Abstract
In human hepatocellular carcinoma (HCC) and many other cancers, somatic point mutations are highly prevalent, yet the mechanisms critical in their generation remain poorly understood. S-nitrosoglutathione reductase (GSNOR), a key regulator of protein S-nitrosylation, is frequently deficient in human HCC. Targeted deletion of the GSNOR gene in mice can reduce the activity of the DNA repair protein O 6-alkylguanine-DNA alkyltransferase (AGT) and promote both carcinogen-induced and spontaneous HCC. In this study, we report that following exposure to the environmental carcinogen diethylnitrosamine, the mutation frequency of a transgenic reporter in the liver of GSNOR-deficient mice (GSNOR−/−) is significantly higher than that in wild-type control. In wild-type mice, diethylnitrosamine treatment does not significantly increase the frequency of the transition from G:C to A:T, a mutation deriving from diethylnitrosamine-induced O 6-ethylguanines that are normally repaired by AGT. In contrast, the frequency of this transition from diethylnitrosamine is increased ~20 times in GSNOR−/− mice. GSNOR deficiency also significantly increases the frequency of the transversion from A:T to T:A, a mutation not affected by AGT. GSNOR deficiency in our experiments does not significantly affect either the frequencies of the other diethylnitrosamine-induced point mutations or hepatocyte proliferation. Thus, GSNOR deficiency, through both AGT-dependent and AGT-independent pathways, significantly raises the rates of specific types of DNA mutations. Our results demonstrate a critical role for GSNOR in maintaining genomic integrity in mice and support the hypothesis that GSNOR deficiency is an important cause of the widespread mutations in human HCC.
Introduction
DNA sequencing of cancer genomes has revealed remarkably large numbers of somatically acquired mutations in many human cancers (1). These mutations, occurring in both protein-coding and other regions throughout the cancer genomes, are mostly single-nucleotide substitutions. There are for instance over 11 000 somatic substitutions in a typical hepatocellular carcinoma (HCC) (2). The genes affected by the somatic mutations are highly heterogeneous (2–4), and mutational heterogeneity is also substantial within individual tumors (1). The characteristic abundance and substantial heterogeneity of somatic mutations in cancer genomes have profound implications in both our perspective of carcinogenesis and therapeutic approaches to cancer. However, defects in DNA repair systems have only been described in limited cancer incidences, and the causes and mechanisms of the somatic mutations remain largely unknown in HCC and many other cancers.
Human HCC, the third leading cause of cancer deaths worldwide, develops mostly in the context of chronic viral hepatitis (5). Inducible nitric oxide synthase (iNOS), a key mediator of innate immune response and inflammation, is often increased both in HCC cells and in the hepatocytes of patients with chronic viral hepatitis and other diseases that predispose to HCC (6–9). Nitric oxide (NO) affects functions of a wide range of proteins—including many important to tumorigenesis—through S-nitrosylation, the covalent modification of cysteine thiols (10). S-nitrosylation is not only influenced by NOS activities but is also prominently regulated by S-nitrosoglutathione reductase (GSNOR), a major denitrosylase (11–13). The human GSNOR gene (ADH5) is located at ~4q23, a region in which chromosomal deletion occurs most frequently in HCC (14–17). We showed that the abundance and activity of GSNOR were significantly decreased in cancer samples from ~50% of patients with HCC (18). Interestingly, gene-expression profiling showed that both GSNOR (ADH5) deficiency and iNOS overexpression in the liver are closely associated with de novo hepatocarcinogenesis after tumor resection and a poor prognosis in HCC patients (19). Thus, excessive S-nitrosylation from GSNOR deficiency and concurrent iNOS overexpression in the liver may contribute critically to human HCC.
