Abstract
The mitogen-activated protein kinase kinase 1 and 2 signaling pathway is a major component of the RAS (Rat sarcoma)/RAF (Radpidly accelerated fibrosarcoma)/MEK (mitogen-activated protein kinase kinase)/ERKs (Extracellular signal-regulated kinases) signaling axis that regulates tumorigenesis and cancer cell growth. MEK is frequently activated in various cancers that have mutations in the KRAS and BRAF oncogenes. Therefore, MEK has been suggested as a therapeutic target for inhibitor development against tumors that are dependent on the activating mutations in mitogen-activated protein kinase signaling. Herein, we report the discovery of three novel MEK inhibitors, herein referred to as CInQ-01, CInQ-03 and CInQ-06. All three inhibitors were highly effective in suppressing MEK1 and MEK2 in vitro kinase activity as well as anchorage-dependent and anchorage-independent cell growth. The inhibitory activity was associated with markedly reduced phosphorylation of ERKs and ribosomal S6 kinases. Furthermore, administration of CInQ-03 inhibited colon cancer cell growth in an in vivo xenograft mouse model and showed no skin toxicity. Overall, these results suggest that these novel MEK inhibitors might be used for chemotherapy or prevention.
Introduction
The activation of v-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog (KRAS), phosphoinositide-3-kinase catalytic alpha polypeptide (PIK3CA) and v-raf murine sarcoma viral oncogene homolog B1 (BRAF) oncogenes and the inactivation of adenomatous polyposis coli, SMAD family member 4 (SMAD4) and p53 tumor suppressor genes are observed in the progression of tumor development from benign epithelium to colorectal cancers (1,2). KRAS is the most frequently mutated gene occurring in about 50% of colorectal tumors and induces the activation of the RAF family Ser/Thr kinases. This overactivation leads to sequential phosphorylation and activation of mitogen-activated protein kinase kinase 1/2 (MEK1/2) and its direct downstream substrates, the extracellular signal-regulated kinases 1/2 (ERK1/2) (3). The p90 ribosomal S6 kinases (RSK) are directly activated by ERK1/2 and promote tumorigenesis, cell proliferation and cell survival (4,5). Thus, therapeutic approaches for inhibiting the Ras signaling pathway will be useful for treating colorectal cancer. However, the efforts to develop effective anti-Ras therapies have been challenging with limited success (2,3). Previous studies showed that ectopic expression of Ras or MEK induces cell transformation in a ERKs-activation-dependent manner (6–8). Cancers displaying an activating BRAF mutation were shown to respond to MEK inhibition (9). T-LAK cell–originated protein kinase (TOPK) is a serine–threonine kinase that is a member of mitogen-activated protein kinase kinase (MAPKK) family (10) and positive feedback between TOPK and ERK2 promotes colorectal cancer formation (11). These studies provide strong rationale for the development of MAPKK family inhibitors for chemotherapeutic intervention in colorectal cancer (12).
MEK1 and 2 show 85% amino acid identity and are required for cell proliferation mediated through cell cycle regulatory events (13). In contrast, the differences between MEK1 and 2 include a higher catalytical activity of MEK2 (14) and MEK2 knockout mice are fully viable, whereas MEK1 knockout is embryonic lethal (15,16).
The adenosine triphosphate (ATP)-binding pocket is highly conserved among different kinase proteins. Therefore, highly selective MEK inhibitors that are non-ATP competitive have been reported and intensely studied pre-clinically (17–19). PD98059 as a first small-molecule MEK inhibitor and U0126 as an allosteric MEK inhibitor have been reported but had pharmaceutical limitations (20,21). Furthermore, second-generation MEK inhibitors, CI-1040 (Pfizer) and PD0325901 (Pfizer), were identified and showed strong antitumor activity in vivo (22,23). However, treatment of patients with these inhibitors showed insufficient antitumor activity or severe toxicity, including blurred vision and neurotoxicity. AZD6244 (ARRY-142886; AstraZeneca) is another second-generation MEK inhibitor that was developed based on the PD184352 structure. It is highly selective for MEK and binds non-competitively with ATP. The benzimidazole derivative AZD6244 suppressed tumor growth in vivo and entered clinical trials (24–26). Unfortunately, MEK inhibitor-associated diarrhea and skin disorders such as rash have been observed (27). Recently, dermatologic side effects associated with AZD6244 were reported and corresponded highly with epidermal growth factor (EGF) receptor inhibitor treatment. Investigators indicated that 77% of patients treated with AZD6244 developed an acute papulopustular rash within 6 weeks. Chronic skin toxicities induced by AZD6244 treatment over 6 weeks included 35% with xerosis cutis and 12% with paronychia, hair abnormalities (e.g. non-scarring alopecia), changes in pigment and skin aging (28). Additionally, Ki-67 expression as a keratinocytic proliferation marker protein induced by AZD6244 was not different in treated compared with matched untreated controls and the proliferation rate was also similar in both groups. Authors also suggested that basal keratinocyte proliferation is distinct from increasing suprabasal proliferation and basal keratinocyte proliferation might be affected by MEK inhibition. Therefore, the location of Ki-67 expression significantly separates the suprabasal keratinocyte layer from the basal layer in AZD6244-treated patients (28,29).
Herein, we report newly discovered potent allosteric and specific MEK inhibitors that exert anticancer activity both in vitro and in vivo. Overall, these findings should be useful for preventing cancer development.
Materials and methods
Reagents
CInQ-01 (N-(12-cyanindolizino[2,3-b]quinoxalin-3-yl)-4-fluoroben zamide, purity: 95%), CInQ-02 (N-(12-cyanindolizino[2,3-b]quinoxalin-3-yl)-2- thiophenecaboxamide, purity: 95%), CInQ-03 (2-chloro-N-(12-cyanindolizino [2,3-b]quinoxalin-2-yl)benzamide, purity: 95%), CInQ-04 (N-(12-cyanindolizino[2,3-b]quinoxalin-2-yl)-4-methylbenzenesulfonamide, purity: 95%), CInQ-05 (2-(1,1-dimethylethyl)-indolizino[2,3-b]quinoxaline-12-carbonitrile, purity: 95%) and CInQ-06 (N-(12-cyanindolizino[2,3-b]quinoxalin-2-yl)-4-fluorobenzamide, purity: 95%) were purchased from InterBioScreen (Moscow, Russia). Active MEK1, inactive ERK2 (MEK substrate) and histone H2AX (TOPK substrate) human recombinant proteins for kinase assays were purchased from Millipore (Temecula, CA). Active MEK2 and active TOPK human recombinant proteins for kinase assays were purchased from SignalChem (Richmond, British Columbia). Antibodies to detect total MEK, phosphorylated MEK, total ERKs, phosphorylated ERKs, total RSK and phosphorylated RSK were purchased from Cell Signaling Technology (Beverly, MA). Antibodies against total MEK1, total MEK2 and β-actin were purchased from Santa Cruz Biotechnology (Santa Cruz, CA).
