Abstract
Bacteriophage Sf6 DNA packaging series initiate at many locations across a 2 kbp region. Our in vivo studies that show that Sf6 small terminase subunit (TerS) protein recognizes a specific packaging (pac) site near the center of this region, that this site lies within the portion of the Sf6 gene that encodes the DNA-binding domain of TerS protein, that this domain of the TerS protein is responsible for the imprecision in Sf6 packaging initiation, and that the DNA-binding domain of TerS must be covalently attached to the domain that interacts with the rest of the packaging motor. The TerS DNA-binding domain is self-contained in that it apparently does not interact closely with the rest of the motor and it binds to a recognition site that lies within the DNA that encodes the domain. This arrangement has allowed the horizontal exchange of terS genes among phages to be very successful.
Keywords: bacteriophage Sf6, DNA packaging, bacteriophage P22, small terminase subunit, TerS
Introduction
The virions of tailed bacteriophages and other large dsDNA viruses contain a highly compacted nucleic acid molecule (Casjens, 1997). During the assembly of such virions, an ATP cleavage powered protein nanomotor pumps the DNA into a preformed capsid protein shell called a procapsid (reviewed in Casjens, 2011; Feiss and Rao, 2012). This DNA translocating ATPase is called the large terminase subunit (TerL), and it moves the dsDNA through a dodecameric ring of portal protein subunits that is present at one icosahedral vertex of the procapsid. Terminase protein's name derives from the fact that many of large dsDNA viruses replicate DNA into overlength concatemers of the genome sequence, and in these cases TerL also carries a nuclease activity that cuts this long DNA to virion size, thus creating the termini of the mature viral DNA chromosome. Recent single-particle optical tweezer experiments have produced new information that has allowed the building of rather detailed mechanistic models for the mechanism of action of this translocase, and these models are being tested (Duffy and Feiss, 2002; Kondabagil, Zhang, and Rao, 2006; Oliveira, Henriques, and Tavares, 2006; Tsay et al., 2009 and 2010; Casjens, 2011; Feiss and Rao, 2012). In a number of such viruses a second protein, called the small terminase subunit (TerS), has been implicated in the initiation of DNA packaging and in the choice of DNA to be packaged in the virion. In these cases TerS proteins recognize a specific site in the phage DNA, but their detailed mechanism of action is very poorly understood (Jackson, Laski, and Andres, 1982; Shinder and Gold, 1988; Casjens et al., 1992a; Chai, Kruft, and Alonso, 1994).
The tailed phage packaging motor usually has three protein components, TerS, TerL and the portal protein which forms the hole through which DNA enters the procapsid. Portal proteins may in some cases also have roles in sensing when the capsid shell is full of DNA (Casjens et al., 1992b; Tavares et al., 1992), in controlling the shape of the coat protein shell (Camacho et al., 1977; Black, Showe, and Steven, 1994) and in controlling the conformational change (expansion) that capsid shells undergo during maturation (Ray et al., 2009). TerL interacts with the portal protein ring of the procapsid in the cases where this interaction is understood, and genetic studies with phages λ and T3 suggest that the C-terminal portion of TerL interacts with portal protein (Frackman, Siegele, and Feiss, 1984; Sippy and Feiss, 1992; Morita, Tasaka, and Fujisawa, 1995; Yeo and Feiss, 1995a and 1995b), but other sequences have also been implicated in phage T4 TerL binding (Lin, Rao, and Black, 1999; Gao and Rao, 2011; Hegde et al., 2012). TerL and TerS also often interact in solution (Poteete and Botstein, 1979; Maluf, Yang and Catalano, 2005), although perhaps not in all phages (Al-Zahrani et al., 2009). In phage λ the C-terminal region of TerS is thought to bind to TerL (Frackman, Siegele, and Feiss, 1985; Yang et al., 1999b). Although atomic structures have recently been determined for several examples of each of the three motor protein subunits, the structure of the assembled, functioning motor is not yet understood.
Although the DNA packaging motor proteins appear to be evolutionarily conserved in spite of having a huge extant diversity in the different tailed phages that have been studied, portal and TerL proteins are universally encoded by tailed phages and are their most highly conserved proteins (Casjens, 2003 and 2008). Portal proteins are always found as dodecameric rings that replace five coat protein subunits at the tail vertex of phage heads, and the x-ray structures of portal rings from the very distantly related phages ϕ29, SPP1 and P22 show that they all have the same basic central fold, in spite of the fact that their amino acid sequences are not convincingly similar (Simpson et al., 2000; Lebedev et al., 2007; Olia et al., 2011). They are quite variable in size and different phage portals can have different “accessory” domains (Tang et al., 2011). The TerL proteins are monomeric when not part of the motor, and existing evidence suggests that four or five TerL molecules participate in the assembled motor (reviewed in Casjens, 2011; Nemecek et al., 2007; Feiss and Rao, 2012). TerS sequence diversity is even larger than that seen among the TerL or portal proteins; BLASTp searches of the sequence database identify a number of apparently unrelated TerS protein families (Casjens and Thuman-Commike, 2011; S. Casjens, unpublished). Crystal structures of octamers of Shigella flexneri phage Sf6 TerS (Zhao et al., 2010), nonamers of Salmonella enterica phage P22 TerS (Roy et al., 2012), nonamers and decamers of Bacillus subtilis phage SF6 TerS (Buttner et al., 2012), and 11- and 12-mers of a fragment of Escherichia coli phage 44RR2 TerS (Sun et al., 2012), as well as the NMR structure of a dimer of a fragment of E. coli phage λ TerS (de Beer et al., 2002) have shown the following: (i) The oligomeric state of the purified TerS proteins varies among the tailed phages, but its assembly state in the complete motor is not known in any case. Thus, either the different motors can accommodate a variable number of TerS subunits, or the oligomeric structure of TerS in the functioning motor may be different from that of the purified TerS proteins. (ii) The TerS C-terminus, where its structure has been determined (in Sf6, SF6 and in part in P22), forms a tubular β-barrel that contains one peptide strand from each subunit. (iii) The Sf6 and SF6 TerS proteins have rather similar overall folds; these two phages are only extremely distantly related in spite of their unfortunately similar names. The P22 TerS fold is partly similar but not identical to these two proteins (see below). The relationships between these more complete structures to the fragment structures of λ and phage 44RR2 TerS structures are less clear (but see Gao and Rao, 2011). Nonetheless, in all of the TerS structures the N-terminal domain is largely helical and includes a helix-turn-helix motif that may be the DNA-binding portion of these proteins (Buttner et al., 2012; Roy et al., 2012; Zhao et al., 2012; Sun et al., 2012). Analysis of mutations of Sf6 TerS have indicated that its N-terminal domain is responsible for binding DNA nonspecifically in vitro (Zhao et al., 2010 and 2012). In apparent contradiction to this view, removal of twenty C-terminal amino acids, which make up most of the C-terminal tubular β-barrel domain of the P22 TerS, does not impair its oligomerization but does block its DNA binding capability (Nemecek et al., 2008; Roy et al., 2012). T4 TerS is dispensable in a T4 in vitro DNA packaging system (Al-Zahrani et al., 2009; Zhang et al., 2011), while in P22 and SPP1 TerS protein is required but specific recognition of the packaging target site is not required in vitro (Poteete and Botstein, 1979; Schmieger, 1984; Schmieger and Koch, 1987; Oliveira, Alonso, and Tavares, 2005). Thus much remains to be understood regarding small terminase subunit function and its role in DNA packaging motor initiation.
The well-studied phage P22 and its relative Sf6 are both members of the “P22-like” tailed phage group, and twelve different weakly homologous proteins build their very similar virions (Casjens et al., 2004; Casjens and Thuman-Commike, 2011; Parent et al., 2012). A major functional difference between these two phages lies in the initiation of DNA packaging. P22 recognizes a specific 22 bp pac site that programs the initiation of processive series of packaging events (Tye, Huberman, and Botstein, 1974; Jackson, Jackson, and Deans, 1978; Casjens et al., 1992a; Wu et al., 2002), and the DNA cleavage that initiates such a series occurs over a 120 bp region that surrounds the pac site (Casjens and Huang, 1982; Casjens et al., 1992a). TerL protein has nonspecific nuclease activity, and it is thought to make these cleavages (Nemecek et al., 2007; Roy and Cingolani, 2012). On the other hand, we found that Sf6 makes its initiation cleavages over a much larger approximately 1600 bp region (Casjens et al., 2004). The reason for this difference was unknown. We report here the localization of the Sf6 pac site near the center of the region in which packaging initiation ends are generated and that the Sf6 TerS subunit is responsible for the large spread in initiation cleavage sites. In addition, genetic analysis of the Sf6 TerS protein supports the notion that this TerS protein has substantial flexibility and interacts with DNA through its N-terminal domain and with TerL through is C-terminal domain.
Results
Horizontal transfer of terS genes among the P22-like phages
There were, as of December 1, 2012, 152 available complete or nearly complete genome sequences of P22-like phages and prophages (this group of phages is defined as in Casjens and Thuman-Commike, 2011). The coat proteins of these phages fall into three major sequence types that are typified by Salmonella phage P22, Shigella phage Sf6 and E. coli phage CUS-3 (see figure 4 of Casjens and Thuman-Commike, 2011). On the other hand, there are six very different TerS “sequence types” that are mostly not convincingly related to one another in amino acid sequence. Figure 1 shows a ClustalW (Larkin et al., 2007) amino acid sequence neighbor-joining tree the of representatives of these TerS types. All of the sequences in figure 1 except the putative ϕSG1 TerS have amino acid sequences that are weakly related to known TerS proteins. (The putative ϕSG1 TerS has no recognizable homology to any protein in the database, but it was included in this analysis because its gene lies in the position at which all other P22-like phages carry their terS genes.) Comparisons of the six types of P22-like TerS proteins typically show less than 15% amino acid sequence identity between types, and simple BLASTp (Altschul et al., 1997) searches with one type often do not find matches in the other groups. The correlation between coat types and TerS types in these genomes is poor; for example different phages with the P22 type coat protein encode TerS proteins of the P22, Ugan1 or CUS-3 types, and different phages with CUS-3 type coat protein have CUS-3, P22, Sf6 or Ugan1 type TerS proteins. Clearly there has been extensive horizontal exchange of terS genes relative to coat protein genes within the P22-like phage group.
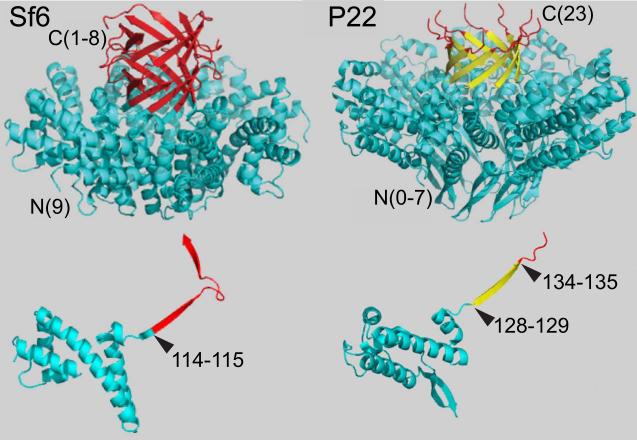
Figure 4. Structures of the Sf6 and P22 TerS proteins.
Phage Sf6 (left) and P22 (right) TerS protein structures are shown as ribbon diagrams (Protein Data Bank ID codes PDB 3HEF and 3P9A, respectively). The native octamer for Sf6 and nonamer for P22 are shown above and single subunits are shown below (Zhao et al., 2010; Roy et al., 2012). The locations of the N- and C-termini in the structures are indicated by “N” and “C”, respectively, and the numbers in parentheses indicate the numbers of flexible terminal amino acids that were not seen in the structures even though they were present in the crystals (different subunits have different numbers of such “missing” amino acids at the Sf6 C-terminus and the P22 N-terminus). In the P22 terS Sf6-hybA and -hybB hybrid phages, the portion of the Sf6 protein shown in blue replaces the P22 blue or blue plus yellow sections, respectively; in the P22 single subunit, the fusion points for P22 Sf6-hybA and Sf6-hybB are shown by color changes and indications of the P22 amino acids of the hybrid proteins between which the fusion took place.
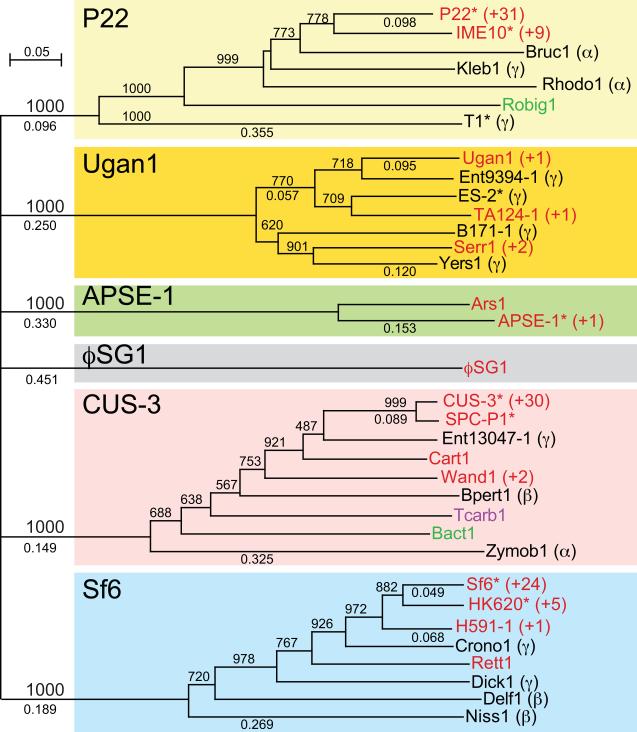
Figure 1. Neighbor-joining tree of TerS proteins of the P22-like phages.
The amino acid sequences of the P22-like TerS proteins were limited to the N-terminal DNA binding domain, and did not contain their short C-terminal domain (such a comparison is dominated by the much larger N-terminal domain and the tree shown is very similar to the tree of the whole proteins; see text). Trees were constructed by Clustal X2 in a Macintosh computer (Larkin et al., 2007), with horizontal branch lengths (numbers between 0 and 1) indicating the fractional amino acid sequence difference, and bootstrap support out of 1000 trials indicated by numbers between 1 and 1000. The very weakly supported deep branching order of the six major TerS types was collapsed, so that these six branches emanate from a single point. The major branches (highlighted with different colored boxes) are depicted on a tree for ease of discussion, in spite of the fact that the six branches may in fact not be homologous; the name of each branch (derived from a typical member) is shown in the upper left corner of the box. The name of the phage or prophage that carries each TerS protein is indicated to the right of each branch tip; functional phages are marked with an asterisk (*). TerS sequences that are nearly identical to those named on the right were collapsed to make the tree more legible, and the number removed is indicated after a plus sign (+) to the right of the phage or prophage name. The colors of the names indicate the following: Red, P22-like phages that infect members of the Enterobacteriaceae bacterial family; Black, non-P22-like phages that infect members of the Proteobacteria phylum, and the host's class within this phylum is indicated in parentheses (e.g., α for Alphaproteobacteria, β for Betaproteobacteria, etc.); Green, non-P22-like prophages whose hosts are members of the Bacteroidetes phylum; Purple, non-P22-like prophage whose host is a member of the Firmicutes phylum. The species of the host and GenBank locus_tag of the TerS proteins in the figure are given in Tables S1 and S2.
