Abstract
Carcinoid tumors are rare neuroendocrine tumors (NETs) that are increasing in incidence. Mutation and altered expression of Wnt/β-catenin signaling components have been described in many tumors but have not been well-studied in NETs. Here, we observed accumulation of β-catenin in the cytoplasm and/or nucleus in 25% of clinical NET tissues. By mutational analysis, the mutations of β-catenin (I35S) and APC (E1317Q, T1493T) were identified in NET cells and the tissues. Expression of representative Wnt inhibitors was absent or markedly decreased in BON, a human pancreatic carcinoid cell line; treatment with 5-aza-2′-deoxycytidine (5-aza-CdR) increased expression levels of the Wnt inhibitors. Methylation analyses demonstrated that CpG islands of SFRP-1 and Axin-2 were methylated, whereas the promoters of DKK-1, DKK-3 and WIF-1 were unmethylated in four NET cells. Aberrant methylation of SFRP-1 was particularly observed in most of clinical NET tissues. In addition, the repression of these unmethylated genes was associated with histone H3 lysine 9 dimethylation (H3K9me2) in BON cells. Together, 5-aza-CdR treatment inhibited cell proliferation and decreased the protein levels of H3K9me2 and G9a. Moreover, a novel G9a inhibitor, UNC0638, suppressed BON cell proliferation through inhibition of Wnt/β-catenin pathway. Overexpression of the inhibitory genes, particularly SFRP-1 and WIF-1 in BON cells, resulted in suppression of anchorage-independent growth and inhibition of tumor growth in mice. Our findings suggest that aberrant Wnt/β-catenin signaling, through either mutations or epigenetic silencing of Wnt antagonists, contributes to the pathogenesis and growth of NETs and have important clinical implications for the prognosis and treatment of NETs.
Introduction
Carcinoid tumors are relatively uncommon, slow growing neuroendocrine tumors (NETs) arising from enterochromaffin cells and predominantly found in the gastrointestinal (GI) tract (1,2). The overall incidence of carcinoid tumors is approximately 2 cases per 100000 people, but the actual incidence is thought to be higher (3,4). Moreover, in contrast to many cancers that have been decreasing over the last decade, the incidence appears to be increasing (3–5). Although the cornerstone of treatment for carcinoid tumors is surgery, oncogenic pathways such as various growth factor receptors and mammalian target of rapamycin (mTOR) have been implicated in the pathogenesis of carcinoid tumors and recently evaluated as therapeutic targets (2,6).
The Wnt/β-catenin signaling involved in early development and tissue maintenance of adults is accomplished via regulation of a specific set of genes implicated in cell proliferation and differentiation (7,8). This signaling leads to stabilization of β-catenin, which activates T-cell factor/lymphoid enhancer-binding factor (TCF/LEF)-dependent transcription of its target genes (9). Hyperactivation of this pathway has been implicated in numerous malignancies (7,9). Adenomatous Polyposis Coli (APC) and β-catenin mutations, as well as altered expression of the pathway components, represent important mechanisms underlying activation of Wnt/β-catenin signaling (8–10).
Wnt antagonists, including members of the ‘destruction complex’ for β-catenin (APC and Axin-2), are commonly mutated and demonstrate a high frequency of aberrant promoter methylation (10–13). In addition, epigenetic silencing of extracellular Wnt inhibitors, such as secreted Frizzled-related proteins (SFRPs), Wnt inhibitory factor-1 (WIF-1) and DICKKOPFs (DKKs) may contribute to the stabilization and accumulation of β-catenin in cancers with or without mutational activation of Wnt/β-catenin signaling (14–21).
Similar to promoter methylation, histone modifications also play critical roles in transcriptional regulation (22). While histone acetylation and arginine methylation of histone H3 and H4 are related to transcriptional activation, methylation of histones may be associated with active or repressive transcriptional activity depending on the methylation site (22,23). Among them, methylation of histone H3 lysine 9 is a well-characterized modification (24,25), which is regulated by histone lysine methyltransferases including G9a and results in repression of the tumor suppressors (e.g. DSC-3 and MASPIN) in breast cancer (26) and RUNX-3 in gastric cancer (27).
Although a high frequency of cytoplasmic accumulation and/or nuclear translocation of β-catenin has been described previously in carcinoid tumors, its correlation with mutational activation of proteins in this cascade, as well as with localization of β-catenin, is controversial (28,29). In our current study, we analyzed accumulation of β-catenin in the cytoplasm and/or nucleus and identified mutations of β-catenin and APC in NET cell lines and clinical tissues. We also demonstrated that transcriptional silencing of typical negative regulators of the Wnt/β-catenin pathway occurred by DNA methylation or histone modification of their promoters in NET cell lines, mainly BON that was established and characterized in our laboratory and has been widely used as a unique model of carcinoid cell biology (30). Finally, we showed that restoration of the Wnt inhibitory genes such as SFRP-1 and WIF-1 resulted in tumor suppressor functions both in vitro and in vivo.
Materials and methods
Materials
Expression vectors (pcDNA3.1His, Invitrogen, Carlsbad, CA) for SFRP-1 and DKK-1 (18) were kindly provided by Dr Hiromu Suzuki (Department of Biochemistry, Sapporo Medical University, Japan). Other vectors in this study included TOPA TA cloning vector from Invitrogen, TCF/LEF-responsive reporter (TOPFlash) and Renilla luciferase reporter from Promega (Madison, WI). All kinds of DNA and RNA isolation kit and HotStarTaq DNA Polymerase were purchased from Qiagen (Valencia, CA). MethylCode™ Bisulfite Conversion Kit, Platinum® Pfx DNA Polymerase, Lipofectamine™ 2000 and NuPAGE® 4–12% Bis-Tris gels were obtained from Invitrogen. A High-Capacity cDNA Reverse Transcription Kit was acquired from Applied Biosystems (Foster City, CA) and iQ SYBR-green Supermix for quantitative reverse transcription–polymerase chain reaction (qRT–PCR) and Sequi-Blot™ PVDF membranes for western blotting were purchased from Bio-Rad (Irvine, CA). A β-catenin antibody was obtained from BD Biosciences (San Jose, CA). The antibody for β-actin and cell lysis buffer were purchased from Cell Signaling (Danvers, MA). Antibodies against active-β-catenin, histone H3, dimethylated at lysine 9 (H3K9me2) and G9a and Chromatin Immunoprecipitation (ChIP) Assay Kit were acquired from Millipore (Bedford, MA). The anti-c-Myc and anti-DKK-1 antibodies were purchased from Epitomics (Burlingame, CA) and Santa Cruz Biotechnology (Santa Cruz, CA), respectively. All other antibodies were obtained from Abcam (Cambridge, MA). ECL™ western blotting detection reagents were purchased from GE Healthcare (Buckinghamshire, UK). Cell Death Detection ELISAplus kit for apoptosis assays was acquired from Roche (Indianapolis, IN) and Dual-Luciferase® Reporter Assay System for luciferase assay was purchased from Promega. UNC0638, a potent G9a inhibitor (31), was obtained from Cayman Chemical (Ann Arbor, MI). All other reagents including 5-aza-2′-deoxycytidine (5-aza-CdR) were purchased from Sigma (St Louis, MO).