Cell culture
All cell lines were purchased from American Type Culture Collection and were cytogenetically tested and authenticated before the cells were frozen. Each vial of frozen cells was thawed and maintained in culture for a maximum of 8 weeks. Enough frozen vials were available for each cell line to ensure that all cell-based experiments were conducted on cells that had been tested and in culture for 8 weeks or less. HCT116 human colon cancer cells were cultured in McCoy’s 5A medium supplemented with 10% fetal bovine serum (FBS; Atlanta Biologicals, Lawrenceville, GA) and 1% antibiotic–antimycotic. HCT15 human colon cancer cells were cultured in RPMI1640 medium supplemented with 10% FBS (Atlanta Biologicals) and 1% antibiotic–antimycotic. HaCaT (human keratinocyte) cells were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% FBS (Atlanta Biologicals) and 1% antibiotic–antimycotic.
In vitro kinase assay
The kinase assay was performed in accordance with instructions provided by Upstate Biotechnology (Billerica, MA). Briefly, the reaction was carried out in the presence of 10 μCi of [γ-32P] ATP with each compound in 40 μl of reaction buffer containing 20mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid (pH 7.4), 10mM MgCl2, 10mM MnCl2 and 1mM dithiothreitol. After incubation at room temperature for 30min, the reaction was stopped by adding 10 μl of protein loading buffer and the mixture was separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis. Each experiment was repeated twice and the relative amounts of incorporated radioactivity were assessed by autoradiography.
Western blot analysis
Cell lysates were prepared with Radio-immunoprecipitation assay (RIPA) buffer (50mM Tris–HCl pH 7.4, 1% NP-40, 0.25% sodium deoxycholate, 0.1% sodium dodecyl sulfate, 150mM NaCl, 1mM ethylenediaminetetraacetic acid, 1× protease inhibitor tablet). Equal amounts of protein were determined using the bicinchoninic acid assay (Pierce, Rockford, IL). Proteins were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis and transferred to polyvinylidene difluoride membranes (Amersham Pharmacia Biotech). Membranes were blocked with 5% non-fat dry milk for 1h at room temperature and incubated with appropriate primary antibodies overnight at 4°C. After washing with phosphate-buffered saline containing 0.1% Tween 20, the membrane was incubated with a horseradish peroxidase-conjugated secondary antibody at a 1:5000 dilution and the signal was detected with a chemiluminescence reagent (Amersham Biosciences Corp.).
Molecular modeling
The crystal structures of MEK1 and MEK2 were obtained from the RCSB Protein Data Bank (PDB entry: 1S9J and 1S9I) (30). The crystal structures were prepared using the Protein Preparation Wizard in Maestro v9.2. Hydrogens were added consistent with a pH of 7. All water molecules were removed and then the structure was minimized with an RMSD cutoff value of 0.3 Å. Three compounds were prepared using LigPrep v2.5 and then assigned AMSOL partial atom charge. The program Glide v5.7 (31) was used for ligand docking. The receptor grid was created with the centroid of the crystal ligand as the center of the grid. Flexible docking was performed with extra precision mode. The number of poses per ligand was set to 10 in postdocking minimization and at most 5 poses would be output. The other parameters were kept as default.
Cell proliferation assay
Cells were seeded (1×103 cells per well) in 96-well plates and incubated for 24h and then treated with different doses of each compound. After incubation for 1, 2 or 3 days, 20 μl of CellTiter96 Aqueous One Solution (3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium [MTS]; Promega) were added and then cells were incubated for 1h at 37°C in a 5% CO2 incubator. Absorbance was measured at 492nm.
Anchorage-independent cell growth
Colon cancer cells (8×103 cells per well) suspended in complete growth medium (McCoy’s 5A or RPMI1640 supplemented with 10% FBS and 1% antibiotics) were added to 0.3% agar with or without different doses of each compound in a top layer over a base layer of 0.6% agar with or without different doses of each compound. HaCaT keratinocytes (8×103 cells per well) suspended in complete growth medium (Dulbecco's modified Eagle's medium supplemented with 10% FBS and 1% antibiotics) were added to 0.3% agar with EGF alone or EGF with different doses of each compound in a top layer over a base layer of 0.6% agar with EGF alone or EGF with different doses of each compound. The cultures were maintained at 37°C in a 5% CO2 incubator for 3 weeks and then colonies were counted under a microscope using the Image-Pro Plus software (v. 4) program (Media Cybernetics).
Luciferase assay for AP-1 reporter activity
Transient transfection was conducted using JetPEI (Qbiogene, Carlsbad, CA), and assays to determine firefly luciferase and Renilla activities were performed according to the manufacturer’s manual (Promega, Madison, WI). Cells (1×104 cells per well) were seeded the day before transfection into 12-well culture plates. Cells were co-transfected with the AP-1 reporter plasmid (250ng) and an internal control (CMV-Renilla, 50ng) and incubated for 24h. Colon cancer cells were treated with individual CInQ inhibitors or U0126 for 2 days. Keratinocytes were treated with CInQ inhibitors or U0126 for 2h before EGF treatment for 12 or 24h. Cells were harvested in Promega lysis buffer. The AP-1 and Renilla luciferase activities were measured using substrates provided in the reporter assay system (Promega). The luciferase activity was normalized to Renilla luciferase activity.
Lentiviral infection
The lentiviral expression vectors, including Gipz-shMEK1 or shMEK2 and packaging vectors, including pMD2.0G and psPAX, were purchased from Addgene (Cambridge, MA). To prepare MEK1/2 viral particles, each viral vector and packaging vectors (pMD2.0G and psPAX) were transfected into HEK293T cells using JetPEI following the manufacturer’s suggested protocols. The transfection medium was changed at 4h after transfection and then cells were cultured for 36h. The viral particles were harvested by filtration using a 0.45mm sodium acetate syringe filter, then combined with 8 µg/ml of polybrane (Millipore, Billerica, MA) and infected into 60% confluent HCT116 cells overnight. The cell culture medium was replaced with fresh complete growth medium for 24h and then cells were selected with puromycine for 36h (1.5 µg/ml of puromycine). The selected cells were used for experiments.
Hematoxylin and eosin staining and immunohistochemistry
Tumor and skin tissues from mice were embedded in paraffin blocks and subjected to hematoxylin and eosin (H&E) staining and immunohistochemistry (IHC). Tissue sections were deparaffinized and hydrated and then permeabilized with 0.5% Triton X-100/1× phosphate-buffered saline for 10min. After developing with 3,3′-diaminobenzidine, the sections were counterstained with H&E. For IHC, sections were hybridized with the primary antibody (1:500) and horseradish peroxidase-conjugated goat antirabbit or mouse IgG antibody was used as the secondary antibody. All sections were observed by microscope and the Image-Pro Plus software (v. 4) program (Media Cybernetics).
Xenograft mouse model
Athymic mice (Cr:NIH(S), NIH Swiss nude, 6–9 week old) were obtained from Charles River and maintained under ‘specific pathogen-free’ conditions based on the guidelines established by the University of Minnesota Institutional Animal Care and Use Committee. Mice were divided into four groups of 10 animals: (i) untreated vehicle group (n = 10); (ii) 1mg CInQ-03/kg body wt (n = 10); (iii) 5mg CInQ-03/kg body wt (n = 10) and (iv) no cells and 5mg CInQ-03/kg body wt (n = 10). HCT116 cells (1.5×106 cells/100 µl) were suspended in serum-free McCoy’s 5A medium and inoculated subcutaneously into the right flank of each mouse. CInQ-03 or vehicle (5% dimethyl sulfoxide in 10% Tween 20) was injected three times per week for 11 days. Tumor volume was calculated from measurements of two diameters of the individual tumor base using the following formula: tumor volume (mm3) = (length × width × height × 0.52). Mice were monitored until tumors reached 1cm3 total volume, at which time mice were euthanized and tumors were extracted.