Recent studies using a mouse line with targeted deletion of the GSNOR gene demonstrated that S-nitrosylation from GSNOR deficiency inactivates the key DNA repair protein O 6-alkylguanine-DNA alkyltransferase (AGT; also known as O 6-methylguanine-DNA-methyltransferase) and promotes HCC (18,20). O 6-alkylguanines in DNA are produced by alkylating N-nitroso compounds, including dialkylnitrosamines, which are widely present in the environment and can also be formed endogenously (21–23). O 6-alkylguanines are mispaired by DNA polymerases to thymine during DNA replication, and the O 6-alkylguanine:T mispairing, through a further round of DNA replication, can result in G:C to A:T mutations (22). Mutagenic O 6-alkylguanines are repaired primarily by AGT (24) although repair of O 6-alkylguanines might be affected by nucleotide excision repair (25). AGT is important for protection against HCC (26–28). We showed that during inflammatory responses following intraperitoneal injection of diethylnitrosamine (DEN) or lipopolysaccharide (LPS), GSNOR deficiency resulted in S-nitrosylation, ubiquitination and proteosomal degradation of AGT, leading to its significant reduction in livers of GSNOR−/− mice (18). Consequently, the repair of carcinogenic O 6-ethylguanines in the livers of DEN-challenged GSNOR−/− mice was impaired. GSNOR−/− mice were found to be very susceptible to both spontaneous and DEN-induced HCC. Predisposition to HCC, S-nitrosylation and depletion of AGT, and accumulation of O 6-ethylguanines due to GSNOR deficiency were remarkably all abolished by concurrent deletion of the iNOS gene in GSNOR−/−iNOS−/− mice, further underscoring the critical role of iNOS-derived S-nitrosylation in AGT inactivation and liver carcinogenesis in GSNOR−/− mice (18). Moreover, hepatocyte-specific deletion of GSNOR caused nitrosative inactivation of liver AGT and increased mortality from DEN (20), demonstrating the importance of GSNOR regulation of S-nitrosylation in liver parenchymal cells. S-nitrosylation may affect DNA repair pathways other than AGT (29), and dialkylnitrosamines can cause a number of mutagenic DNA alkylations in addition to O 6-alkylguanines (30). NO and related reactive nitrogen species at high levels may also damage DNA directly (31). However, it is unknown whether GSNOR deficiency in vivo affects the rate or spectrum of DNA mutation.
To examine the impact of GSNOR deficiency on DNA mutations, we employed Big Blue transgenic mice, a well-established system for detecting DNA mutations in vivo (32), and measured the rates of both spontaneous and DEN-induced mutations in livers of GSNOR−/− Big Blue double-transgenic mice. We found that after DEN treatment, GSNOR deficiency significantly raises the rate of a specific subset of single-nucleotide substitutions, demonstrating the importance of GSNOR to genomic integrity. Our findings suggest that GSNOR deficiency may provide a novel mechanism for abundant point mutations in HCC.
Materials and methods
Animals
GSNOR−/− mice (12) were crossed with Big Blue mice (Agilent, Santa Clara, CA) to obtain GSNOR−/−Big Blue double-transgenic mice. All mice were in a C57BL/6 background. The mice were maintained on normal mouse chow (5058 PicoLab Mouse Diet 20) in a specific pathogen-free facility at the University of California at San Francisco (UCSF). The experimental protocol was approved by the Institutional Animal Care and Use Committee of UCSF.
Genotyping
Genomic DNA was isolated from ear punctures using the Extract-N-Amp Tissue PCR Kit (Sigma–Aldrich, St Louis, MO). cII was detected by PCR using the lambda select-cII sequencing primers (5′-CCACACCTATGGTGTATG-3′ and 5′-CCTCTGCCGAAGTTGAGTAT-3′). The GSNOR knockout allele was detected by PCR using 5′-CCTGAAGCAGCTACTCCCACTACCAC-3′ and 5′-TCTTGACGAGTTCTTCTGAGG-3′ primers, whereas the GSNOR wild-type allele was detected using 5′- GGCATGTCTTCATTTAGCTCAC-3′ and 5′-TCAAGAGGTGAGGCTACAAGTT-3′ primers. PCR was performed using RedExtract-N-Amp PCR ReadyMix (Sigma–Aldrich) in a 10 μl reaction volume. Cycling parameters for both cII and GSNOR alleles included a 5min denaturation at 95°C, followed by 30 cycles of 30 s at 94°C, 45 s at 54°C and 1min at 72°C, with a final extension of 10min at 72°C.
