Abstract
To explore the possibility that underlying genetic susceptibility influences radiation-induced lung injury, we studied pulmonary damage after radiation therapy for lung cancer and single nucleotide polymorphisms implicated in radiation sensitivity. An objective radiologic method (perfusion single photon emission computed tomography) was used to assess radiation sensitivity. An association between single nucleotide polymorphisms in XRCC1 and BRCA1 and radiation sensitivity of the lungs was noted.
Background
The primary objective of this study was to evaluate the association between radiation sensitivity of the lungs and candidate single nucleotide polymorphisms (SNP) in genes implicated in radiation-induced toxicity.
Methods
Patients with lung cancer who received radiation therapy (RT) had pre-RT and serial post-RT single photon emission computed tomography (SPECT) lung perfusion scans. RT-induced changes in regional perfusion were related to regional dose, which generated patient-specific dose-response curves (DRC). The slope of the DRC is independent of total dose and the irradiated volume, and is taken as a reflection of the patient’s inherent sensitivity to RT. DNA was extracted from blood samples obtained at baseline. SNPs were determined by using a combination of high-resolution melting, TaqMan assays, and direct sequencing. Genotypes from 33 SNPs in 22 genes were compared against the slope of the DRC by using the Kruskal-Wallis test for ordered alternatives.
Results
Thirty-nine self-reported Caucasian patients with pre-RT and ≥6 month post-RT SPECTs, and blood samples were identified. An association between genotype and increasing slope of the DRC was noted in G(1301) A in XRCC1 (rs25487) (P = .01) and G(3748) A in BRCA1 (rs16942) (P = .03).
Conclusions
By using an objective radiologic assessment, polymorphisms within genes involved in repair of DNA damage (XRCC1 and BRCA1) were associated with radiation sensitivity of the lungs.
Keywords: Genetics, Lung toxicity, Radiation sensitivity, Single photon emission computed tomography, Single-nucleotide polymorphism
Introduction
Radiation therapy (RT) is an important treatment modality for patients with cancer, and approximately 50% to 60% of cancer patients will receive RT at some point during their illness. The adverse effects of RT depend principally on the site being treated, the volume of normal tissue irradiated, the dose per fraction, the total dose administered, and whether chemotherapy is also being administered. However, it is also clinically apparent that some cancer patients are more sensitive to the adverse effects of RT than are others, even when all of the factors listed above are relatively constant. Rare hereditary disorders (eg, ataxia-telangiectasia, Nijmegen Breakage syndrome, Fanconi anemia) suggest that genetic differences in key genes may influence radiation sensitivity.1
Single nucleotide polymorphisms (SNPs) are DNA sequence variations, in either coding or noncoding regions, in which a single nucleotide (adenine, guanine, thymine, and cytosine) varies among a population. To identify genetic factors that may be contributing to radiation-induced toxicity, SNPs in genes associated with DNA repair pathways and other radiation-related processes have been studied. By using this candidate gene approach, some studies, but not all,2–4 have demonstrated an association between certain SNPs and acute and long-term adverse effects of RT. These genes include TGFB1,5–11 SOD2,12,13 XRCC1,11–14 XRCC3,13,15,16 XRCC6,16 MSH2,15 MSH3,15 ATM,17,18 p53,19 FSHR,20 ABCA1,21 IL12RB2,21 LIG4,22 and RAD51.23
Most of the endpoints studied to date have been subjective signs or symptoms reported by the patient or provider (eg, erectile dysfunction or breast fibrosis). More objective outcomes may be preferred when evaluating potential genetic contributors of radiation sensitivity. As part of institutional review board–approved prospective studies at Duke University, patients with lung cancer have undergone serial perfusion single photon emission computed tomographies (SPECT) before and after thoracic RT. Dose-dependent changes in SPECT lung perfusion defects after treatment provide an objective assessment of inherent radiation sensitivity.24,25 Most of these same patients had blood samples collected and stored for correlative studies.
We recently reported an association between a polymorphism in the promoter of the transforming growth factor β1 gene (TGFB1) and radiation sensitivity assessed by using radiation-induced SPECT changes.26 Herein, we assess for the possible association between 12 additional SNPs that have been associated with radiation sensitivity in previous studies, in addition to numerous other SNPs within genes known to be involved in DNA damage recognition and repair, which may contribute to radiation sensitivity.
Materials and Methods
Patient Population
As part of institutional review board–approved prospective clinical trials at Duke University, patients with lung cancer who were receiving definitive RT underwent a pretreatment perfusion SPECT scan as well as serial posttreatment scans to assess radiation-induced lung injury. Blood was drawn on several patients at baseline for correlative studies and was stored at −80°C. Patients were included in the present analysis if they (1) underwent a pre-RT SPECT, (2) underwent a ≥6-month post-RT SPECT, (3) had at least 1 banked blood sample, and (4) were Caucasian. Changes in SPECT perfusion after RT largely develop within the first 6 months after treatment with minimal changes thereafter.25,27 Only self-reported Caucasian patients were studied because the relative allelic frequency of SNPs may differ among ancestries.
RT Planning
Patients underwent computed tomography (CT) based 3-dimensional treatment planning by using Plan University of North Carolina software (PLUNC). The patients either received conventionally fractionated RT (1.8–2 Gy every day to 40–70 Gy) or accelerated RT by using a concomitant boost.28 For the latter, the patients received 1.25 Gy twice a day to the clinical target volume, including the primary tumor and mediastinum, usually with anteroposterior/posteroanterior fields. The gross tumor volume received a concurrent boost of 35 cGy twice a day with off-cord fields, thus delivering 1.6 Gy twice a day to the gross tumor volume. After the initial 57.6 Gy, the gross tumor volume received an additional dose at 1.6 Gy twice a day to a total dose of 73.6–86.4 Gy). Chemotherapy was administered at the discretion of the multimodality team.