Immunohistochemistry
Paraffin-embedded tissue blocks of carcinoid tumors from colon (n = 1), small bowel (n = 11), lung (n = 6) and thymus (n = 2) were sectioned (5 μm) and deparaffinized in xylene and rehydrated in a descending ethanol series. Immunostaining with β-catenin antibody was performed as described previously (32). Assessment of the stained slides was performed blindly by an experienced pathologist.
Cell culture
The human pancreatic carcinoid cell line BON (30) is cultured in Dulbecco’s modified Eagle’s medium and F12K in a 1:1 ratio supplemented with 5% fetal bovine serum. QGP-1 (pancreatic somatostatinoma cell line) purchased from Japan Health Sciences Foundation (Osaka, Japan) and NCI-H727 and UMC-11 cells (lung carcinoid tumor cell lines) obtained from ATCC (Manassas, VA) are maintained in RPMI 1640 medium with 10% fetal bovine serum.
Direct DNA sequence analysis
To screen for mutations in exon 3 of β-catenin or in a mutation cluster region in exon 15 of APC, genomic DNA extracted from cell lines or paraffin-embedded tissues was amplified by PCR. For APC, primer sets designed for screening codons 1231–1623 of APC were used. The primer sequences for β-catenin and APC are listed in Supplementary Table 1, available at Carcinogenesis Online. The PCR products were purified from agarose gels and were confirmed by direct sequence analyses using both forward and reverse primers.
RNA isolation, reverse transcription–polymerase chain reaction and qRT–PCR analysis
Total RNA was isolated from cultured cells using RNeasy kits (Qiagen) according to the manufacturer’s instructions. Reverse transcription–polymerase chain reaction (RT–PCR) analysis of SFRP-1, Axin-2, DKK-1, DKK-3 and WIF-1 expression was performed using cDNA synthesized from 1 µg of total RNA and the primers listed in Supplementary Table 1, available at Carcinogenesis Online. The PCR products were analyzed on a 2% agarose gel. qRT–PCR was performed using iCycler with the iQ SYBR-green Supermix (Bio-Rad) and the same gene-specific primers; β-actin was used as an internal control (33). The relative quantity of the transcripts was calculated by the formula 2−ΔΔCt, where ΔCt was determined by subtracting the average β-actin Ct value from the average target Ct value.
Methylation analysis
Methylation of 5′ regions of Wnt inhibitor genes was analyzed using methylation-specific PCR (MSP) and bisulfite sequencing analysis. Briefly, PCR was performed using bisulfite-modified genomic DNA and the primers listed in Supplementary Table 1, available at Carcinogenesis Online. For MSP, primer sets that distinguish between unmethylated (U) and methylated (M) DNA were used. The PCR products were visualized by 2% agarose gel. For bisulfite sequencing, PCR products were cloned into the TOPO TA cloning vector (Invitrogen) and the plasmids from individual bacterial colonies were sequenced using both side primers.
Chromatin immunoprecipitation analysis
ChIP analysis was performed according to the manufacturer’s protocol (Millipore). Purified DNA from cells incubated with 5-aza-CdR was then amplified across the DKK-1, DKK-3 and WIF-1 promoter region (Supplementary Table 1, available at Carcinogenesis Online). The relative quantity of the transcripts for DKK-1 and WIF-1 by real-time PCR was calculated by the formula 2−ΔΔCt, where ΔCt was determined by subtracting the average β-globin Ct value from the average target Ct value. The quantitation of DKK-3 was performed using gel-based PCR on a 2% agarose gel.
Western blot, cell proliferation and apoptosis analyses
Western blot, cell proliferation and apoptosis analyses were performed with the materials in this study as described previously (34).
Luciferase reporter assays
BON cells were plated in 24-well plates and transiently transfected with the TOPFlash reporter (0.4 μg) and the Renilla luciferase reporter (0.05 μg) with or without pcDNA3.1His vectors containing Wnt inhibitors (0.4 μg each) used previously (18). For construction of the expression vector for WIF-1, the complete open reading was produced by PCR from the WIF-1 cDNA clone (OriGene Technologies, Rockville, MD) using specified primers (Supplementary Table 1, available at Carcinogenesis Online). Transfections were performed using Lipofectamine™ 2000 according to the manufacturer’s instructions (Invitrogen). For UNC0638 treatment, varying concentrations of UNC0638 were treated into BON cells after 1 day. The cells were harvested and luciferase activity was measured 2 days after transfection.
Colony formation assay
BON cells (1×106 cells) were transfected with 10 μg of each Wnt inhibitor or empty vector by electroporation using GenePulser XCell (Bio-Rad). Cells were then plated in 6-well plates and selected for 2–3 weeks with G418 (0.8mg/ml). Colonies were then stained with crystal violet solution and imaged using the Alpha Innotech Imaging system and counted using the AlphaEaseFC™ software (Alpha Innotech Corporation, San Leandro, CA).
Soft agar assay
To measure anchorage-independent growth, BON cells were plated in growth medium containing 0.4% agarose in 6-well plates at a concentration of 2.5×105 cells per well onto a bottom layer of solidified 0.8% agarose. After incubation for 4 weeks, colonies were stained with crystal violet solution and quantified using AlphaEaseFC™ software. We also assessed colony size and dispersion using a Nikon TE2000 inverted microscope and NIS Elements AR3.10 software (Nikon, Melville, NY).
Tumor growth in nude mice
BON cells (5×106) stably transfected with empty vector or SFRP-1 were collected in 100 μl of sterile phosphate-buffered saline and inoculated subcutaneously into 6-week-old male athymic nude mice (n = 5 per group). After 3 weeks, the tumor size was measured with a vernier caliper and the tumor volume was defined as (longest diameter) × (shortest diameter)2/2. All animal procedures were performed in the nude mouse facility using protocols approved by the Institutional Animal Care and Use Committee.