Figure 1 also shows that four of the above TerS types include proteins encoded by phages outside of the P22-like group. For example, the TerS type exemplified by phage CUS-3 (pink box in figure 1) includes TerS proteins encoded by prophage Bpert1 in a Bordetella pertussis genome (a Betaproteobacteria), prophage Zymob1 in a Zymomonas mobilis genome (an Alphaproteobacteria), prophage Tcarb1 in a Thermosinus carboxydivorans genome, and prophage Bact1 in a Bacteroides species genome (see Tables S1 and S2 for details of these bacterial hosts and prophage terS genes). The first two of these bacterial hosts are in the Proteobacteria phylum but reside in different taxonomic classes from the Gamma-Proteobacteria hosts of the P22-like phages, the third is in the Firmicutes phylum, and the last is in the Bacteriodetes phylum. These four prophages are not P22-like in either the organization of their virion assembly genes or the sequence of the encoded proteins (not shown). Similarly, bone fide tailed phages T1 and ES-2 have TerS proteins that reside in the P22 and Ugan1 families, respectively. T1 infects E. coli and is a member of the Siphoviridae with a long, noncontractile tail (German, Misra, and Kropinski, 2006), and ES-2 infects Cronobacter sakazakii and is a member of the Myoviridae with a long contractile tail (Lee, Park, and Chang, 2011). The fact that these proteins encoded by other phage types are present inside four of the branches of the P22-like TerS tree means that no matter where the root of the tree actually lies, such “outsiders” are present inside at least three of the P22-like branches.
Not all phage genes are exchanged at this frequency. Sub-nanometer resolution 3-dimensional cryoelectron microscopic reconstructions of virions have been determined for three phages that typify the three types of coat proteins in the P22-like group, P22 (Jiang et al., 2003; Parent et al., 2010; Tang et al., 2011), Sf6 (Parent et al., 2012) and CUS-3 (K. Parent, T. Baker, E. Gilcrease and S. Casjens, unpublished). These structures show that all three coat proteins have a phage HK97 coat protein polypeptide fold (Wikoff et al., 2000) that is embellished with an “extra” telokin-like domain at the same location in the protein in all three cases. This domain is not present in this location in other phage coat proteins whose structures are known, indicating that the P22-like coat proteins have not been subject to horizontal exchange from outside the P22-like group and have diverged within this group from a common ancestor (Parent et al., 2012). In addition, portal and scaffolding proteins are, like coat protein, present as three major types in the P22-like phages, but these types correlate perfectly with the coat protein types; thus, coat, portal and scaffolding protein genes have not been shuffled by evolutionary exchange (Casjens and Thuman-Commike, 2011). We conclude that horizontal exchange of terS genes must have occurred among the different P22-like phages, as well as between this group and other tailed phage types, while the procapsid assembly (coat, portal and scaffolding protein) genes have not enjoyed such free exchange.
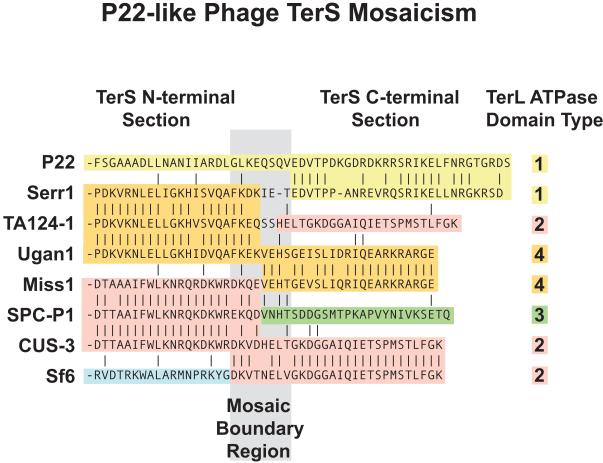
The tailed phages are well known for having mosaic genomes, and while analyzing a much smaller number of the P22-like phage terS genes, we discovered that the locations of the boundaries between “mosaic sections” that were formed during the exchanges of terS genes are correlated with protein domain boundaries. Mosaic boundaries near the C-terminus, but within the terS gene could be identified in several P22-like phages (Casjens and Thuman-Commike, 2011). There are now nearly three times as many P22-like phage genome sequences known, and among these we identify eight different situations in which the location of the TerS mosaic boundary can be quite accurately located (other sequence types exist but the mosaic boundary cannot be located because no relatives with shuffled TerS domains have been found to date). Figure 2 shows that these eight different mosaic boundaries are all present within a small region, and that four different N-terminal TerS domains have been extensively shuffled relative to four different C-terminal domains at these boundaries. The sizes of these small C-terminal domains range from 23 amino acids in Ugan1 to 28 in P22. We also note that in all of the >150 P22-like phage genomes examined to date, the different C-terminal TerS protein section types correlate perfectly with several different TerL N-terminal domain sequence types (right column, figure 2), suggesting that evolutionary shuffling events that create phages with different combinations of TerS C-terminal section and TerL protein types have not survived. Since P22 TerS and TerL are known to form a mixed oligomeric protein complex (Poteete and Botstein, 1979; Roy et al., 2012), this failure to survive is most easily explained if the C-terminal domain of TerS is important for the interaction between TerS and TerL (Casjens and Thuman-Commike, 2011).
Figure 2. TerS C-terminal amino acid sequence relationships.
The amino acid sequence relationships surrounding the C-terminal and N-terminal mosaic sectional boundaries of the TerS proteins of eight P22-like phages are shown. The same background color denotes similar sequences; each sequence ends on the right at the C-terminus of the protein (the sequence relationships of the remainder of the N-terminal sections are similar to the portions shown). The numbers in the right column represent the sequence type of the TerL N-terminal ATPase domain of each phage (Casjens and Thuman-Commike, 2011; S. Casjens, unpublished). Table S2 gives the Genbank locus_tags for these terS genes.
TerS functional domains
In order to begin to test the idea, derived from the above comparative genomic analysis, that the C-terminal domain of TerS is responsible for its interaction with TerL, we created new phage genome constructs in which the N-terminal TerS domain from phage Sf6 replaces the parallel domain of phage P22 and tested their functionality. Phage Sf6 TerS was chosen because its amino acid sequence is essentially unrelated to that of P22 TerS (the two proteins are only 11.7% identical and the few identities are scattered throughout the protein alignment), and its crystal structure has been solved (Zhao et al., 2010). This combination of N-terminal TerS domain (blue background in figure 2) and terL (yellow background in figure 2) has not been found in nature. We performed these replacements in a P22 prophage, since virion assembly is not required for maintenance of a prophage, and therefore mutations lethal for virion assembly or function can be constructed. The resulting prophages can be induced to lytic growth to test for successful DNA packaging and virion assembly. We previously constructed a P22 sieA−Δ1, 15−ΔS302∷KanR, 13−amH101 in which the three mutations allow efficient tailspike gene expression after induction, kanamycin selection for lysogens, and control of lysis, respectively (Cortines et al., 2011; Padilla-Meier et al., 2012). This prophage, present in sup° S. enterica serotype Typhimurium LT2 strain UB-1791 (all bacterial and phage strains are listed in table 1), produces fully-tailed, plaque-forming virions after induction, and it was used in all genetic manipulations of the terS gene described here. In short, we first replaced the native bacterial galK gene with a tetracycline resistance cassette (TetRA) (Karlinsey, 2007). Then, galK recombineering (Warming et al., 2005) was used to replace part of the terS gene of the P22 prophage with the E. coli galK gene expression cassette from plasmid pGalK (Warming et al., 2005), and this galK cassette was in turn replaced by the desired part of the Sf6 terS gene (details in Materials and Methods).
Table 1.
Bacteria and bacteriophage strains used in this study
| Name | Genotypea | Source |
|---|---|---|
| Salmonella enterica serovar Typhimurium LT2 | ||
| UB-0002 | (DB7004) leuA am414, supE | Winston, Botstein, and Miller, 1979 |
| UB-0020 | (MS1868) leuA am414, Fels2−, r−, m+, sup°; from K. Hughes | Youderian et al., 1983 |
| UB-0134 | leuA−am414, Fels2−, cob−ΔCRR299 (P22 sieA−44, ant−am222, ΔAp68 [tpfr49 a1−, 9−, c2+, mnt+]); from J. Roth | Youderian, Chadwick, and Susskind, 1982 |
| UB-1760 | (TT23216) LT2 terY2∷CamR; from J. Roth | Kulesus et al., 2008 |
| UB-1766 | (TT25401) LT2 CRR2061(zfa-9223∷kan,zfa-9228∷TetRA Peut) eut-38∷MudA; from J. Roth | Pimkin, Pimkina, and Markham, 2009 |
| UB-1790 | UB-0020 galK∷TetRA-1 (P22 13-amH101, 15-Δsc302∷KanR, sieA-Δ1) | Padilla-Meier et al., 2012 |
| UB-1958 | UB-0020 galK∷TetRA-1 (P22 13-amH101, 15-Δsc302∷KanR, 3∷Sf6-hybG, sieA-Δ1) | This report |
| UB-1960 | UB-0020 galK∷TetRA-1 (P22 13-amH101, 15-Δsc302∷KanR, 3∷Sf6-hybJ, sieA-Δ1) | This report |
| UB-1961 | UB-0020 galK∷TetRA-1 (P22 13-amH101, 15-Δsc302∷KanR, 3∷galK-1, sieA-Δ1)/pKD46 | This report |
| UB-1982 | UB-0020 galK+, CamR-1, TetRA-2 | This report |
| UB-1985 | UB-0020 galK∷P22pacL, CamR-1, TetRA-2 | This report |
| UB-1988 | UB-0020 galK∷P22pacL, CamR-1, TetRA-2 (P22 13-amH101, 15-Δsc302∷KanR, sieA-Δ1) | This report |
| UB-1991 | UB-0020 galK∷P22pacR, CamR-1, TetRA-2 | This report |
| UB-2019 | UB-0020 galK∷TetRA-1 (P22 13-amH101, 15-Δsc302∷KanR, 3∷Sf6-hybA, sieA-Δ1) | This report |
| UB-2021 | UB-0020 galK∷TetRA-1 (P22 13-amH101, 15-Δsc302∷KanR, 3∷Sf6-hybD, sieA-Δ1) | This report |
| UB-2022 | UB-0020 galK∷TetRA-1 (P22 13-amH101, 15-Δsc302∷KanR, 3∷Sf6-hybE, sieA-Δ1) | This report |
| UB-2023 | UB-0020 galK∷TetRA-1 (P22 13-amH101, 15-Δsc302∷KanR, 3∷Sf6-hybF, sieA-Δ1) | This report |
| UB-2024 | UB-0020 galK∷TetRA-1 (P22 13-amH101, 15-Δsc302∷KanR, 3∷Sf6-hybC, sieA-Δ1) | This report |
| UB-2033 | UB-0020 galK∷TetRA-1 (P22 13-amH101, 15-Δsc302∷KanR, 3∷Sf6-hybI, sieA-Δ1) | This report |
| UB-2040 | UB-0020 galK∷TetRA-1 (P22 13-amH101, 15-Δsc302∷KanR, 3∷Sf6-hybB, sieA-Δ1) | This report |
| UB-2041 | UB-0020 galK∷TetRA-1 (P22 13-amH101, 15-Δsc302∷KanR, 3∷Sf6-hybK 3- D115A, V116A, T117A, sieA-Δ1) | This report |
| UB-2042 | UB-0020 galK∷TetRA-1 (P22 13-amH101, 15-Δsc302∷KanR, 3∷Sf6-hybL 3- P118A, D119a, K120A, sieA-Δ1) | This report |
| UB-2043 | UB-0020 galK∷TetRA-1 (P22 13-amH101, 15-Δsc302∷KanR, 3∷Sf6-hybM 3- G121A, D122A, R123A, sieA-Δ1) | This report |
| UB-2044 | UB-0020 galK∷TetRA-1 (P22 13-amH101, 15-Δsc302∷KanR, 3∷Sf6-hybN 3- D124A, K125A, R126A, sieA-Δ1) | This report |
| UB-2045 | UB-0020 galK∷TetRA-1 (P22 13-amH101, 15-Δsc302∷KanR, 3∷Sf6-hybO 3- R127A, S128A, R129A, sieA-Δ1) | This report |
| UB-2046 | UB-0020 galK∷TetRA-1 (P22 13-amH101, 15-Δsc302∷KanR, 3∷Sf6-hybP 3- I130A, K131A, E132A, sieA-Δ1) | This report |
| UB-2047 | UB-0020 galK-Δ1, CamR-1, TetRA-2 | This report |
| UB-2071 | UB-0020 galK∷TetRA-1 (P22 13-amH101, 15-Δsc302∷KanR, 3∷Sf6-hybQ 3- L133A, F134A, N135A, sieA-Δ1) | This report |
| UB-2072 | UB-0020 galK∷TetrA-1 (P22 13-amH101, 15-Δsc302∷KanR, 3∷Sf6-HybR 3- D113A, K114A, sieA-Δ1) | This report |
| UB-2073 | UB-0020 galK∷TetRA-1 (P22 13-amH101, 15-Δsc302∷KanR, 3∷Sf6-HybS 3- E115A, V116A, sieA-Δ1) | This report |
| UB-2074 | UB-0020 galK∷TetrA-1 (P22 13-amH101, 15-Δsc302∷KanR, 3∷Sf6-hybT SGSG inserted between K114 and E115, sieA-Δ1) | This report |
| UB-2075 | UB-0020 galK∷TetRA-1 (P22 13-amH101, 15-Δsc302∷KanR, 3∷Sf6-hybU SGSGSGSG inserted between K114 and E115, sieA-Δ1) | This report |
| UB-2093 | UB-0020 galK+, CamR-1, TetRA-2 (P22 13-amH101, 15-Δsc302∷KanR, 3∷Sf6-hybA, sieA-Δ1) | This report |
| UB-2094 | UB-0020 galK∷P22pacR, CamR-1, TetRA-2 (P22 13-amH101, 15-Δsc302∷KanR, 3∷Sf6-hybA, sieA-Δ1) | This report |
| UB-2095 | UB-0020 galK∷Sf6pac3L, CamR-1, TetRA-2 (P22 13-amH101, 15-Δsc302∷KanR, 3∷Sf6-hybA, sieA-Δ1) | This report |
| UB-2099 | UB-0020 galK-Δ1, CamR-1, TetRA-2 (P22 13-amH101, 15-Δsc302∷KanR, 3∷Sf6-hybA, sieA-Δ1) | This report |
| UB-2100 | UB-0020 galK:Sf6pac5R, CamR-1, TetRA-2 (P22 13-amH101, 15-Δsc302∷KanR, 3∷Sf6-hybA, sieA-Δ1) | This report |
| UB-2101 | UB-0020 galK∷Sf6pAc1L, CamR-1, TetRA-2 (P22 13-amH101, 15-Δsc302∷KanR, 3∷Sf6-hybA, sieA-Δ1) | This report |
| UB-2102 | UB-0020 galK∷Sf6pac4R, CamR-1, TetRA-2 (P22 13-amH101, 15s-Δsc302∷KanR, 3∷Sf6-hybA, sieA-Δ1) | This report |
| UB-2103 | UB-0020 galK∷Sf6pac3R, CamR-1, TetRA-2 (P22 13-amH101, 15-Δsc302∷KanR, 3∷Sf6-hybA, sieA-Δ1) | This report |
| UB-2104 | UB-0020 galK∷Sf6pac1R, CamR-1, TetRA-2 (P22 13-amH101, 15-Δsc302∷KanR, 3∷Sf6-hybA, sieA-Δ1) | This report |
| UB-2105 | UB-0020 galK∷Sf6pac2R, CamR-1, TetRA-2 (P22 13-amH101, 15-Δsc302∷KanR, 3∷Sf6-hybA, sieA-Δ1) | This report |
| UB-2106 | UB-0020 galK∷Sf6frag7, CamR-1, TetRA-2 (P22 13-amH101, 15-Δsc302∷KanR, 3∷Sf6-hybA, sieA-Δ1) | This report |
| UB-2118 | UB-0020 galK-Δ1, CamR-1, TetRA-2 (P22 13-amH101, 15-Δsc302∷KanR, sieA-Δ1) | This report |
| UB-2119 | UB-0020 galK+, CamR-1, TetrA-2, (P22 13-amH101, 15-Δsc302∷KanR, sieA-Δ1) | This report |
| UB-2120 | UB-0020 galK∷P22pacR, CamR-1, TetRA-2 (P22 13-amH101, 15-Δsc302∷KanR, sieA-Δ1) | This report |
| UB-2121 | UB-0020 galK∷Sf6pac1L, CamR-1, TetRA-2 (P22 13-amH101, 15-Δsc302∷KanR, sieA-Δ1) | This report |
| UB-2123 | UB-0020 galK∷Sf6pac2R, CamR-1, TetRA-2 (P22 13-amH101, 15-Δsc302∷KanR, sieA-Δ1) | This report |
| UB-2124 | UB-0020 galK∷Sf6pac1R, CamR-1, TetRA-2 (P22 13-amH101, 15-Δsc302∷KanR, sieA-Δ1) | This report |
| Escherichia coli K-12 | ||
| UB-0049 | (NF1829) araD-139, Δ7679(araABOIC-,leu-), galUK-, lac-ΔX74, rspL-, thi- / F' lac Iq1, lac Z∷Tn5(KanR), lacY+ | Shultz et al., 1982 |
| Shigella flexneri | ||
| UB-1458 | PE577; gift of R. Morona | Casjens et al., 2004 |
| UB-1469 | PE577 (Sf6); gift of R. Morona | This report |
| UB-1564 | PE577 (Sf6 62∷KanR) | This report |
| Bacteriophages | ||
| UC-911 | P22 13-amH101, 15-Δsc302∷KanR, sieA-Δ1 | Padilla-Meier et al., 2012 |
| UC-920 | Sf6 63∷pac'0 (plasmid pPP311 XhoI-NcoI cloning region with no insert; see text) | This report |
| UC-921 | Sf6 63∷pac'A (Sf6 bp 1–200) | This report |
| UC-922 | Sf6 63∷pac'B (Sf6 bp 149–424) | This report |
| UC-923 | Sf6 63∷pac'C (Sf6 bp 200–424) | This report |
| UC-924 | Sf6 63∷pac'D (Sf6 bp 140 to 210) | This report |
| UC-925 | Sf6 63∷pac'E (Sf6 (bp 149–225) | This report |
| UC-929 | P22 13-amH101, 15-Δsc302∷KanR, 3∷Sf6-hybA, sieA-Δ1 | This report |
| UC-930 | P22 13-amH101, 15-Δsc302∷KanR, 3∷Sf6-hybB, sieA-Δ1 | This report |
Strain names in parentheses are the names used in the laboratory from which the strain was obtained. Strain name UB-0020 in middle column indicates that the strain also carries the UB-0020 alleles. Amino acid numbers refer to the amino acids of the Sf6-hybB TerS protein.