Perfusion SPECTs
SPECT lung perfusion scans were obtained after the intravenous injection of technetium-99m–labeled macroaggregated albumin as previously described.24,29,30 The pre- and postradiation SPECT lung images were registered to each other and to the radiation treatment planning scan (and hence the 3-dimensional dose distribution), largely manually with the assistance of some automatic image registration tools. The SPECT images were translated and rotated (in 6 degrees of freedom) until the “edges” of the SPECT-defined perfusion (the area of rapid gradient in counts per cubic centimeter) were aligned with the CT-defined lung borders, also considering the presence of the tumor and regions of emphysema that influence the SPECT images.31 We recognize that a perfect registration is not possible for an elastic organ in a breathing patient. Every attempt was made to have all of the scans and the radiation treatment delivered with the patient in a similar position. In all cases, the registration was performed by an experienced physicist.
This multi-image registration facilitated the analysis relating changes in regional perfusion (comparison of pre- and post-radiation SPECTs) to the regional radiation dose (from the planning CT). After registration, the quantitative SPECT data were resampled by tri-linear interpolation to match the spatial sampling of the planning CT data set. Within each lung pixel, the change in regional perfusion was quantified by comparing pre- and post-radiation SPECTs.27 For each patient, and at each dose level (D), the reduction in the percentage of SPECT counts (compared with the pre-RT scan) was calculated as percent reductionD = 100 × (1 − postD)/preD; in which postD and preD are the percentage SPECT counts on the post- and pre-RT scans in the region that received D Gy, respectively.27
Changes in regional perfusion after RT were related to regional radiation doses (via image fusion), which yielded a patient-specific dose-response curve (DRC). The DRC slope is independent of irradiated volume and is taken as a reflection of the patient’s inherent sensitivity to radiation. The DRC was obtained from the nontumor-bearing contralateral lung to avoid issues related to reperfusion after treatment of central tumors. Because SPECTs provide only relative perfusion information, the DRC was “normalized” by assuming that absolute perfusion is unchanged in “control” regions that receive very low doses (typically <2.5 Gy).27 The normalized percent reduction in regional perfusion, R, at dose D, is thus32:
In which the 0 subscripts refer to the lung regions at <2.5 Gy. We recognize that normalization may be imprecise because function in the low-dose region may increase due to compensation. Nevertheless, this is the method that has been used in the 2 centers that have generated DRCs, and we know of no way to correct for this possibility.
Studied SNPs
Genes of interest were selected based on peer-reviewed publications and knowledge of DNA damage repair pathways. Each gene was reviewed on the National Center for Biotechnology Information database. SNPs within genes with a minor allele frequency more than 0.10 in a Caucasian population were selected for analysis. SNPs within coding regions that resulted in a synonymous mutation (no change in amino acid) were not included. Hardy-Weinberg equilibrium was assessed for all SNPs.
Genotyping Methods
DNA extraction was performed by using the QIAamp DNA Blood Mini Kit (Qiagen, Germantown, MD) according to the manufacturer’s instructions. DNA was found to be of high quality, and no samples had to be discarded secondary to impurity or degradation. SNP genotypes were determined by using a combination of high-resolution melting (HRM) assays, TaqMan assays, and direct DNA sequencing. Primer pairs for each SNP were designed by using the National Center for Biotechnology Information–Primer Blast program to create approximately 125 bp amplicons that contain each SNP. HRM assays were performed on an Applied Biosystems 7500 Fast Real-Time polymerase chain reaction system according to the manufacturer’s instructions and were analyzed with Applied Biosystems HRM v2.0 software (Applied Biosystems, Foster, CA). Based on the HRM curves, selected samples were DNA sequenced to confirm the SNP genotype by using the Duke University DNA Analysis Facility shared resource. Predefined or custom-designed TaqMan SNP genotyping assays were obtained from Applied Biosystems and were performed on a 7500 Fast Real-Time polymerase chain reaction instrument according to the manufacturer’s standard protocol by using 20-ng human genomic DNA per 20 μL assay. Data analysis was performed by using Applied Biosystems 7500 Software v2.0.4.
Statistical Analysis
All statistical summaries and analyses were produced by using the R statistical environment.33 The SNPs were tested for the Hardy-Weinberg equilibrium by using an exact test34 provided by the R extension package genetics. SNPs not in Hardy-Weinberg equilibrium were not further assessed. The association between genotype and the slope of the DRC was tested for each SNP by using the Kruskal-Wallis equilibrium powered for ordered alternatives.35 The implementation provided by the R extension package coin36 was used for this purpose. The analyses for all SNPs in this study were a priori powered for additive genetic effects. Because this was an exploratory study, the results were not adjusted for multiple testing to minimize the risk of false negatives.
Results
Patient Characteristics
Forty-five unrelated patients with a baseline blood sample and pre-RT and ≥6-month post-RT SPECTs, and with usable DNA, were identified. Patients who did not self-report as Caucasian were excluded (n = 6), which left 39 for the present analysis. The majority had a history of smoking (92%). Patients received either conventionally fractionated RT (n = 29) or accelerated hyperfractionated RT (n = 10), with chemotherapy administered in the majority (82%). Chemotherapy consisted of a platinum-based doublet in all patients (96% carboplatin and 4% cisplatin). The median total radiation dose was 66 Gy (range, 40–86.4 Gy). Patient and treatment characteristics can be found in Table 1.
Table 1.
Patient Characteristics (n = 39)
| Characteristic | |
|---|---|
| Median Age (Range), y | 63 (46–87) |
| Sex, No. (%) | |
| Men | 20 (51) |
| Women | 19 (49) |
| Stage, No. (%) | |
| I | 3 (8) |
| II | 2 (5) |
| III | 27 (69) |
| IV | 3 (8) |
| X (locally) | 4 (10) |
| Histology, No. (%) | |
| Squamous | 5 (13) |
| Adenocarcinoma | 6 (15) |
| Non–small-cell not otherwise specified | 24 (62) |
| Small cell | 4 (10) |
| Smoking Status, No. (%) | |
| Current | 12 (31) |
| Former | 24 (61) |
| Never | 3 (8) |
| Treatment, no. (%) | |
| Sequential ChT/RT | 22 (56) |
| Concurrent ChT/RTa | 10 (26) |
| RT alone | 7 (18) |
| Median Radiation Dose (range), Gy | 66 (40–86.4) |
| Mean Lung Dose, Median (range), Gy | 18.5 (5–29) |
| Median V20 (range) | 33 (6–51) |
Abbreviations: ChT = chemotherapy; RT = radiation therapy; V20 = volume of lung receiving 20 Gy or more.