Statistical analysis
Descriptive statistics including mean and standard deviation (SD) were calculated and bar graphs generated to summarize the results for respective experiments. Within each experiment, comparisons between two groups were performed using two-sample t-tests, whereas comparisons across groups were accomplished using one-way analysis of variance models and test for linear trend of increasing dose levels or pairwise comparisons with control group were subsequently performed using contrast statements. Estimates of mutation rates in cell lines and carcinoid tissues were calculated along with exact 95% confidence intervals, whereas tests whether mutation proportions were significantly different from the null value of 0 were performed using the binomial test. Normality assumptions of the analysis of variance and two-sample t-tests for each outcome were assessed. P-values < 0.05 were considered statistically significant.
Results
Analysis of β-catenin localization and mutation of β-catenin and APC in NETs
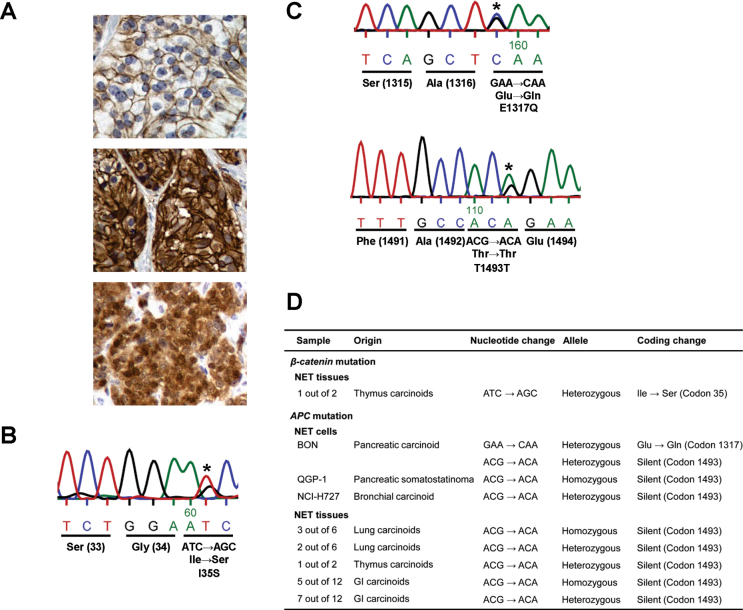
To examine Wnt/β-catenin signaling status in NETs, we first assessed β-catenin localization in clinical NET tissues (i.e. lung, thymus and GI carcinoids) since it is well known that activation of this pathway results in the localization of β-catenin in the cytoplasm and/or nucleus. In the normal epithelium of the colon, ileum and other organs, β-catenin staining was mainly localized to the cell membrane as reported previously (28) (data not shown). In 75% of the clinical samples (11 of 12 GI carcinoids and 4 of 6 lung carcinoids), there was membranous but not cytoplasmic or nuclear staining, similar to that of normal epithelium (Figure 1A, upper). In 15% cases (1 of 12 GI carcinoids and 2 of 6 lung carcinoids), cytoplasmic and membranous staining was detected (Figure 1A, middle), whereas nuclear and cytoplasmic staining was observed in two carcinoid tumors of the thymus (Figure 1A, bottom). Activation of Wnt/β-catenin signaling was confirmed in the NET samples demonstrating both cytoplasmic + membranous and nuclear + cytoplasmic staining, although there was variability in the degree of activation.
Fig. 1.
Analysis of β-catenin localization in clinical NET samples and direct DNA sequence analysis of β-catenin and APC in NET cell lines and clinical specimens. (A) Immunohistochemical analysis of β-catenin in clinical NET samples. Membranous staining of β-catenin without cytoplasmic and nuclear staining in a small bowel carcinoid tumor (upper). Cytoplasmic and membranous staining of β-catenin without nuclear staining in another small bowel carcinoid tumor (middle). Strong nuclear and cytoplasmic staining of β-catenin in a thymus carcinoid tumor (bottom). (B) I35S mutation in β-catenin (ATC to AGC substitution in nucleotide) in a thymus carcinoid tissue. (C) E1317Q mutation in APC (GAA to CAA substitution in nucleotide) in BON cells (upper). T1493T silent mutation in APC (ACG to ACA substitution in nucleotide) in BON cells (bottom). Asterisk indicates base substitution sites. (D) Summary of β-catenin and APC mutations in NET cell lines and clinical samples.
Next, we examined mutations in exon 3 of β-catenin and a mutation cluster region in exon 15 of APC in 4 NET cell lines and the 20 clinical NET samples. As shown in Figure 1B, a missense mutation (I35S) of β-catenin was identified in a thymus carcinoid tumor, which resulted in replacement of the Ile (ATC) at codon 35 by Ser (AGC); β-catenin accumulation in the cytoplasm and nucleus was observed (Figure 1A, bottom), and there appeared to be a correlation between accumulation of β-catenin and β-catenin mutation. In contrast, no β-catenin mutations were detected in the other NET cells and clinical samples. Moreover, a missense mutation containing a Glu (GAA) to Gln (CAA) substitution at codon 1317 (E1317Q) in the APC was identified in BON cells (Figure 1C, upper) but was not noted in the other samples. We also found a silent mutation (T1493T) of APC where the ACG at codon 1493 was replaced by ACA in BON cells (Figure 1C, bottom). The same silent mutation of APC was noted in QGP-1 and NCI-H727 cells and in NET tissue samples including all GI carcinoid tumors (Figure 1D). Estimates of silent mutation rates and 95% exact binomial confidence intervals for NET cell lines and clinical samples were 0.75 (3/4) and 95% CI: 0.19, 0.99 and 0.9 (18/20) and 95% CI: 0.68, 0.99, respectively. Based on the binomial test, each P-value for both specimens testing whether the mutation proportion is significantly different from 0 was noted to be statistically significant (P < 0.0001).
Negative regulators of the Wnt/β-catenin pathway are re-expressed by 5-aza-CdR treatment in BON cells
In addition to mutations of APC and β-catenin, the aberrant activation of Wnt/β-catenin signaling can also be achieved by silencing of inhibitory genes for the pathway (11,13–21). To determine whether the silencing of negative regulators of the pathway was due to methylation, we treated BON, the most extensively utilized NET cells, with the DNA methyltransferase inhibitor, 5-aza-CdR, and examined the expression of representative Wnt/β-catenin signaling inhibitors, SFRP-1, Axin-2, DKK-1, DKK-3 and WIF-1 using RT–PCR (Figure 2A). We found that treatment with 5-aza-CdR (either 0.5 or 5 µM) restored the expression of Wnt inhibitor genes. To confirm these results, mRNA expression levels were also investigated by qRT–PCR (Figure 2B). Incubation of cells with 5-aza-CdR (0.5 μM) resulted in a significant induction of all tested genes (6.2-fold for SFRP-1; 5.2-fold for Axin-2; 11.4-fold for DKK-1; 2.7-fold for DKK-3 and 4.7-fold for WIF-1). The effects were more pronounced after treatment with 5 μM 5-aza-CdR, suggesting that Wnt inhibitor genes were the targets of epigenetic silencing through methylation in BON cells.