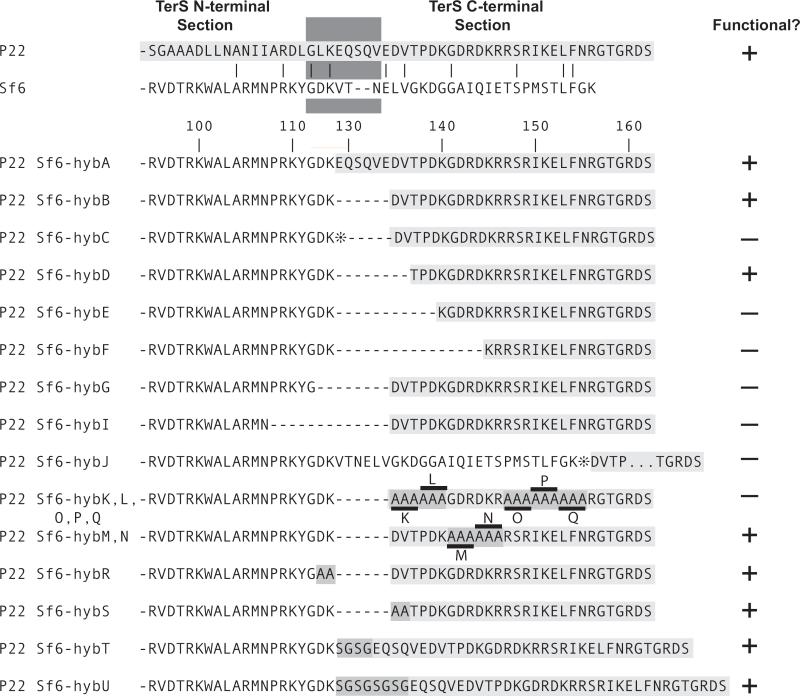
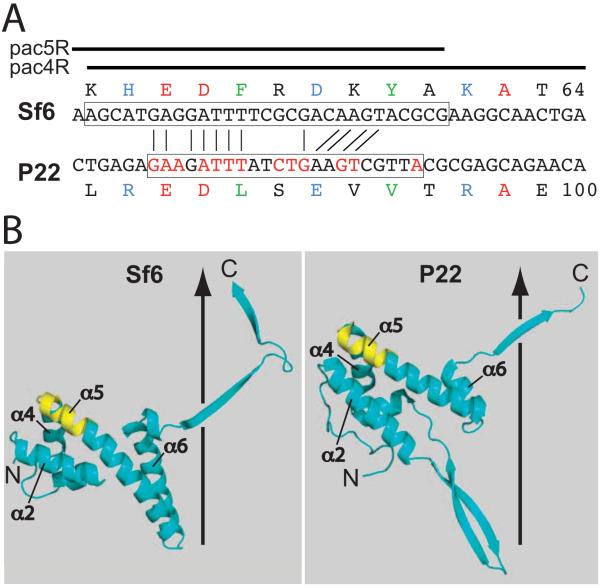
Two prophage constructs with hybrid terS genes, P22 Sf6-hybA and P22 Sf6-hybB, whose hybrid junctions are shown in figure 3, gave yields of plaque-forming phages upon induction to lytic growth with Mitomycin C that were very similar to the parental P22 prophage with its fully P22 terS gene. In these two hybrid phages Sf6 terS codons 1–114 replace codons 1–128 or 1–134 of P22 terS; in both cases the Sf6 terS sequence is fused translationally in-frame to the remaining C-terminal P22 terS sequences. The observation that these two hybrid phages are functional is perhaps somewhat surprising, since the isolated P22 TerS protein is a nonamer ring (Nemecek et al., 2008; Roy et al., 2012), and the Sf6 TerS forms an octamer ring (Zhao et al., 2010). Figure 4 shows ribbon diagrams of both oligomers and single subunits of the oligomers; these structures show that the mosaic junction of the Sf6-hybA TerS protein correspond precisely to the boundary between the N-terminal globular domain and the C-terminal β-barrel domain of both proteins. The oligomeric state of the hybrid proteins has not been determined. Nonetheless, the functionality of these two hybrid phages allows two initial conclusions to be drawn. (i) Sf6 TerS amino acids 1–114 are sufficient, when fused to amino acids 137–162 of P22 TerS, to supply TerS function to phage P22. It seems very likely that, like P22 itself, all the P22-like phages including Sf6 are dependent upon a pac site that is recognized by TerS. If so, (ii) the Sf6 pac site must lie within the first 114 codons of the Sf6 terS gene (see below).
Figure 3. Phage P22-Sf6 hybrid TerS proteins.
The upper two amino acid sequences are the C-termini of the TerS proteins of phages P22 (light gray background) and Sf6 (white background). The dark gray box above marks the approximate division between the two functional domains of the TerS proteins (see text). Below, the junctions are shown for the various P22-Sf6 hybrid TerS proteins discussed in the text; the numbers above indicate the amino acids of the Sf6 protein (left of junction) and P22 protein (right of junction). Medium gray backgrounds indicate amino acid changes or insertions, and asterisks (①) denote UGA stop codons. The rightmost column indicates the functionality of the hybrid TerS proteins as determined by the phage titer on Salmonella UB-0002 of the lysate 3 hr after induction by the addition of 1.5 μg/ml carbadox of each prophage containing strain at a cell density of 2×108/ml; a yield of 1×1010-2×1011 phage/ml indicated a functional protein (+) and yields <105/ml indicated a nonfunctional protein (−).
In order to test whether the covalent connection between the N-terminal Sf6 domain and the C-terminal P22 domain is essential, a UGA stop codon was engineered between the two TerS domains of prophage P22 terS Sf6-hybB to create P22 terS Sf6-hybC (figure 3). In addition, a prophage carrying the TerS Sf6-hybJ was constructed in which the entire 140-codon Sf6 terS gene is present with a stop codon separating it from the P22 C-terminal region. In both of these the progeny yield upon induction is >106-fold lower than the P22 terS Sf6-hybB parent. The nonfunctionality of these two phages indicates that neither the free Sf6 TerS N-terminal domain nor the full-length Sf6 TerS protein can supply TerS function to the P22 packaging apparatus, and that the covalent connection between the N-terminal Sf6 TerS domain and the C-terminal P22 TerS domain is essential. These observations support the idea that the TerS C-terminal domain is required to attach the TerS N-terminal domain to the rest of the DNA packaging apparatus.
The Sf6-hybB terS gene was trimmed from the Sf6-P22 junction point to determine how much of the Sf6 N-terminal domain and of the P22 C-terminal domain are required at the fusion point to generate a functional hybrid TerS protein. Deletion of two of the P22 amino acids (P22 D135 and V136 of Sf6-hybD TerS in UB-2021) from the C-terminal side of the hybrid junction resulted in a functional phage, but removal of three (P22 T137, P138 and D139 in Sf6-hybE TerS in UB-2022) or eight (Sf6-hybF TerS in UB-2023) additional P22 amino acids resulted in nonfunctional phages (amino acid numbers are as shown in figure 3). Removal of two amino acids, D113 and K114, from the Sf6 (N-terminal) side of the Sf6-hybB junction resulted in the nonfunctional phage P22 terS Sf6-hybG (UB-1958), as did removal of seven amino acids (P22 terS Sf6-hybI in UB-2033) (figure 3). Comparison of Sf6-hybK and -hybS TerS proteins (below) indicates that the third amino acid of the P22 part of Sf6-hybB TerS, T137, is important. Thus, removal or alteration of two or more amino acids from the Sf6 side or three or more amino acids from the P22 side of the Sf6-hybB junction drastically reduces the in vivo activity of TerS. To determine what parts of the P22 portion of the functional Sf6-hybB protein might be most important for its function, triple alanine substitutions were made that replace sets of three adjacent codons between P22 codons 135 and 155 of the Sf6-hybB terS gene (Sf6-hybK through hybQ; similar changes could not be easily made in amino acids 156–162 because their codons overlap the terL gene). Of these mutants, changes from P22 TerS amino acids 138 to 143 did not inactivate the TerS protein, while changes in the 135 to 137 or 144 to 155 amino acid intervals did have a strong negative effect on phage yield after induction (figure 3). These results suggest that protein-protein (most likely TerS-TerL, above) contacts are not critical in the middle portion of the C-terminal domain of the Sf6-hybB TerS protein.
Since P22 Sf6-hybA and -hybB TerS proteins are both functional, it appeared that the length of the connection between the N-terminal DNA-binding domain and the essential parts of the C-terminal domain is not critical. We tested this by inserting four or eight “linker” amino acids between the Sf6 and P22 sequences of Sf6-hybA TerS protein to create P22 terS Sf6-hybT and -hybU (prophages of strains UB-2074 and -2075, respectively) (figure 3). Both of these changes result in plaque-forming phages, indicating that these TerS proteins with extended inter-domain linkers are functional. On the other hand the connection between the two TerS domains can apparently be too short. In P22 terS Sf6-hybG deletion of Sf6 TerS amino acids D113-K114 inactivates TerS, but in P22 terS Sf6-hybR (figure 3; strain UB-2072) replacement of D113-K114 with two alanine residues is functional. Thus, it appears that the inter-domain linker length rather than specific D113 and K114 side chain structure is required for TerS function. Comparison of the TerS proteins in P22 terS Sf6-hybD (two amino acids shorter than Sf6-hybB) and Sf6-hybU (fourteen amino acids longer than Sf6-hybB) show that the linker between the two domains can vary in length by at least 16 amino acids without inactivating the protein. We conclude that the precise spatial juxtaposition between the two TerS domains is much less important than the presence of a physical peptide backbone connection between them.
The Sf6 pac site
The hybrid phage experiments above suggest that the Sf6 pac site should lie within the first 112 codons of the Sf6 terS gene (bp 1–336 of the Sf6 genome; accession No. AF547987), since this is the only Sf6 sequence in the smallest functional hybrid P22 terS Sf6-hybR. To test this idea directly, we engineered inverted, nonadjacent duplications of sections of this region into a nonessential location of the Sf6 genome and tested their ability to program the initiation of packaging series. Figure 5 describes this strategy, which is patterned after our previous genetic analysis of the phage P22 pac site (Wu et al., 2002). Packaging series initiation DNA cleavage events can be visualized in agarose gels as the restriction fragment from the initiating end of the DNA of the first packaging event in a series; this fragment is called the “pac fragment,” and a packaging initiation cleavage near the pac site forms one end of the fragment and restriction endonuclease cleavage forms the other end (see Jackson, Jackson, and Deans, 1978; Casjens et al., 1992a; Wu et al., 2002). Thus, by analyzing whether or not a pac fragment is generated by a potential pac site, the packaging series initiating activity of such a site can be determined.