With or without induction and/or consolidation ChT.
All the patients underwent pre- and post-radiation SPECTs. The median time between completing treatment and follow-up SPECT was 8 months (range, 6–17 months). The median DRC slope was 0.38 (ie, 0.38% reduction in perfusion per Gy), with a range of −1.14 to 1.12. There was no correlation between DRC slope and the time interval of follow-up SPECT, consistent with our prior analyses for time intervals >6 months.25,27
Forty-three SNPs in 23 genes were initially screened. Eight SNPs had a relative minor allele frequency of zero and were not evaluated further (rs28897686, rs28897687, and rs28897688 in BRCA1; rs2229033 in ATR; rs3730017 in NOS2; rs75521089 in HIF1A; rs28897729 in BRCA2; and rs80233386 in RAD51). Genotyping results from 1 SNP (rs1650697 in MSH3) were not thought to be reliable, and the results were discarded. One SNP was not in Hardy-Weinberg equilibrium and was not assessed further (rs1801321 in RAD51). This left 33 SNPs in 22 genes for the present analysis (Table 2).
Table 2.
Genes and SNPs Studied
| SNP | Gene | Location | Position | Base Substitution | Amino Acid Position | Amino Acid Substitution |
|---|---|---|---|---|---|---|
| rs2071746 | HMOX1 | Promoter | −413 | A → T | NA | NA |
| rs2071747 | HMOX1 | Exon 1 | 99 | G → C | 7 | Asp → His |
| rs2779249 | NOS2A | Promoter | 974 | C → A | NA | NA |
| rs2297518 | NOS2A | Exon 16 | 2087 | G → A | 608 | Ser → Leu |
| rs11549465 | HIF1A | Exon 12 | 2148 | C → T | 582 | Pro → Ser |
| rs1801320 | RAD51 | Promoter | −135 | G → C | NA | NA |
| rs11226 | RAD52 | Promoter | −2259 | C → T | NA | NA |
| rs1042522 | TP53 | Exon 4 | 412 | C → G | 72 | Pro → Arg |
| rs1805388 | LIG4 | Exon 2 | 261 | C → T | 9 | Thr → Ile |
| rs1062557 | GRP | Exon 1 | 108 | C → A | 4 | Arg → Ser |
| rs1801516 | ATM | Exon 37 | 5942 | G → A | 1853 | Asp → Asn |
| rs16941 | BRCA1 | Exon 10 | 3345 | A → G | 1038 | Glu → Gly |
| rs16942 | BRCA1 | Exon 10 | 3780 | G → A | 1183 | Arg → Lys |
| rs144848 | BRCA2 | Exon 10 | 1341 | T → G | 372 | Asn → His |
| rs1799782 | XRCC1 | Exon 6 | 700 | C → T | 194 | Arg → Trp |
| rs25489 | XRCC1 | Exon 9 | 959 | G → A | 280 | Arg → His |
| rs25487 | XRCC1 | Exon 10 | 1316 | G → A | 399 | Arg → Gln |
| rs3218536 | XRCC2 | Exon 3 | 649 | G → A | 188 | Arg → His |
| rs861539 | XRCC3 | Unknown | 1045 | C → T | 241 | Thr → Met |
| rs4880 | SOD2 | Unknown | 201 | T → C | 16 | Val → Ala |
| rs13181 | ERCC2 | Exon 23 | 2298 | G → T | 751 | Lys → Gln |
| rs2227928 | ATR | Exon 4 | 754 | C → T | 211 | Thr → Met |
| rs2229032 | ATR | Exon 43 | 7396 | G → A | 2425 | Arg → Gln |
| rs1130409 | APEX1 | Exon 5 | 776 | G → T | 148 | Glu → Asp |
| rs26279 | MSH3 | Unknown | 3386 | G → A | 1045 | Ala → Thr |
| rs184967 | MSH3 | Unknown | 3099 | G → A | 949 | Arg → Gln |
| rs1799977 | MLH1 | Unknown | 715 | A → G | 219 | Ile → Val |
| rs3790566 | IL12RB2 | Intron | NA | T → C | NA | NA |
| rs3790567 | IL12RB2 | Intron | NA | A → G | NA | NA |
| rs3790568 | IL12RB2 | Intron | NA | G → A | NA | NA |
| rs2230806 | ABCA1 | Unknown | 969 | G → A | 219 | Arg → Lys |
| rs2253304 | ABCA1 | Intron | NA | A → G | NA | NA |
| rs2487058 | ABCA1 | Intron | NA | C → T | NA | NA |
Abbreviations: A = adenine; C = cytosine; G = guanine; NA = not applicable; SNP = single nucleotide polymorphism; T = thymine.
Among the 33 SNPs studied, 2 were associated with increasing slope of the SPECT DRC (Table 3). These included rs16942 (a SNP in exon 10 of BRCA1) (P = .03) and rs25487 (a SNP in exon 10 of XRCC1) (P = .01) (Figure 1). None of the other studied SNPs were statistically significant (Table 3). As both XRCC1 and BRCA1 are directly involved or regulate base excision repair, a simple parametric linear SNP-SNP association analysis, with both SNPs coded additively, was performed. The asymptotic P value for the interaction was .67. Due to the small sample size, the resulting sparseness in the data may render this analysis of limited use.
Table 3.