Fig. 2.
Expression analysis of Wnt/β-catenin inhibitor genes in BON cells after 5-aza-CdR treatment. (A) RT–PCR analysis of SFRP-1, Axin-2, DKK-1, DKK-3, WIF-1 and β-actin expression in BON cells with or without 5-aza-CdR treatment. Cells were treated with varying concentrations of 5-aza-CdR for 96h with media changed every 24h. Total RNA was isolated from BON cells treated with 0 [dimethyl sulfoxide (DMSO)], 0.5 or 5 μM 5-aza-CdR and cDNA was synthesized from 1 µg of total RNA. The reaction was performed using HotStarTaq DNA Polymerase (Qiagen) and the primers listed in Supplementary Table 1, available at Carcinogenesis Online. The PCR products were visualized by 2% agarose gel. (B) qRT–PCR analysis confirmed that treatment with 5-aza-CdR induced the restoration of the Wnt/β-catenin pathway inhibitor genes in BON cells. The reaction was performed using iCycler with the iQ SYBR-green Supermix (Bio-Rad), cDNA and the same primers. Each gene expression was assessed by evaluating threshold cycle (Ct) values. The relative amount of mRNA expression was calculated by the comparative ΔΔCt method. Columns represent triplicate data points; error bars, SD; *P < 0.05 versus DMSO treatment; β-actin was used as internal control in both analyses.
The CpG islands in the 5′ regions of SFRP-1 and Axin-2 are hypermethylated in NET cells and the methylation of SFRP-1 is highly associated with in clinical NET tissues
To determine whether the induction of expression of the aforementioned Wnt/β-catenin signaling pathway inhibitors by 5-aza-CdR was due to CpG island methylation, we examined the methylation status of the 5′ regions of these genes using MSP analysis and bisulfite sequencing in four NET cell lines. MSP analysis using two MSP primer pairs revealed methylation of the 5′ region of SFRP-1 in all of the tested cell lines (Figure 3A). The methylation profile of CpG sites in the 5′ region of SFRP-1 was further analyzed in BON and QGP-1 cells by bisulfite sequencing (Figure 3B). Whereas complete methylation was noted in QGP-1 cells, the CpG islands of SFRP-1 were partially methylated in BON cells. Moreover, the promoter of Axin-2 was also methylated in these four NET cells as noted by MSP analysis (Figure 3A). Bisulfite sequencing of BON and QGP-1 cells confirmed hypermethylation of the CpG islands consistent with the MSP data (Figure 3C). Based on the above findings, we further investigated CpG island methylation of SFRP-1 and Axin-2 in the same NET patients’ samples used in immunohistochemical and mutational analyses. Aberrant CpG site methylation of SFRP-1 was observed in almost all NET samples by MSP using the two primer pairs but in only one GI carcinoid (p12), the methylation of Axin-2 was shown (Figure 3D). These data demonstrated that silencing of SFRP-1 and Axin-2 expression was linked with DNA methylation in NET cell lines and methylation of SFRP-1 was strongly associated with clinical NET tissues especially.
Fig. 3.
DNA methylation analyses of the 5′ region of the SFRP-1 and Axin-2 in NET cells and tissues. (A) MSP analysis of CpG islands of SFRP-1 and Axin-2 with respective two primer pairs (MSP-A and -B) and primers specific for the methylated (M) and unmethylated (U) DNA in four NET cells. NS represents non-specific bands or primer dimers for PCR products. (B) Bisulfite genomic sequencing analysis of two 5′ regions of SFRP-1 in BON and QGP-1 cells. (C) Bisulfite genomic sequencing analysis of Axin-2 CpG islands in BON and QGP-1 cells. Each row of circles represents the DNA sequence of an individual clone; closed and open circles indicate methylated and unmethylated CpG sites, respectively. (D) MSP analysis of the upstream region of the SFRP-1 and Axin-2 with the same primers described above in clinical NET samples (patient numbers).
Repression of unmethylated Wnt inhibitor genes is associated with the level of H3K9me2
The CpG island methylation status of the DKK-1, DKK-3 and WIF-1 was also tested using both MSP analysis (Figure 4A and Supplementary Figure 1A, available at Carcinogenesis Online) and bisulfite sequencing (Supplementary Figure 2, available at Carcinogenesis Online). Contrary to SFRP-1 and Axin-2 and unlike in most tumor cells in which the Wnt inhibitors are silenced by DNA methylation, the CpG sites of DKK-1 and DKK-3 in the four NET cell lines were found to be unmethylated by both analyses (Figure 4A and Supplementary Figures 1A and 2A, available at Carcinogenesis Online). Similar to DKKs, the CpG islands of WIF-1 were also unmethylated in the cell lines (Figure 4A and Supplementary Figure 2B, available at Carcinogenesis Online). In addition to DNA demethylation, 5-aza-CdR is also associated with H3K9 demethylation in the regulatory regions of silenced genes (26,35,36). In particular, induction of the silenced genes such as two antimetastatic tumor suppressor genes, DSC-3 and MASPIN, in breast cancer (26) and RUNX-3 in gastric cancer (27) by treatment with 5-aza-CdR was closely associated with significant decreases of H3K9me2 in the promoters. To examine whether H3K9me2 was involved in expression of the above genes, we performed ChIP analyses with an anti-H3K9me2 antibody. Interestingly, the level of H3K9me2 in the promoter of DKK-1 (Figure 4B), DKK-3 (Supplementary Figure 1B, available at Carcinogenesis Online) and WIF-1 (Figure 4C) decreased significantly by 5-aza-CdR in a dose-dependent manner. These data suggest that treatment of 5-aza-CdR results in strong H3K9me2 reduction in the gene promoters, which leads to increased expression of the inhibitors. Together, silencing of DKK-1, DKK-3 and WIF-1 expression in BON cells was not associated with DNA methylation but with histone modification. We next determined by cell counting and western blotting whether 5-aza-CdR treatment affected cell proliferation and the protein levels of above silenced Wnt inhibitors, H3K9me2 and G9a, the histone methyltransferase responsible for mono methylation and dimethylation of histone H3 lysine 9 (37), respectively. Treatment with 5-aza-CdR augmented the protein levels of silenced Wnt antagonists in BON cells (Supplementary Figure 3A, available at Carcinogenesis Online) consistent with RT–PCR analysis (Figure 2) and led to a pronounced inhibition of BON cell proliferation (Supplementary Figure 3B, available at Carcinogenesis Online) resulting from reactivation of silenced tumor suppressor genes including the Wnt inhibitors. Previously, it was reported that 5-aza-CdR treatment in human breast cancer cells reduced the global levels of H3K9me2 and the level of G9a was also decreased by treating with 5-aza-CdR although the exact mechanism is not known (26). Consistent with the report, treatment of 5-aza-CdR decreased the level of H3K9me2 and G9a protein expression in BON cells (Figure 4D). In order to further verify whether G9a-mediated H3K9me2 activates Wnt signaling by inhibiting the transcription of Wnt antagonists, BON cells were treated with varying concentrations of UNC0638, a novel G9a inhibitor (31). Cell proliferation (Supplementary Figure 4A, available at Carcinogenesis Online) and TOPFlash activity (Figure 4E) were significantly decreased by UNC0638 in a dose-dependent manner. Moreover, treatment with UNC0638 resulted in decrease in H3K9me2, leading to increased levels of DKK-1, DKK-3 and WIF-1 and reduced levels of c-Myc, a target of Wnt/β-catenin pathway (Figure 4F). These data indicated that H3K9me2 and G9a directly regulate the silencing of the Wnt inhibitors in BON cells. In addition, BON cells treated with UNC0638 demonstrated significantly increased sensitivity to serum-starvation-induced apoptosis (Supplementary Figure 4B, available at Carcinogenesis Online).
