Abstract
Purpose.
Lacrimal gland (LG) morphogenesis and repair are regulated by a complex interplay of intrinsic factors (e.g., transcription factors) and extrinsic signals (e.g., soluble growth/signaling factors). Many of these interconnections remain poorly characterized. Runt-related (Runx) factors belong to a small family of heterodimeric transcription factors known to regulate lineage-specific proliferation and differentiation of stem cells. The purpose of this study was to define the expression pattern and the role of Runx proteins in LG development and regeneration.
Methods.
Expression of epithelial-restricted transcription factors in murine LG was examined using immunostaining, qRT-PCR, and RT2Profiler PCR microarrays. The role of Runx transcription factors in LG morphogenesis was studied using siRNA and ex vivo LG cultures. Expression of Runx transcription factors during LG regeneration was assessed using in vivo model of LG regeneration.
Results.
We found that Runx factors are expressed in the epithelial compartment of the LG; in particular, Runx1 was restricted to the epithelium with highest level of expression in ductal and centroacinar cells. Downregulation of Runx1 to 3 expression using Runx-specific siRNAs abolished LG growth and branching and our data suggest that Runx1, 2, and 3 are partially redundant in LG development. In siRNA-treated LG, reduction of branching correlated with reduction of epithelial proliferation, as well as expression of cyclin D1 and the putative epithelial progenitor cell marker cytokeratin-5. Runx1, Runx3, and cytokeratin-5 expression increased significantly in regenerating LG and there was modest increase in Runx2 expression during LG differentiation.
Conclusions.
Runx1 and 2 are new markers of the LG epithelial lineage and Runx factors are important for normal LG morphogenesis and regeneration.
Keywords: lacrimal gland regeneration, stem and progenitor cells, Runx1, Runx2, qRT PCR microarrays, Runx3
The purpose of this study was to define the expression pattern and the role of Runx proteins in lacrimal gland development and regeneration.
Introduction
Runt-related (Runx) factors belong to a small family of heterodimeric transcription factors that control stem cell fate and regulate lineage-specific proliferation and differentiation.1 All Runx factors bear a conserved DNA-binding and heterodimerization domain2; however, they have different expression patterns and roles in tissue morphogenesis and homeostasis. Mouse genetic studies show that Runx1 (also known as AML1/Cbfa2) is essential for hematopoiesis,3,4 Runx2 (also known as AML3/Cbfa1) is required for osteogenesis,5 and Runx3 (also known as AML2/Cbfa3) is involved in gut development, neurogenesis, and lung alveolar differentiation.6–9 Runx proteins can act as repressors or activators depending on the cellular context.10,11 Runx proteins also contribute significantly to the transduction of fibroblast growth factor (FGF), Notch, transforming growth factor β, and Wnt signals,12–15 which control stem cell function and tissue regeneration.
Recent reports indicate that Runx proteins play a role in regulation of stem cells in epithelial derivatives.16–18 In particular, Runx1 to 3 are expressed in hair follicles where they regulate morphogenesis and stem cell survival.19–22 Runx1 is involved in regulation of epithelial cell adhesion, migration, and epithelial-mesenchymal cross talk.16,23
The expression and/or role of Runx proteins in development and regeneration of major ocular glands, lacrimal gland (LG) and meibomian gland (MG), has not been previously studied. LG is an exocrine-type gland that accounts for the bulk of the aqueous portion of the preocular tear film.24 Murine LG development starts at approximately E13.5 as an invagination of conjunctival epithelium into the surrounding mesenchyme. Subsequently, epithelial ducts form an elaborate network through a process known as branching morphogenesis.25,26 The branching pattern and function of the LG is regulated by epidermal growth factor, FGFs, bone morphogenetic proteins (BMPs), Wnts, and numerous transcription factors.27–32 Recent studies indicate that, similar to other exocrine glands (pancreas, salivary, mammary),33–36 the LG has a high regenerative potential and is able to repair itself even after substantial damage.37 During LG regeneration, the epithelial component of the gland undergoes epithelial-mesenchymal transition.24,38 In this process, epithelial cells lose cell junctions, polarity, and epithelial-specific markers, and acquire migratory phenotype and expression of mesenchymal markers.37 When gland remodeling is completed, cells return to an epithelial phenotype and form new LG ductal and acinar structures.37 There is also evidence for a population of proliferating nestin-positive stem cells that expand during LG regeneration; a subset of these cells bear markers of myoepithelial cells, suggesting a common progenitor for myoepithelial and epithelial lineages.37 It is possible that LG regeneration involves both dedifferentiation of mature epithelial cells, and activation, proliferation, and migration of epithelial stem cells.
Inflammation of the lacrimal gland, such as in Sjögren's syndrome, graft versus host disease, or other pathological conditions, can induce LG destruction. Unfortunately, these pathological conditions are also associated with a decline in LG regenerative ability. Inflammation may impact the stem cell niche, or change expression of important regulators of LG repair.24,38 Defining the mechanism(s) and factors that control LG morphogenesis and regeneration is important for developing new strategies to treat LG pathologies. Currently, we have limited understanding of the factors that induce progenitor cell proliferation and differentiation, and maintain the differentiated state once regeneration is completed.
In this study, we identified several transcription factors with a specific pattern of expression in the epithelial or mesenchymal cell lineage of the LG. We found that Runx1 and Runx2 were restricted to the LG epithelial compartment and Runx3 was weakly expressed in both epithelial and mesenchymal compartments. Downregulation of Runx1 to 3 expression in LG organ cultures significantly reduced LG growth, branching, and epithelial proliferation; thus Runx factors are necessary for normal LG morphogenesis. Runx1 and 3 expression increased during LG regeneration/remodeling and correlated with upregulation of cytokeratin 5 (Krt5), while Runx2 expression remained largely unchanged. We conclude that Runx proteins are LG epithelial cell lineage markers and regulate LG morphogenesis. Moreover, Runx1 and 3 may be important for LG regeneration.
Materials and Methods
Mice
Imprinting control region (ICR) (CD-1) out-bred timed-pregnant mice (Harlan Laboratories, Hayward, CA) were used for isolation of embryonic LGs for organ cultures and quantitative (qRT-PCR) arrays. Reactivatable Runx1 knockout mice (Runx1Re/Re) have been previously described39,40 and were used for studies of Runx1 expression and function. Female BALB/c mice (Taconic, Germantown, NY) were used for regeneration studies and qRT-PCR arrays. All experiments were in accordance with the ARVO Statement for the Use of Animals in Ophthalmic and Vision Research and were approved by The Scripps Research Institute and Tufts Medical Center Institutional Animal Care and Use Committees. ICR mice were sacrificed and embryos were kept in dissection medium (high-glucose minimum essential media [MEM] supplemented with 10% fetal bovine serum [FBS] and antibiotic/antimycotic) on ice prior to LG isolation. BALB/c mice were anesthetized and the exorbital LGs were left untreated (control) or were injected with 2 μL volume of recombinant human IL-1α (1 μg, a generous gift from the BRB Preclinical Repository of the National Cancer Institute), as previously described.41 Mice were sacrificed at various times postinjection and LGs removed and processed for RNA extraction.
Culture Media
LG dissection was performed in high-glucose MEM supplemented with 10% FBS and antibiotic-antimycotic (cat.# 15240062; Invitrogen, Carlsbad, CA).
LG organ cultures were maintained in CMRL Medium-1066 (cat.# 11530037; Invitrogen) supplemented with Insulin-Transferrin-Selenium-Supplement (cat.# 51300044; Invitrogen), human transferring-40 μg/mL (cat.# 0030124SA; Invitrogen), 0.1% Albumax I (cat.# 11020021; Invitrogen), and 150 μg/mL ascorbic acid (prepared fresh) (cat.# 07157; STEMCELL Technologies Inc., Vancouver, Canada).