Genotype Frequencies and Association With Slope of DRCa
| SNP | Gene | Genotypes | P Value |
|---|---|---|---|
| rs2071746 | HMOX1 | 9 AA, 22 AT, 8 TT | .59 |
| rs2071747 | HMOX1 | 36 GG, 3 GC | .67 |
| rs2779249 | NOS2A | 4 AA, 23 AC, 12 CC | .54 |
| rs2297518 | NOS2A | 3 AA, 14 AG, 22 GG | .73 |
| rs11549465 | HIF1A | 27 CC, 11 CT, 1 TT | .52 |
| rs1801320 | RAD51 | 34 GG, 5 CG | .21 |
| rs11226 | RAD52 | 14 CC, 18 CT, 7 TT | .21 |
| rs1042522 | TP53 | 22 GG, 17 GC | .97 |
| rs1805388 | LIG4 | 25 CC, 13 CT, 1 TT | .93 |
| rs1062557 | GRP | 25 AA, 12 AC, 2 CC | .18 |
| rs1801516 | ATM | 1 AA, 11 AG, 27 GG | .58 |
| rs16941 | BRCA1 | 19 AA, 15 AG, 5 GG | .47 |
| rs16942 | BRCA1 | 19 AA, 13 AG, 7 GG | .03 |
| rs144848 | BRCA2 | 2 GG, 15, GT, 22 TT | .09 |
| rs1799782 | XRCC1 | 33 CC, 6 CT | .24 |
| rs25489 | XRCC1 | 35 GG, 4 AG | .98 |
| rs25487 | XRCC1 | 6 AA, 20 AG, 13 GG | .01 |
| rs3218536 | XRCC2 | 1 AA, 6 AG, 32 GG | .83 |
| rs861539 | XRCC3 | 14 CC, 18 CT, 7 TT | .09 |
| rs4880 | SOD2 | 11 CC, 16 CT, 12 TT | .98 |
| rs13181 | ERCC2 | 3 GG, 23 GT, 13 TT | .66 |
| rs2227928 | ATR | 15 CC, 19 CT, 5 TT | .44 |
| rs2229032 | ATR | 29 GG, 10 GA | .16 |
| rs1130409 | APEX1 | 12 GG, 21 GT, 6 TT | .27 |
| rs26279 | MSH3 | 17 AA, 19 AG, 3 GG | .20 |
| rs184967 | MSH3 | 29 GG, 9 GA, 1 AA | .30 |
| rs1799977 | MLH1 | 12 AA, 23 AG, 4 GG | .15 |
| rs3790566 | IL12RB2 | 22 CC, 17 CT | .33 |
| rs3790567 | IL12RB2 | 22GG, 17 GA | .32 |
| rs3790568 | IL12RB2 | 33 GG, 6 GA | .89 |
| rs2230806 | ABCA1 | 13 CC, 21 CT, 5 TT | .60 |
| rs2253304 | ABCA1 | 12 GG, 21 GA, 6 AA | .97 |
| rs2487058 | ABCA1 | 15 CC, 19 CT, 5 TT | .98 |
Bold indicates statistically significant associations.
Abbreviations: A = adenine; C = cytosine; DRC = dose-response curve; G = guanine; SNP = single nucleotide polymorphism.
DRC of perfusion single photon emission computed tomography after radiation therapy.
Figure 1.
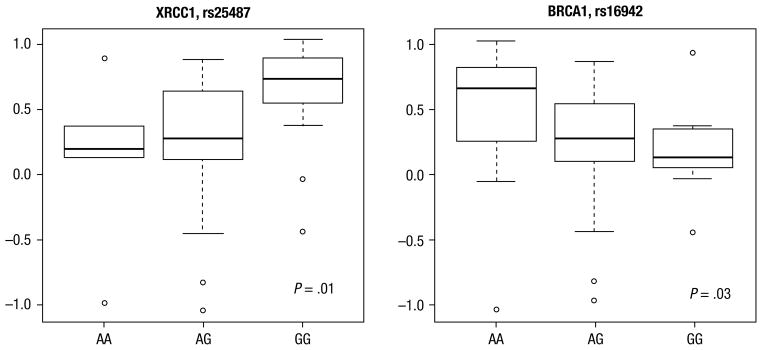
Boxplots for rs25487 (XRCC1) and rs16942 (BRCA1), Produced by Using the R Boxplot Function With Its Default Settings, Comparing Genotype With Slope of the Single Photon Emission Computed Tomography Dose-Response Curve. The Line in the Middle Is the Median (Q2). The Upper and Lower Edges of the Box Are the 25th (Q1) and 75th (Q3) Percentiles. The Height of the Box is the Interquartile Range (IQR). The Whiskers Are Computed as Follows: Exclude All Data Points That Are Either 1.5 × IQR Below Q1 or 1.5 × IQR Above Q3. The Whiskers Correspond to the Range of This Reduced Data Set. Any Data Points (outliers) From the Original Sample Below or Above the Whiskers Are Also Plotted
Discussion
By using this unique database, an objective radiologic endpoint was available to assess radiation sensitivity of the lungs. Radiographic abnormalities, indicative of injury, are noted in the lung almost immediately after thoracic RT.27,37 The radiographic findings most extensively studied are increased density on CT and decreased pulmonary perfusion on SPECT. Radiation dose is the single most important contributor to the development of radiographic abnormalities.38,39 An optimal endpoint to study radiation sensitivity would be both objective and clinically relevant. SPECT abnormalities, admittedly, are only weakly correlated with clinical outcomes, such as change in pulmonary function or development of symptoms.40 However, radiation pneumonitis, which has also been studied in the context of SNPs,5,14,19,22,23,41–43 is notoriously difficult to accurately identify and grade because it is a clinical diagnosis.44 Further, although clinical symptoms are dependent on dosimetric parameters (the volume of lung that receives a certain radiation dose), the SPECT DRC is independent of volume. Thus, both approaches should be viewed as complementary when studying genetic susceptibility to RT.
In this study, when using the slope of the SPECT DRC, we observed that 2 single nucleotide polymorphisms, one in XRCC1 and one in BRCA1, were associated with radiation sensitivity of the lungs assessed with SPECT. In a prior analysis, the −509 promoter in the TGFB1 gene was also associated with radiation sensitivity.26 Multiple other studies have also observed an association among the −509 SNP in TGFB1 and radiation toxicities, including altered breast appearance,7 breast fibrosis,8,9 erectile dysfunction,6 rectal bleeding, and miscellaneous severe complications.10 However, several other studies have not observed such relationships.2,45,46
The protein product of XRCC1 is involved in the base excision repair pathway in which single-strand breaks, generated after exposure to ionizing radiation or other harmful stimuli, are repaired. An association between radiation sensitivity and an SNP in exon 10 of XRCC1 was observed in our study (rs25487). This polymorphism substitutes an A (adenine) for G (guanine) at messenger RNA position 1316, which leads to an amino acid substitution at position 399 (glutamine for arginine). Patients with the ancestral allele (G) were found to be more radiosensitive.