Fig. 4.
Repression of DKK-1 and WIF-1 is associated with the level of H3K9me2 in BON cells. (A) MSP analysis with primers specific for the methylated (M) and unmethylated (U) DKK-1 and WIF-1 CpG islands in four NET cell lines. ChIP analysis demonstrated reduction of H3K9me2 within the DKK-1 (B) and WIF-1 (C) promoter in BON cells following 5-aza-CdR treatment for 96h. The amount of recruitment of H3K9me2 to the DKK-1 and WIF-1 promoter was quantified by real-time PCR and plotted as the H3K9me2 binding ratio of DKK-1 or WIF-1 to β-globin and then expressed relative to DMSO-treated BON cells. Columns represent triplicate data points; error bars, SD; *P < 0.05 versus DMSO treatment. (D) BON cells were treated with 5-aza-CdR for 96h and protein extracts were analyzed by western blotting with the indicated antibodies; β-actin was used as an internal control for protein loading. (E) Relative luciferase activity obtained using a TOPFlash and, as a control, a Renilla luciferase reporter in BON cells treated with indicated concentrations of UNC0638. Columns represent triplicate data points; error bars, SD; *P < 0.05 versus DMSO treatment. (F) Western blotting analysis of UNC0638 effects on expression of silenced proteins and its related proteins. BON cells were treated with 0 or 2.5 μM UNC0638 for 1 day and protein extracts were analyzed with the indicated antibodies.
Suppression of TCF/LEF transcriptional activity and cell growth by Wnt inhibitor gene expression in BON cells
To examine the relationship between the silencing of Wnt inhibitors and the Wnt/β-catenin signaling activity, we transiently cotransfected BON cells with a TOPFlash and the Wnt inhibitor genes showing repressed expression. TCF/LEF activity was significantly suppressed by overexpression of the respective genes (0.4-fold for SFRP-1, 0.6-fold for WIF-1 and 0.3-fold for DKK-1) compared with empty vector (Figure 5A). Furthermore, in order to determine the effect of these genes in regulating growth, we expressed the Wnt inhibitors in BON cells and then assessed for colony formation (Figure 5B). After transfection and selection with G418, we found that introduction of these inhibitors markedly suppressed colony formation (51% for SFRP-1, 57% for WIF-1 and 41% for DKK-1) (Figure 5C). Overexpression of DKK-3 also diminished the TCF/LEF activity (0.8-fold) and colony formation (37%) compared with empty vector (Supplementary Figure 5, available at Carcinogenesis Online). Taken together, these results demonstrate that Wnt inhibitor genes, silenced in BON cells, can inhibit Wnt/β-catenin signaling activity and suppress tumor cell growth.
Fig. 5.
Tumor suppressive effects of Wnt inhibitor genes in BON cells. (A) Relative luciferase activity obtained using a TOPFlash and Renilla luciferase reporter in BON cells transiently transfected with control (empty vector) or the indicated Wnt inhibitor genes. Columns represent triplicate data points; error bars, SD; *P < 0.05 versus control. (B) Representative results from a colony formation assay carried out in BON cells transfected with control (empty vector) or the respective Wnt inhibitor gene constructs. (C) The relative number of colonies compared with the control; error bars, SD; *P < 0.05 versus control from triplicate analyses.
Tumor suppressor effects of SFRP-1 and WIF-1 in BON cells
Next, we established and selected BON cell clones demonstrating high levels of SFRP-1 (Figure 6) or WIF-1 (Supplementary Figure 6, available at Carcinogenesis Online) following G418 selection. Reporter assays using TOPFlash showed significant reduction of the transcriptional activity in both SFRP-1 stable cells compared with control cells stably transfected with the empty vector (Figure 6A). Expression of SFRP-1 decreased the levels of active-β-catenin, which recognizes the active, dephosphorylated form of β-catenin on Ser37 or Thr41, and c-Myc (Figure 6B). Additionally, to determine the effect of SFRP-1 expression on anchorage-independent growth, we performed soft agar assays using the stable transfectants. The numbers of colonies in cells stably expressing SFRP-1 were significantly lower than those of control cells (Figure 6C). Moreover, the size of colonies in the SFRP-1 clones was relatively small compared with those of control cells (Figure 6D). Finally, to investigate the effect of SFRP-1 on tumor growth in vivo, we injected the stable BON cell clones subcutaneously into the flanks of athymic nude mice and assessed growth. After 3 weeks, tumor growth was significantly reduced in mice injected with the SFRP-1-overexpressing BON cell clones (Figure 6E). Similarly, reduction of reporter activity and expression of the active form of β-catenin and c-Myc and suppression of anchorage-independent growth were shown in WIF-1-overexpressing BON cells (Supplementary Figure 6, available at Carcinogenesis Online). Collectively, these results demonstrate that restoration of Wnt antagonists, particularly SFRP-1 and WIF-1, reduces TCF/LEF activity, expression of active-β-catenin and c-Myc and tumor growth in vitro and in vivo implying a tumor suppressor function for the Wnt inhibitor genes in NET cells.