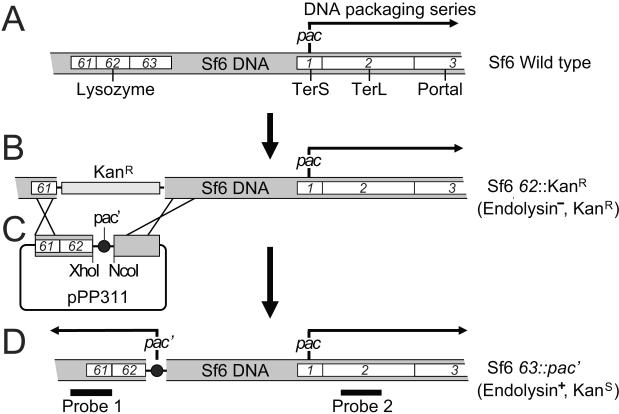
Figure 5. Building phage Sf6 genomes with two pac sites.
(A) The section of the Sf6 genome containing genes 61(holin)-62 (endolysin)-63 (lambda Rz homologue) and 1 (small terminase)-2 (large terminase)-3 (portal protein). (B) The same Sf6 genome section as in part A with a kanamycin resistance cassette replacing genes 62 and 63 (prophage of Shigella strain UB-1564). (C) Plasmid pPP311 carrying a sequence (represented by a solid black circle) to be tested for Sf6 pac site activity; see text for its construction. (D) Sf6 63∷pac' in which pac tester sequences from pPP311 derivatives have replaced the KanR cassette of the phage in part B. The black arrows indicates the direction of DNA packaging from an active pac' site replacing Sf6 gene 63 and from the native pac site in gene 1 (see text), and Southern probes 1 and 2 are defined in the text.
To generate Sf6 phages with duplicated sequences for such a test, we first constructed a defective Sf6 prophage in which a KanR cassette replaces the essential endolysin-encoding gene 62 (figure 5B and Materials and Methods). Second, a plasmid, pPP311, was constructed which carries a complete 62 gene, a multicloning site transcriptionally downstream of gene 62 into which a sequence to be tested for pac activity can be inserted, and additional Sf6 homology downstream of gene 62 (figure 5C). When this plasmid carrying a potential pac site in its multicloning site is transformed into a S. flexneri cell carrying the above Sf6 62∷KanR defective prophage, and the prophage is induced to lytic growth with mitomycin C, the only plaque-forming phages produced must have their KanR gene insertion replaced by the intact 62 gene and the adjacent potential pac site by homologous recombination with the plasmid; note that integration of the whole plasmid into the Sf6 genome makes it too large to be completely packaged. The orientation of such a potential pac site in the Sf6 genome is determined by its orientation in plasmid pPP311, and in the constructs studied here this orientation was designed to program any packaging series leftward on Sf6 DNA (figure 5D).
Sequences spanning various parts of the Sf6 terS gene were amplified using primers with XhoI and NcoI site-containing 3'-tails, and the resulting DNAs were inserted between the XhoI and NcoI sites of plasmid pPP311 so that their orientation was opposite to their native orientation in the terS gene (figure 5). These sequences were then moved into the Sf6 gene 63 region as described above. The resulting phages (strains UC-920 through UC-925; table 1) form plaques, so the inserted second pac site (called pac' hereafter) does not interfere with lytic growth (as is also the case with phage P22; Wu et al., 2002). DNA was isolated from the virions thus produced, cut with restriction endonuclease PmeI or BsrGI, and the fragments displayed by 0.8% agarose gel electrophoresis. The gel was stained with ethidium bromide and Southern probed with probe 1 (figure 6; probe 1 DNA was PCR amplified from the bp 35981–36507 region of the Sf6 genome, so that it hybridizes to any pac' fragments extending leftward from the XhoI-NcoI cloning site). The PmeI and BsrGI enzymes were chosen because they produce pac' fragments in the 1–4 kbp size range, where analysis of the width of the diffuse pac' fragment band is most accurately performed, and because their overlapping true restriction fragments are much larger than their pac' fragments and so will not interfere with the analysis. Figure 6A shows that, as expected, if nothing is inserted into the pPP311 cloning site, Southern analysis with probe 1 of the DNA from the resulting phage (UC-920) shows no leftward-extending pac' fragment, indicating that packaging series are not initiated on the DNA present between the XhoI and NcoI sites of pPP311 (or at other fortuitous sites in this region). On the other hand when Sf6 sequence A (Sf6 pac'A bp 1–200; figure 6A) was inserted between the XhoI and NcoI sites (phage UC-921), a diffuse band in the Southern autoradiograph is centered at about 1.7 kbp for the BsrGI digest and 3.5 kbp for the PmeI digest (figure 6A). The BsrGI-PmeI double digest diffuse band is identical to that in the BsrGI digest, showing that the variable end of this family of fragments is the right end and thus is where packaging initiated. The diffuse PmeI and BsrGI pac' bands extend from about 2.5 to 4.4 kbp and 3.0 to 0.9 kbp in the electrophoresis gel, respectively, and so both have measured widths of 2.0±0.1 kbp (figure 6). Probing BsrGI cut DNA with probe 2 (amplified from bp 761–1408 of Sf6 chromosome) showed only the diffuse pac fragment band (at ~6000 bp) initiating from the native pac site in the terS gene region (data not shown; see also Casjens et al., 2004). Insertion of Sf6 DNA fragment B (Sf6 pac'B bp 200–424) into the XhoI-NcoI insertion site did not show any pac' fragment (data not shown). This finding indicates that packaging series initiation is sequence-specific in that any Sf6 sequence at this position is not sufficient to cause initiation.
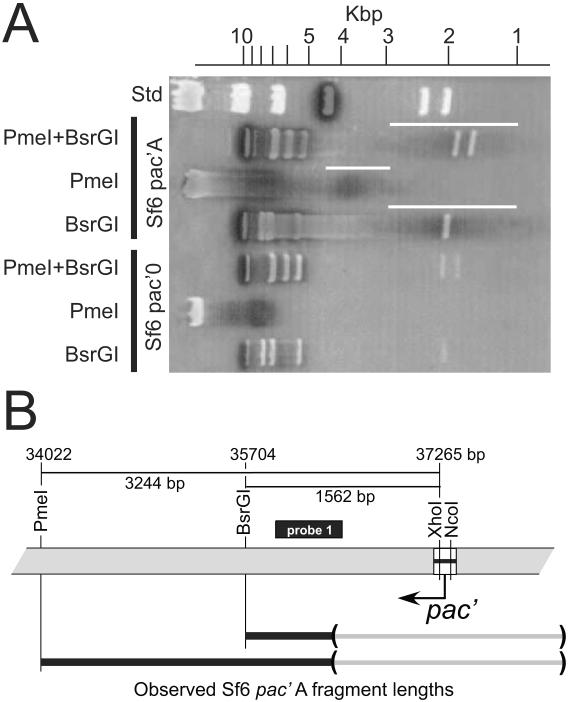
Figure 6. Pac fragments generated by a second pac site in phage Sf6.
(A) A 0.8% agarose electrophoresis gel of Sf6 pac'0 (strain UC-920) and Sf6 pac'A (UC-921) DNA cleaved with the indicated restriction enzymes (phage λ DNA cleaved with HindIII serve as size standards in the rightmost lane). White bands are the DNA fragments stained with ethidium bromide. Superimposed on the stained gel is an autoradiogram of the same gel probed with probe 1, a PCR amplified DNA fragment that extends from Sf6 bp 35981 to 36520 (hybridizing DNA shown in black; the Sf6 probe cross-reacts with one lambda DNA band as expected since the Sf6 and lambda holin genes are nearly identical). The white vertical lines to the right of the Sf6 pac'A lanes mark the width of the pac' fragment bands. (B) Above, a map of the gene 62-pac' region (see figure 5) of the Sf6 63::pac0 (UC-920) genome is shown with distances between relevant restriction sites. The location of Southern probe 1 is indicated by a black bar. Below, horizontal lines represent the observed pac fragments in part A that were generated by packaging initiation in the pac'A sequence. The left ends of these lines are anchored to the locations of the restriction sites that were cleaved to generate the fragments, and the parentheses at the right ends enclose the regions in which the right ends of these fragments occur.
The directionality of packaging from the pac'A sequence above is the same as we previously reported for the native Sf6 packaging, and its pac' fragment band width is similar to what was previously reported for normal packaging series initiation by wild type phage Sf6 (Casjens et al., 2004). The Sf6 pac site was located more precisely through the analysis of the ability of smaller Sf6 DNA regions to generate pac' fragments. These DNA fragments are shown in figure 7. Fragment C (Sf6 bp 149–424) gave rise to a pac' fragment of similar intensity to that from fragment A (data not shown). Since these findings suggested a region with pac site activity in the overlap between fragments A and C, shorter fragments D (bp 140 to 210) and E (bp 149–225) from this overlap region (figure 7) were also inserted at the XhoI-NcoI insertion site as described above to create phages UC-924 and UC-925, respectively. Both cause the generation of diffuse pac' fragments (data not shown). These observations strongly support the idea that these DNA sequences at the pac' site are programming packaging series initiation in a fashion that accurately reflects wild type initiation. We conclude from these experiments that the information for the Sf6 pac site lies in the bp 149–210 interval within the Sf6 terS gene.
Figure 7. DNA fragments tested for Sf6 pac activity.
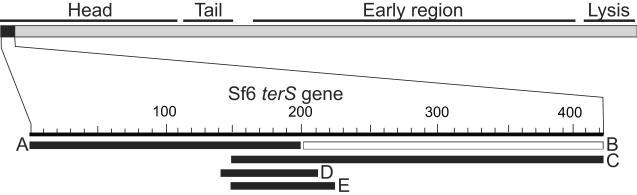
A map of the Sf6 genome is shown with its functional regions indicated above and the terS gene (gene 1) shown in black. Below, the DNA fragments (A through E, see text) that were test for Sf6 pac activity are indicated as horizontal bars for which black and white indicate activity and no activity, respectively (see text).
Because of technical difficulties in accurately analyzing such highly diffuse DNA “bands” in electrophoresis gels, we devised a second method for genetic identification of the Sf6 pac site. P22 that is growing lytically can initiate packaging on induced, defective prophages in the Salmonella chromosome (Weaver and Levine, 1978; Youderian et al., 1988); however, the sequences required for such initiation were not analyzed in detail. It therefore seemed possible that a pac site inserted into the host bacterium's chromosome would program a high frequency of transduction of markers near that site. As a proof of principle we first inserted a short sequence containing the previously characterized P22 pac site (Wu et al., 2002) into the Salmonella chromosome and tested its ability to cause an increase in transduction frequency of nearby markers by phage P22. To more easily monitor transduction, we used recombineering methods to construct Salmonella strain UB-1982 in which tetracycline (TetRA) and chloramphenicol (CamR) resistance genes are inserted 10 kbp on either side of the native galK gene in the chromosome (Figure 8A); 10 kbp is long enough that the effects of any exonucleolytic nibbling at DNA ends that might occur during transduction packaging events that initiate at pac sites at the galK location (see below) should be minimized, and the drug resistance genes would be packaged in the first headful of any packaging series that might initiate from such a pac site (details of strain constructions are in Materials and Methods). The galK gene of UB-1982 was then replaced by a 40 bp P22 sequence containing the pac site in either of the two possible orientations, and the resulting strains were lysogenized by P22 UC-911 (table 1). The prophages of these strains (UB-1988 and UB-2120), as well as control strains in which the galK gene was present (UB-2119) or neatly deleted (UB-2118) were induced, and the frequencies of transduction of CamR and TetRA by the resulting lysates were measured as described in Materials and Methods. Figure 8C line 3 shows that the inserted sequence P22 pacL stimulates CamR transduction about 100-fold over the control strains UB-2118 and UB-2119 (lines 1 and 2) that have no inserted pac site. This stimulation depends on the orientation of the pac site, and the orientation of P22 pacL1 should program packaging only in the direction of the CamR marker (Jackson, Jackson, and Deans, 1978; Casjens et al., 1992a; Wu et al., 2002). The P22 pacR site oriented in the opposite direction does not confer a stimulation of CamR transduction. Inserted pac site stimulation of transduction of the TetRA cassette by P22 is less striking because of a rather high background transduction when no inserted pac site is present (”none” or “galK” in figure 8C lines 1 and 2). This is likely due to a natural pac-like site in Salmonella DNA that programs this transduction; nonetheless, at least a 3-fold increase of TetRA transduction was consistently observed when the P22 pac site was oriented so that it directs packaging towards the TetRA cassette (P22 pacR). We conclude that this 40 bp sequence is sufficient to program P22 packaging series initiation on the Salmonella chromosome. We also note that the previous experimental characterization of the P22 pac site had only shown that this region is necessary for packaging initiation (Wu et al., 2002), and this is the first demonstration that it is also sufficient for initiation. Thus, there are likely no other sequences in the P22 genome that are required in cis for packaging in addition to the known pac site.
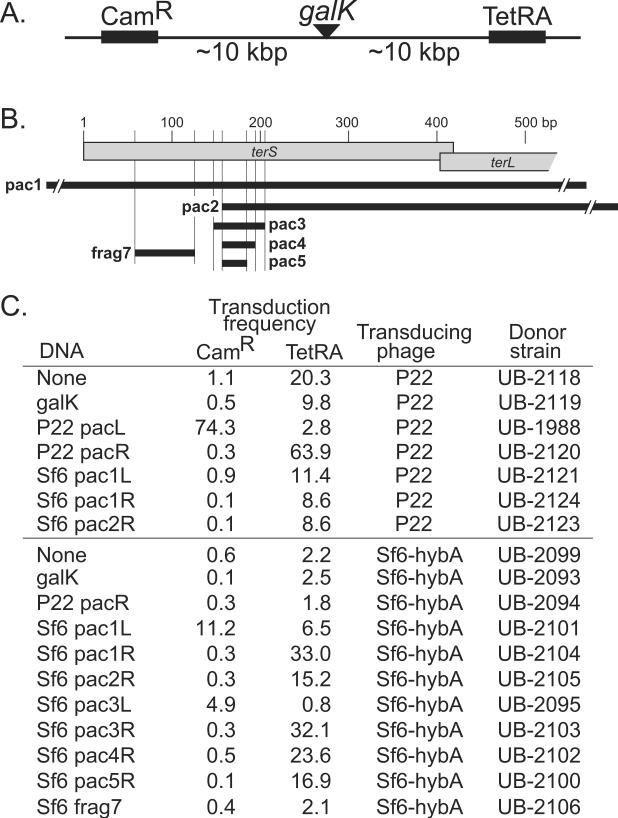
Figure 8. Transductional measurement of Sf6 pac activity.
(A) Chloramphenicol and tetracycline resistance cassettes in the Salmonella strain UB-1982 chromosome. (B) The locations of the Sf6 DNA fragments that were used to replace the galK gene in the Salmonella chromosome are indicated by horizontal black bars (exact coordinates given in Materials and Methods). (C) Transduction frequencies are presented transduced colonies per plaque-forming phage particle (×10−6), measured as described in Materials and Methods. The leftmost column indicates the DNA that was present in the host donor chromosome, where L and R indicate that the pac sites were oriented to program DNA packaging towards CamR or TetRA from the insertion site at galK. Three or more replicates were performed for each transduction that gave similar results, and a representative set of results is shown.