Yin et al14 observed that the XRCC1 Q399R AA genotype, after correcting for potential confounding factors, was associated with a reduced risk of radiation pneumonitis in patients with non–small-cell lung cancer (0.48) (P = .04). Andreassen et al13 observed an association between G/G homozygotes and radiation-induced subcutaneous fibrosis after treatment for breast cancer, with heterozygotes at intermediate risk compared with AA homozygotes. Similarly, in a study of patients with nasopharyngeal cancer, Alsbeih et al11 noted an association between the G allele and grade 2 to 3 late fibrosis after head-and-neck RT for nasopharyngeal carcinoma. Chang-Claude et al47 observed a trend for a higher risk of acute adverse effects in breast cancer patients with normal weight and with this same genetic change, although the difference was not statistically significant.
However, Giotopoulos et al9 studied patients with breast cancer who received either postlumpectomy or postmastectomy RT (without an electron boost) and evaluated high-grade telangiectasias and fibrosis. SOMA score 2 to 4 telangiectasias developed in 2% of patients with GG, 11% of patients with AG, and 5% of patients with AA. It was reported that heterozygosity was more likely to develop telangiectasias (P = .01). Because AA homozygotes had a lower risk than heterozygotes, the biologic significance of this is ambiguous. Burri et al12 did not observe an association between this polymorphism and complications after brachytherapy, with or without external-beam RT (rectal bleeding, erectile dysfunction, urinary morbidity). Other investigators have also not found an association between this polymorphism and subjective endpoints of radiation sensitivity, including breast fibrosis,48 altered breast appearance after RT,2,7 and acute skin reactions.49
A pooled analysis of several studies (differing endpoints, scoring systems, patient populations) by Langsenlehner et al50 did not observe an effect of the XRCC1 R399Q polymorphism on risk of late radiation-induced toxicity. There was an association with XRCC1 R280H, with an odds ratio of 0.6. We did not observe an association with this polymorphism (P =.98), but there were only 4 heterozygotes in our population.
This is the first study, to our knowledge, that has demonstrated a possible association between the G(3780)A SNP in BRCA1 and radiation sensitivity of the lungs. BRCA1, typically associated with inherited breast cancer, encodes a protein that joins other proteins involved in DNA damage recognition and repair to form a complex known as the BRCA1-associated genome surveillance complex. BRCA1 plays a central role in repairing DNA double-strand breaks. A lower risk of toxicity was noted in patients with the AG or GG genotype. Few studies have evaluated BRCA1 polymorphisms and their possible relationship to radiation toxicity. Damaraju et al51 did not observe an association among any BRCA1 SNPs, including position 3780, and bladder and/or rectal toxicity after pelvic RT. We did not observe an association among many SNPs that had previously been associated with RT toxicity (Table 4). This may be due to a lack of statistical power, lack of true causation, or organ and/or tissue specific differences.
Table 4.
SNPs Associated With Radiation Sensitivity
| Gene | Name | SNP | Endpointa |
|---|---|---|---|
| TGFB1 | Transforming growth factor β1 | rs1800469; rs1800470; rs1800471 | Radiation pneumonitis,5 altered breast appearance,7 breast fibrosis,7,8 erectile dysfunction,6 rectal bleeding,6 miscellaneous severe complications,10 neck fibrosis,11 perfusion SPECT defects26 |
| SOD2 | Superoxide dismutase 2 | rs4880 | Rectal bleeding,12 subcutaneous fibrosis13 |
| XRCC1 | X-ray repair cross- complementing protein 1 | rs25489; rs25487 | Erectile dysfunction,12 subcutaneous fibrosis,11,13 rectal/bladder toxicity,50 pneumonitis14 |
| XRCC3 | X-ray repair cross- complementing protein 3 | rs861539 | Acute skin reaction,15 subcutaneous fibrosis and telangiectasia,13 severe dysphagia16 |
| MSH2 | mutS homolog 2 | rs2303428 | Acute skin reaction15 |
| MSH3 | mutS homolog 3 | rs26279 | Acute skin reaction15 |
| ATM | Ataxia telangiectasia mutated | Multiple | Rectal bleeding,17 erectile dysfunction,17 urinary morbidity,17 chest wall fibrosis,18 subcutaneous late effects,52 pneumonitis,43 miscellaneous adverse reactions,53 late effects54 |
| MC1R | Melanocortin 1 receptor | Multiple | Severe acute adverse effects55 |
| FSHR | Follicle-stimulating hormone receptor | rs2268363 | Erectile dysfunction20 |
| P53 | Tumor protein p53 | rs1042522 | Radiation pneumonitis19 |
| ABCA1 | ATP-binding cassette, subfamily A, member 1 | rs2230806; rs2253304; rs2487058 | Radiation dermatitis21 |
| IL12RB2 | Interleukin-12 receptor, beta 2 | rs379056; rs3790566; rs3790568 | Radiation dermatitis21 |
| RAD51 | RAD51 homolog | rs1801320 | Pneumonitis23 |
| LIG4 | Ligase IV, DNA, ATP-dependent | rs1805388 | Pneumonitis22 |
Abbreviations: ATP = adenosine triphosphate; SNP = single nucleotide polymorphism; SPECT = single photon emission computed tomography.
Not all SNPs are associated with all endpoints.
To date, most studies that evaluated radiation toxicity have used relatively subjective endpoints. This is understandable because most toxicities that patients experience are not readily quantifiable. Our study was unique because we used an objective measure of RT sensitivity, namely dose-dependent changes in regional perfusion in the lung after thoracic RT. We have observed possible associations between SNPs in TGFβ1, XRCC1, and BRCA1, and a greater decline in pulmonary perfusion after RT. Large, genome-wide association studies may ultimately be required to discover which polymorphisms are most responsible for interpatient differences in radiation sensitivity. Ideally, objective and clinically relevant endpoints of radiation sensitivity will be used in these endeavors.