Fig. 6.
Tumor suppressive properties of SFRP-1 in BON cells. (A) Relative luciferase activity obtained using a TOPFlash and a Renilla in SFRP-1 BON cell clones (S1-2 and S1-4). Columns represent triplicate data points; error bars, SD; *P < 0.05 versus control (empty vector). (B) Western blot analysis showing expression of Wnt/β-catenin pathway-related proteins regulated by SFRP-1. Overexpression of SFRP-1 decreases the level of active-β-catenin and c-Myc, a target of Wnt/β-catenin signaling pathway. (C) The relative number of colonies compared with the control in soft agar assay. Colony formation of the BON cell clones was assessed over a period of 4 weeks. Columns represent triplicate data points; error bars, SD; *P < 0.05 versus control. (D) Representative results from a soft agar assay carried out in two SFRP-1 and control BON cell clones. (E) Overexpression of SFRP-1 suppresses growth of BON cell xenografts in vivo. Athymic nude mice were inoculated subcutaneously with the stable BON cell clones. Five mice were used in each group, and the cells were inoculated at one site in the flank of each mouse. Size of the tumors was measured after 3 weeks. Each value represents mean ± SD of tumor volume (mm3) (*P < 0.05 versus control).
Discussion
Accumulation of β-catenin in the cytoplasm and/or nucleus activates the transcription of Wnt target genes associated with cell proliferation and survival and is linked to human diseases including a variety of cancers (7,8). β-Catenin accumulation has been observed in GI carcinoid tumors (28,29), but the precise molecular mechanisms and functional consequences of Wnt/β-catenin pathway alterations have not been delineated. In the present study, we found that gene mutations (β-catenin and APC) and/or repression of Wnt inhibitors through epigenetic silencing by promoter methylation (SFRP-1 and Axin-2) or histone modification, H3K9me2 (DKK-1, DKK-3 and WIF-1), lead to aberrant Wnt/β-catenin signaling in NET samples, providing multiple mechanisms for the deregulation of this pathway in NETs.
First, we determined β-catenin localization in 20 NET clinical specimens and assessed mutational status of β-catenin and APC in the NET cells and tissue samples. Compared with previous reports (28,29), the frequency of cytoplasmic and/or nuclear localization of β-catenin in GI carcinoids was low (8.3%; 1 out of 12). In addition, only membranous β-catenin expression has been reported previously in non-GI carcinoid tumors (29); however, in our study, two of six lung and all thymus carcinoids demonstrated cytoplasmic and/or nuclear staining of β-catenin. The missense mutation (I35S) of β-catenin detected in a thymus carcinoid abrogates the phosphorylation-dependent interaction of β-catenin, resulting in activation of the Wnt/β-catenin signaling pathway (38). Previously, this mutation was noted to be infrequent and mainly identified in hepatocellular carcinoma (39). Besides, in BON cells, we found an APC missense mutation (E1317Q) which, although controversial, has been reported as a colorectal cancer risk associated allele (40–42). A silent mutation (T1493T) of APC was also detected and confirmed to be statistically significant in 75% of NET cell lines and 90% of the clinical samples. This silent alteration has been detected previously as a frequent neutral polymorphism identified in numerous types of tumors (43,44). Together, alteration in Wnt/β-catenin signaling could be detected in both GI and non-GI carcinoid tumors and mutations of β-catenin and APC are infrequent but may be a factor in the pathogenesis of NETs, albeit in a relatively small sample size.
It is becoming increasingly evident that the aberrant activation of Wnt/β-catenin signaling can also be achieved by the epigenetic silencing of the Wnt inhibitors that are also regulated by β-catenin/TCF complex as components of a negative feedback loop (7,9). We demonstrated minimal to no expression of the Wnt inhibitors, SFRP-1, Axin-2, DKK-1, DKK-3 and WIF-1 in the BON cell line. However, expression of these Wnt inhibitor genes increased after treatment with 5-aza-CdR, indicating that Wnt inhibitor genes are the targets of epigenetic silencing through methylation. The mechanism for downregulation of the Wnt antagonists in cancers is predominately due to silencing by promoter hypermethylation (10); indeed, we demonstrated that silencing of SFRP-1 and Axin-2 was associated with their promoter hypermethylation in all NET cell lines and that the methylation of SFRP-1 was strongly associated with the majority of carcinoid tumor tissues although cytoplasmic and/or nuclear staining of β-catenin was not detected in all of the tumors. Promoter methylation of Wnt inhibitors provides a reliable assessment of prognosis in a number of cancers. For example, aberrant promoter methylation of SFRP-1 was associated with an overall shorter survival in patients with breast cancers (45), WIF-1 methylation is a poor prognostic factor for patients with acute myeloid leukemia (AML) (46) and DKK-3 methylation is an independent prognostic marker for gastric cancers (20). Therefore, methylation of SFRP-1 alone may not be sufficient to deregulation of Wnt/β-catenin signaling but portend a prognosis in patients with NETs.
Whereas DKK-1, DKK-3 and WIF-1 are silenced by DNA methylation in most tumor cells, the promoters of these genes were found to be unmethylated despite increased expression with 5-aza-CdR treatment in NET cells. Recently, it has been shown that treatment with 5-aza-CdR is associated with histone H3K9 demethylation in some silenced genes (26,35,36). Moreover, histone modification, including H3K9me2, occurred with silencing of a tumor suppressor gene p16 INK4a (47). Also, epigenetic repression of DACT3, which is another negative Wnt regulator in colon cancer, is associated with histone modifications instead of promoter hypermethylation (48). Here, we show that, in the case of DKK-1, DKK-3 and WIF-1 in NET cells, epigenetic silencing occurs through repressive histone modification, H3K9me2, and does not involve or require methylation of the promoter. Using a novel G9a inhibitor, UNC0638, we further elucidated that the silencing of the Wnt inhibitors is allowed by H3K9me2 and inhibition of proliferation by UNC0638 is closely related with reduction of Wnt/β-catenin signaling and induction of apoptosis in BON cells. Previously, G9a was reported to be required for the maintenance of the malignant phenotype and that elevated expression of G9a, which is associated with poor prognosis, promotes invasion and metastasis in lung cancer (49). Moreover, knockdown of G9a induces cell apoptosis; overexpression of this protein is noted in all types of cancers analyzed (50). G9a attenuates the expression (26) and activity (50) of tumor suppressor genes, suggesting this protein may promote tumor cell properties. In line with the above evidences, our findings suggest that G9a may also be involved in NET tumorigenesis by regulating H3K9me2.