Antibodies
The following primary antibodies were used for immunostaining: mouse monoclonal E-cadherin antibodies (Clone 36/E-cadherin; BD Pharmingen, San Jose, CA; or clone DECMA-1; Millipore, Billerica, MA), mouse monoclonal antibody to Pecam (CD31, clone MEC 13.3; BD Pharmingen), rat monoclonal antibody against Ser28 phosphohistone-H3 (P-H3) (Clone HTA28; Sigma-Aldrich, St. Louis, MO), laminin antibody produced in rabbit (cat.# L 9393; Sigma-Aldrich), and cytokeratin 5 (Krt5) rabbit polyclonal antibody (cat.# AF138; Covance, San Diego, CA). Complimentary secondary antibodies were obtained from Invitrogen.
X-Gal Staining
Tissue was fixed with 1.25% paraformaldehyde (PFA) and 1.25% glutaraldehyde solution and processed for X-Gal staining as described in the Supplementary Methods.
Immunoblotting
Total protein was prepared from small interfering RNAs (siRNAs)-treated LG organ cultures (approximately 10–15 LGs per each condition) using radioimmunoprecipitation assay (RIPA) lysis buffer and sonicated. Equal aliquots of protein were resolved by SDS-PAGE, transferred to polyvinylidene difluoride membrane (PDVF), and probed with antibody to Cyclin D1 (cat.# 2922; Cell Signaling, Boston, MA). β-actin antibody (cat.# 4970; Cell Signaling) was used as a loading control. Bands were quantified by densitometry, and the cyclin-D1/β-actin ratio calculated. Two experiments were performed, and the results from Runx1, 2, and 3 siRNA-treated cultures were normalized to the values from the control siRNA-treated cultures.
Isolation of LG Epithelium and Mesenchyme
Epithelial and mesenchymal components of E16.5 LGs were separated as previously described.31,42,43 Briefly, LGs were dissected and treated with trypsin/pancreatin solution on ice for 40 minutes, and the epithelial structure was mechanically separated from the mesenchyme. To prepare trypsin/pancreatin solution, trypsin (cat.# 85450C-25G; Invitrogen) and pancreatin (P3292; Sigma) were dissolved in Tyrode solution (see Supplementary Methods) to a final concentration of 2.25% wt/vol and 0.75% wt/vol respectively. Solution was filter sterilized.
qRT-PCR and qRT-PCR Microarray
Three independent experiments were performed to obtain triplicates for each microarray. For each experiment, LGs were isolated from at least two litters of embryos. RNA was isolated using RNeasy Mini kit (cat.# 74104; SABiosciences, Qiagen, Valencia, CA); cDNA was made using RT2 First Strand Kit (cat.# 330401; SABiosciences, Qiagen). Pathway-specific PCR arrays or individual primer PCR assays were performed using RT2 SYBR Green qPCR Mastermix (SABiosciences, Invitrogen) (see Supplementary Methods for primers and arrays). Quantitative RT-PCR was performed on the ABI 7300 system (Life Technologies, Grand Island, NY) and data analyzed using online normalization and analysis tools (provided in the public domain, http://sabiosciences.com/pcrarraydataanalysis.php).
RNA Interference
The siRNAs targeting Runx1, Runx2, and Runx3, as well as negative control (scrambled) and penetration control (CY3-conjugated) siRNAs were from Ambion, Invitrogen. RNA interference (RNAi) was tested using at least two different siRNAs for each of Runx1, Runx2, or Runx3. Control and Runx siRNAs were delivered using Lipofectamine RNAiMAX Transfection Reagent (Invitrogen). Transfection efficiency was determined to be 95% to 98% using the Cy3-labelled siRNA. Details of RNAi and qRT-PCR are described in the Supplementary Methods. Gene expression profiles were generated by normalizing the Ct (threshold cycle) of genes of interest with a housekeeping gene 29S and then comparing gene expression in LGs treated with control siRNA and combinations of Runx1, 2, and 3 siRNAs. Approximately 10 to 15 LGs per each condition were used. Each experiment was repeated at least three times.
Statistical Analysis
For each experiment, approximately 10 to 15 LGs per condition/treatment were used. For gene or protein expression studies, LG tissue samples were pooled together. All experiments were repeated two to three times. Data are expressed as means ± SD from two to three experiments. Paired Student's t-test was used to determine the statistical significance of differences between experimental (Runx siRNA-treated) and control (control siRNA-treated) LGs. P values less than 0.05 were considered statistically significant.
Results
Expression of Stem Cell Markers in Separated Epithelial and Mesenchymal Components of Developing LG
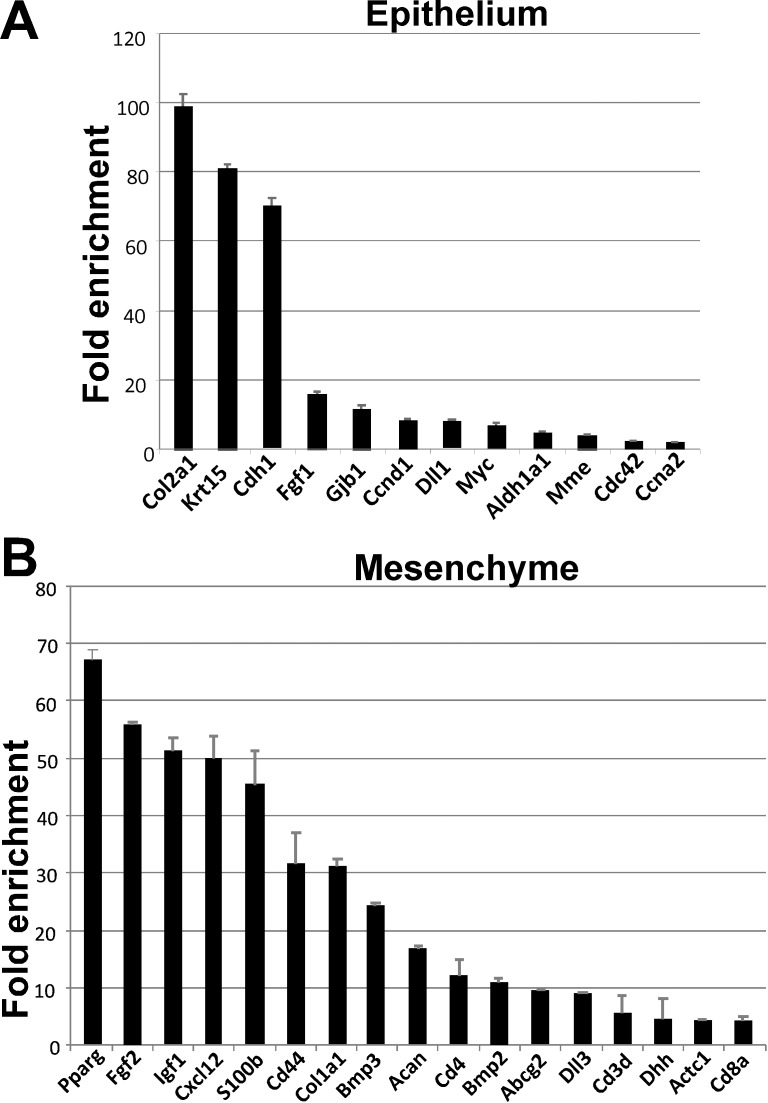
Remodeling of tissue after injury recapitulates many of the cellular events that govern embryonic development, including stem/progenitor cell activation.44 Thus, to begin to identify novel markers of LG progenitors that may be involved in regeneration, we first examined stem cell–associated signaling pathways and regulators in embryonic LG using stem cells and stem cell transcription factors qRT-PCR arrays. To identify markers specific for the epithelial and mesenchymal lineages, we prepared RNA from enzymatically separated E16.5 LG epithelial and mesenchymal components. Complete separation of epithelial and mesenchymal cells was confirmed by measuring expression of Barx2 (which is expressed only in LG epithelium) and Barx1 (which is expressed only in LG mesenchyme).28 Comparison of epithelial and mesenchymal cells using the Stem Cell Array (PAMM-405A; SABiosciences, Qiagen), showed that epithelial cells were enriched for cell cycle regulators, such as Cyclin D1 (CcnD1), cell division cycle 42 homolog (Cdc42), Cyclin A2 (Ccna2), and regulator of cell proliferation myelocytomatosis oncogene (Myc) (Fig. 1). These markers are consistent with our observation that epithelium is more proliferative than mesenchyme (see Supplementary Fig. S1). LG mesenchyme preferentially expressed growth factors such as FGF2 and FGF3; BMP1, 2, and 3; growth differentiation factor 3 (GDF3); and insulin-like growth factor-1. Mesenchyme was also enriched in hematopoietic cell lineage markers (glycoproteins, cluster differentiation 4 and 8) CD4 and CD8a, mesenchymal progenitor cell markers, such as Acan (Agc1), and various adhesion and matrix proteins (Fig. 1). These findings suggest that mesenchyme of developing LG undergoes substantial remodeling and is a source of growth factors important for epithelial morphogenesis.30,31
Figure 1.