DEN treatment
DEN (Sigma–Aldrich) was prepared in phosphate-buffered saline without calcium or magnesium. Male pups were given a single intraperitoneal injection of DEN (25 µg/g body wt) or saline control at postnatal day 15. Mice were euthanized at postnatal day 45 and liver samples were collected.
cII mutant detection
cII mutants were analyzed with the lambda Select-cII Mutation Detection System for Big Blue Rodents (Agilent). Liver samples (60mg) from Big Blue mice were homogenized with a Wheaton Dounce tissue grinder, digested with proteinase K and dialyzed in Tris-EDTA buffer (pH 7.5) at room temperature for 48h. High molecular weight genomic DNA was extracted from the samples using the RecoverEase DNA Isolation Kit (Agilent). The lambda phage genome in the mouse genomic DNA was recovered and packaged in vitro using the Transpak reagent (Agilent). The packaged phages were used to infect Escherichia coli strain G1250 and subsequently mixed with top agar and plated on 100mm agar plates. The total number of plaque-forming phages in the packaged sample was estimated by incubating the phage-infected G1250 cells in titer plates overnight at 37°C (non-selective condition). Lambda phages containing mutant cII genes were selected by culture for 40–48h at 24°C (selective condition). The putative cII mutants were isolated and used to infect G1250 cells to confirm the mutant phenotype by replating under the selective growth condition. Only mutants with the confirmed phenotype were used for calculating mutant frequency and further analysis.
DNA sequence analysis
The cII gene in mutant phages was amplified by PCR using the cII sequencing primers described above and performed with the following conditions: 5 μl of plaque sample DNA, 0.4 μM of each primer and RedExtract-N-Amp PCR ReadyMix (Sigma) in a 20 μl reaction volume. The PCR product was purified by the Qiaquik PCR purification kit (Qiagen), eluted in 30 μl of sterile water, and 1 μl run on a 1.5% agarose gel to verify the presence of a specific 432 base-pair band representing the cII region. The purified DNA products and cII sequencing primers were sent to sequencing companies (SeqWright, Houston TX and Sequetech, CA) and the sequence returned compared in a multiple sequence alignment to the wild-type cII Genbank reference sequence (J02459) using ClustalX 2.0.11. Sequence chromatograms were analyzed for sequence quality and verification of mutations.
Determination of mutation frequency and statistical analysis
Mutant frequency, defined as the fraction of phages carrying mutations in cII, was determined by dividing the number of verified mutant plaques by the total number of plaques screened. The frequency of independent mutations was determined by correction for recurrent mutations within a single mouse. Mutant and mutation frequencies were analyzed statistically by analysis of variance (33).