Since this in vivo assay for pac site activity behaves as predicted for phage P22, we replaced the Salmonella UC-1982 galK gene (figure 8A) with several sections of the phage Sf6 genome and determined their ability to support initiation of packaging (i.e., increased transduction frequency) during lytic growth of P22 terS Sf6-hybA (UC-929). These inserted sequences are diagrammed in figure 8B (see Materials and Methods for precise endpoints). They were chosen from the Sf6 terS gene and environs, since the experiments above indicated that this region harbors the Sf6 pac site. Each of these strains was lysogenized with P22 terS Sf6-hybA, and the transduction of the CamR and TetRA markers was measured after induction of the prophage to lytic growth. Six different overlapping Sf6 DNA sequences (Sf6 pac1-pac5 and frag7; figure 8B) were tested for P22 terS Sf6-hybA pac activity, several of them in both orientations. The frag7 sequence (UB-2106) did not show an increase in transduction over strains that carried the native galK gene (UB-2093) or a neat deletion of the galK gene (UB-2099) indicating that it does not contain an Sf6 pac site. The five remaining inserted Sf6 sequences all mediate unidirectional transduction increases (figure 8C lines 11–17). For example, in one orientation the Sf6 pac3R fragment (Sf6 bp 145–203; figure 8C line 15) stimulates transduction of TetRA about 15-fold over strains with no pac site, but does not stimulate transduction of CamR; in the other orientation the same sequence (Sf6 pac3L; line 14) stimulates CamR transduction about 10-fold but has no effect on TetRA transduction. The direction of these transduction increases agrees perfectly in all cases with our previous determination of the direction of native Sf6 DNA packaging (Casjens et al., 2004) and with the results from our Southern analysis of phages with duplicated pac sites above. The difference in background transduction frequency of CamR and TetRA without inserted pac sites could be due to several factors, but (as with P22 above) the presence of an Sf6 pac-like site in the native Salmonella chromosome sequence that programs some TetRA transduction by P22 Sf6-hybA is a likely explanation.
Replacement of the galK gene by Sf6 pac1R, pac2R, pac3R, pac4R or pac5R sequences all gave rather similar 10- to 20-fold increases in TetRA transduction (figure 8C). The smallest Sf6 sequence tested, pac5R, is sufficient for the bulk of this effect, and thus at least most of the Sf6 pac site lies within Sf6 bp 154–183. We also found that P22 TerS does not utilize the Sf6 pac site (compare UB-2121, -2123 and -2124 with UB-2118 and UB-2119 in figure 8C), and the Sf6-hybA TerS does not utilize the P22 pac site (UB-2094 vs. UB-2093 and UB-2099 in figure 8C). This data strongly supports the idea that the N-terminal globular TerS domain of Sf6 is responsible for pac site recognition in vivo. All of the above experiments support the idea that the phage Sf6 pac site lies inside the Sf6 terS gene between bp 154 and 183, and that packaging proceeds rightwards on the Sf6 genome from recognition events at that site. A more detailed analysis of the Sf6 pac site will be the subject of future studies.
TerS controls the location of packaging initiation DNA cleavages
As was described above, Sf6 generates packaging series initiation DNA ends over a large approximately 2000 bp region. This distribution can be visualized in agarose electrophoresis gels as the pac DNA fragment(s) (the restriction fragment from the initiation end of the DNA of the first member of a packaging series, above). In order to determine whether the TerS source affects the distribution of packaging initiation DNA cleavages, we analyzed the pac fragments of the functional TerS hybrid phages P22 terS Sf6-hybA (UC-921) and Sf6-hybB (UC-922) in comparison to that of P22 (UC-911). Figure 9 shows that after NdeI cleavage P22 DNA has a rather sharp ~3 kbp long pac fragment band as was previously known (Jackson, Jackson, and Deans, 1978; Casjens et al., 1992a); however, P22 terS Sf6-hybA DNA has a broad and diffuse NdeI pac fragment band that is about 2 kbp wide. Southern analysis was used to visualize these bands to avoid confusion regarding the source of the DNA bands. As above, restriction enzymes were chosen to display the pac fragments in the 2–5 kbp size range and to ensure that the overlapping true restriction fragments do not migrate at the same position as the pac fragments in the electrophoresis gel. Restriction enzyme analysis again showed that the packaging initiation (non-restriction enzyme)-generated ends of the diffuse pac fragment band were centered on the terS gene and that packaging proceeds rightward from the pac site (figure 9B). Parallel analysis of P22 terS Sf6-hybB DNA gave identical results (data not shown). The approximately 2 kbp band width is similar to the approximately 1.6 kbp distribution measured for phage Sf6 itself (Casjens et al., 2004). The difference between the 2 kbp measured here and the 1.6 kbp reported previously is almost certainly due to the rather arbitrary nature of determining the positions of the outer edges of such very diffuse gel “bands”; re-analysis of our previous data (Casjens et al., 2004) indicates that the pac fragment band width in those experiments was slightly underestimated due to the use of more conservative criteria for determining the band's outer edges and is in fact indistinguishable from the band widths measured here. Since the N-terminal domain of the Sf6 terS gene is the only Sf6 genetic information present in the hybrid phages analyzed in this way, we conclude that this domain of TerS is responsible for the difference between the P22 packaging initiation cleavage-containing region of 120 bp and the much larger Sf6 region (see Discussion for possible mechanisms).
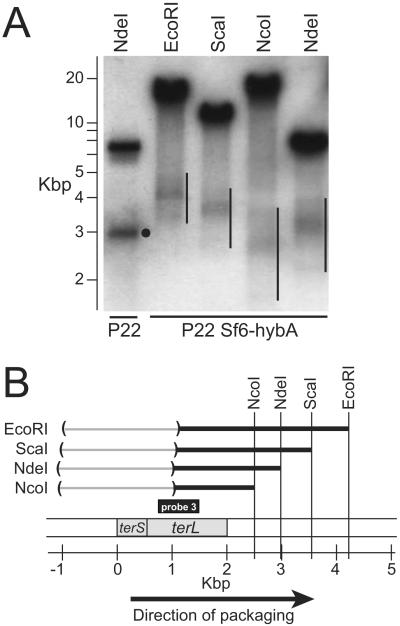
Figure 9. Pac fragments of P22 Sf6-hybA.
(A) Southern analysis of P22 Sf6-hybA pac fragments. DNA from P22 3∷Sf6-hybA, 13−amH101, 15−Δsc302∷KanR, sieA−Δ1 DNA was cleaved with the restriction endonuclease indicated above and separated by electrophoresis in 1.0% agarose. The gel was probed with 32P-labeled probe 3 (PCR amplified from bp 732–1497 of P22 DNA) and exposed to x-ray film. The black circle marks the P22 NdeI pac fragment location, and vertical black lines mark the widths of the diffuse pac fragments of P22 Sf6-hybA DNA; the bands higher in the gel are the true restriction fragments (restriction cuts at both ends) that are generated from packaging events other than the first one in a series. (B) A map of the packaging series initiation region of P22 Sf6-hybA is shown with a scale in kbp below it; the locations of the terS and terL genes are noted on the map. Horizontal black lines represent the pac fragments in part A that were generated by initiation of packaging series on P22 Sf6-hybA DNA. The right ends of these lines are anchored to the locations of the restriction sites that were cleaved to generate the pac fragments, and the parentheses enclose the regions in which the left (packaging initiation) ends of these fragments occur.
Discussion
The role of phage Sf6 TerS in DNA packaging
Analysis of the genome sequences of extant natural variants among the P22-like phages led us to the predictions that the C-terminal 23–28 amino acids of their TerS proteins are responsible for the interaction of TerS with the rest of the DNA packaging machinery and that their N-terminal domains recognize specific pac sites to choose the DNA molecules to be packaged. To test these ideas we constructed a fusion of the N-terminal domain of the phage Sf6 TerS with the C-terminal portion of P22 TerS in an otherwise completely phage P22 context, a combination that has not yet been found in nature. Several conclusions can be drawn from the fact that such hybrid phages are functional and from our genetic analysis of this hybrid TerS protein.
-
(1)
The N-terminal domain of TerS is responsible for the sequence specificity of pac site recognition in vivo, since the hybrid TerS with an Sf6 N-terminal domain and P22 C-terminal domain utilizes the Sf6 pac site and not the P22 pac site. Similarly, as expected from our previous analysis of its pac site (Wu et al., 2002), we found that phage P22 TerS cannot utilize the Sf6 pac site. However, we (Nemecek et al., 2008) and Roy et al. (2012) have found that removal of twenty or more C-terminal residues from P22 TerS results in a protein that no longer binds DNA nonspecifically in vitro. This observation is in apparent conflict with the idea that the N-terminal domain of TerS is solely responsible for DNA binding and could indicate that the C-terminus might also participate in some way.
-
(2)
Since the pac recognition specificities Sf6 and P22 TerS proteins are different (neither utilizes the other's site), the functional P22 terS Sf6-hybA and -hybB phages should be utilizing an Sf6 pac site that resides within the DNA that encodes the N-terminal domain of Sf6 TerS. Our experimental analysis showed that phage Sf6 does indeed carry its pac site near the center of this region.
-
(3)
Purified P22 TerS protein forms a complex with TerL (Poteete and Botstein, 1979; Roy et al., 2012; our unpublished results). The genetic findings presented here strongly suggest that the C-terminal domain of TerS is responsible for this binding, and while this work was underway Roy et al. (2012) showed that removal of the C-terminal 22 amino acids from P22 TerS abrogates its ability to bind TerL but not its ability to form nonamers. The evolutionary co-segregation of the C-terminal domain of TerS with the N-terminal half of TerL strongly suggests an interaction between these domains. Genetic studies with phages lambda and T4 TerS proteins indicate that their C-terminal regions also participate in binding to their cognate TerL's (Frackman, Siegele, and Feiss, 1985; Yang et al., 1999a; Gao and Rao, 2011). Since these three terminases are extremely different, having essentially no recognizable sequence similarity beyond an ATPase motif in TerL, this appears to be a very ancient interaction strategy that has been preserved in spite of the long evolutionary divergence between these two terminases.
-
(4)
The N-terminal domains of P22 and Sf6 TerS have essentially no amino acid similarity and yet the Sf6 domain can function in an otherwise completely P22 context. It thus seems very unlikely that this domain has important intimate interactions with the rest of the DNA packaging motor. If such interactions existed, the many differences between the Sf6 and P22 TerS proteins would make the interaction very unlikely in the hybrid phages studied here.
-
(5)
The fact that the connecting “linker” region between the essential parts of the N- and C-terminal domains of the TerS protein can vary in length by at least 16 amino acids and still be functional further suggests a very flexible connection between the N-terminal domain of TerS and the rest of the packaging machinery. The apparent flexibility of this connection is somewhat surprising, since it might have been expected that the different components of a DNA translocating molecular motor would have to occupy very specific spatial positions in the motor. However, there are other indications of TerS flexibility which include a mutant of P22 whose isolated TerS is present as decamers instead of nonamers is functional in vivo (Nemecek et al., 2008), P22 TerS has 23 C-terminal amino acids that are not ordered in crystals (Roy et al., 2012), and analyses of alternate crystal structures of each of the SF6, T4 and Sf6 TerS proteins indicate that they have considerable structural flexibility (Buttner et al., 2012; Sun et al., 2012; Zhao et al., 2012).
-
(6)
We also found that the N-terminal domain of TerS is responsible for the distribution of packaging series initiation DNA cleavages, since a P22 that carries only the DNA-binding domain of Sf6 TerS has a packaging initiation end distribution similar to that of phage Sf6 and not to that of phage P22. This could be explained by terminase moving along the DNA between pac recognition and DNA cleavage (see below), and if so this putative sliding/rolling would be due to movement of TerS or the TerS-TerL complex on the DNA and not, for example, after the DNA might be handed off from TerS to TerL.
Unlike the cohesive end generating terminases, the terminases of the headful packaging phages that have been examined do not generate precise DNA ends at the start of packaging series (summarized in Wu et al., 2002 and Casjens et al., 2005). We previously showed that two such tailed phages, Sf6 and ES18, generate packaging initiation ends over large regions of about 2000 and 500 bp, respectively (Casjens et al., 2004 and 2005). The frequency of such ends across the Sf6 DNA indicates that the frequency of cleavage decreases with increasing distance from near the center of the region (Casjens et al., 2004). We report here the localization of the phage Sf6 pac site to a short sequence near the center of this region, which is consistent with models that have a decreasing probability of cleavage with increased distance from the pac site.
The mechanism by which such a multiplicity of packaging initiation ends might be generated from targeted pac site recognition events is not known, but several models can potentially account for this observation. (i) The region that contains the packaging series initiation ends contains a number of recognition (pac) sites, any of which can be used to initiate packaging series. Alternatively, a single terminase complex could bind pac and either (ii) cut nearby DNA that happens to bend or loop into contact with it, (iii) recruit additional adjacent terminase complexes to bind nonspecifically and create a patch of terminase-covered DNA in which any terminase could generate the initiation end for a given packaging series, or (iv) move in either direction along the DNA away from the pac site before cleaving the DNA, perhaps by “sliding” or “rolling” of the multimer? Finally, (v) ends created at pac by a terminase cleavage could be attacked by an exonuclease before actual packaging starts. The localization of the Sf6 pac site to a small region rules out models that depend on the presence of many pac sites. Furthermore, the “patch of bound terminases” model seems rather unlikely since the terminase subunits, especially TerL, are not made in large amounts (Casjens and King, 1974; Poteete and Botstein, 1979), and because in our unpublished experiments substantially lowering the amount of P22 TerS during infection did not cause the initiation cleavage events to become less frequent farther from the pac site in that phage. If terminase bound to the pac site cleaves nearby DNA that has looped into contact with it, the nuclease active site should be separate from the DNA binding site and DNA cleavage should not occur within the pac site itself; however, in P22 cleavages are made inside the pac site (Casjens et al., 1992a; Wu et al., 2002) which would make this model less tenable (but multimeric TerL structures in which only one subunit is bound to pac could obviate this argument). Finally, although terminases have endonuclease activity, there is no evidence for exonuclease participation in packaging in any phage system, and such a model demands initial cleavage far upstream from the pac site (in the packaging directionality sense), and it is difficult to imagine how the ends could then be most frequent near the pac site. Thus, by default, current evidence suggests that the sliding/rolling model is most likely, although it has not been demonstrated directly. Understanding the mechanism of any such movement must await further experimentation.
Whatever the true mechanism, the four headful phages whose packaging initiation DNA cleavages and pac site have been previously precisely characterized, P22, Mu, P1 and SPP1, generate a number of alternate packaging initiation ends within approximately 120, 150, 12 and 7 bp regions, respectively, and all have a pac recognition site near or within the region within which the ends are generated (Deichelbohrer, Messer, and Trautner, 1982; Groenen and van de Putte, 1985; Sternberg and Coulby, 1987; Harel et al., 1990; Casjens et al., 1992a; Chai, Lurz, and Alonso, 1995). Phage T4 likely uses this strategy as well, but it is less well characterized in this regard (Wu and Black, 1995; Wu, Lin, and Black, 1995). In phage P22 the recognition site, pac, is a 22 bp sequence that lies approximately in the center of the 120 bp region where the DNA ends are generated (Wu et al., 2002), so this relationship is similar in P22 and Sf6 although the cleavage sites extend much further away from the pac site in Sf6 than they do in P22, suggesting a possible underlying mechanistic similarity between them. On the other hand we also note that tailed phages are extremely diverse and variations on any theme are not unexpected. It is known, for example, that phage Mu terminase binds its pac site near the end of the phage genome (which is integrated into the host chromosome) and only makes DNA cleavages in one direction from the pac site, in the host DNA that is adjacent to the integrated phage DNA. Nonetheless, the lack of precise DNA cleavage during initiation of packaging appears to be a common feature among headful packaging phages.