Finally, we examined whether restoration of Wnt antagonists, which are silenced in NET cells, would have tumor suppressive effects in the BON cell line. Silencing of various Wnt inhibitor genes and downregulation of their expression have been reported in many cancer types; induction of their expression leads to tumor suppression (11,13–21). In this regard, we also confirmed that induction of Wnt antagonists exerts a tumor suppressive effect in BON cells further indicating that epigenetic silencing of these Wnt inhibitors contribute to NET tumorigenesis. Further work is required to elucidate the correlation or connection between the deregulated Wnt/β-catenin pathway and genetic abnormalities for the components of other pathways such as PI3K/Akt/mTOR and Ras/Raf/MAPK signaling, which are also important in NET functions.
In summary, our findings suggest that the aberrant Wnt/β-catenin signaling through epigenetic silencing of Wnt negative regulatory genes or infrequent gene mutation contributes to NET pathogenesis and growth. Assessment of the methylation status of Wnt inhibitors, especially SFRP-1, may be used as a prognostic indicator and to identify patients with NETs who would most likely benefit from a treatment regimen to include Wnt inhibitors or G9a inhibitors.
Supplementary material
Supplementary Table 1 and Figures 1–6 can be found online at http://carcin.oxfordjournals.org/
Funding
National Institute of Diabetes and Digestive and Kidney Diseases (5R01DK048498-18 to B.M.E.) and the National Institute on Aging (5R37AG010885-21 to B.M.E.).
Supplementary Material
Acknowledgement
We thank Hiromu Suzuki (Department of Biochemistry, Sapporo Medical University, Japan) for providing expression vectors, Tianyan Gao and Cynthia Long for helpful suggestions and technical assistance and Catherine E.Anthony, Heather N.Russell-Simmons, Hope Johnson and Donna A.Gilbreath for manuscript preparation.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations:
- 5-aza-CdR
5-aza-2ʹ-deoxycytidine
- ChIP
chromatin immunoprecipitation
- DKK
DICKKOPF
- GI
gastrointestinal
- H3K9me2
dimethylation of histone H3 at lysine 9
- MSP
methylation-specific PCR
- NET
neuroendocrine tumor
- (q)RT-PCR
(quantitative) reverse transcripton- polymerase chain reaction
- SD
standard deviation
- SFRP
secreted Frizzled-related protein
- TCF/LEF
T-cell factor/lymphoid enhancer-binding factor
- WIF-1
Wnt inhibitory factor-1.
References
- 1. Modlin I.M., et al. (2005). Current status of gastrointestinal carcinoids. Gastroenterology, 128, 1717–1751 [DOI] [PubMed] [Google Scholar]
- 2. Valentino J., et al. (2011). Recent advances in the diagnosis and treatment of gastrointestinal carcinoids. Adv. Surg., 45, 285–300 [DOI] [PubMed] [Google Scholar]
- 3. Modlin I.M., et al. (2003). A 5-decade analysis of 13,715 carcinoid tumors. Cancer, 97, 934–959 [DOI] [PubMed] [Google Scholar]
- 4. Maggard M.A., et al. (2004). Updated population-based review of carcinoid tumors. Ann. Surg., 240, 117–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kohler B.A., et al. (2011). Annual report to the nation on the status of cancer, 1975–2007, featuring tumors of the brain and other nervous system. J. Natl Cancer Inst., 103, 714–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bowen K.A., et al. (2009). An analysis of trends and growth factor receptor expression of GI carcinoid tumors. J. Gastrointest. Surg., 13, 1773–1780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Clevers H. (2006). Wnt/beta-catenin signaling in development and disease. Cell, 127, 469–480 [DOI] [PubMed] [Google Scholar]
- 8. Klaus A., et al. (2008). Wnt signalling and its impact on development and cancer. Nat. Rev. Cancer, 8, 387–398 [DOI] [PubMed] [Google Scholar]
- 9. MacDonald B.T., et al. (2009). Wnt/beta-catenin signaling: components, mechanisms, and diseases. Dev. Cell, 17, 9–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ying Y., et al. (2009). Epigenetic disruption of the WNT/beta-catenin signaling pathway in human cancers. Epigenetics, 4, 307–312 [DOI] [PubMed] [Google Scholar]
- 11. Esteller M., et al. (2000). Analysis of adenomatous polyposis coli promoter hypermethylation in human cancer. Cancer Res., 60, 4366–4371 [PubMed] [Google Scholar]
- 12. Koch A., et al. (2004). Mutations and elevated transcriptional activity of conductin (AXIN2) in hepatoblastomas. J. Pathol., 204, 546–554 [DOI] [PubMed] [Google Scholar]
- 13. Tseng R.C., et al. (2008). Epigenetic silencing of AXIN2/betaTrCP and deregulation of p53-mediated control lead to wild-type beta-catenin nuclear accumulation in lung tumorigenesis. Oncogene, 27, 4488–4496 [DOI] [PubMed] [Google Scholar]
- 14. Suzuki H., et al. (2004). Epigenetic inactivation of SFRP genes allows constitutive WNT signaling in colorectal cancer. Nat. Genet., 36, 417–422 [DOI] [PubMed] [Google Scholar]
- 15. Mazieres J., et al. (2004). Wnt inhibitory factor-1 is silenced by promoter hypermethylation in human lung cancer. Cancer Res., 64, 4717–4720 [DOI] [PubMed] [Google Scholar]
- 16. Aguilera O., et al. (2006). Epigenetic inactivation of the Wnt antagonist DICKKOPF-1 (DKK-1) gene in human colorectal cancer. Oncogene, 25, 4116–4121 [DOI] [PubMed] [Google Scholar]
- 17. Urakami S., et al. (2006). Combination analysis of hypermethylated Wnt-antagonist family genes as a novel epigenetic biomarker panel for bladder cancer detection. Clin. Cancer Res., 12, 2109–2116 [DOI] [PubMed] [Google Scholar]
- 18. Sato H., et al. (2007). Frequent epigenetic inactivation of DICKKOPF family genes in human gastrointestinal tumors. Carcinogenesis, 28, 2459–2466 [DOI] [PubMed] [Google Scholar]
- 19. Nojima M., et al. (2007). Frequent epigenetic inactivation of SFRP genes and constitutive activation of Wnt signaling in gastric cancer. Oncogene, 26, 4699–4713 [DOI] [PubMed] [Google Scholar]
- 20. Yu J., et al. (2009). Promoter methylation of the Wnt/beta-catenin signaling antagonist Dkk-3 is associated with poor survival in gastric cancer. Cancer, 115, 49–60 [DOI] [PubMed] [Google Scholar]