Results
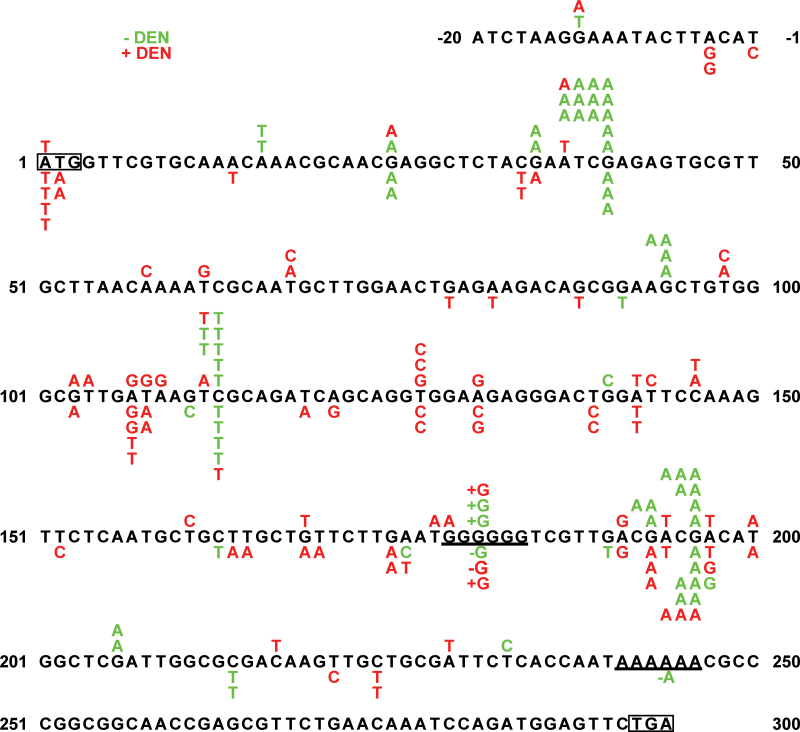
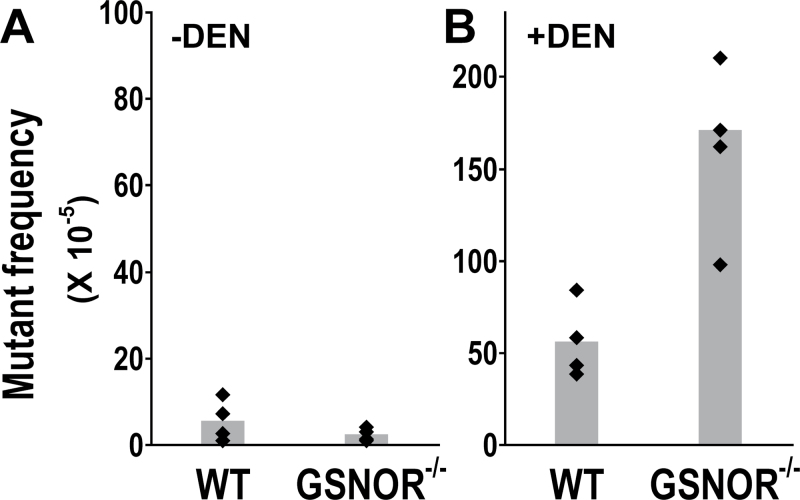
To investigate the effects of GSNOR deficiency on DNA mutation in vivo, we crossed GSNOR−/− mice with Big Blue transgenic reporter mice containing in a single locus with ~30 copies of the lambda bacteriophage genome in recoverable shuttle vectors (34). Direct selection of the recovered phages containing mutations in the lambda cII transgene, a widely used assay for point mutations in Big Blue mice (35), was employed to analyze genomic DNA from the livers of wild-type and GSNOR−/− Big Blue mice. To determine the frequency of spontaneous mutations, we screened 2 million plaques each from 45-day-old unchallenged wild-type and GSNOR−/− Big Blue mice (Table I). Mutants obtained from the primary screen were verified for the mutant phenotype by replating under the selective condition. We found that the frequencies of total verified cII mutants in phages from four GSNOR−/− mice did not statistically differ from those of four wild-type mice (Figures 1 and 2A). This mutant frequency was comparable with that reported previously in wild-type mice (32). To determine the type of mutational changes that occurred, we sequenced the cII gene in the mutants (Table I and Figure 1). DNA sequence analysis identified one change of a single nucleotide in each of the sequenced mutants of both GSNOR−/− and control mice, indicating no false-positive identification of the cII mutants by our phage-based assay. The majority of mutations in both mice is G:C to A:T transitions, predominantly at CpG sites (Figure 1 and Supplementary Figure S1, available at Carcinogenesis Online). We detected mutations at nucleotide positions 40, 113 and 196, which occur in two to four of both the GSNOR−/− and control mice (Figure 1). These positions were previously reported hotspots for spontaneous mutations in the cII transgene (36,37). Highly recurrent (4–8 times) mutations detected at hotspots 40, 113 and 196 in two wild-type mice may result from independent mutational events in the mice or largely from clonal expansion of the mutant cells in early development of the animals. Mutant frequencies in these mice thus might overestimate independent mutations. The frequency of verified independent mutations, obtained by subtracting all recurrent mutations within a single mouse, was comparable between the GSNOR−/− and wild-type mice (Supplementary Figure S2A, available at Carcinogenesis Online). Overall, these results suggest that spontaneous mutations occur comparably in the liver of GSNOR−/− and wild-type mice at an early age.
Table I.