TerS and pac site evolution and horizontal exchange
The shortest Sf6 sequence that we tested that has pac site activity was the 30 bp fragment pac5R (figure 8), and its sequence is related to the P22 pac site. Nine of the 13 bp that are known to be important in P22 pac site recognition (Wu et al., 2002) are present in this Sf6 sequence if a two bp deletion in P22 is allowed (7 of 13 if not; figure 10A). Although the Sf6 and P22 TerS proteins do not have convincing sequence similarity, they do have partly similar folds. They both contain a cluster of five α-helices that have the same connectivity and similar but not identical spatial arrangements (figure 10B and figure 2 of Roy et al., 2012). A major difference between these two protein structures is the replacement of the short connection between α-helices 2 and 3 in Sf6 by a rather long β-hairpin in P22 (figure 10B). Although the minimal regions currently known to contain the P22 and Sf6 pac sites encode amino acids 89–96 and 51–61 of the two TerS proteins, respectively, it is striking that these two regions actually lie in precisely the same spatial location in the two proteins. Figure 10B shows that both reside at the N-terminus of α-helix 5. It is also interesting to note that this region is at the outer rim of the TerS multimer and that this region of the Sf6 octamer likely contacts DNA in its in vitro non-sequence-specific DNA-binding activity; in particular, Sf6 TerS Lys59, which is encoded within the pac site region identified here, is likely in contact with bound DNA (Zhao et al., 2010 and 2012). Of the seven amino acids encoded by the P22 pac site, there are two identities and three similarities in the parallel Sf6 sequence (figure 10A). If, as seems likely, the site on TerS that binds DNA nonspecifically in vitro is also involved in specific pac site recognition in vivo, then the pac site DNA itself encodes at least some of the amino acids that participate directly in recognition of the pac site DNA. This would represent a very striking example of molecular efficiency and evolutionary ingenuity. Such an arrangement would ensure that the pac site cannot be separated from the protein region that recognizes it during horizontal exchange of genetic material among different phages. It thus seems likely that the P22 and Sf6 TerS proteins are ancient homologues that have diverged a great deal, but have retained common DNA binding features.
Figure 10. P22 and Sf6 pac sites.
(A) The nucleotide sequences surrounding the Sf6 and P22 pac site regions are compared. In both cases packaging proceeds rightward from the pac site as shown here; red nucleotides indicate those bp in which mutations are known that largely abolish P22 pac site activity (Wu et al., 2002). The nucleotides shown are Sf6 bp 154–194 (Acc. No. AF547987) and P22 nucleotides 41722–41724 and 1–38 (Acc. No. BK00583; this sequence crosses the position at which the circular sequence is opened in the GenBank annotation). The minimal regions that have pac activity are boxed. The amino acids encoded by these sequences are shown above (Sf6) and below (P22) the nucleotide sequences with the number of the last amino acid shown on the right. Identical amino acids in the two phages are shown in red, similar amino acids are shown in blue (same charge) and green (hydrophobic). Above, the horizontal black bars indicate the bps present in the pac4R and pac5R fragments tested for pac activity in the transduction assay (see text and figure 8B). (B) Phage Sf6 and P22 TerS single subunit protein ribbon diagrams are shown (Protein Data Bank ID codes 3P9A and 3HEF, respectively). The N- and C-termini are indicated, as are α–helices 2, 4, 5 and 6 (Zhao et al., 2010; Roy et al., 2012). The yellow portions mark amino acids 89–97 encoded by the pac site of P22 and amino acids 51–61 encoded by the shortest Sf6 pac-containing fragment tested (pac5R) in this report. The locations of the central channels of the TerS multimer rings (see text) are indicated by vertical black arrows.
We showed here that the phage Sf6 pac site lies within its terS gene; however, not all tailed phage packaging initiation sites lie within the gene that encodes the TerS protein. For example in phages like lambda whose intravirion (mature) DNAs have sequence-specific, single-strand protruding cohesive ends, the recognition site (called cos in these phages) is typically immediately upstream of the adjacent terS and terL genes (e.g. lambda, N15 and HK97 and many other phages, but there are exceptions, for example phage, E. coli phage P2 (Ziermann and Calendar, 1990; Linderoth et al., 1991) and mycophage Giles (Hatfull, 2012) where the cos sites are far from the terS gene). These cos sites are typically more complex than the pac sites of headful packaging phages and contain dyad symmetry (Feiss and Rao, 2012), so it may be difficult to integrate them into the terS gene in these cases. The pac sites of five headful packaging phages have been studied (see above). The pac sites of P22 (Wu et al., 2002), P1 (Sternberg and Coulby, 1987; Lobocka et al., 2004), SPP1 (Chai, Lurz, and Alonso, 1995) and Sf6 (shown here) lie inside the terS gene, and less direct evidence has indicated a possible site within the T4 terS gene (Wu and Black, 1995; Wu, Lin, and Black, 1995). However in one less well-studied headful packaging phage, T1, the pac site appears to be in the early region and not within the terS gene (Liebeschuetz and Ritchie, 1986; Roberts, Martin, and Kropinski, 2004). Similarly, the phage Mu (a different sort of headful packager, above) pac site is nearly half the genome away from the TerS gene (Groenen and van de Putte, 1985; Harel et al., 1990; Morgan et al., 2002). Thus, although it is not universal, it is at least very common for the pac sites of headful packaging phages to lie with the terS gene.
Our findings thus support a picture in which the DNA that encodes the N-terminal domain of TerS is a self-contained, exchangeable unit that contains the pac site. It encodes the protein domain than binds the pac site and can function to recognize DNA for phage packaging as long as it can bind to TerL in the packaging apparatus through its C-terminal domain. This arrangement has allowed the horizontal exchange of terS genes among phages to be very successful, but the nature of the evolutionary advantage that might be gained by such exchanges remains mysterious.
Materials and Methods
Phage and bacterial strains
All bacterial and phage strains used in this study are listed in Table 1. S. enterica UB-0002 was used to propagate phage P22 strains and S. flexneri PE577 (UB-1458) was used to propagate phage Sf6 strains. E. coli NF1829 was used to carry plasmids and as transformation recipient during plasmid construction. All plasmid and phage constructs were confirmed by determination of the sequence of the modified region.
Construction of Sf6 phages with two pac sites
In order to genetically move sequences of choice into the phage Sf6 genome, a plasmid into which such sequences can be inserted and an Sf6 phage that accepts these sequences from the plasmid were constructed. The DNA accepting prophage, Sf6 62∷KanR, was constructed as follows: Plasmid pPP309 was constructed by ligating the following three polymerase chain reaction (PCR) amplified DNA fragments into plasmid pUC18 (Yanisch-Perron, Vieira, and Messing, 1985) that was opened with restriction enzymes HindIII and EcoRI, transforming into E. coli, and selecting for ampicillin resistance: (i) Sf6 bp 35805–36769 (Casjens et al., 2004; accession No. AF547987) with HindIII and BamHI site containing primer tails at the two ends, respectively, cut with these two enzymes; (ii) kanamycin resistance cassette amplified from bp 1823 to 2741 of plasmid pACYC177 (Rose, 1988) with BamHI and NdeI site containing primer tails, respectively, cut with these two enzymes; and (iii) Sf6 bp 37247 to 38249 with NdeI and EcoRI site containing tails, respectively, cut with these two enzymes. The resulting plasmid insert contains Sf6 bp 35805–38249 except the KanR cassette replaces bp 36770 and 37246. Plasmid pPP309 was electroporated into S. flexneri strain PE577 that carried a wild type Sf6 prophage. This strain (UB-1469) was induced to lytic growth with 1 μg/ml mitomycin C, phage particles from the resulting lysate were used to infect S. flexneri strain PE577, and the surviving cells were selected for growth in the presence of 50 μg/ml kanamycin and screened for cells that did not carry ampicillin resistance of the pPP309 plasmid. The resulting KanR AmpS bacterial strain (UB-1564) carries a prophage, Sf6 62∷KanR, in which the KanR cassette replaces gene the essential gene 62 (phage encoded endolysin). It does not release plaque-forming phages upon induction with mitomycin C.
A plasmid designed to carry the sequences to be placed in the Sf6 genome was constructed as follows: (i) Sf6 bp 36472 to 37247 were PCR amplified with HindIII and XhoI site containing primer tails, respectively, cut with HindIII and XhoI, and ligated to HindII+SalI cut pUC18 plasmid (New England Biosciences, Ipswich, MA); and (ii) Sf6 bp 37606 to 38628 were PCR amplified with BglII-XhoI-KpnI-NcoI and EcoRI site containing primer tails, respectively, and ligated into the plasmid from step (i) that was cut with BamHI and EcoRI. The resulting plasmid, pPP311, contains Sf6 bp 36472–38628 in which 36 bps (5'-GTCGACTCTAGAGGATCTCTCGAGGGTACCCCATGG) that contain unique XhoI and NcoI sites (underlined) replace Sf6 bp 37247–37606 (figure 5). The latter replacement inactivates gene 63; this spanin-encoding Sf6 gene is a homologue of phage λ gene Rz and phage P22 gene 15 and is not essential in those phages under low divalent cation growth conditions (Casjens et al., 1989; Summer et al., 2007). This work showed that Sf6 gene 63 is also required for efficient plaque formation in the presence of divalent cations, so all gene 63 defective phages were grown in the presence of 10 mM NaCitrate (Casjens et al., 1989).
DNA sequences to be tested for pac activity were inserted between the XhoI-NcoI sites of pPP311 and then moved into the phage Sf6 genome as follows: Oligonucleotide primers with 3'-tails containing XhoI and NcoI cleavage sites were used to PCR amplify sequences from the Sf6 genome, or double-stranded oligonucleotides with near terminal XhoI and NcoI sites were synthesized. These DNA fragments were cleaved with XhoI and NcoI and ligated into similarly cleaved plasmid pPP311. The ligation reaction mixture was transformed into E. coli NF1829 (UB-0049), and the resulting plasmid structures were confirmed by restriction mapping and sequence determination. These plasmids were moved into S. flexneri PE577 (Sf6 62∷KanR) UB-1564 by electroporation. These ampicillin and kanamycin resistant cells were grown to 2×108/ml in LB broth and induced with 1 μg/ml mitomycin C (Sigma, St. Louis, MO). After about 4 hours of shaking at 37°C, cell lysis was completed by shaking with a few drops of chloroform, and cell debris was removed by centrifugation. The resulting phages, UC-920 through UC-925 (table 1), all of which form plaques on S. flexneri PE577 (UB-1458), must have acquired a functional endolysin gene (gene 62) by homologous recombination with the plasmid. Integration of the whole plasmid into the Sf6 62∷KanR prophage makes the genome too large to be packaged, so the plaque-forming phages must have also enjoyed a second homologous recombination event in the bp 37606–38623 region that causes a neat replacement of the KanR cassette of Sf6 62∷KanR by the intact endolysin gene and the sequences cloned into the XhoI-NcoI sites of pPP311 (figure 5).
P22 prophage recombineering
P22 prophages with Sf6-P22 hybrid terS genes were constructed by recombineering as follows: Primer oligonucleotides A and B (Table S3) for PCR amplification were used that amplify the E. coli galK gene expression cassette from plasmid pGalK as described by Warming et al. (2005). These primers had 50 nt 3'-tails that correspond to P22 sequence immediately 5' of the gene 3 (terS) start codon and 3' of its codon 134. This amplified DNA was electroporated into UB-1790 cells (this strain is galK− and carries a P22 13−amH101, 15−Δsc302∷KanR, sieA−Δ1 (UC-911) prophage (Padilla-Meier et al., 2012); table 1), and colonies were selected that utilize galactose as the sole carbon source. The resulting strain (UB-1961) carries a galK expression cassette that replaces gene 3 codons 1–133 of the resident prophage. Here and below, in each recombineering step the phage lambda Red expression plasmid pKD46 (Datsenko and Wanner, 2000; Karlinsey, 2007) was present to stimulate recombinational replacement and was removed by growth overnight at 41°C before the strain was used further. DNA containing the first 114 codons of phage Sf6 gene 1 (terS) was then PCR amplified from plasmid pET28a-gp1 (Zhao et al., 2010) with primers C and D (Table S3). Primer C contains the 50 nucleotides of sequence immediately upstream of P22 gene 3 and 20 nucleotides that correspond to the first 20 nucleotides of the Sf6 gene 1 (terS) at its 3'-end; primer D contains 50 nucleotides that correspond to P22 gene 3 codons 129–146 and 18 nucleotides that correspond to Sf6 gene 1 codons 109–114 at its 3'-end. This amplified DNA was electroporated into strain UB-1961, and galK− cells resistant to 2-deoxygalactose (Sigma, St. Louis, MO) were selected as described by Warming et al. (2005). This recombinational replacement of the galK gene of UB-1961 resulted in a strain (UB-2019) whose P22 prophage contains a hybrid terS gene (called terS Sf6-hybA) in which Sf6 gene 1 codons 1–114 neatly replace codons 1–134 of P22 gene 3. Oligonucleotide C and variants of D were used in the same manner to construct prophages with terS genes that have different Sf6-P22 fusion points and junction sequences (hybA-hybJ and hybU-hybT; table 1 and figure 3). Salmonella strains carrying P22 prophages with amino acid changes to alanine in the Sf6-P22 hybB terS gene were made as follows: The terS Sf6-hybB gene along with 224 upstream and 104 downstream bp was amplified from strain UB-2040 DNA using primers E and F (Table S3), and the resulting fragment was cleaved by restriction enzymes XbaI and HindIII and cloned into similarly cleaved plasmid pBLSK (Stratagene, La Jolla, CA). This plasmid, pPP465, was modified by QUICKCHANGE methodology (Qiagen, Valencia, CA) to make the bp changes in terS hybrids hybK-hybS (table 1 and figure 3). The modified plasmid phage DNA inserts were PCR amplified and used to recombinationally replace the galK gene of UB-1961 as described above.