Stem cell PCR array revealed genes differentially expressed in LG epithelium and mesenchyme. Total RNA was prepared from E16.6 enzymatically separated LG epithelium and mesenchyme. Relative expression was calculated using the household genes provided in the array. Bars represent means ± SEM of two experiments. LG epithelium was enriched with expression of collagen, type II, alpha 1 (Col2a1), keratin 15 (Krt5), cadherin 1 (Cdh1), fibroblast growth factor 1 (Fgf1), gap junction protein, beta 1 (Gjb1), Cyclin D1 (CcnD1), delta-like 1 (Dll1), myelocytomatosis oncogene (Myc), aldehyde dehydrogenase family 1, subfamily A1 (Aldh1a1), membrane metalloendopeptidase (Mme), cell division cycle 42 homolog (Cdc42), Cyclin A2 (Ccna2). LG mesenchyme was enriched with expression of peroxisome proliferator activated receptor gamma (Pparg), fibroblast growth factor 2 (Fgf2), insulin-like growth factor 1 (Igf1), chemokine (C-X-C motif) ligand 12 (Cxcl12), S100 protein, beta polypeptide (S100b), CD44 antigen (CD44), aldehyde dehydrogenase family 1, subfamily A1 (Aldh1a1), bone morphogenetic protein 3 (Bmp3), aggrecan (Acan), CD4 antigen (CD4), bone morphogenetic protein 2 (Bmp2), ATP-binding cassette, subfamily G (WHITE), member 2 (Abcg2), delta-like 3, (Dll3), CD3 antigen, delta polypeptide (CD3d), desert hedgehog (Dhh), actin, alpha, cardiac muscle 1 (Actc1), CD8 antigen, alpha chain (CD8a).
To define a transcription factor signature for LG epithelium, we compared epithelial and mesenchymal cells using the Stem Cell Transcription Factor PCR Array (PAMM-501Z; SABiosciences, Qiagen). Sox6 and Sox9 (SRY-box containing transcription factors 6 and 9, stem cell markers), Foxp1 (forkhead transcription factor-1, regulator of epithelial injury response), Six2 (Sine oculis-related homeobox), Runx1 (runt family transcription factor), and Pax6 (paired homeobox factor) were exclusively expressed in LG epithelium (Table 1). Specificity of Pax6 for the LG epithelial compartment has been previously reported.27,28 Relative to mesenchyme, LG epithelium was also enriched in Myc, and the epigenetic regulators DnmtB (DNA methyltransferase 3B), and Tert (Telomerase reverse transcriptase) (Tables 1, 2).
Table 1. .
Genes Expressed in the LG Epithelium at E16.5
|
Lacrimal Gland Epithelium |
E16.5 | |
|
Gene |
Fold-Up Regulation |
P
Value |
| Dnmt3b | 6.306 | 0.00309 |
| Foxp1 | 7.336 | 0.000959 |
| Myc | 6.786 | 0.000038 |
| Pax6 | 17.530 | 0.000005 |
| Runx1 | 11.926 | 0.001072 |
| 62 | 17.441 | 0.003939 |
| Sox6 | 7.988 | 0.000005 |
| Sox9 | 4.957 | 0.004121 |
| Tert | 2.266 | 0.004974 |
| Vdr | 7.369 | 0.000321 |
LGs were isolated at E16.5 and epithelium and mesenchyme were separated and gene expression in the epithelium and mesenchyme was compared in qRT-PCR stem cell transcription factors array.
Table 2. .
Genes Expressed in the LG Mesenchyme at E16.5
|
Lacrimal Gland Mesenchyme |
E16.5 | |
|
Gene |
Fold-Up Regulation |
P
Value |
| Foxp2 | 7.672 | 0.007084 |
| Gata6 | 40.564 | 0.000308 |
| Klf2 | 8.576 | 0.021912 |
| Klf4 | 6.208 | 0.000546 |
| Nfatc1 | 6.294 | 0.002887 |
| Nr2f2 | 9.021 | 0.000207 |
| Pax1 | 14.392 | 0.001801 |
| Pitx2 | 48.605 | 0.00389 |
| Pparg | 38.927 | 0.000666 |
| Zfpm2 | 13.236 | 0.02511 |
| Dlx1 | 14.540 | 0.028748 |
| Esr1 | 23.755 | 0.011947 |
| Gata1 | 5.480 | 0.029089 |
LGs were isolated at E16.5 and epithelium and mesenchyme were separated and gene expression in the epithelium and mesenchyme was compared in qRT-PCR stem cell transcription factors array.
Several transcriptional regulators were restricted to LG mesenchyme (Table 2) or enriched in mesenchyme relative to epithelium, including Foxp2, which is also found early in developing conjunctiva,45 Pax1, and Kruppel-like factors, Klf2 and Klf4. The latter factors are involved in trafficking and differentiation of CD8+ and CD4+ T cells, which are important players in animal models of autoimmunity.46
Runx1 Is a New Marker of the LG Epithelial Cell Lineage
Our study provides the first evidence that Runx1 expression is restricted to the LG epithelium. In other tissues, Runx1 is involved in stem cell specification, proliferation, and differentiation,21,23,46 and influences tissue homeostasis.46 Runx1 was recently shown to be involved in differentiation of the epithelial secretory cell lineage in colon.46 Moreover, absence of Runx1 was associated with global changes in expression of inflammation-related genes.3,46,47 Thus, we decided to further investigate Runx expression and function during LG development.