Statistical analysis
All quantitative results are expressed as mean ± SD or SE. Statistically significant differences were obtained using the Student’s t-test or by one-way analysis of variance. A P < 0.05 was considered to be statistically significant.
Results
CInQ inhibitors suppress MEK1 and MEK2 kinase activities
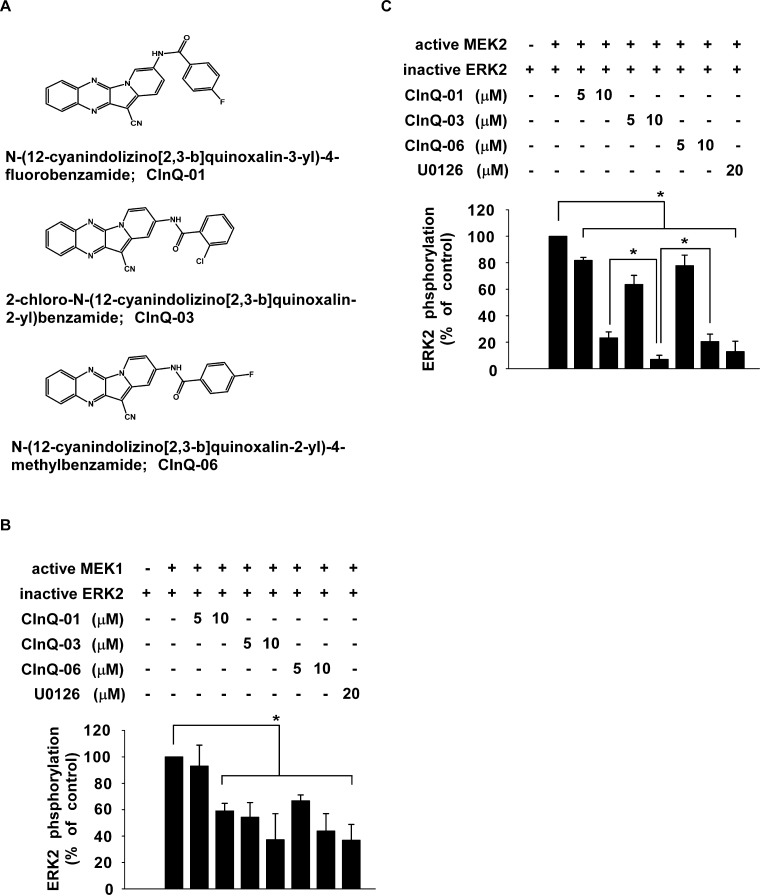
Previously, we reported the identification of a novel TOPK inhibitor (HI-TOPK-032). To identify a more potent TOPK inhibitor, we screened analogs of HI-TOPK-032 (Supplementary Figure 1A, available at Carcinogenesis Online; CInQ-01 to CInQ-06) using an in vitro TOPK kinase assay. Interestingly, these analogs had little effect on TOPK activity (30 µM; Supplementary Figure 1B, available at Carcinogenesis Online). Because TOPK is a MAPKK family member, we tested the effect of the analogs (30 µM) on MEK activity and results showed that MEK1 kinase activity was strongly inhibited by CInQ-01, -03 or -06. U0126 is a well-known MEK inhibitor and was used as a positive control. These data suggested that CInQ-01, -03 and -06 are potent MEK inhibitors (Supplementary Figure 1C, available at Carcinogenesis Online). Therefore, these three compounds (Figure 1A; 5 and 10 µM) were selected and evaluated further by in vitro MEK1 and MEK2 kinase assays. Results showed that CInQ-01, -03 and -06 significantly suppressed MEK1 and MEK2 kinase activities in a dose-dependent manner (Figure 1B and C). Additionally, inhibition of MEK2 kinase activity by CInQ-03 (10 µM) was stronger than the others. These results suggested that CInQ-01, -03 and -06 are specific and potent MEK inhibitors.
Fig. 1.
CInQ inhibitors suppress MEK kinase activity. (A) Respective chemical structures of CInQ-01, -03 and -06. CInQ inhibitors (CInQ-01, -03 and -06) significantly suppress (B) MEK1 and (C) MEK2 kinase activities in a dose-dependent manner. The effect of CInQ inhibitors or U0126, a well-known MEK inhibitor, on MEK activity was assessed by an in vitro kinase assay using MEK1 (active, 250ng) or MEK2 (active, 500ng) and inactive ERK2 (MEK1 or 2 substrate, 500ng) proteins with [γ-32P] ATP. For B and C, all data are represented as means ± SD of values from three independent experiments. Band density was measured using the Image J (NIH) software program and is illustrated as percent of control (no inhibitors, second column). The asterisk (*) indicates a significant (P < 0.05) decrease induced by CInQ-01, -03 or -06 compared with untreated control.
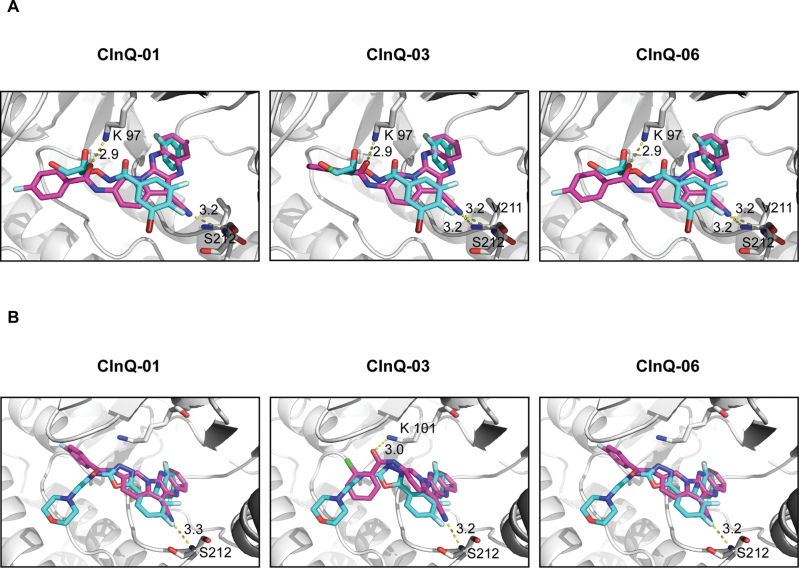
Computer modeling of the MEK and CInQ inhibitor complexes
We performed molecular docking of the individual CInQ inhibitors and MEK in order to determine the binding orientation of the inhibitors with MEK. In the docked structures of MEK1 with the individual CInQ inhibitors, all compounds formed hydrogen bonds with Lys97 and Ser212 and the docked poses overlap very well with the crystal ligand PD318088 (Figure 2A). In the crystal structure of MEK1, PD318088 also forms a hydrogen bond with Lys97. The docking score of MEK1 with PD318088 is −9.45, and the docking scores of MEK1 with CInQ-01, -03 and -06 are −7.84, −7.29 and −7.83, respectively. Additionally, in the docked structure of MEK2 with the CInQ inhibitors, all compounds formed a hydrogen bond with Ser216 (Figure 2B). CInQ-03 forms an additional hydrogen bond with Lys101, which also forms a hydrogen bond with PD334581 in the crystal structure of MEK2. The docking score of MEK2 with PD334581 is −8.92, and the docking scores of MEK2 with CInQ-01, -03 and -06 are −8.54, −6.49, and −8.58, respectively. These scores suggest a binding affinity that is similar to the known inhibitor.