We acknowledge the limitations of our analysis. The number of patients is small, and a verification cohort is not available. We were constrained to use a candidate gene approach as opposed to a genome-wide association study, given the small number of patients with available SPECT data to perform our analysis. We also did not correct for multiple hypotheses given the small number of patients. Thus, the associations observed could have been due to chance alone. Finally, multivariate analyses to correct for possible confounding factors (chemotherapy, smoking status, pulmonary function, etc) were not deemed statistically feasible in our population. These same factors did not have a dramatic effect on the DRC in our prior analyses.39 The fundamental and unique strength of our study, as opposed to almost all other studies that evaluated radiation toxicity, is the objective radiologic endpoint of radiation sensitivity that was used. In this study, in which an objective radiologic assessment of radiation-induced lung injury was used, polymorphisms within genes involved in repair of DNA damage (XRCC1 and BRCA1) were associated with radiation sensitivity of the lungs.
Clinical Practice Points.
Dosimetric factors predict the risk of radiation-induced lung injury but have limitations.
Underlying genetic factors, including SNPs, may contribute to risk.
Further work is necessary to evaluate whether genetic factors are contributory and how this should change clinical treatment recommendations.
Acknowledgments
The study was supported in part by NIH grants CA69579 (L. Marks) and CA115748 (S. Das).
Footnotes
Disclosure
The authors have stated that they have no conflicts of interest.
Presented at the 14th World Conference on Lung Cancer, Amsterdam, Netherlands, July 3–7, 2011
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Pollard JM, Gatti RA. Clinical radiation sensitivity with DNA repair disorders: an overview. Int J Radiat Oncol Biol Phys. 2009;74:1323–31. doi: 10.1016/j.ijrobp.2009.02.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barnett GC, Coles CE, Elliott RM, et al. Independent validation of genes and polymorphisms reported to be associated with radiation toxicity: a prospective analysis study. Lancet Oncol. 2012;13:65–77. doi: 10.1016/S1470-2045(11)70302-3. [DOI] [PubMed] [Google Scholar]
- 3.Alsner J, Andreassen CN, Overgaard J. Genetic markers for prediction of normal tissue toxicity after radiotherapy. Semin Radiat Oncol. 2008;18:126–35. doi: 10.1016/j.semradonc.2007.10.004. [DOI] [PubMed] [Google Scholar]
- 4.Andreassen CN, Alsner J. Genetic variants and normal tissue toxicity after radiotherapy: a systematic review. Radiother Oncol. 2009;92:299–309. doi: 10.1016/j.radonc.2009.06.015. [DOI] [PubMed] [Google Scholar]
- 5.Yuan X, Liao Z, Liu Z, et al. Single nucleotide polymorphism at rs1982073:T869C of the TGFbeta 1 gene is associated with the risk of radiation pneumonitis in patients with non-small-cell lung cancer treated with definitive radiotherapy. J Clin Oncol. 2009;27:3370–8. doi: 10.1200/JCO.2008.20.6763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peters CA, Stock RG, Cesaretti JA, et al. TGFB1 single nucleotide polymorphisms are associated with adverse quality of life in prostate cancer patients treated with radiotherapy. Int J Radiat Oncol Biol Phys. 2008;70:752–9. doi: 10.1016/j.ijrobp.2007.05.023. [DOI] [PubMed] [Google Scholar]
- 7.Andreassen CN, Alsner J, Overgaard J, et al. TGFB1 polymorphisms are associated with risk of late normal tissue complications in the breast after radiotherapy for early breast cancer. Radiother Oncol. 2005;75:18–21. doi: 10.1016/j.radonc.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 8.Quarmby S, Fakhoury H, Levine E, et al. Association of transforming growth factor beta-1 single nucleotide polymorphisms with radiation-induced damage to normal tissues in breast cancer patients. Int J Radiat Biol. 2003;79:137–43. [PubMed] [Google Scholar]
- 9.Giotopoulos G, Symonds RP, Foweraker K, et al. The late radiotherapy normal tissue injury phenotypes of telangiectasia, fibrosis and atrophy in breast cancer patients have distinct genotype-dependent causes. Br J Cancer. 2007;96:1001–7. doi: 10.1038/sj.bjc.6603637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Ruyck K, Van Eijkeren M, Claes K, et al. TGFbeta1 polymorphisms and late clinical radiosensitivity in patients treated for gynecologic tumors. Int J Radiat Oncol Biol Phys. 2006;65:1240–8. doi: 10.1016/j.ijrobp.2006.03.047. [DOI] [PubMed] [Google Scholar]
- 11.Alsbeih G, Al-Harbi N, Al-Hadyan K, et al. Association between normal tissue complications after radiotherapy and polymorphic variations in TGFB1 and XRCC1 genes. Radiat Res. 2010;173:505–11. doi: 10.1667/RR1769.1. [DOI] [PubMed] [Google Scholar]
- 12.Burri RJ, Stock RG, Cesaretti JA, et al. Association of single nucleotide polymorphisms in SOD2, XRCC1 and XRCC3 with susceptibility for the development of adverse effects resulting from radiotherapy for prostate cancer. Radiat Res. 2008;170:49–59. doi: 10.1667/RR1219.1. [DOI] [PubMed] [Google Scholar]
- 13.Andreassen CN, Alsner J, Overgaard M, et al. Prediction of normal tissue radiosensitivity from polymorphisms in candidate genes. Radiother Oncol. 2003;69:127–35. doi: 10.1016/j.radonc.2003.09.010. [DOI] [PubMed] [Google Scholar]