Analysis of lambda cII transgene in wild-type and GSNOR–/– Big Blue mice
| Samplea | Plaques screened | Mutants identifiedb | Mutants sequenced |
|---|---|---|---|
| WT − DEN | 2.1E+06 | 161 | 46 |
| KO − DEN | 2.2E+06 | 44 | 25 |
| WT + DEN | 3.2E+05 | 202 | 37 |
| KO + DEN | 4.3E+05 | 488 | 58 |
aLiver samples from wild-type (WT) and GSNOR−/− (KO) mice treated with DEN (+DEN) or saline (−DEN).
bMutants verified by replating under the selective condition.
Fig. 1.
Mutational spectra at the cII locus in the livers of wild-type and GSNOR−/− Big Blue mice. Shown are the nucleotide sequence changes in the untreated (green) and DEN-treated (red) wild-type (above the reference cII sequence) or GSNOR−/− (below the sequence) mice. Translational start and stop codons are outlined. +G and −G indicate single-nucleotide insertion and deletion, respectively, in the underlined G repeats; −A indicates single nucleotide deletion in the underlined A repeats.
Fig. 2.
Overall mutation frequency from DEN treatment is increased by GSNOR deficiency. (A) The frequency of spontaneous cII mutants in the livers of four untreated GSNOR−/− mice is not significantly different from that in four wild-type (WT) controls (P = 0.2). (B) Mutant frequency in the livers of four GSNOR−/− mice is significantly higher than that in four wild-type controls after DEN challenge (P = 0.006). The mutant frequencies of the DEN-treated wild-type and GSNOR−/− mice are significantly higher than those in the unchallenged wild-type (P = 0.003) and GSNOR−/− (P = 0.001) mice, respectively. Each dot represents the data from a single mouse; bar represents the group mean.
To determine the effects of GSNOR deficiency on mutagenesis induced by environmental mutagens, we injected DEN intraperitoneally into 15-day-old GSNOR−/− (n = 4) and wild-type (n = 4) Big Blue mice. DNA mutations were analyzed 30 days after DEN injection (postnatal day 45) to permit DNA adduct formation and cell proliferation required for fixation of mutations. We collected genomic DNA from the livers of the mice and screened for DNA mutations in the cII transgene using the phage-based assay. Mutant frequency, as expected, was significantly increased in both wild-type and GSNOR−/− mice by DEN treatment (Figure 2). Importantly, however, the mutant frequency in livers of DEN-treated GSNOR−/− mice was significantly higher than that in the wild-type control (Figure 2B). The mean mutant frequency of GSNOR−/− mice was about three times of the wild-type control; the mutant frequency in each of the four GSNOR−/− mice was higher than that in any of the four wild-type controls (Figure 2B). DNA sequence analysis identified one nucleotide change in each of the sequenced mutants of DEN-treated GSNOR−/− and wild-type mice (Table I and Figure 1), indicating no false-positive identification of cII mutants by the phage-based assay. In addition, no mutation was detected more than twice in the sequenced mutants of any individual DEN-treated mouse, suggesting that mutant frequencies obtained in this study were unlikely compromised by any ‘jackpot’ mutation from clonal expansion. We also found that the frequency of independent mutations, obtained by subtracting recurrent mutations, was significantly higher in DEN-treated GSNOR−/− mice than in the wild-type control (Figure S2B). Thus, the true mutation frequency of independent mutational events from DEN, lying likely between the unadjusted and adjusted mutant frequencies, was significantly increased in the GSNOR−/− mice.