Fig. 2.
Computer models of CInQ inhibitors docked with MEK. (A) Docking models of MEK1 with CInQ-01, -03 or -06. Carbons of PD318088, the crystal ligand, are shown in cyan. (B) Docking models of MEK2 with CInQ-01, -03 or -06. Carbons of PD334581, the crystal ligand, are shown in cyan. Carbons of MEK1 or 2 are shown in green and carbons of individual compounds are shown in magenta. Nitrogen is shown in blue, oxygen in red, fluorine in light cyan, chlorine in light green, bromine in ruby and iodine in light purple.
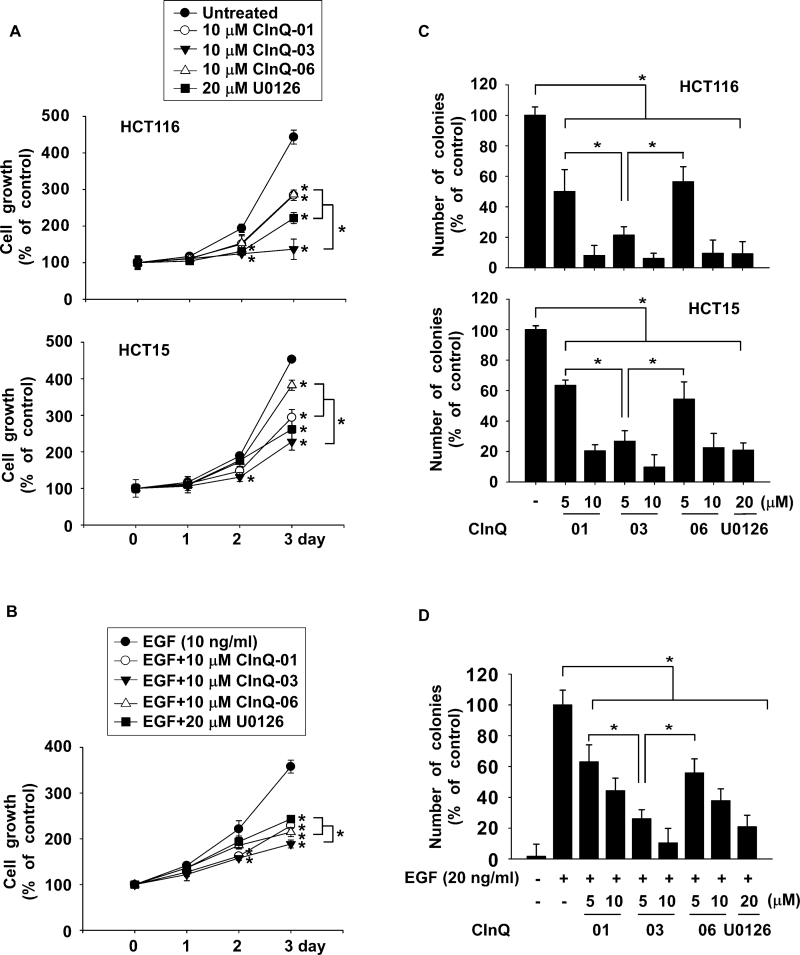
CInQ inhibitors suppress anchorage-dependent and anchorage-independent cell growth
We examined the effect of the CInQ inhibitors on colon cancer cell growth and on the growth of EGF-induced HaCaT keratinocytes. Cell growth was measured using the MTS assay at 1, 2 or 3 days after treatment with EGF alone or EGF and individual CInQ inhibitors. Results indicated that colon cancer cell growth was significantly decreased by the respective CInQ inhibitors (Figure 3A). EGF-induced HaCaT cell growth was also strongly suppressed by CInQ inhibitors (Figure 3B).
Fig. 3.
CInQ-01, -03 or -06 exerts anticancer activity. (A) CInQ inhibitors dose dependently inhibit colon cancer cell growth. Cells were treated with the individual CInQ inhibitor or U0126 for 1, 2 or 3 days. (B) Effect of CInQ inhibitors on EGF-induced growth of human keratinocytes (HaCaT cells). Cells were co-treated with EGF and CInQ inhibitor or U0126 for 1, 2 or 3 days and growth was analyzed by MTS assay. For A and B, all data are shown as means ± SD of values from three independent experiments with triplicate samples. The asterisk (*) indicates a significant (P < 0.05) difference in growth of cells treated with inhibitor compared with untreated or EGF-only-treated control. (C) CInQ inhibitors dose dependently suppress anchorage-independent colon cancer cell growth. Cells were treated with individual CInQ inhibitors or U0126 in 0.3% agar and incubated for 3 weeks. (D) Effect of individual CInQ inhibitors or U0126 on EGF-induced transformation of HaCaT cells. Cells were co-treated with EGF and CInQ inhibitors or U0126 in 0.3% agar and incubated for 2 weeks. Colonies were counted using a microscope and the Image-Pro Plus (v. 6) computer software program. For C and D, all data are shown as means ± SD of values from three independent experiments each with triplicate samples. The asterisk (*) indicates a significant (P < 0.05) difference in colony formation in cells treated with CInQ inhibitors or U0126 compared with untreated or EGF-only-treated control cells.
Next, to determine the effect of the CInQ inhibitors on anchorage-independent growth and EGF-induced transformation, cells were seeded with EGF alone or EGF with individual CInQ inhibitors in 0.3% agar and incubated for 2 or 3 weeks. Data showed that anchorage-independent cancer cell growth was strongly suppressed by CInQ inhibitors in a dose-dependent manner (Figure 3C). EGF-induced HaCaT cell transformation was also significantly inhibited by the respective CInQ inhibitors (Figure 3D).