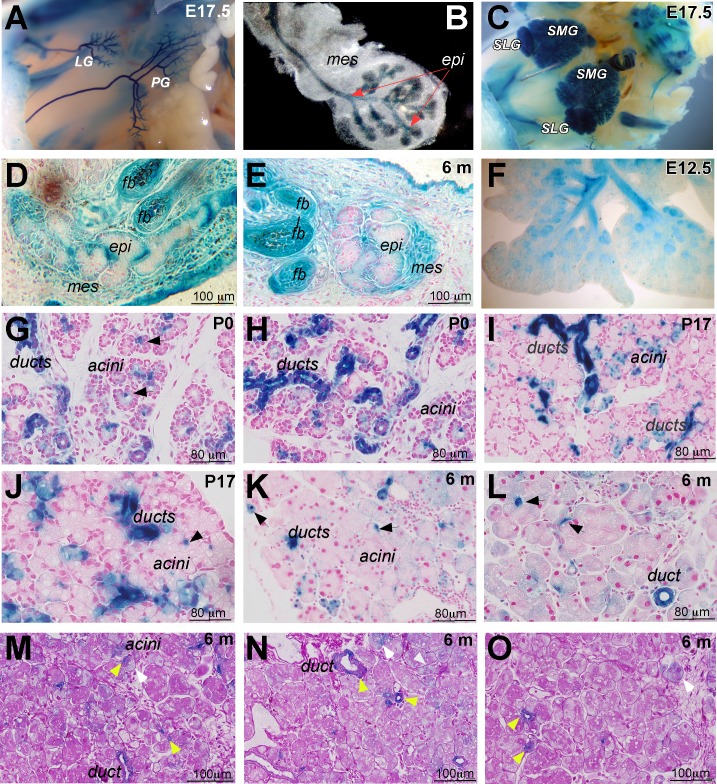
A reversible Runx1 knockin-LacZ mouse was used to examine Runx1 expression and function in LG morphogenesis.39,40 During embryonic development, Runx1 was expressed throughout the epithelium of LG, parotid glands (PG), submandibular (SMG), and sublingual glands (SLG) (Figs. 2A–C). Runx1-LacZ was also expressed in ductal and mesenchymal components of the MG, in the follicle bulge region (fb) of eyelashes (Figs. 2D, 2E), and in other branched organs, such as lung (Fig. 2F) and mammary gland (not shown). In early postnatal (P0–P17) and adult (6-month-old) mice, Runx1 expression remained restricted to the epithelial component of the LG with high level of expression in ductal epithelium in a subset of cells within the acini called centroacinar cells (Figs. 2G–J, black arrows), which represent the extension of the intercalated duct cells. Centroacinar cells in other branched glands are reported to be progenitor-like and involved in tissue regeneration.48,49 Runx1-LacZ also marked cells in ducts that morphologically and positionally resemble myoepithelial cells (MECs) (not shown). The expression of Runx1 within LG ducts, centroacinar cells, and MECs suggests a specific function in epithelial progenitor populations. Adult Runx1-LacZ LG sections were stained using the Periodic Acid-Schiff (PAS) reaction, which identifies a subset of acinar and ductal cells.50 PAS-positive (PAS+) epithelial cells (pink, see Figs. 2M–O) were seen in main and intercalated ducts and within acini (Figs. 2M, 2N). Runx-LacZ marked all PAS+ ducts and centroacinar cells (Figs. 2M–O, yellow arrows) and some PAS−acinar cells (Figs. 2M, 2N, white arrows).
Figure 2.
Analysis of Runx1 expression in the lacrimal gland of heterozygous Runx1 knockin-lacZ mice. (A, B) Whole-mount X-Gal staining of mouse embryo at E17.0. Runx1 expression is restricted to the epithelium of the LG and PG; (B) shows an excised LG. Red arrows mark epithelial component of the lacrimal gland. (C) Runx1-lacZ is expressed in the SLG and SMG. (D, E) Runx1 is also expressed in the epithelial and mesenchymal components of the meibomian gland (mei) and lung epithelium (F). (G–L) Expression pattern of Runx1 in the LGs at postnatal development day 0 (P0; [G, H]), P17 (I, J), 6-month-old (K, L) old mice. Runx1 expression is restricted to the ducts and centroacinar cells (black arrows) within the secretory acini. LGs obtained from Runx1 knockin-lacZ mice were stained with X-Gal and paraffin sections were prepared. Nuclei were stained with fast red to visualize LG morphology. (M–O) PAS reaction was also used for staining of sections of Runx1-LacZ LGs. Strong LacZ staining was seen within all PAS+ ducts, including a main duct (m. duct) and centroacinar cells (yellow arrows) and within some PAS– acini/cells (white arrows).
Role of Runx1 Gene Function in LG Development
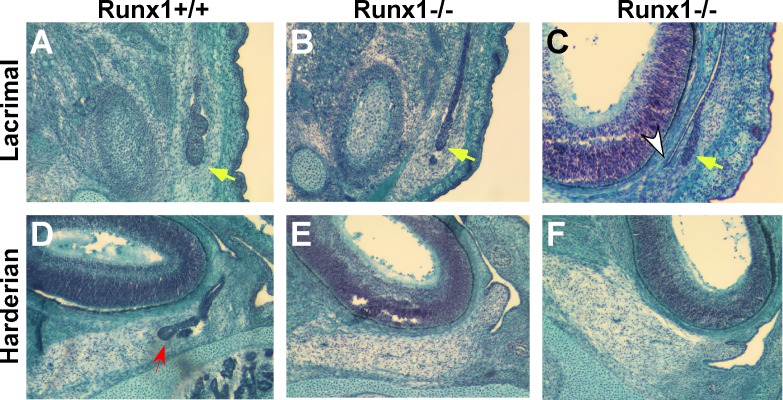
Runx1-null mice die at E11.0–12.5 due to impaired endothelial and hematopoietic stem cell function. In Tie2-Cre;Runx1Fl°x:lacZ/Fl°x:lacZ embryos, Cre recombinase is expressed under the control of the endothelial/hematopoietic-specific Tie2 promoter,51–53 resulting in selective reactivation of Runx1 in hematopoietic cells. The gross appearance of rescued Tie2-Cre;Runx1Fl°x:lacZ/Fl°x:lacZ embryos was indistinguishable from that of wild-type (WT) embryos. Serial paraffin sections of E16.5–17.0 embryos showed only slight difference between Cre;Runx1Fl°x:lacZ/Fl°x:lacZ and WT glands: WT LG started to branch and had several small buds (Fig. 3A), while many Cre;Runx1Fl°x:lacZ/Fl°x:lacZ LGs contained only one bud (Figs. 3B, 3C) that was short and close to conjunctiva (Fig. 3C). At E16.5, harderian glands (HG) emerge from the nasal part of conjunctival epithelium; HG were evident in WT embryos (Fig. 3D, red arrow), but absent in Cre;Runx1Fl°x:lacZ/Fl°x:lacZ embryos (Figs. 3E, 3F). Despite the embryonic delays, adult Cre;Runx1Fl°x:lacZ/Fl°x:lacZ LG and HG morphology was essentially normal with only slight variation in gland sizes (not shown). Thus Runx1 controls timing of primary bud outgrowth, but may be redundant in later development.
Figure 3.
Mutation in the Runx1 gene reduces LG budding and perturbs HG development in embryos at E16.5. (A–C) LG; (D–F) HG development in WT (Runx+/+) (A, D) and mutant (Runx–/–) (B, C, E, F) embryos. Yellow arrows indicate LG budding, white arrow conjunctiva, and red arrow HG epithelium component in WT embryos.
Role of Runx Genes in LG Development
To examine possible redundancy of Runx genes, Runx 1, 2, and 3 expression was measured by qRT-PCR in adult ocular glands and other branched organs (lung and submandibular gland) (Table 3). All three Runx genes were expressed in LG and HG; Runx1 and Runx2 were restricted to the epithelial component of the glands, whereas Runx3 mRNA was detected in both epithelium and mesenchyme (Table 4).
Table 3. .
Analysis of Gene Expression in Ocular Glands and Other Branched Organs
|
Expression |
Lacrimal Gland |
Harderian Gland |
Meibomian Gland |
Lung |
Submandibular Gland |
| Runx1 | +++ | +++ | +++ | +++ | +++ |
| Runx2 | +++ | +++ | +++ | ++ | +++ |
| Runx3 | ++ | + | + | + | + |
| Barx1 | ++ | +++ | + | N/A | +++ |
| Barx2 | +++ | +++ | ++ | + | +++ |
| Pax6 | +++ | +/– | +++ | + | – |
+++, highly expressed; ++, intermediate level of expression; +, low level of expression; +/–, very low expression; N/A, not determined; –, no expression.
Table 4. .
Analysis of Runx1 to 3 Expression in Separated LG Epithelium and Mesenchyme
| Expression |
LG Epithelium |
LG Mesenchyme |
| Runx1 | +++ | – |
| Runx2 | ++ | – |
| Runx3 | + | + |
| Barx1 | – | +++ |
| Barx2 | +++ | – |
+++, highly expressed; ++, intermediate level of expression; +, low level of expression; –, no expression.