- 21. Licchesi J.D., et al. (2010). Transcriptional regulation of Wnt inhibitory factor-1 by Miz-1/c-Myc. Oncogene, 29, 5923–5934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Shilatifard A. (2006). Chromatin modifications by methylation and ubiquitination: implications in the regulation of gene expression. Annu. Rev. Biochem., 75, 243–269 [DOI] [PubMed] [Google Scholar]
- 23. Zhang Y., et al. (2001). Transcription regulation by histone methylation: interplay between different covalent modifications of the core histone tails. Genes Dev., 15, 2343–2360 [DOI] [PubMed] [Google Scholar]
- 24. Richards E.J., et al. (2002). Epigenetic codes for heterochromatin formation and silencing: rounding up the usual suspects. Cell, 108, 489–500 [DOI] [PubMed] [Google Scholar]
- 25. Fuks F. (2005). DNA methylation and histone modifications: teaming up to silence genes. Curr. Opin. Genet. Dev., 15, 490–495 [DOI] [PubMed] [Google Scholar]
- 26. Wozniak R.J., et al. (2007). 5-Aza-2′-deoxycytidine-mediated reductions in G9A histone methyltransferase and histone H3 K9 di-methylation levels are linked to tumor suppressor gene reactivation. Oncogene, 26, 77–90 [DOI] [PubMed] [Google Scholar]
- 27. Lee S.H., et al. (2009). Hypoxic silencing of tumor suppressor RUNX3 by histone modification in gastric cancer cells. Oncogene, 28, 184–194 [DOI] [PubMed] [Google Scholar]
- 28. Fujimori M., et al. (2001). Accumulation of beta-catenin protein and mutations in exon 3 of beta-catenin gene in gastrointestinal carcinoid tumor. Cancer Res., 61, 6656–6659 [PubMed] [Google Scholar]
- 29. Su M.C., et al. (2006). Nuclear translocation of beta-catenin protein but absence of beta-catenin and APC mutation in gastrointestinal carcinoid tumor. Ann. Surg. Oncol., 13, 1604–1609 [DOI] [PubMed] [Google Scholar]
- 30. Evers B.M., et al. (1991). Establishment and characterization of a human carcinoid in nude mice and effect of various agents on tumor growth. Gastroenterology, 101, 303–311 [DOI] [PubMed] [Google Scholar]
- 31. Vedadi M., et al. (2011). A chemical probe selectively inhibits G9a and GLP methyltransferase activity in cells. Nat. Chem. Biol., 7, 566–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yu T., et al. (2012). Regulation of the potential marker for intestinal cells, Bmi1, by β-catenin and the zinc finger protein KLF4: implications for colon cancer. J. Biol. Chem., 287, 3760–3768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Deng G., et al. (1999). Methylation of CpG in a small region of the hMLH1 promoter invariably correlates with the absence of gene expression. Cancer Res., 59, 2029–2033 [PubMed] [Google Scholar]
- 34. Gulhati P., et al. (2012). Sorafenib enhances the therapeutic efficacy of rapamycin in colorectal cancers harboring oncogenic Kras and Pik3ca. Carcinogenesis, 33, 1782–1790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Fahrner J.A., et al. (2002). Dependence of histone modifications and gene expression on DNA hypermethylation in cancer. Cancer Res., 62, 7213–7218 [PubMed] [Google Scholar]
- 36. Nguyen C.T., et al. (2002). Histone H3-lysine 9 methylation is associated with aberrant gene silencing in cancer cells and is rapidly reversed by 5-aza-2′-deoxycytidine. Cancer Res., 62, 6456–6461 [PubMed] [Google Scholar]
- 37. Rice J.C., et al. (2003). Histone methyltransferases direct different degrees of methylation to define distinct chromatin domains. Mol. Cell, 12, 1591–1598 [DOI] [PubMed] [Google Scholar]
- 38. Kikuchi A. (2003). Tumor formation by genetic mutations in the components of the Wnt signaling pathway. Cancer Sci., 94, 225–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Austinat M., et al. (2008). Correlation between beta-catenin mutations and expression of Wnt-signaling target genes in hepatocellular carcinoma. Mol. Cancer, 7, 21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hahnloser D., et al. (2003). The APC E1317Q variant in adenomatous polyps and colorectal cancers. Cancer Epidemiol. Biomarkers Prev., 12, 1023–1028 [PubMed] [Google Scholar]
- 41. Rozek L.S., et al. (2006). APC E1317Q is not associated with colorectal cancer in a population-based case-control study in Northern Israel. Cancer Epidemiol. Biomarkers Prev., 15, 2325–2327 [DOI] [PubMed] [Google Scholar]
- 42. Hall M.J., et al. (2009). Risk of colorectal neoplasia associated with the adenomatous polyposis coli E1317Q variant. Ann. Oncol., 20, 1517–1521 [DOI] [PubMed] [Google Scholar]
- 43. Gayther S.A., et al. (1995). Rapid detection of rare variants and common polymorphisms in the APC gene by PCR-SSCP for presymptomatic diagnosis and showing allele loss. J. Med. Genet., 32, 568–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Davies S.M., et al. (1994). Frequent polymorphism in exon 15 of the adenomatous polyposis coli gene. Hum. Genet., 93, 329–330 [DOI] [PubMed] [Google Scholar]
- 45. Veeck J., et al. (2006). Aberrant methylation of the Wnt antagonist SFRP1 in breast cancer is associated with unfavourable prognosis. Oncogene, 25, 3479–3488 [DOI] [PubMed] [Google Scholar]
- 46. Chim C.S., et al. (2006). Preferential methylation of Wnt inhibitory factor-1 in acute promyelocytic leukemia: an independent poor prognostic factor. Leukemia, 20, 907–909 [DOI] [PubMed] [Google Scholar]
- 47. Bachman K.E., et al. (2003). Histone modifications and silencing prior to DNA methylation of a tumor suppressor gene. Cancer Cell, 3, 89–95 [DOI] [PubMed] [Google Scholar]
- 48. Jiang X., et al. (2008). DACT3 is an epigenetic regulator of Wnt/beta-catenin signaling in colorectal cancer and is a therapeutic target of histone modifications. Cancer Cell, 13, 529–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Chen M.W., et al. (2010). H3K9 histone methyltransferase G9a promotes lung cancer invasion and metastasis by silencing the cell adhesion molecule Ep-CAM. Cancer Res., 70, 7830–7840 [DOI] [PubMed] [Google Scholar]
- 50. Huang J., et al. (2010). G9a and Glp methylate lysine 373 in the tumor suppressor p53. J. Biol. Chem., 285, 9636–9641 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.