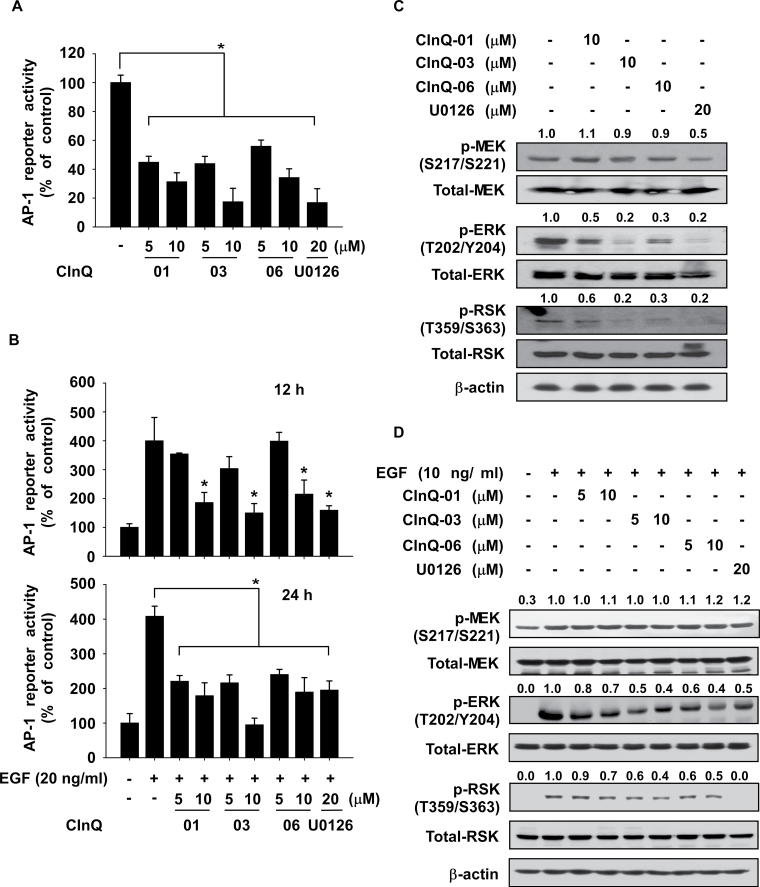
CInQ inhibitors suppress activator protein-1 activity
We then determined whether CInQ inhibitors had an effect on activator protein-1 (AP-1) reporter activity in HCT116 or HaCaT cells stimulated with EGF. HCT116 cells were treated with individual CInQ inhibitors for 48h and HaCaT cells were treated with CInQ inhibitor for 2h before stimulation with EGF for 12 or 24h. AP-1 reporter activity was strongly suppressed by CInQ inhibitors in colon cancer cells (Figure 4A) and in EGF-treated HaCaT cells (Figure 4B). We also examined the effect of these inhibitors on downstream signaling of MEK in colon cancer cells and EGF-induced HaCaT cells. Results showed that each CInQ inhibitor suppressed phosphorylation of ERK1/2 and RSK (Figure 4C and D). However, the CInQ inhibitors had little effect on the phosphorylation of MEK.
Fig. 4.
Effect of CInQ-01, -03 or -06 on AP-1 promoter activity. (A) CInQ inhibitors suppress AP-1 reporter activity in colon cancer cells. Cells were transfected with the AP-1-luciferase reporter and CMV-Renilla plasmids. At 1 day after transfection, cells were treated with individual CInQ inhibitors or U0126 and incubated for 2 days. The AP-1 reporter activity was assessed. (B) Effect of CInQ inhibitors on EGF-induced AP-1 reporter activity in HaCaT cells. Cells were transfected with the AP-1-luciferase reporter and CMV-Renilla plasmids for 1 day. Cells were then treated with CInQ inhibitors or U0126 for 2h before treatment with EGF (10ng/ml) for 12 or 24h. For A and B, all data are shown as means ± SD of values from three independent experiments each with triplicate samples. The asterisk (*) indicates a significant (P < 0.05) difference in AP-1 reporter activity in cells treated with CInQ inhibitors or U0126 compared with untreated or EGF-only-treated control cells. (C) CInQ inhibitors suppress AP-1 signaling in colon cancer cells. Colon cancer cells were treated with individual CInQ inhibitors or U0126 for 2 days. (D) Effect of CInQ inhibitors on EGF-induced AP-1 signaling in HaCaT cells. Cells were treated with CInQ inhibitor or U0126 for 2h before treatment with EGF for 15min. The expression of MEK downstream proteins was determined by western blotting. β-Actin was used to verify equal protein loading. Band density was measured using the Image J (NIH) software program. Similar results were obtained from three independent experiments and representative blots are shown.
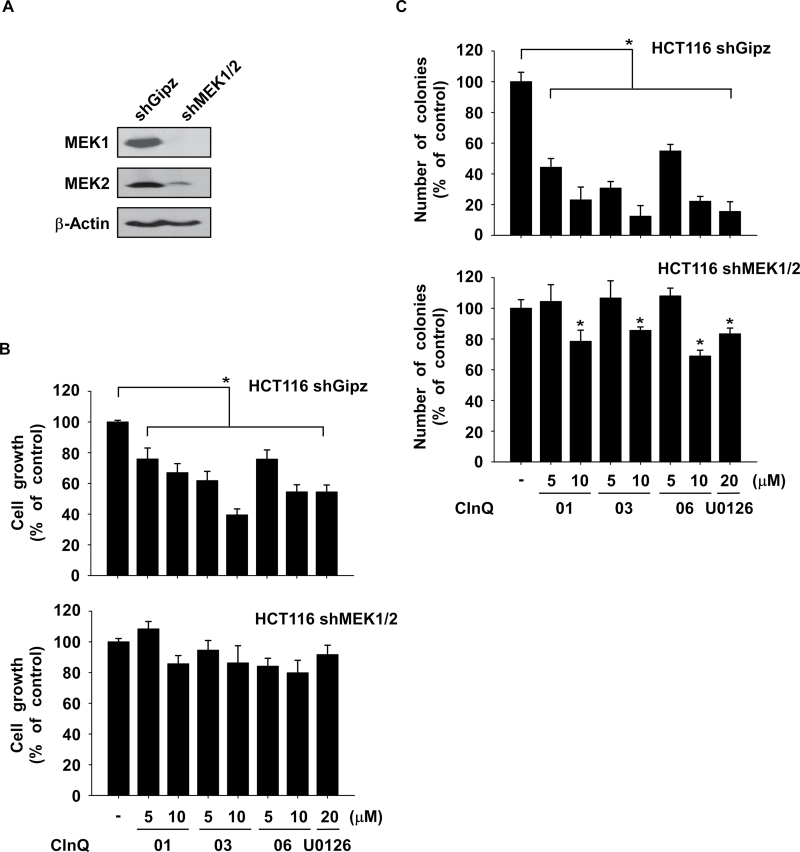
The anticancer effects of CInQ inhibitors are dependent on MEK expression
To further study the anticancer effects of the CInQ inhibitors, we established HCT116 cells stably expressing mock (shGipz) or knockdown of MEK1/2 (shMEK1/2) and analyzed MEK expression by western blot (Supplementary Figure 2A and B, available at Carcinogenesis Online; Figure 5A). The effect of inhibitors on growth of shGipz or shMEK1/2 colon cancer cells was assessed by MTS assay at 72h. Results indicated that cells expressing shMEK1/2 were resistant to the antigrowth effect of the CInQ inhibitors compared with shGipz-expressing cells (Figure 5B). Furthermore, we examined the effect of CInQ inhibitors on anchorage-independent colon cancer cell growth. Results showed that the inhibition of anchorage-independent colon cancer cell growth induced by the CInQ inhibitors was less effective in shMEK cells compared with shGipz cells (Figure 5C). These results indicated that the anticancer activity induced by CInQ inhibitors is dependent on MEK expression.
Fig. 5.
The anticancer activity exerted by CInQ inhibitors is dependent on MEK expression. (A) Colon cancer cells stably expressing knockdown of MEK1/2 were established using lentiviral infection. The expression of MEK1 and 2 was determined by western blotting. β-Actin was used to verify equal protein loading. Band density was measured using the Image J (NIH) software program. (B) The effect of CInQ inhibitors or U0126 on cell growth was not as efficient in knockdown MEK1/2 cells compared with control cells. Cells were treated with CInQ inhibitors or U0126 for 3 days and cell growth was analyzed by MTS assay. (C) CInQ inhibitors or U0126 were not as effective to inhibit anchorage-independent cell growth in knockdown MEK1/2 cells compared with control cells. Cells were treated with CInQ inhibitors or U0126 in 0.3% agar and incubated for 3 weeks at 37°C/5% CO2. For B and C, all data are shown as means ± SD of values from three independent experiments each with triplicate samples. The asterisk (*) indicates a significant (P < 0.05) decrease in colony formation induced by CInQ inhibitors or U0126 compared with untreated control cells.