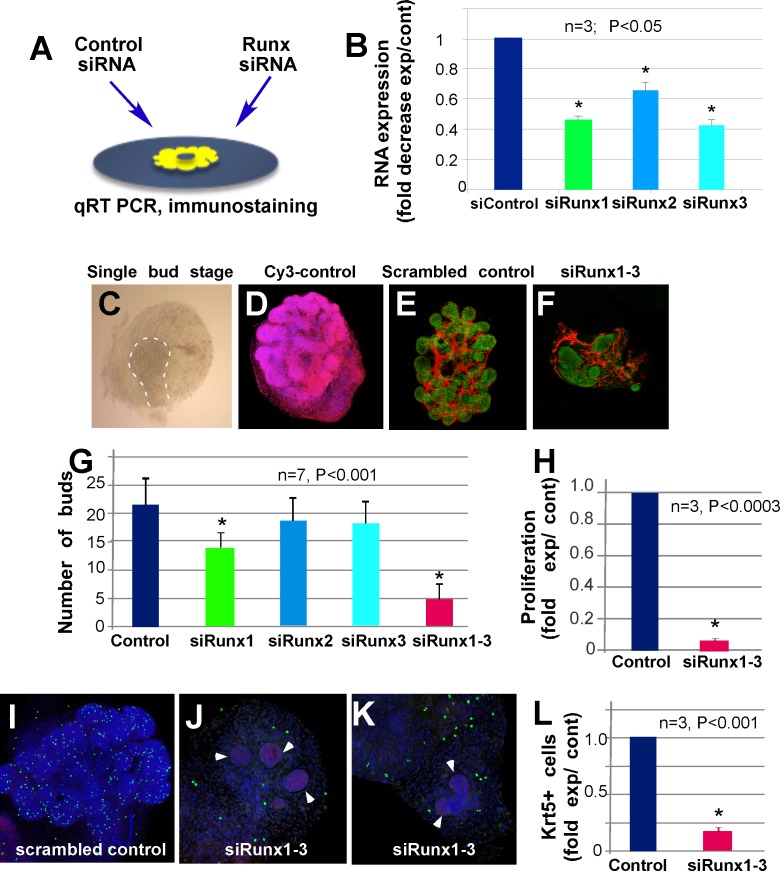
To further define the function of Runx genes in LG development, we inhibited their expression using either single siRNAs targeted against each Runx isoform or a combination of siRNAs to target all isoforms. The Runx-targeted siRNAs were applied to LG organ cultures prepared at E15 (single-bud stage, Figs. 4A, 4C). Scrambled siRNA acted as a control and CY3-conjugated control siRNA was used to evaluate tissue penetration (Figs. 4D, 4E). Labeled siRNA (penetration control) was found in all cells of treated LGs (Fig. 4D). Reduction of Runx RNA was confirmed by qRT-PCR (Fig. 4B). Application of Runx1 siRNA alone induced a small but statistically significant decrease in LG branching (two-tailed P0 = 0.0007) (Fig. 4G), while application of Runx2 (two-tailed P0 = 0.4251) or Runx3 siRNA (two-tailed P0 = 0.5423) had no effect (Fig. 4G). However, LG treated with combined Runx1 and Runx2 siRNAs (two-tailed P0 < 0.0001) or Runx1, Runx2, and Runx3 siRNAs (two-tailed P0 = 0.0001) showed greatly reduced branching and irregular bud shape relative to control siRNA treated LGs (Figs. 4E–G).
Figure 4.
Knockdown of Runx1, 2, and 3 reduces LG epithelial growth, cell proliferation, and number of Krt5-expressing progenitor cells. (A) Scheme of ex vivo siRNA treatment. LG were isolated at one bud stage (also see [C]) at E15.5 and cultured on a filter floating on the surface of culture medium. Control and Runx siRNAs were delivered using RNAiMAX (Invitrogen). (B) Runx1, 2, and 3 RNAs fold decrease after specific siRNA application is shown relative to control-treated LGs. (C) Example of LG at one bud stage used for siRNA application. (D) CY-3–labeled control RNA is found in all cells of the LG. (E) Scrambled control siRNA application does not affect LG branching. (F) Combined specific Runx1, 2, and 3 siRNA applications strongly reduce LG growth/branching. (G) Quantification of buds in siRNA-treated glands shows significant inhibition of branching after application of Runx1, or combinations of Runx1 and 2 or Runx1, 2, and 3 siRNAs (E-cadherin is green, PECAM is red). (H–K) The number of proliferating cells is decreased in Runx-siRNA treated LGs (H) quantification of H-3P antibody-stained proliferating cells in control and Runx1, 2, and 3 siRNA-treated LG buds. Control (I) and Runx1, 2, and 3 siRNA-treated (J–K) LG were stained with H-3P antibody (green), E-cadherin (red), and 4′,6-diamidino-2-phenylindole (blue) (white arrowheads show epithelium). LGs treated with Runx1, 2, and 3 siRNA show almost no cell proliferation in the epithelial component of the gland, while proliferating cells are still present in mesenchyme. (L) Number of Krt5-expressing cells is strongly reduced in LGs treated with Runx1, 2, and 3 siRNA. Asterisks denote statistical significance in the results compared to the control.
Runx proteins are reported to regulate stem cell proliferation.23 To assess cell proliferation in LGs treated with Runx siRNAs, we used an phospho-histone-3 antibody (Ser28-H3-P, referred to here as H3-P) that detects phosphorylated histones at the onset of mitosis.54 Combined application of siRNA to all three Runx genes almost completely abolished proliferation in the epithelial component of the gland (Figs. 4H–K), although proliferating cells were still detected in the mesenchyme (Figs. 4J, 4K). Control siRNAs had no effect on cell proliferation (Figs. 4I, 4H). We hypothesize that perturbation of Runx1, 2, and 3 during LG morphogenesis decreases the number of proliferating progenitor cells (either epithelial stem cells or MECs). Consistent with this, expression of cytokeratin-5 (Krt5), a reported marker of epithelial progenitors in several tissues,55–57 was strongly reduced in Runx-siRNA–treated LG (Fig. 4L). To explore the mechanism of Runx regulation of cell proliferation, we measured CyclinD1 protein (a highly conserved cyclin protein family member expressed in the epithelium [see Fig. 1]) in siRNA-treated LG by immunoblotting. CyclinD1 was modestly downregulated by Runx1 siRNA treatment and strongly downregulated by application of all three Runx siRNAs; application of single Runx2 or Runx3 siRNAs had no significant effect on CyclinD1 levels (Fig. 5).
Figure 5.
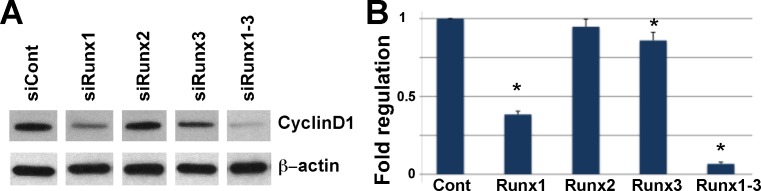
Knockdown of Runx1, 2, and 3 expression results in decreased CyclinD1 protein. (A) Protein was prepared from siRNA-treated LG organ cultures, resolved by SDS-PAGE, and probed with the cyclin-D1 or β-actin antibodies. (B) Graphic representation of densitometric analyses of Western blots from two independent experiments (mean ± SD of Optical Density [OD] of bands). For analysis of the Western blot images ImageJ 1.46 (National Institutes of Health, Bethesda, MD) has been used. The relative densities of the loading-control bands and the sample bands were calculated. Adjusted density for each sample lane was calculated by dividing the sample relative density by the loading-control relative density for each lane separately. Asterisks mark significant statistical difference in the level of CyclinD1 of experimental siRNA treated lacrimal glands compared to control treated glands.