Insertion of pac sites into the Salmonella chromosome
Strain UB-1982 was constructed by sequentially inserting chloramphenicol and tetracycline resistance cassettes immediately 5' of bp 827343 and bp 849069, respectively, into the S. enterica LT2 strain UB-0020 genome (LT2 Acc. No. AE006468; recombineering details described above). Oligonucleotide pairs G/H and I/J (Table S3) were used with template DNA from strains UB-1760 and UB-1766, respectively, to PCR amplify fragments that contain the CamR and TetRA resistance cassettes with 40 bp of identity to the S. enterica LT2 chromosome at both ends. This placed the CamR-1 and TetRA-2 resistance cassettes about 10 kbp on either side of the galK gene which lies at bp 838373–839521. The CamR-1 insert lies between Salmonella open reading frames STM764 and STM765, and the TetRA-2 insert lies between STM784 and STM785; none of these reading frames has a known function, and we did not observe a growth phenotype for either insertion. In strain UB-2047 the galK gene was neatly deleted from UB-1982 by replacement it with synthetic oligonucleotide K (Table S3) annealed to its complementary oligonucleotide. The galK gene was replaced by the phage P22 pac site (P22 pacL and pacR in figure 8C) with recombineering replacement of galK in UB-1982 by annealed oligonucleotides L and M or N and O, respectively (Table S3). The resulting strains, UB-1985 and UB-1991, contain P22 nucleotides bp 41716–41724/1–31 in opposite orientations (this sequence crosses the opening of the circular P22 sequence in its GenBank annotation; Acc. No. BK000583). Strains in which the UB-1982 S. enterica LT2 bp galK gene is replaced by phage Sf6 sequences (UB-2093–2095, 2099 and 2100–2106) to be tested for pac site activity were created in a manner analogous to the construction of strain UB-1991. DNA fragments containing Sf6 bp 61–120 (called “fragN”), 388891–690 (Sf6 pac1), 155–821 (Sf6 pac2), 145–203 (Sf6 pac3), 155–194 (Sf6 pac4), and 154–183 (Sf6 pac5), with 40 bp tails for replacing galK were prepared by PCR templated by Sf6 DNA or were made synthetically. Note that the Sf6 pac1 sequence crosses the opening of the circular Sf6 sequence in its GenBank annotation (Accession No. AF547987). Several of these were made with the Sf6 sequences in different orientations; L and R (as in Sf6 pac1L or pac2R) indicate an orientation for which the final construct packaging should proceed towards the CamR or TetR cassette, respectively. For use in transduction experiments, the strains in this paragraph were lysogenized by phage P22 13−amH101, 15−Δsc302∷kanR, sieA−Δ1 (UC-911) or P22 13−amH101, 15−Δsc302∷kanR, 3∷Sf6-hybA, sieA−Δ1 (UC-929) by infecting and selecting for kanamycin resistance. Figure 8C and table 1 give the strain names and genotypes of these constructs.
Generalized transduction measurements
Transduction frequency measurements were performed as follows: The strains indicated in figure 8C were grown in L broth to 2×108/ml at 37°C, induced by the addition of 1.5 μg/ml carbadox (Sigma, St. Louis, MO), and lysed with chloroform after 3 hrs of shaking at 37°C (the strains each carry a P22 sieA−Δ1, 13−amH101, 15−Δsc302∷KanR or a P22, sieA−Δ1, 13−amH101, 15−Δsc302∷KanR, 3∷Sf6-hybA prophage). The resulting lysate was titered on UB-0002 to determine the phage yield. CamR or TetR carrying transducing particles were measured by infecting strain UB-0134 (Youderian, Chadwick, and Susskind, 1982) or UB-0020 freshly grown to 2×108 cells/ml in LB broth at a multiplicity of infection of 0.5 with phage from the above lysates for 90 min at 37°C, and the infected cells were plated on selective medium containing tetracycline or chloramphenicol to determine the number of drug resistant transduced colonies. Transduction frequencies are shown as transduced colonies/plaque-forming phage particle (×10−6) (Schmieger, 1972).
Southern analysis
Probes were labeled with 32P and Southern analysis was carried out as previously described (Casjens and Huang, 1993; Casjens et al., 1995).
Supplementary Material
Highlights
We identify the bacteriophage Sf6 DNA packaging recognition (pac) site.
We show that the N-terminal domain of Sf6 TerS protein recognizes the pac site.
The C-terminal TerS domain binds to the rest of the packaging apparatus.
The "linker" between the TerS N- and C-terminal domains can be very flexible.
We show that the P22-like TerS genes have undergone extensive horizontal exchange.
Acknowledgements
This work was supported by NIH grant RO1 AI074825 to SRC. We thank Gino Cingolani for access to the P22 TerS protein structure before its publication, Miriam Susskind, John Roth, Eric Kofoid and Anthony Poteete for Salmonella strains, Renato Morona for Shigella strains, Surabhi Kasera for help with hybrid phage construction and Liang Tang for plasmid pET28a-gp1.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Al-Zahrani AS, Kondabagil K, Gao S, Kelly N, Ghosh-Kumar M, Rao VB. The small terminase, gp16, of bacteriophage T4 is a regulator of the DNA packaging motor. J. Biol. Chem. 2009;284:24490–500. doi: 10.1074/jbc.M109.025007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black LW, Showe M, Steven AC. Morphogenesis of the T4 head. In: Karam J, editor. Molecular biology of bacteriophage T4. ASM Press; Washington, DC: 1994. pp. 218–258. [Google Scholar]
- Buttner CR, Chechik M, Ortiz-Lombardia M, Smits C, Ebong IO, Chechik V, Jeschke G, Dykeman E, Benini S, Robinson CV, Alonso JC, Antson AA. Structural basis for DNA recognition and loading into a viral packaging motor. Proc. Natl. Acad. Sci. U S A. 2012;109:811–6. doi: 10.1073/pnas.1110270109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho A, Jimenez F, De La Torre J, Carrascosa JL, Mellado RP, Vasquez C, Vinuela E, Salas M. Assembly of Bacillus subtilis phage ϕ29. 1. Mutants in the cistrons coding for the structural proteins. Eur. J. Biochem. 1977;73:39–55. doi: 10.1111/j.1432-1033.1977.tb11290.x. [DOI] [PubMed] [Google Scholar]
- Casjens S. Principles of virion structure, function and assembly. In: Chiu W, Burnett R, Garcea R, editors. Structural Biology of Viruses. Oxford University Press; Oxford: 1997. pp. 3–37. [Google Scholar]
- Casjens S. Prophages and bacterial genomics: what have we learned so far? Molec. Microbiol. 2003;49:277–300. doi: 10.1046/j.1365-2958.2003.03580.x. [DOI] [PubMed] [Google Scholar]
- Casjens S. Diversity among the tailed-bacteriophages that infect the Enterobacteriaceae. Res. Microbiol. 2008;159:340–348. doi: 10.1016/j.resmic.2008.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casjens S, Delange M, Ley HL, Rosa P, Huang WM. Linear chromosomes of Lyme disease agent spirochetes: genetic diversity and conservation of gene order. J. Bacteriol. 1995;177:2769–80. doi: 10.1128/jb.177.10.2769-2780.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casjens S, Eppler K, Parr R, Poteete AR. Nucleotide sequence of the bacteriophage P22 gene 19 to 3 region: identification of a new gene required for lysis. Virology. 1989;171:588–98. doi: 10.1016/0042-6822(89)90628-4. [DOI] [PubMed] [Google Scholar]
- Casjens S, Huang WM. Initiation of sequential packaging of bacteriophage P22 DNA. J. Mol. Biol. 1982;157:287–98. doi: 10.1016/0022-2836(82)90235-2. [DOI] [PubMed] [Google Scholar]
- Casjens S, Huang WM. Linear chromosomal physical and genetic map of Borrelia burgdorferi, the Lyme disease agent. Molec. Microbiol. 1993;8:967–80. doi: 10.1111/j.1365-2958.1993.tb01641.x. [DOI] [PubMed] [Google Scholar]
- Casjens S, King J. P22 morphogenesis. I: Catalytic scaffolding protein in capsid assembly. J. Supramol. Struct. 1974;2:202–24. doi: 10.1002/jss.400020215. [DOI] [PubMed] [Google Scholar]
- Casjens S, Sampson L, Randall S, Eppler K, Wu H, Petri JB, Schmieger H. Molecular genetic analysis of bacteriophage P22 gene 3 product, a protein involved in the initiation of headful DNA packaging. J. Mol. Biol. 1992a;227:1086–99. doi: 10.1016/0022-2836(92)90523-m. [DOI] [PubMed] [Google Scholar]
- Casjens S, Winn-Stapley D, Gilcrease E, Moreno R, Kühlewein C, Chua JE, Manning PA, Inwood W, Clark AJ. The chromosome of Shigella flexneri bacteriophage Sf6: complete nucleotide sequence, genetic mosaicism, and DNA packaging. J. Mol. Biol. 2004;339:379–394. doi: 10.1016/j.jmb.2004.03.068. [DOI] [PubMed] [Google Scholar]
- Casjens S, Wyckoff E, Hayden M, Sampson L, Eppler K, Randall S, Moreno E, Serwer P. Bacteriophage P22 portal protein is part of the gauge that regulates packing density of intravirion DNA. J. Mol. Biol. 1992b;224:1055–74. doi: 10.1016/0022-2836(92)90469-z. [DOI] [PubMed] [Google Scholar]
- Casjens SR. The DNA-packaging nanomotor of tailed bacteriophages. Nat. Rev. Microbiol. 2011;9:647–57. doi: 10.1038/nrmicro2632. [DOI] [PubMed] [Google Scholar]
- Casjens SR, Gilcrease EB, Winn-Stapley DA, Schicklmaier P, Schmieger H, Pedulla ML, Ford ME, Houtz JM, Hatfull GF, Hendrix RW. The generalized transducing Salmonella bacteriophage ES18: complete genome sequence and DNA packaging strategy. J. Bacteriol. 2005;187:1091–104. doi: 10.1128/JB.187.3.1091-1104.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casjens SR, Thuman-Commike PA. Evolution of mosaically related tailed bacteriophage genomes seen through the lens of phage P22 virion assembly. Virology. 2011;411:393–415. doi: 10.1016/j.virol.2010.12.046. [DOI] [PubMed] [Google Scholar]
- Chai S, Kruft V, Alonso JC. Analysis of the Bacillus subtilis bacteriophages SPP1 and SF6 gene 1 product: a protein involved in the initiation of headful packaging. Virology. 1994;202:930–9. doi: 10.1006/viro.1994.1415. [DOI] [PubMed] [Google Scholar]
- Chai S, Lurz R, Alonso JC. The small subunit of the terminase enzyme of Bacillus subtilis bacteriophage SPP1 forms a specialized nucleoprotein complex with the packaging initiation region. J. Mol. Biol. 1995;252:386–98. doi: 10.1006/jmbi.1995.0505. [DOI] [PubMed] [Google Scholar]
- Cortines JR, Weigele PR, Gilcrease EB, Casjens SR, Teschke CM. Decoding bacteriophage P22 assembly: identification of two charged residues in scaffolding protein responsible for coat protein interaction. Virology. 2011;421:1–11. doi: 10.1016/j.virol.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U S A. 2000;97:6640–5. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Beer T, Fang J, Ortega M, Yang Q, Maes L, Duffy C, Berton N, Sippy J, Overduin M, Feiss M, Catalano CE. Insights into specific DNA recognition during the assembly of a viral genome packaging machine. Mol. Cell. 2002;9:981–91. doi: 10.1016/s1097-2765(02)00537-3. [DOI] [PubMed] [Google Scholar]
- Deichelbohrer I, Messer W, Trautner TA. Genome of Bacillus subtilis Bacteriophage SPP1: Structure and Nucleotide Sequence of pac, the Origin of DNA Packaging. J. Virol. 1982;42:83–90. doi: 10.1128/jvi.42.1.83-90.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy C, Feiss M. The large subunit of bacteriophage lambda's terminase plays a role in DNA translocation and packaging termination. J. Mol. Biol. 2002;316:547–61. doi: 10.1006/jmbi.2001.5368. [DOI] [PubMed] [Google Scholar]
- Feiss M, Rao VB. The bacteriophage DNA packaging machine. Adv Exp. Med. Biol. 2012;726:489–509. doi: 10.1007/978-1-4614-0980-9_22. [DOI] [PubMed] [Google Scholar]
- Frackman S, Siegele DA, Feiss M. A functional domain of bacteriophage lambda terminase for prohead binding. J. Mol. Biol. 1984;180:283–300. doi: 10.1016/s0022-2836(84)80005-4. [DOI] [PubMed] [Google Scholar]
- Frackman S, Siegele DA, Feiss M. The terminase of bacteriophage lambda. Functional domains for cosB binding and multimer assembly. J. Mol. Biol. 1985;183:225–38. doi: 10.1016/0022-2836(85)90215-3. [DOI] [PubMed] [Google Scholar]
- Gao S, Rao V. Specificity of interactions among the DNA-packaging machine components of T4-related bacteriophages. J. Biol. Chem. 2011;286:3944–56. doi: 10.1074/jbc.M110.196907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- German G, Misra R, Kropinski A. The T1-like bacteriophages. In: Calendar R, editor. The Bacteriophages Second Edition. Oxford University Press; Oxford: 2006. pp. 211–224. [Google Scholar]
- Groenen MA, van de Putte P. Mapping of a site for packaging of bacteriophage Mu DNA. Virology. 1985;144:520–2. doi: 10.1016/0042-6822(85)90292-2. [DOI] [PubMed] [Google Scholar]
- Harel J, Duplessis L, Kahn JS, DuBow MS. The cis-acting DNA sequences required in vivo for bacteriophage Mu helper-mediated transposition and packaging. Arch. Microbiol. 1990;154:67–72. doi: 10.1007/BF00249180. [DOI] [PubMed] [Google Scholar]
- Hatfull GF. The secret lives of mycobacteriophages. Adv. Virus Res. 2012;82:179–288. doi: 10.1016/B978-0-12-394621-8.00015-7. [DOI] [PubMed] [Google Scholar]
- Hegde S, Padilla-Sanchez V, Draper B, Rao VB. Portal-large terminase interactions of the bacteriophage T4 DNA packaging machine implicate a molecular lever mechanism for coupling ATPase to DNA translocation. J. Virol. 2012;86:4046–57. doi: 10.1128/JVI.07197-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson EN, Jackson DA, Deans RJ. EcoRI analysis of bacteriophage P22 DNA packaging. J. Mol. Biol. 1978;118:365–88. doi: 10.1016/0022-2836(78)90234-6. [DOI] [PubMed] [Google Scholar]
- Jackson EN, Laski F, Andres C. Bacteriophage P22 mutants that alter the specificity of DNA packaging. J. Mol. Biol. 1982;154:551–63. doi: 10.1016/s0022-2836(82)80014-4. [DOI] [PubMed] [Google Scholar]
- Jiang W, Li Z, Zhang Z, Baker ML, Prevelige PE, Jr., Chiu W. Coat protein fold and maturation transition of bacteriophage P22 seen at subnanometer resolutions. Nat. Struct. Biol. 2003;10:131–5. doi: 10.1038/nsb891. [DOI] [PubMed] [Google Scholar]
- Karlinsey JE. Lambda-Red genetic engineering in Salmonella enterica serovar Typhimurium. Methods Enzymol. 2007;421:199–209. doi: 10.1016/S0076-6879(06)21016-4. [DOI] [PubMed] [Google Scholar]
- Kondabagil KR, Zhang Z, Rao VB. The DNA translocating ATPase of bacteriophage T4 packaging motor. J. Mol. Biol. 2006;363:786–99. doi: 10.1016/j.jmb.2006.08.054. [DOI] [PubMed] [Google Scholar]