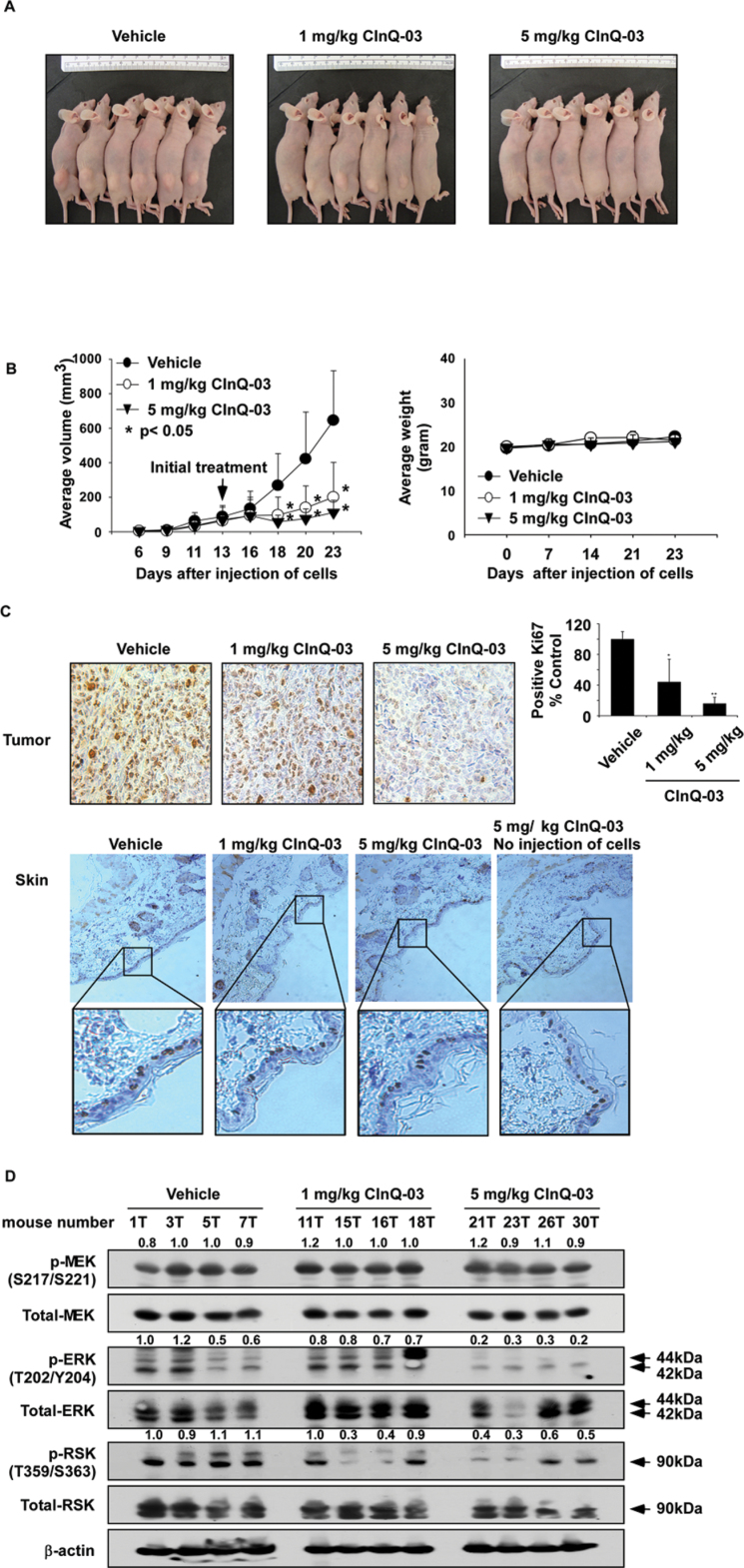
CInQ-03 inhibits colon cancer tumor growth in a xenograft mouse model
Based on the results described above, CInQ-03 showed a stronger anticancer effect compared with the other inhibitors. Therefore, we selected CInQ-03 as the most potent MEK inhibitor for further study in vivo. HCT116 colon cancer cells were injected into the flank of athymic nude mice and mice were treated with CInQ-03 at 1 or 5mg/kg or vehicle three times a week over a period of 11 days after the average tumor volume grew to about 70mm3. Treatment of mice with 1 or 5mg/kg of CInQ-03 strongly suppressed HCT116 tumor growth by over 70% relative to the vehicle-treated group (Figure 6A and B, left panel; P < 0.05). Additionally, mice seemed to tolerate treatment with CInQ-03 without overt signs of toxicity or significant loss of body weight similar to the vehicle-treated group (Figure 6B, right panel). Furthermore, the effects of CInQ-03 on a tumor proliferation marker were evaluated by IHC and H&E staining of HCT116 tumor and skin tissues after 11 days of treatment. The expression of Ki-67 was markedly decreased by treatment with CInQ-03 (Figure 6C, upper panel). However, Ki-67 expression in skin tissues in CInQ-03-treated tissues was similar to the vehicle-treated group (Figure 6C, lower panel).
Fig. 6.
CInQ-03 prevents xenograft tumor growth. (A) Representative photographs of tumor-bearing athymic nude mouse treated or not treated with CInQ-03. (B, left panel) CInQ-03 suppresses colon tumor growth. HCT116 colon cancer cells were injected subcutaneously into the dorsal right flank of mice. Micewere injected with CInQ-03 or vehicle three times a week for 11 days. Mice were monitored until tumors reached 1cm3 total volume, at which time mice were euthanized. Tumors and skin tissues were harvested. Tumor volume was calculated from measurements of two diameters of the individual tumor based on the following formula: tumor volume (mm3) = (length × width × height × 0.52). Data are shown as means ± SE of values obtained from the experiments. The asterisk (*) indicates a significant difference between tumors from untreated and treated mice as determined by t-test (P < 0.05). (B, right panel) CInQ-03 has no effect on mouse body weight. Body weights from treated or untreated groups of mice were obtained once a week for 23 days. (C) H&E staining and IHC analysis of tumor and skin tissues. Treated or untreated groups of mice were euthanized and then tumors and skin tissues were harvested. Colon tumor (upper panel) and skin tissue (lower panel) slides were prepared from paraffin sections after fixation with formalin and then stained with H&E or anti-Ki-67. Expression of Ki-67 was visualized by light microscope (×200). The number of Ki-67-stained cells (right panel in C) was counted from IHC results (N = 3; *P < 0.05, **P < 0.01). The fourth upper panel in C is from mice not injected with cells but with compound only—no tumors developed. (D) CInQ-03 inhibits MEK-target protein expression in HCT116 colon tumor tissues. Tumor tissues from groups treated with vehicle, 1 or 5mg CInQ-03 per kg body wt were immunoblotted with antibodies to detect total MEK, phosphorylated MEK, total ERKs, phosphorylated ERKs, total RSK, phosphorylated RSK and β-actin. β-Actin was used to verify equal protein loading. Band density was measured using the Image J (NIH) software program.