Runx1 and 3 Are Upregulated During Early Stages of LG Regeneration
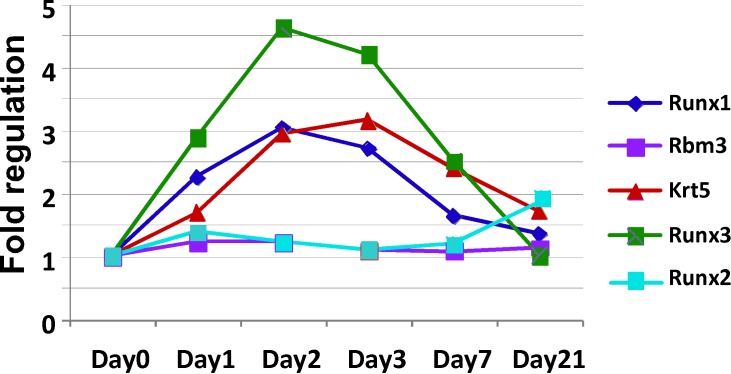
To elicit LG regeneration, we used the established IL-1α injection method. IL-1α induces a strong immune response, extensive inflammation, and LG damage, followed by a regenerative phase that restores morphology by 7 to 10 days after injury.37 If Runx genes are expressed in epithelial progenitor cells or cells undergoing active remodeling during LG regeneration, we predicted that Runx expression would increase during the regeneration phase. LGs were injected with IL-1α or saline (vehicle control), and collected at day 1, 2, 3, 7, or 21 after injection for RNA extraction. Levels of Runx1 to 3, Krt5 (epithelial progenitor marker), RBM3 (member of the glycine-rich RNA-binding protein family, ubiquitously expressed in the LG), and PrL27 (noncoding RNA, poly[A]-bearing) and PrS13 (16S ribosomal RNA gene) housekeeping mRNAs were determined by qRT-PCR (Table 5, Fig. 6, graphic representation of Table 5). Runx1 and Runx3 were upregulated in regenerating LG. Runx2 mRNA remained largely unchanged during the period of LG remodeling, but was slightly increased during epithelial differentiation/maturation (Fig. 6). As expected, Rbm3 expression was largely unchanged. Significantly, changes in Runx1 and Runx3 expression during LG regeneration correlated with changes in expression of Krt5, peaking during the first 3 days and declining from 7 to 21 days back to the baseline level in uninjured glands. Thus, Runx1 and Runx3 are induced during LG regeneration and may be involved in expansion of epithelial progenitor cells.
Table 5. .
Changes in Gene Expression During Lacrimal Gland Regeneration
|
|
Day 1 |
Day 2 |
Day 3 |
Day 7 |
Day 21 |
|||||
|
Fold Regulation |
P0 Value |
Fold Regulation |
P0 Value |
Fold Regulation |
P0 Value |
Fold Regulation |
P0 Value |
Fold Regulation |
P0 Value |
|
| Runx1 | 2.2342 | 0.000174 | 3.0275 | 0.000005 | 2.7089 | 0.000054 | 1.6359 | 0.00017 | 1.358 | 0.019599 |
| Runx2 | 0.931 | 0.130836 | 1.2128 | 0.212677 | 0.9869 | 0.555818 | 1.1843 | 0.282192 | 2.1496 | 0.003509 |
| Runx3 | 2.8746 | 0.000221 | 4.6128 | 0.00001 | 4.1988 | 0.000004 | 2.5115 | 0.00028 | 1.006 | 0.430019 |
| Krt5 | 1.6831 | 0.006834 | 2.9337 | 0.000223 | 3.1363 | 0.000064 | 2.3936 | 0.005639 | 1.7249 | 0.029249 |
| RBM3 | 1.1965 | 0.353178 | 1.2148 | 0.199725 | 0.9289 | 0.542553 | 1.0698 | 0.726245 | 0.9809 | 0.466395 |
Expression of Runx1, Runx2, Runx3, Krt5, and Rbm3 in IL-1 injected and control LGs was studied by qRT-PCR. Fold regulation was calculated by comparing of the level of expression of each gene in IL-1–treated glands with its expression in saline-injected glands. Gene expression was normalized to PrL27 and PrS13 housekeeping genes. The experiment was performed in triplicate. The ΔΔCt-based fold regulation and P0 value calculations were performed using the RT2 Profiler PCR Array Data Analysis software (SABiosciences, Qiagen). Statistically significant changes and corresponding P0 values are shown in bold.
Figure 6.
Graphic representation of Table 5. LGs were injected with IL-1 or saline (vehicle control) and collected at day 1, 2, 3, 7, or 21 after injection for RNA extraction. Levels of Runx1, Runx2, Runx3, Krt5, RBM3, and PrL27 and PrS13 mRNAs were determined by qRT-PCR. Increase in Runx1 and Runx3 expression correlates with increase in expression of epithelial progenitor cell marker Krt5, while expression of Runx2 and ubiquitously expressed Rbm3 remains unchanged during regeneration.
Discussion
Here we studied the expression and function of Runx factors during LG development and regeneration. We identified Runx1 and Runx2 as novel specific epithelial markers of the LG, and showed that Runx3 is expressed in both LG epithelium and mesenchyme. Consistent with our data, recent publications show that Runx proteins are expressed in epithelial cells in salivary and mammary glands, prostate, intestine, and lung.16,18,19,23,46,58 Recent lineage tracing studies of skin epithelia using Runx1-CreER knockin mice reported that embryonic Runx1+ placode cells show long-term contribution to multiple hair follicle epithelial lineages,21 suggesting a role for Runx1 in multipotential epithelial stem cells. The elevated level of Runx1 expression in LG centroacinar and ductal cells, upregulation of Runx1 expression during LG regeneration, and the reported contribution of Runx1 in controlling multiple stem cell lineages, suggests that high level of Runx1 expression might mark LG epithelial progenitor cell population(s). However, currently, the identity of LG stem/progenitor cells remains unclear and it is unknown whether the ductal, acinar, and MEC lineages originate from a common multipotent stem cell or multiple lineage-specific stem/progenitor cells.
The dramatic impairment of LG epithelial proliferation and branching by blockage of Runx1 to 3 correlates with other reports that Runx proteins regulate lineage-specific cell proliferation, differentiation, and cell migration.59–63 RUNX functions are also associated with several cancers and other human pathologies; Runx1 and Runx2 proteins may act as oncogenes by directly promoting cell proliferation62,64 and Runx1 has been implicated in promoting cell cycle progression through induction of cyclin D proteins.65 Indeed, we show that Runx proteins regulate cyclin D1 expression at least during LG development. Interestingly cyclin D1 and CDK4 were recently reported to induce Runx2 and Runx3 phosphorylation, ubiquitylation, and proteasomal degradation, suggesting complex interaction between Runx and cell cycle proteins.66
Runx proteins have been implicated in regulation of multiple developmental and regenerative processes, including cell adhesion and matrix production, cell movement, and cytoskeletal organization. For example, Runx1 directly regulates E-cadherin and matrix metalloproteinase 9 (MMP9) expression.67 Downregulation of E-cadherin and upregulation of MMP9 is closely linked with epithelial mesenchymal transition (EMT)38 during LG regeneration. Although knockdown of Runx expression during LG morphogenesis only modestly affected E-cadherin expression (not shown), changes in adhesion and matrix remodeling may be part of the mechanism by which Runx proteins control morphogenesis and regeneration.
Defining new biomarkers of the LG epithelial lineage and of potential stem/progenitor cells is critical to understanding the LG regenerative process, and may aid the development of cell-based interventions for LG degeneration.
Supplementary Material
Acknowledgments
Supported by National Eye Institute/National Institutes of Health Grants 1 R21 EY021292 (HPM) and RO1-EY12383 (DZ) and Sjögren's syndrome Foundation (HPM).