- 14.Yin M, Liao Z, Liu Z, et al. Functional polymorphisms of base excision repair genes XRCC1 and APEX1 predict risk of radiation pneumonitis in patients with non-small cell lung cancer treated with definitive radiation therapy. Int J Radiat Oncol Biol Phys. 2011;81:e67–73. doi: 10.1016/j.ijrobp.2010.11.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mangoni M, Bisanzi S, Carozzi F, et al. Association between genetic polymorphisms in the XRCC1, XRCC3, XPD, GSTM1, GSTT1, MSH2, MLH1, MSH3, and MGMT genes and radiosensitivity in breast cancer patients. Int J Radiat Oncol Biol Phys. 2011;81:52–8. doi: 10.1016/j.ijrobp.2010.04.023. [DOI] [PubMed] [Google Scholar]
- 16.Werbrouck J, De Ruyck K, Duprez F, et al. Acute normal tissue reactions in head-and-neck cancer patients treated with IMRT: influence of dose and association with genetic polymorphisms in DNA DSB repair genes. Int J Radiat Oncol Biol Phys. 2009;73:1187–95. doi: 10.1016/j.ijrobp.2008.08.073. [DOI] [PubMed] [Google Scholar]
- 17.Cesaretti JA, Stock RG, Lehrer S, et al. ATM sequence variants are predictive of adverse radiotherapy response among patients treated for prostate cancer. Int J Radiat Oncol Biol Phys. 2005;61:196–202. doi: 10.1016/j.ijrobp.2004.09.031. [DOI] [PubMed] [Google Scholar]
- 18.Andreassen CN, Overgaard J, Alsner J, et al. ATM sequence variants and risk of radiation-induced subcutaneous fibrosis after postmastectomy radiotherapy. Int J Radiat Oncol Biol Phys. 2006;64:776–83. doi: 10.1016/j.ijrobp.2005.09.014. [DOI] [PubMed] [Google Scholar]
- 19.Yang M, Zhang L, Bi N, et al. Association of P53 and ATM polymorphisms with risk of radiation-induced pneumonitis in lung cancer patients treated with radiotherapy. Int J Radiat Oncol Biol Phys. 2011;79:1402–7. doi: 10.1016/j.ijrobp.2009.12.042. [DOI] [PubMed] [Google Scholar]
- 20.Kerns SL, Ostrer H, Stock R, et al. Genome-wide association study to identify single nucleotide polymorphisms (SNPs) associated with the development of erectile dysfunction in African-American men after radiotherapy for prostate cancer. Int J Radiat Oncol Biol Phys. 2010;78:1292–300. doi: 10.1016/j.ijrobp.2010.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Isomura M, Oya N, Tachiiri S, et al. IL12RB2 and ABCA1 genes are associated with susceptibility to radiation dermatitis. Clin Cancer Res. 2008;14:6683–9. doi: 10.1158/1078-0432.CCR-07-4389. [DOI] [PubMed] [Google Scholar]
- 22.Yin M, Liao Z, Liu Z, et al. Genetic variants of the nonhomologous end joining gene LIG4 and severe radiation pneumonitis in nonsmall cell lung cancer patients treated with definitive radiotherapy. Cancer. 2012;118:528–35. doi: 10.1002/cncr.26214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yin M, Liao Z, Huang YJ, et al. Polymorphisms of homologous recombination genes and clinical outcomes of non-small cell lung cancer patients treated with definitive radiotherapy. PLoS ONE. 2011;6:e20055. doi: 10.1371/journal.pone.0020055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marks LB, Spencer DP, Bentel GC, et al. The utility of SPECT lung perfusion scans in minimizing and assessing the physiologic consequences of thoracic irradiation. Int J Radiat Oncol Biol Phys. 1993;26:659–68. doi: 10.1016/0360-3016(93)90285-4. [DOI] [PubMed] [Google Scholar]
- 25.Zhang J, Ma J, Zhou S, et al. Radiation-induced reductions in regional lung perfusion: 0. 1–12 year data from a prospective clinical study. Int J Radiat Oncol Biol Phys. 2010;76:425–32. doi: 10.1016/j.ijrobp.2009.02.005. [DOI] [PubMed] [Google Scholar]
- 26.Kelsey CR, Jackson L, Langdon S, et al. A polymorphism within the promoter of the TGFbeta1 gene is associated with radiation sensitivity using an objective radiologic endpoint. Int J Radiat Oncol Biol Phys. 2012;82:e247–55. doi: 10.1016/j.ijrobp.2011.02.064. [DOI] [PubMed] [Google Scholar]
- 27.Woel RT, Munley MT, Hollis D, et al. The time course of radiation therapy-induced reductions in regional perfusion: a prospective study with >5 years of follow-up. Int J Radiat Oncol Biol Phys. 2002;52:58–67. doi: 10.1016/s0360-3016(01)01809-0. [DOI] [PubMed] [Google Scholar]
- 28.Maguire PD, Marks LB, Sibley GS, et al. 73. 6 Gy and beyond: hyperfractionated, accelerated radiotherapy for non-small-cell lung cancer. J Clin Oncol. 2001;19:705–11. doi: 10.1200/JCO.2001.19.3.705. [DOI] [PubMed] [Google Scholar]
- 29.Marks LB, Munley MT, Spencer DP, et al. Quantification of radiation-induced regional lung injury with perfusion imaging. Int J Radiat Oncol Biol Phys. 1997;38:399–409. doi: 10.1016/s0360-3016(97)00013-8. [DOI] [PubMed] [Google Scholar]
- 30.Marks LB, Fan M, Clough R, et al. Radiation-induced pulmonary injury: symptomatic versus subclinical endpoints. Int J Radiat Biol. 2000;76:469–75. doi: 10.1080/095530000138466. [DOI] [PubMed] [Google Scholar]
- 31.Marks LB, Spencer DP, Sherouse GW, et al. The role of three dimensional functional lung imaging in radiation treatment planning: the functional dose-volume histogram. Int J Radiat Oncol Biol Phys. 1995;33:65–75. doi: 10.1016/0360-3016(95)00091-C. [DOI] [PubMed] [Google Scholar]
- 32.Levinson B, Marks LB, Munley MT, et al. Regional dose response to pulmonary irradiation using a manual method. Radiother Oncol. 1998;48:53–60. doi: 10.1016/s0167-8140(98)00057-7. [DOI] [PubMed] [Google Scholar]