We then examined the effect of CInQ-03 on MEK downstream signaling in tumor tissues. Expression of MEK-targeted proteins was analyzed by western blot. Results indicated that phosphorylation of ERKs and RSK was strongly suppressed by CInQ-03 treatment (Figure 6D). These findings indicated that HCT116 colon tumor growth was suppressed by CInQ-03 through its targeting of the MEK signaling pathway.
Discussion
The anticancer activities of a highly selective non-ATP competitive inhibitor of MEK1/2 have been reported in in vitro and in vivo studies (17,18,32–34). Recently, phase I and II clinical evaluation of several MEK inhibitors, including PD0325901, AZD6244 and XL518, which share a common core structure, have been conducted (24,35,36). Despite many efforts to identify MEK inhibitors and provide effective preclinical results using second-generation MEK inhibitors, which are currently in clinical trials for various solid malignancies, none has yet been approved. Current inhibitors are associated with diarrhea and dermatologic toxicities including rash (28,35). Although subtle structural diversities suggest biological differences, a similar toxicity by these MEK inhibitors has been observed. Therefore, identification of MEK inhibitors that are structurally different and less toxic than current MEK inhibitors is needed. To characterize the most potent MEK inhibitors, computer docking and scoring were conducted to compare the CInQs and current MEK inhibitors. Interestingly, although the CInQs occupy a similar docking pose with MEKs, the CInQs have a distinct chemical structure compared with current MEK inhibitors, such as PD318088, PD334581, PD184352, PD0325901 and AZD6244. On the other hand, several questions have arisen regarding an explanation for the differential inhibitory mechanism and skin toxicity between CInQs and current MEK inhibitors. In addition, pharmacokinetic and pharmacodynamic values for CInQs are also needed. These questions will be addressed in future studies.
Results of the MEK1 docking study showed that the CInQ compounds have similar docking scores and display almost the same binding mode. However, in the MEK2 docking study, CInQ-03 displayed a higher (i.e. poorer) docking score than the other two, but exhibited a better binding mode. Notably, the CInQ compounds all bind deeply into the binding pocket in a manner similar to the crystal ligand. Overall, the docking and experimental data suggest that the CInQ compounds display an ability to inhibit MEK1/2 and CInQ-03 is more potent than the other two.
We found that expression of Ki-67 was strongly inhibited by treatment with CInQ-03 in tumor tissues (Figure 6C, upper panels). Next, to examine potential skin toxicity that might be induced by CInQ-03, H&E-stained skin tissues were analyzed. Previous reports suggested that dermatologic side effects induced by AZD6244 were associated with a shift in Ki-67 positive expression from the basal layer to the suprabasal keratinocyte layers (28,29). Interestingly, expression in the basal layer and the suprabasal keratinocyte layers of skin tissues treated with CInQ-03 was similar to the vehicle-treated group (Figure 6C, lower panel). In contrast, EGF-induced growth of keratinocytes was significantly inhibited by CInQs (Figure 3B). To maintain keratinocyte homeostasis, MEK/ERK/RSK signaling is required for proliferation (37,38). Our group reported that TOPK phosphorylated ERKs and that TOPK was phosphorylated by ERK2 (11). Additionally, our preliminary data showed that U0126 could inhibit TOPK activity (data not shown). Apoptosis of keratinocytes might be increased if MEK- or TOPK-mediated ERKs activation is completely blocked by current MEK inhibitors. No one has studied the effect of MEK inhibitors derived from benzhydroxamate or benzimidazole derivatives on MAPKK family members, especially TOPK. CInQ-03 as a specific MEK inhibitor can block MEK-mediated ERKs activation but has no effect on TOPK-mediated ERKs activation.
In this study, to identify a specific MEK inhibitor, we examined the effect of candidate compounds on MEK family kinase activities by using in vitro MEK and TOPK kinase assays. We found that CInQ inhibitors are specific and potent MEK inhibitors in in vitro and cell-based assays. Furthermore, results from a xenograft mouse model indicated that administration of CInQ-03 at 1 or 5mg/kg body wt for 11 days significantly suppressed colon cancer cell growth and was not toxic (Figure 6B). Furthermore, suppression of phosphorylation of ERKs by CInQ-03 in a cell-based assay was highly correlated with the in vivo animal results (Figure 4C and D; Figure 6D).
Based on these findings, we suggest that CInQ-03 is a novel and specific MEK inhibitor both in vitro and in vivo. These results should be useful for development of novel MEK inhibitors. Future studies will investigate the efficacy of CInQ-03 and pharmacological characterization and detailed examination of dermatologic toxicities.
Supplementary material
Supplementary Figures 1 and 2 can be found at http://carcin.oxfordjournals.org/
Funding
The Hormel Foundation and National Institutes of Health grants CA120388, CA027502, R37CA081064 and ES016548 and the Korea Research Council of Fundamental Science and Technology (KRCF) Research Fellowship for Young Scientists.
Supplementary Material
Acknowledgements
We thank Tonya Poorman for secretarial assistance in submitting this manuscript. Current address for Dr. Hua Xie is State Key Laboratory of Drug Research, Shanghai Institute of Materia Medica, Chinese Academy of Sciences, Beijing, China.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations:
- ATP
adenosine triphosphate
- CInQ-01
N-(12-cyanindo lizino[2,3-b]quinoxalin-3-yl)-4-fluorobenzamide
- CInQ-03
2-chloro-N-(12- cyanindolizino[2,3-b]quinoxalin-2-yl)benzamide
- EGF
epidermal growth factor
- FBS
fetal bovine serum
- H&E
hematoxylin and eosin
- MAPK
mitogen-activated protein kinase
- MAPKK
mitogen-activated protein kinase kinase
- MEK1/2
mitogen-activated protein kinase kinase 1 and 2
- MTS
3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium
- RSK
ribosomal S6 kinases
- TOPK
T-LAK cell–originated protein kinase.