Disclosure: D. Voronov, None; A. Gromova, None; D. Liu, None; D. Zoukhri, None; A. Medvinsky, None; R. Meech, None; H.P. Makarenkova, None
References
- 1. van Wijnen AJ, Stein GS, Gergen JP, et al. Nomenclature for Runt-related (RUNX) proteins. Oncogene. 2004; 23: 4209–4210 [DOI] [PubMed] [Google Scholar]
- 2. Wang CQ, Jacob B, Nah GS, Osato M. Runx family genes, niche, and stem cell quiescence. Blood Cells Mol Dis. 2010; 44: 275–286 [DOI] [PubMed] [Google Scholar]
- 3. Mukai K, Benbarak MJ, Tachibana M, et al. Critical role of P1-Runx1 in mouse basophil development. Blood. 2012; 120: 76–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Growney JD, Shigematsu H, Li Z, et al. Loss of Runx1 perturbs adult hematopoiesis and is associated with a myeloproliferative phenotype. Blood. 2005; 106: 494–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Maruyama Z, Yoshida CA, Furuichi T, et al. Runx2 determines bone maturity and turnover rate in postnatal bone development and is involved in bone loss in estrogen deficiency. Dev Dyn. 2007; 236: 1876–1890 [DOI] [PubMed] [Google Scholar]
- 6. Friedrich MJ, Rad R, Langer R, et al. Lack of RUNX3 regulation in human gastric cancer. J Pathol. 2006; 210: 141–146 [DOI] [PubMed] [Google Scholar]
- 7. Reis BS, Rogoz A, Costa-Pinto FA, Taniuchi I, Mucida D. Mutual expression of the transcription factors Runx3 and ThPOK regulates intestinal CD4(+) T cell immunity. Nat Immunol. 2013; 14: 271–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Levanon D, Bettoun D, Harris-Cerruti C, et al. The Runx3 transcription factor regulates development and survival of TrkC dorsal root ganglia neurons. EMBO J. 2002; 21: 3454–3463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lee JM, Shin JO, Cho KW, et al. Runx3 is a crucial regulator of alveolar differentiation and lung tumorigenesis in mice. Differentiation. 2011; 81: 261–268 [DOI] [PubMed] [Google Scholar]
- 10. Uchida H, Zhang J, Nimer SD. AML1A and AML1B can transactivate the human IL-3 promoter. J Immunol. 1997; 158: 2251–2258 [PubMed] [Google Scholar]
- 11. Taniuchi I, Osato M, Egawa T, et al. Differential requirements for Runx proteins in CD4 repression and epigenetic silencing during T lymphocyte development. Cell. 2002; 111: 621–633 [DOI] [PubMed] [Google Scholar]
- 12. Burns CE, Traver D, Mayhall E, Shepard JL, Zon LI. Hematopoietic stem cell fate is established by the Notch-Runx pathway. Genes Dev. 2005; 19: 2331–2342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kagoshima H, Shigesada K, Kohara Y. RUNX regulates stem cell proliferation and differentiation: insights from studies of C. elegans. J Cell Biochem. 2007; 100: 1119–1130 [DOI] [PubMed] [Google Scholar]
- 14. Reinhold MI, Naski MC. Direct interactions of Runx2 and canonical Wnt signaling induce FGF18. J Biol Chem. 2007; 282: 3653–3663 [DOI] [PubMed] [Google Scholar]
- 15. Selvamurugan N, Kwok S, Alliston T, Reiss M, Partridge NC. Transforming growth factor-beta 1 regulation of collagenase-3 expression in osteoblastic cells by cross-talk between the Smad and MAPK signaling pathways and their components, Smad2 and Runx2. J Biol Chem. 2004; 279: 19327–19334 [DOI] [PubMed] [Google Scholar]
- 16. Chimge NO, Frenkel B. The RUNX family in breast cancer: relationships with estrogen signaling. Oncogene. 2013; 32: 2121–2130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lim M, Zhong C, Yang S, Bell AM, Cohen MB, Roy-Burman P. Runx2 regulates survivin expression in prostate cancer cells. Lab Invest. 2010; 90: 222–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fowler M, Borazanci E, McGhee L, et al. RUNX1 (AML-1) and RUNX2 (AML-3) cooperate with prostate-derived Ets factor to activate transcription from the PSA upstream regulatory region. J Cell Biochem. 2006; 97: 1–17 [DOI] [PubMed] [Google Scholar]
- 19. Raveh E, Cohen S, Levanon D, Groner Y, Gat U. Runx3 is involved in hair shape determination. Dev Dyn. 2005; 233: 1478–1487 [DOI] [PubMed] [Google Scholar]
- 20. Osorio KM, Lee SE, McDermitt DJ, et al. Runx1 modulates developmental, but not injury-driven, hair follicle stem cell activation. Development. 2008; 135: 1059–1068 [DOI] [PubMed] [Google Scholar]
- 21. Osorio KM, Lilja KC, Tumbar T. Runx1 modulates adult hair follicle stem cell emergence and maintenance from distinct embryonic skin compartments. J Cell Biol. 2011; 193: 235–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Glotzer DJ, Zelzer E, Olsen BR. Impaired skin and hair follicle development in Runx2 deficient mice. Dev Biol. 2008; 315: 459–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hoi CS, Lee SE, Lu SY, et al. Runx1 directly promotes proliferation of hair follicle stem cells and epithelial tumor formation in mouse skin. Mol Cell Biol. 2010; 30: 2518–2536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zoukhri D. Mechanisms involved in injury and repair of the murine lacrimal gland: role of programmed cell death and mesenchymal stem cells. Ocul Surf. 2010; 8: 60–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hogan BL, Grindley J, Bellusci S, Dunn NR, Emoto H, Itoh N. Branching morphogenesis of the lung: new models for a classical problem. Cold Spring Harb Symp Quant Biol. 1997; 62: 249–256 [PubMed] [Google Scholar]
- 26. Horowitz A, Simons M. Branching morphogenesis. Circ Res. 2008; 103: 784–795 [DOI] [PubMed] [Google Scholar]
- 27. Makarenkova HP, Ito M, Govindarajan V, et al. FGF10 is an inducer and Pax6 a competence factor for lacrimal gland development. Development. 2000; 127: 2563–2572 [DOI] [PubMed] [Google Scholar]
- 28. Tsau C, Ito M, Gromova A, Hoffman MP, Meech R, Makarenkova HP. Barx2 and Fgf10 regulate ocular glands branching morphogenesis by controlling extracellular matrix remodeling. Development. 2011; 138: 3307–3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Marechal H, Jammes H, Rossignol B, Mauduit P. EGF precursor mRNA and membrane-associated EGF precursor protein in rat exorbital lacrimal gland. Am J Physiol. 1999; 276: C734–C746 [DOI] [PubMed] [Google Scholar]
- 30. Mattiske D, Sommer P, Kidson SH, Hogan BL. The role of the forkhead transcription factor, Foxc1, in the development of the mouse lacrimal gland. Dev Dyn. 2006; 235: 1074–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Dean C, Ito M, Makarenkova HP, Faber SC, Lang RA. Bmp7 regulates branching morphogenesis of the lacrimal gland by promoting mesenchymal proliferation and condensation. Development. 2004; 131: 4155–4165 [DOI] [PubMed] [Google Scholar]
- 32. Dean CH, Miller LA, Smith AN, Dufort D, Lang RA, Niswander LA. Canonical Wnt signaling negatively regulates branching morphogenesis of the lung and lacrimal gland. Dev Biol. 2005; 286: 270–286 [DOI] [PubMed] [Google Scholar]
- 33. Li WC, Rukstalis JM, Nishimura W, et al. Activation of pancreatic-duct-derived progenitor cells during pancreas regeneration in adult rats. J Cell Sci. 2010; 123: 2792–2802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Burgess KL, Dardick I, Cummins MM, Burford-Mason AP, Bassett R, Brown DH. Myoepithelial cells actively proliferate during atrophy of rat parotid gland. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 1996; 82: 674–680 [DOI] [PubMed] [Google Scholar]