- Kulesus RR, Diaz-Perez K, Slechta ES, Eto DS, Mulvey MA. Impact of the RNA chaperone Hfq on the fitness and virulence potential of uropathogenic Escherichia coli. Infect. Immun. 2008;76:3019–26. doi: 10.1128/IAI.00022-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–8. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Lebedev AA, Krause MH, Isidro AL, Vagin AA, Orlova EV, Turner J, Dodson EJ, Tavares P, Antson AA. Structural framework for DNA translocation via the viral portal protein. EMBO J. 2007;26:1984–94. doi: 10.1038/sj.emboj.7601643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YD, Park JH, Chang HI. Genomic sequence analysis of virulent Cronobacter sakazakii bacteriophage ES2. Arch. Virol. 2011;156:2105–8. doi: 10.1007/s00705-011-1096-7. [DOI] [PubMed] [Google Scholar]
- Liebeschuetz J, Ritchie DA. Phage T1-mediated transduction of a plasmid containing the T1 pac site. J. Mol. Biol. 1986;192:681–92. doi: 10.1016/0022-2836(86)90021-5. [DOI] [PubMed] [Google Scholar]
- Lin H, Rao VB, Black LW. Analysis of capsid portal protein and terminase functional domains: interaction sites required for DNA packaging in bacteriophage T4. J. Mol. Biol. 1999;289:249–60. doi: 10.1006/jmbi.1999.2781. [DOI] [PubMed] [Google Scholar]
- Linderoth NA, Ziermann R, Haggard-Ljungquist E, Christie GE, Calendar R. Nucleotide sequence of the DNA packaging and capsid synthesis genes of bacteriophage P2. Nucleic Acids Res. 1991;19:7207–14. doi: 10.1093/nar/19.25.7207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobocka MB, Rose DJ, Plunkett G, 3rd, Rusin M, Samojedny A, Lehnherr H, Yarmolinsky MB, Blattner FR. Genome of bacteriophage P1. J. Bacteriol. 2004;186:7032–68. doi: 10.1128/JB.186.21.7032-7068.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maluf N, Yang Q, Catalano C. Self-association properties of the bacteriophage lambda terminase holoenzyme: implications for the DNA packaging motor. J. Mol. Biol. 2005;347:523–42. doi: 10.1016/j.jmb.2005.01.016. [DOI] [PubMed] [Google Scholar]
- Morgan GJ, Hatfull GF, Casjens S, Hendrix RW. Bacteriophage Mu genome sequence: analysis and comparison with Mu-like prophages in Haemophilus, Neisseria and Deinococcus. J Mol Biol. 2002;317:337–59. doi: 10.1006/jmbi.2002.5437. [DOI] [PubMed] [Google Scholar]
- Morita M, Tasaka M, Fujisawa H. Structural and functional domains of the large subunit of the bacteriophage T3 DNA packaging enzyme: importance of the C-terminal region in prohead binding. J. Mol. Biol. 1995;245:635–44. doi: 10.1006/jmbi.1994.0052. [DOI] [PubMed] [Google Scholar]
- Nemecek D, Gilcrease EB, Kang S, Prevelige PE, Jr., Casjens S, Thomas GJ., Jr. Subunit conformations and assembly states of a DNA-translocating motor: the terminase of bacteriophage P22. J. Mol. Biol. 2007;374:817–36. doi: 10.1016/j.jmb.2007.08.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemecek D, Lander GC, Johnson JE, Casjens SR, Thomas GJ., Jr. Assembly architecture and DNA binding of the bacteriophage P22 terminase small subunit. J. Mol. Biol. 2008;383:494–501. doi: 10.1016/j.jmb.2008.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olia AS, Prevelige PE, Jr., Johnson JE, Cingolani G. Three-dimensional structure of a viral genome-delivery portal vertex. Nat. Struct. Mol. Biol. 2011;18:597–603. doi: 10.1038/nsmb.2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira L, Alonso JC, Tavares P. A defined in vitro system for DNA packaging by the bacteriophage SPP1: insights into the headful packaging mechanism. J. Mol. Biol. 2005;353:529–39. doi: 10.1016/j.jmb.2005.08.063. [DOI] [PubMed] [Google Scholar]
- Oliveira L, Henriques AO, Tavares P. Modulation of the viral ATPase activity by the portal protein correlates with DNA packaging efficiency. J. Biol. Chem. 2006;281:21914–23. doi: 10.1074/jbc.M603314200. [DOI] [PubMed] [Google Scholar]
- Padilla-Meier GP, Gilcrease EB, Weigele PR, Cortines JR, Siegel M, Leavitt JC, Teschke CM, Casjens SR. Unraveling the role of the C-terminal helix turn helix of the coat-binding domain of bacteriophage P22 scaffolding protein. J. Biol. Chem. 2012;287:33766–80. doi: 10.1074/jbc.M112.393132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parent KN, Gilcrease EB, Casjens SR, Baker TS. Structural evolution of the P22-like phages: comparison of Sf6 and P22 procapsid and virion architectures. Virology. 2012;427:177–88. doi: 10.1016/j.virol.2012.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parent KN, Khayat R, Tu LH, Suhanovsky MM, Cortines JR, Teschke CM, Johnson JE, Baker TS. P22 coat protein structures reveal a novel mechanism for capsid maturation: stability without auxiliary proteins or chemical crosslinks. Structure. 2010;18:390–401. doi: 10.1016/j.str.2009.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimkin M, Pimkina J, Markham GD. A regulatory role of the Bateman domain of IMP dehydrogenase in adenylate nucleotide biosynthesis. J. Biol. Chem. 2009;284:7960–9. doi: 10.1074/jbc.M808541200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poteete AR, Botstein D. Purification and properties of proteins essential to DNA encapsulation by phage P22. Virology. 1979;95:565–73. doi: 10.1016/0042-6822(79)90509-9. [DOI] [PubMed] [Google Scholar]
- Ray K, Oram M, Ma J, Black LW. Portal control of viral prohead expansion and DNA packaging. Virology. 2009;391:44–50. doi: 10.1016/j.virol.2009.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts MD, Martin NL, Kropinski AM. The genome and proteome of coliphage T1. Virology. 2004;318:245–66. doi: 10.1016/j.virol.2003.09.020. [DOI] [PubMed] [Google Scholar]
- Rose RE. The nucleotide sequence of pACYC177. Nucleic Acids Res. 1988;16:356. doi: 10.1093/nar/16.1.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy A, Bhardwaj A, Datta P, Lander G, Cingolani G. Small terminase couples viral DNA-binding to genome-packaging ATPase activity. Structure. 2012;20:1403–1413. doi: 10.1016/j.str.2012.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy A, Cingolani G. Structure of P22 headful packaging nuclease. J. Biol. Chem. 2012;287:28196–205. doi: 10.1074/jbc.M112.349894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmieger H. Phage P22-mutants with increased or decreased transduction abilities. Mol. Gen. Genet. 1972;119:75–88. doi: 10.1007/BF00270447. [DOI] [PubMed] [Google Scholar]
- Schmieger H. pac sites are indispensable for in vivo packaging of DNA by phage P22. Mol. Gen. Genet. 1984;195:252–5. doi: 10.1007/BF00332755. [DOI] [PubMed] [Google Scholar]
- Schmieger H, Koch E. In vitro assay of packaging protein gp3 of Salmonella phage P22. Intervirology. 1987;28:157–62. doi: 10.1159/000150011. [DOI] [PubMed] [Google Scholar]
- Shinder G, Gold M. The Nul subunit of bacteriophage lambda terminase binds to specific sites in cos DNA. J. Virol. 1988;62:387–92. doi: 10.1128/jvi.62.2.387-392.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shultz J, Silhavy TJ, Berman ML, Fiil N, Emr SD. A previously unidentified gene in the spc operon of Escherichia coli K12 specifies a component of the protein export machinery. Cell. 1982;31:227–35. doi: 10.1016/0092-8674(82)90422-6. [DOI] [PubMed] [Google Scholar]
- Simpson AA, Tao Y, Leiman PG, Badasso MO, He Y, Jardine PJ, Olson NH, Morais MC, Grimes S, Anderson DL, Baker TS, Rossmann MG. Structure of the bacteriophage 29 DNA packaging motor. Nature. 2000;408:745–50. doi: 10.1038/35047129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sippy J, Feiss M. Analysis of a mutation affecting the specificity domain for prohead binding of the bacteriophage lambda terminase. J. Bacteriol. 1992;174:850–6. doi: 10.1128/jb.174.3.850-856.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg N, Coulby J. Recognition and cleavage of the bacteriophage P1 packaging site (pac). II. Functional limits of pac and location of pac cleavage termini. J. Mol. Biol. 1987;194:469–79. doi: 10.1016/0022-2836(87)90675-9. [DOI] [PubMed] [Google Scholar]
- Summer E, Berry J, Tran T, Niu L, Struck D, Young R. Rz/Rz1 lysis gene equivalents in phages of Gram-negative hosts. J. Mol. Biol. 2007;373:1098–112. doi: 10.1016/j.jmb.2007.08.045. [DOI] [PubMed] [Google Scholar]
- Sun S, Gao S, Kondabagil K, Xiang Y, Rossmann MG, Rao VB. Structure and function of the small terminase component of the DNA packaging machine in T4-like bacteriophages. Proc Natl Acad Sci U S A. 2012;109:817–22. doi: 10.1073/pnas.1110224109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J, Lander GC, Olia AS, Li R, Casjens S, Prevelige P, Jr., Cingolani G, Baker TS, Johnson JE. Peering down the barrel of a bacteriophage portal: the genome packaging and release valve in P22. Structure. 2011;19:496–502. doi: 10.1016/j.str.2011.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavares P, Santos MA, Lurz R, Morelli G, de Lencastre H, Trautner TA. Identification of a gene in Bacillus subtilis bacteriophage SPP1 determining the amount of packaged DNA. J. Mol. Biol. 1992;225:81–92. doi: 10.1016/0022-2836(92)91027-m. [DOI] [PubMed] [Google Scholar]
- Tsay JM, Sippy J, DelToro D, Andrews BT, Draper B, Rao V, Catalano CE, Feiss M, Smith DE. Mutations altering a structurally conserved loop-helix-loop region of a viral packaging motor change DNA translocation velocity and processivity. J. Biol. Chem. 2010;285:24282–9. doi: 10.1074/jbc.M110.129395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsay JM, Sippy J, Feiss M, Smith DE. The Q motif of a viral packaging motor governs its force generation and communicates ATP recognition to DNA interaction. Proc. Natl. Acad. Sci. U S A. 2009;106:14355–60. doi: 10.1073/pnas.0904364106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tye BK, Huberman JA, Botstein D. Non-random circular permutation of phage P22 DNA. J. Mol. Biol. 1974;85:501–28. doi: 10.1016/0022-2836(74)90312-x. [DOI] [PubMed] [Google Scholar]
- Warming S, Costantino N, Court DL, Jenkins NA, Copeland NG. Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Res. 2005;33:e36. doi: 10.1093/nar/gni035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver S, Levine M. Replication in situ and DNA encapsulation following induction of an excision-defective lysogen of Salmonella bacteriophage P22. J. Mol. Biol. 1978;118:389–411. doi: 10.1016/0022-2836(78)90235-8. [DOI] [PubMed] [Google Scholar]
- Wikoff WR, Liljas L, Duda RL, Tsuruta H, Hendrix RW, Johnson JE. Topologically linked protein rings in the bacteriophage HK97 capsid. Science. 2000;289:2129–33. doi: 10.1126/science.289.5487.2129. [DOI] [PubMed] [Google Scholar]
- Winston F, Botstein D, Miller JH. Characterization of amber and ochre suppressors in Salmonella typhimurium. J. Bacteriol. 1979;137:433–9. doi: 10.1128/jb.137.1.433-439.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CH, Black LW. Mutational analysis of the sequence-specific recombination box for amplification of gene 17 of bacteriophage T4. J. Mol. Biol. 1995;247:604–17. [PubMed] [Google Scholar]
- Wu CH, Lin H, Black LW. Bacteriophage T4 gene 17 amplification mutants: evidence for initiation by the T4 terminase subunit gp16. J. Mol. Biol. 1995;247:523–8. [PubMed] [Google Scholar]
- Wu H, Sampson L, Parr R, Casjens S. The DNA site utilized by bacteriophage P22 for initiation of DNA packaging. Molec. Microbiol. 2002;45:1631–1646. doi: 10.1046/j.1365-2958.2002.03114.x. [DOI] [PubMed] [Google Scholar]
- Yang Q, Berton N, Manning MC, Catalano CE. Domain structure of gpNu1, a phage lambda DNA packaging protein. Biochemistry. 1999a;38:14238–47. doi: 10.1021/bi991408f. [DOI] [PubMed] [Google Scholar]
- Yang Q, Berton N, Manning MC, Catalano CE. Domain structure of gpNu1, a phage lambda DNA packaging protein. Biochemistry. 1999b;38:14238–47. doi: 10.1021/bi991408f. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–19. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Yeo A, Feiss M. Mutational analysis of the prohead binding domain of the large subunit of terminase, the bacteriophage lambda DNA packaging enzyme. J. Mol. Biol. 1995a;245:126–40. [PubMed] [Google Scholar]
- Yeo A, Feiss M. Specific interaction of terminase, the DNA packaging enzyme of bacteriophage lambda, with the portal protein of the prohead. J. Mol. Biol. 1995b;245:141–50. doi: 10.1006/jmbi.1994.0013. [DOI] [PubMed] [Google Scholar]
- Youderian P, Chadwick SJ, Susskind MM. Autogenous regulation by the bacteriophage P22 arc gene product. J. Mol. Biol. 1982;154:449–64. doi: 10.1016/s0022-2836(82)80006-5. [DOI] [PubMed] [Google Scholar]
- Youderian P, Sugiono P, Brewer KL, Higgins NP, Elliott T. Packaging specific segments of the Salmonella chromosome with locked-in Mud-P22 prophages. Genetics. 1988;118:581–92. doi: 10.1093/genetics/118.4.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youderian P, Vershon A, Bouvier S, Sauer RT, Susskind MM. Changing the DNA-binding specificity of a repressor. Cell. 1983;35:777–83. doi: 10.1016/0092-8674(83)90110-1. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Kottadiel VI, Vafabakhsh R, Dai L, Chemla YR, Ha T, Rao VB. A promiscuous DNA packaging machine from bacteriophage T4. PLoS Biol. 2011;9:e1000592. doi: 10.1371/journal.pbio.1000592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Finch CJ, Sequeira RD, Johnson BA, Johnson JE, Casjens SR, Tang L. Crystal structure of the DNA-recognition component of the bacterial virus Sf6 genome-packaging machine. Proc. Natl. Acad. Sci. USA. 2010;107:1971–6. doi: 10.1073/pnas.0908569107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Kamau YN, Christensen TE, Tang L. Structural and Functional Studies of the Phage Sf6 Terminase Small Subunit Reveal a DNA-Spooling Device Facilitated by Structural Plasticity. J. Mol. Biol. 2012 doi: 10.1016/j.jmb.2012.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziermann R, Calendar R. Characterization of the cos sites of bacteriophages P2 and P4. Gene. 1990;96:9–15. doi: 10.1016/0378-1119(90)90334-n. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.