References
- 1. Schubbert S., et al. (2007). Hyperactive Ras in developmental disorders and cancer. Nat. Rev. Cancer, 7, 295–308 [DOI] [PubMed] [Google Scholar]
- 2. Sjöblom T., et al. (2006). The consensus coding sequences of human breast and colorectal cancers. Science, 314, 268–274 [DOI] [PubMed] [Google Scholar]
- 3. Pearson G., et al. (2001). Mitogen-activated protein (MAP) kinase pathways: regulation and physiological functions. Endocr. Rev., 22, 153–183 [DOI] [PubMed] [Google Scholar]
- 4. Anjum R., et al. (2008). The RSK family of kinases: emerging roles in cellular signalling. Nat. Rev. Mol. Cell Biol., 9, 747–758 [DOI] [PubMed] [Google Scholar]
- 5. Eisinger-Mathason T.S., et al. (2010). RSK in tumorigenesis: connections to steroid signaling. Steroids, 75, 191–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cowley S., et al. (1994). Activation of MAP kinase kinase is necessary and sufficient for PC12 differentiation and for transformation of NIH 3T3 cells. Cell, 77, 841–852 [DOI] [PubMed] [Google Scholar]
- 7. Mansour S.J., et al. (1994). Transformation of mammalian cells by constitutively active MAP kinase kinase. Science, 265, 966–970 [DOI] [PubMed] [Google Scholar]
- 8. Shields J.M., et al. (2000). Understanding Ras: ‘it ain’t over ‘til it’s over’. Trends Cell Biol., 10, 147–154 [DOI] [PubMed] [Google Scholar]
- 9. Estep A.L., et al. (2007). Mutation analysis of BRAF, MEK1 and MEK2 in 15 ovarian cancer cell lines: implications for therapy. PLoS ONE, 2, e1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kim D.J., et al. (2012). Novel TOPK inhibitor HI-TOPK-032 effectively suppresses colon cancer growth. Cancer Res., 72, 3060–3068 [DOI] [PubMed] [Google Scholar]
- 11. Zhu F., et al. (2007). Bidirectional signals transduced by TOPK-ERK interaction increase tumorigenesis of HCT116 colorectal cancer cells. Gastroenterology, 133, 219–231 [DOI] [PubMed] [Google Scholar]
- 12. McCubrey J.A., et al. (2010). Emerging MEK inhibitors. Expert Opin. Emerg. Drugs, 15, 203–223 [DOI] [PubMed] [Google Scholar]
- 13. Liu X., et al. (2004). The MAP kinase pathway is required for entry into mitosis and cell survival. Oncogene, 23, 763–776 [DOI] [PubMed] [Google Scholar]
- 14. Zheng C.F., et al. (1993). Cloning and characterization of two distinct human extracellular signal-regulated kinase activator kinases, MEK1 and MEK2. J. Biol. Chem., 268, 11435–11439 [PubMed] [Google Scholar]
- 15. Bélanger L.F., et al. (2003). Mek2 is dispensable for mouse growth and development. Mol. Cell. Biol., 23, 4778–4787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Giroux S., et al. (1999). Embryonic death of Mek1-deficient mice reveals a role for this kinase in angiogenesis in the labyrinthine region of the placenta. Curr. Biol., 9, 369–372 [DOI] [PubMed] [Google Scholar]
- 17. Davies B.R., et al. (2007). AZD6244 (ARRY-142886), a potent inhibitor of mitogen-activated protein kinase/extracellular signal-regulated kinase kinase ½ kinases: mechanism of action in vivo, pharmacokinetic/pharmacodynamic relationship, and potential for combination in preclinical models. Mol. Cancer Ther., 6, 2209–2219 [DOI] [PubMed] [Google Scholar]
- 18. Lorusso P.M., et al. (2005). Phase I and pharmacodynamic study of the oral MEK inhibitor CI-1040 in patients with advanced malignancies. J. Clin. Oncol., 23, 5281–5293 [DOI] [PubMed] [Google Scholar]
- 19. Menon U., et al. (2005). Prospective study using the risk of ovarian cancer algorithm to screen for ovarian cancer. J. Clin. Oncol., 23, 7919–7926 [DOI] [PubMed] [Google Scholar]
- 20. Dudley D.T., et al. (1995). A synthetic inhibitor of the mitogen-activated protein kinase cascade. Proc. Natl. Acad. Sci. U.S.A., 92, 7686–7689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Favata M.F., et al. (1998). Identification of a novel inhibitor of mitogen-activated protein kinase kinase. J. Biol. Chem., 273, 18623–18632 [DOI] [PubMed] [Google Scholar]
- 22. Sebolt-Leopold J.S., et al. (1999). Blockade of the MAP kinase pathway suppresses growth of colon tumors in vivo . Nat. Med., 5, 810–816 [DOI] [PubMed] [Google Scholar]
- 23. Solit D.B., et al. (2006). BRAF mutation predicts sensitivity to MEK inhibition. Nature, 439, 358–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Adjei A.A., et al. (2008). Phase I pharmacokinetic and pharmacodynamic study of the oral, small-molecule mitogen-activated protein kinase kinase ½ inhibitor AZD6244 (ARRY-142886) in patients with advanced cancers. J. Clin. Oncol., 26, 2139–2146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Huynh H., et al. (2007). Targeted inhibition of the extracellular signal-regulated kinase kinase pathway with AZD6244 (ARRY-142886) in the treatment of hepatocellular carcinoma. Mol. Cancer Ther., 6, 138–146 [DOI] [PubMed] [Google Scholar]
- 26. Yeh T.C., et al. (2007). Biological characterization of ARRY-142886 (AZD6244), a potent, highly selective mitogen-activated protein kinase kinase ½ inhibitor. Clin. Cancer Res., 13, 1576–1583 [DOI] [PubMed] [Google Scholar]
- 27. Messersmith W.A., et al. (2006). Novel targets in solid tumors: MEK inhibitors. Clin. Adv. Hematol. Oncol., 4, 831–836 [PubMed] [Google Scholar]
- 28. Schad K., et al. (2010). Mitogen-activated protein/extracellular signal-regulated kinase kinase inhibition results in biphasic alteration of epidermal homeostasis with keratinocytic apoptosis and pigmentation disorders. Clin. Cancer Res., 16, 1058–1064 [DOI] [PubMed] [Google Scholar]
- 29. Balagula Y., et al. (2011). Dermatologic side effects associated with the MEK ½ inhibitor selumetinib (AZD6244, ARRY-142886). Invest. New Drugs, 29, 1114–1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ohren J.F., et al. (2004). Structures of human MAP kinase kinase 1 (MEK1) and MEK2 describe novel noncompetitive kinase inhibition. Nat. Struct. Mol. Biol., 11, 1192–1197 [DOI] [PubMed] [Google Scholar]
- 31. Friesner R.A., et al. (2006). Extra precision glide: docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. J. Med. Chem., 49, 6177–6196 [DOI] [PubMed] [Google Scholar]
- 32. Daouti S., et al. (2009). Characterization of a novel mitogen-activated protein kinase kinase ½ inhibitor with a unique mechanism of action for cancer therapy. Cancer Res., 69, 1924–1932 [DOI] [PubMed] [Google Scholar]
- 33. Iverson C., et al. (2009). RDEA119/BAY 869766: a potent, selective, allosteric inhibitor of MEK1/2 for the treatment of cancer. Cancer Res., 69, 6839–6847 [DOI] [PubMed] [Google Scholar]
- 34. Yang J.Y., et al. (2010). Activation of FOXO3a is sufficient to reverse mitogen-activated protein/extracellular signal-regulated kinase kinase inhibitor chemoresistance in human cancer. Cancer Res., 70, 4709–4718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wang D., et al. (2007). Clinical experience of MEK inhibitors in cancer therapy. Biochim. Biophys. Acta, 1773, 1248–1255 [DOI] [PubMed] [Google Scholar]
- 36. Wang J.Y., et al. (2007). Recent advances of MEK inhibitors and their clinical progress. Curr. Top. Med. Chem., 7, 1364–1378 [DOI] [PubMed] [Google Scholar]
- 37. Dumesic P.A., et al. (2009). Erk1/2 MAP kinases are required for epidermal G2/M progression. J. Cell Biol., 185, 409–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Scholl F.A., et al. (2009). Mek1/2 gene dosage determines tissue response to oncogenic Ras signaling in the skin. Oncogene, 28, 1485–1495 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.