- 35. Lombaert IM, Knox SM, Hoffman MP. Salivary gland progenitor cell biology provides a rationale for therapeutic salivary gland regeneration. Oral Dis. 2011; 17: 445–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Smith GH. Mammary stem cells come of age, prospectively. Trends Mol Med. 2006; 12: 287–289 [DOI] [PubMed] [Google Scholar]
- 37. Zoukhri D, Fix A, Alroy J, Kublin CL. Mechanisms of murine lacrimal gland repair after experimentally induced inflammation. Invest Ophthalmol Vis Sci. 2008; 49: 4399–4406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. You S, Avidan O, Tariq A, et al. Role of epithelial-mesenchymal transition in repair of the lacrimal gland after experimentally induced injury. Invest Ophthalmol Vis Sci. 2012; 53: 126–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Liakhovitskaia A, Gribi R, Stamateris E, et al. Restoration of Runx1 expression in the Tie2 cell compartment rescues definitive hematopoietic stem cells and extends life of Runx1 knockout animals until birth. Stem Cells. 2009; 27: 1616–1624 [DOI] [PubMed] [Google Scholar]
- 40. Samokhvalov IM, Thomson AM, Lalancette C, Liakhovitskaia A, Ure J, Medvinsky A. Multifunctional reversible knockout/reporter system enabling fully functional reconstitution of the AML1/Runx1 locus and rescue of hematopoiesis. Genesis. 2006; 44: 115–121 [DOI] [PubMed] [Google Scholar]
- 41. Zoukhri D, Ko S, Stark PC, Kublin CL. Roles of caspase 1 and extracellular signal-regulated kinase in inflammation-induced inhibition of lacrimal gland protein secretion. Invest Ophthalmol Vis Sci. 2008; 49: 4392–4398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Weaver M, Dunn NR, Hogan BL. Bmp4 and Fgf10 play opposing roles during lung bud morphogenesis. Development. 2000; 127: 2695–2704 [DOI] [PubMed] [Google Scholar]
- 43. Makarenkova HP, Hoffman MP, Beenken A, et al. Differential interactions of FGFs with heparan sulfate control gradient formation and branching morphogenesis. Sci Signal. 2009; 2: ra55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lombaert IM, Knox SM, Hoffman MP. Salivary gland progenitor cell biology provides a rationale for therapeutic salivary gland regeneration. Oral Dis. 2011; 17: 445–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Gupta D, Harvey SA, Kaminski N, Swamynathan SK. Mouse conjunctival forniceal gene expression during postnatal development and its regulation by Kruppel-like factor 4. Invest Ophthalmol Vis Sci. 2011; 52: 4951–4962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Fijneman RJ, Anderson RA, Richards E, et al. Runx1 is a tumor suppressor gene in the mouse gastrointestinal tract. Cancer Sci. 2012; 103: 593–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wong WF, Kohu K, Nakamura A, et al. Runx1 deficiency in CD4+ T cells causes fatal autoimmune inflammatory lung disease due to spontaneous hyperactivation of cells. J Immunol. 2012; 188: 5408–5420 [DOI] [PubMed] [Google Scholar]
- 48. Nagasao J, Yoshioka K, Amasaki H, Mutoh K. Centroacinar and intercalated duct cells as potential precursors of pancreatic endocrine cells in rats treated with streptozotocin. Ann Anat. 2003; 185: 211–216 [DOI] [PubMed] [Google Scholar]
- 49. Stanger BZ, Stiles B, Lauwers GY, et al. Pten constrains centroacinar cell expansion and malignant transformation in the pancreas. Cancer Cell. 2005; 8: 185–195 [DOI] [PubMed] [Google Scholar]
- 50. Millar TJ, Herok G, Koutavas H, Martin DK, Anderton PJ. Immunohistochemical and histochemical characterisation of epithelial cells of rabbit lacrimal glands in tissue sections and cell cultures. Tissue Cell. 1996; 28: 301–312 [DOI] [PubMed] [Google Scholar]
- 51. Ward NL, Putoczki T, Mearow K, Ivanco TL, Dumont DJ. Vascular-specific growth factor angiopoietin 1 is involved in the organization of neuronal processes. J Comp Neurol. 2005; 482: 244–256 [DOI] [PubMed] [Google Scholar]
- 52. Kisanuki YY, Hammer RE, Miyazaki J, Williams SC, Richardson JA, Yanagisawa M. Tie2-Cre transgenic mice: a new model for endothelial cell-lineage analysis in vivo. Dev Biol. 2001; 230: 230–242 [DOI] [PubMed] [Google Scholar]
- 53. Stifani N, Freitas AR, Liakhovitskaia A, Medvinsky A, Kania A, Stifani S. Suppression of interneuron programs and maintenance of selected spinal motor neuron fates by the transcription factor AML1/Runx1. Proc Natl Acad Sci U S A. 2008; 105: 6451–6456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Dyson MH, Thomson S, Inagaki M, et al. MAP kinase-mediated phosphorylation of distinct pools of histone H3 at S10 or S28 via mitogen- and stress-activated kinase 1/2. J Cell Sci. 2005; 118: 2247–2259 [DOI] [PubMed] [Google Scholar]
- 55. Knox SM, Lombaert IM, Reed X, Vitale-Cross L, Gutkind JS, Hoffman MP. Parasympathetic innervation maintains epithelial progenitor cells during salivary organogenesis. Science. 2010; 329: 1645–1647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Rock JR, Hogan BL. Developmental biology. Branching takes nerve. Science. 2010; 329: 1610–1611 [DOI] [PubMed] [Google Scholar]
- 57. Luo X, Okubo T, Randell S, Hogan BL. Culture of endodermal stem/progenitor cells of the mouse tongue. In Vitro Cell Dev Biol Anim. 2009; 45: 44–54 [DOI] [PubMed] [Google Scholar]
- 58. Haley KJ, Lasky-Su J, Manoli SE, et al. RUNX transcription factors: association with pediatric asthma and modulated by maternal smoking. Am J Physiol Lung Cell Mol Physiol. 2011; 301: L693–L701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zusso M, Methot L, Lo R, Greenhalgh AD, David S, Stifani S. Regulation of postnatal forebrain amoeboid microglial cell proliferation and development by the transcription factor Runx1. J Neurosci. 2012; 32: 11285–11298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Inoue K, Ito Y. Neuroblastoma cell proliferation is sensitive to changes in levels of RUNX1 and RUNX3 protein. Gene. 2011; 487: 151–155 [DOI] [PubMed] [Google Scholar]
- 61. Zagami CJ, Zusso M, Stifani S. Runx transcription factors: lineage-specific regulators of neuronal precursor cell proliferation and post-mitotic neuron subtype development. J Cell Biochem. 2009; 107: 1063–1072 [DOI] [PubMed] [Google Scholar]
- 62. Nevadunsky NS, Barbieri JS, Kwong J, et al. RUNX3 protein is overexpressed in human epithelial ovarian cancer. Gynecol Oncol. 2009; 112: 325–330 [DOI] [PubMed] [Google Scholar]
- 63. Fukamachi H, Ito K. Growth regulation of gastric epithelial cells by Runx3. Oncogene. 2004; 23: 4330–4335 [DOI] [PubMed] [Google Scholar]
- 64. Akech J, Wixted JJ, Bedard K, et al. Runx2 association with progression of prostate cancer in patients: mechanisms mediating bone osteolysis and osteoblastic metastatic lesions. Oncogene. 2009; 29: 811–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Peterson LF, Boyapati A, Ranganathan V, et al. The hematopoietic transcription factor AML1 (RUNX1) is negatively regulated by the cell cycle protein cyclin D3. Mol Cell Biol. 2005; 25: 10205–10219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Zhang M, Xie R, Hou W, et al. PTHrP prevents chondrocyte premature hypertrophy by inducing cyclin-D1-dependent Runx2 and Runx3 phosphorylation, ubiquitylation and proteasomal degradation. J Cell Sci. 2009; 122: 1382–1389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Liu YN, Lee WW, Wang CY, Chao TH, Chen Y, Chen JH. Regulatory mechanisms controlling human E-cadherin gene expression. Oncogene. 2005; 24: 8277–8290 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.