- 33.R Development Core Team. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2010. [Google Scholar]
- 34.Emigh T. A comparison of tests for Hardy-Weinberg equilibrium. Biometrics. 1980;36:627–42. [PubMed] [Google Scholar]
- 35.Hajek J, Sidak Z, Pranab KS. Theory of rank tests. San Diego, CA: Academic Press; 1999. [Google Scholar]
- 36.Hothorn T, Hornik K, van de Wiel MA, et al. A Lego system for conditional inference. Am Stat. 2006;60:257–63. [Google Scholar]
- 37.Skoczylas JZ, Bentzen SM, Overgaard M, et al. Time course of radiological lung density changes after postmastectomy radiotherapy. Acta Oncol. 2000;39:181–7. doi: 10.1080/028418600430743. [DOI] [PubMed] [Google Scholar]
- 38.Ma J, Zhang J, Zhou S, et al. Regional lung density changes after radiation therapy for tumors in and around thorax. Int J Radiat Oncol Biol Phys. 2010;76:116–22. doi: 10.1016/j.ijrobp.2009.01.025. [DOI] [PubMed] [Google Scholar]
- 39.Garipagaoglu M, Munley MT, Hollis D, et al. The effect of patient-specific factors on radiation-induced regional lung injury. Int J Radiat Oncol Biol Phys. 1999;45:331–8. doi: 10.1016/s0360-3016(99)00201-1. [DOI] [PubMed] [Google Scholar]
- 40.Fan M, Marks LB, Hollis D, et al. Can we predict radiation-induced changes in pulmonary function based on the sum of predicted regional dysfunction? J Clin Oncol. 2001;19:543–50. doi: 10.1200/JCO.2001.19.2.543. [DOI] [PubMed] [Google Scholar]
- 41.Tomioka T, Ogawa M, Sawamura S, et al. A case of post-radiation therapy patient with difficulty in intubation unexpected preoperatively [in Japanese with English abstract] Masui. 2003;52:406–8. [PubMed] [Google Scholar]
- 42.Hildebrandt MA, Komaki R, Liao Z, et al. Genetic variants in inflammation-related genes are associated with radiation-induced toxicity following treatment for non-small cell lung cancer. PLoS ONE. 2010;5:e12402. doi: 10.1371/journal.pone.0012402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang L, Yang M, Bi N, et al. ATM polymorphisms are associated with risk of radiation-induced pneumonitis. Int J Radiat Oncol Biol Phys. 2010;77:1360–8. doi: 10.1016/j.ijrobp.2009.07.1675. [DOI] [PubMed] [Google Scholar]
- 44.Kocak Z, Evans ES, Zhou SM, et al. Challenges in defining radiation pneumonitis in patients with lung cancer. Int J Radiat Oncol Biol Phys. 2005;62:635–8. doi: 10.1016/j.ijrobp.2004.12.023. [DOI] [PubMed] [Google Scholar]
- 45.Barnett GC, Coles CE, Burnet NG, et al. No association between SNPs regulating TGF-beta1 secretion and late radiotherapy toxicity to the breast: results from the RAPPER study. Radiother Oncol. 2010;97:9–14. doi: 10.1016/j.radonc.2009.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Martin S, Sydenham M, Haviland J, et al. Test of association between variant tgbeta1 alleles and late adverse effects of breast radiotherapy. Radiother Oncol. 2010;97:15–8. doi: 10.1016/j.radonc.2010.03.019. [DOI] [PubMed] [Google Scholar]
- 47.Chang-Claude J, Popanda O, Tan XL, et al. Association between polymorphisms in the DNA repair genes, XRCC1, APE1, and XPD and acute side effects of radiotherapy in breast cancer patients. Clin Cancer Res. 2005;11:4802–9. doi: 10.1158/1078-0432.CCR-04-2657. [DOI] [PubMed] [Google Scholar]
- 48.Zschenker O, Raabe A, Boeckelmann IK, et al. Association of single nucleotide polymorphisms in ATM, GSTP1, SOD2, TGFB1, XPD and XRCC1 with clinical and cellular radiosensitivity. Radiother Oncol. 2010;97:26–32. doi: 10.1016/j.radonc.2010.01.016. [DOI] [PubMed] [Google Scholar]
- 49.Zhou L, Xia J, Li H, et al. Association of XRCC1 variants with acute skin reaction after radiotherapy in breast cancer patients. Cancer Biother Radiopharm. 2010;25:681–5. doi: 10.1089/cbr.2010.0811. [DOI] [PubMed] [Google Scholar]
- 50.Langsenlehner T, Renner W, Gerger A, et al. Association between single nucleotide polymorphisms in the gene for XRCC1 and radiation-induced late toxicity in prostate cancer patients. Radiother Oncol. 2011;98:387–93. doi: 10.1016/j.radonc.2011.01.021. [DOI] [PubMed] [Google Scholar]
- 51.Damaraju S, Murray D, Dufour J, et al. Association of DNA repair and steroid metabolism gene polymorphisms with clinical late toxicity in patients treated with conformal radiotherapy for prostate cancer. Clin Cancer Res. 2006;12:2545–54. doi: 10.1158/1078-0432.CCR-05-2703. [DOI] [PubMed] [Google Scholar]
- 52.Iannuzzi CM, Atencio DP, Green S, et al. ATM mutations in female breast cancer patients predict for an increase in radiation-induced late effects. Int J Radiat Oncol Biol Phys. 2002;52:606–13. doi: 10.1016/s0360-3016(01)02684-0. [DOI] [PubMed] [Google Scholar]
- 53.Angèle S, Romestaing P, Moullan N, et al. ATM haplotypes and cellular response to DNA damage: association with breast cancer risk and clinical radiosensitivity. Cancer Res. 2003;63:8717–25. [PubMed] [Google Scholar]
- 54.Ho AY, Fan G, Atencio DP, et al. Possession of ATM sequence variants as predictor for late normal tissue responses in breast cancer patients treated with radiotherapy. Int J Radiat Oncol Biol Phys. 2007;69:677–84. doi: 10.1016/j.ijrobp.2007.04.012. [DOI] [PubMed] [Google Scholar]
- 55.Fogarty GB, Muddle R, Sprung CN, et al. Unexpectedly severe acute radiotherapy side effects are associated with single nucleotide polymorphisms of the melanocortin-1 receptor. Int J Radiat Oncol Biol Phys. 2010;77:1486–92. doi: 10.1016/j.ijrobp.2009.07.1690. [DOI] [PubMed] [Google Scholar]