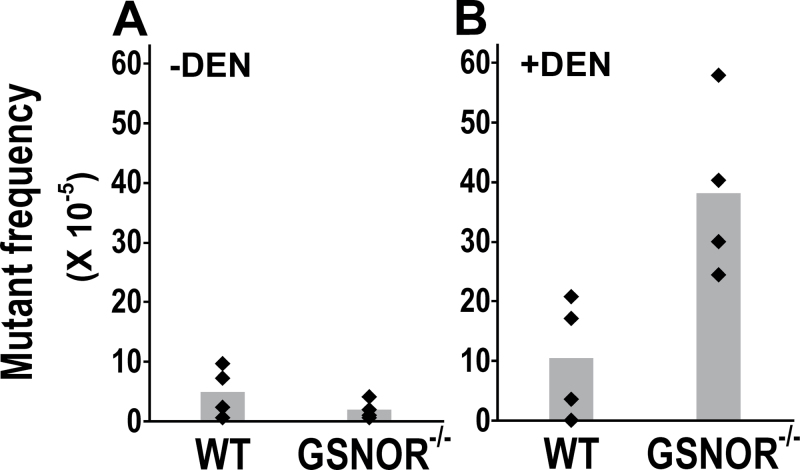
To identify potential mechanisms responsible for increased mutagenesis in DEN-treated GSNOR−/− mice, we determined the frequencies of cII mutants with G:C to A:T transitions. This transition, a major mutation from DEN treatment, results mainly from DEN-induced O 6-ethylguanine, which can occur in both CpG and non-CpG sites and is repaired primarily by AGT. We found that the mutant frequency of this transition was not significantly increased by DEN treatment in wild-type mice (Figure 3; P = 0.4). In sharp contrast, the mutant frequency was increased by DEN about 20 times in GSNOR−/− mice (Figure 3; P = 0.003). Furthermore, the mutant frequency in DEN-treated GSNOR–/– mice was significantly higher than that in DEN-treated wild-type controls (Figure 3B). These results demonstrate that in DEN-challenged mice, GSNOR deficiency substantially increases the frequency of G:C to A:T transitions.
Fig. 3.
GSNOR deficiency increases the frequency of G:C to A:T transitions from DEN treatment. The frequency of cII mutants containing G:C to A:T transitions is from the liver of untreated (A) or DEN-treated (B) wild-type and GSNOR−/− mice (n = 4 in each group). The frequency is significantly higher in DEN-treated GSNOR−/− mice than in DEN-treated wild-type mice (P = 0.02).
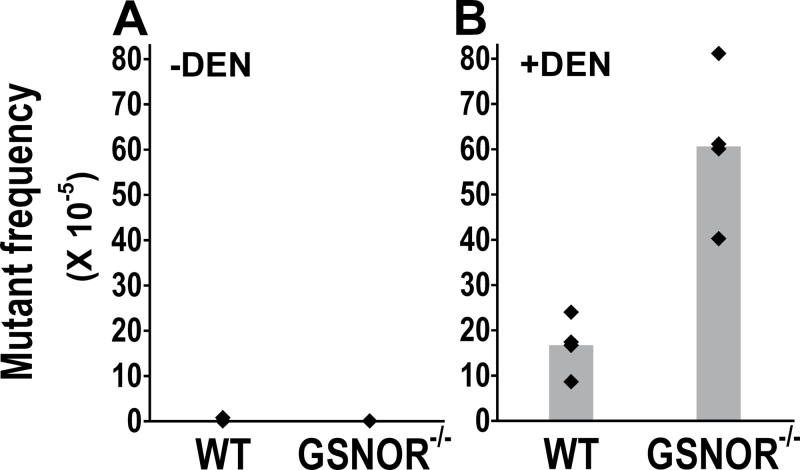
The most common point mutation in wild-type cells after treatment with ethylating agents is A:T to T:A transversion (38–40). We found that while A:T to T:A transversions were almost undetectable in untreated mice, the mutant frequency of this transversion was significantly increased by DEN in both wild-type and GSNOR−/− mice (Figures 1 and 4). Interestingly, the frequency of the transversion was significantly higher in DEN-treated GSNOR−/− mice than that in DEN-treated wild-type mice (Figure 4B). Thus, GSNOR appears to play a protective role against DEN-induced A:T to T:A transversions.
Fig. 4.
GSNOR deficiency increases the frequency of A:T to T:A transversions from DEN treatment. The frequencies of cII mutants containing A:T to T:A transversions are from the livers of untreated (A) or DEN-treated (B) wild-type and GSNOR−/− mice (n = 4 in each group). The frequency is significantly higher in DEN-treated GSNOR−/− mice than in wild-type control (P = 0.003).
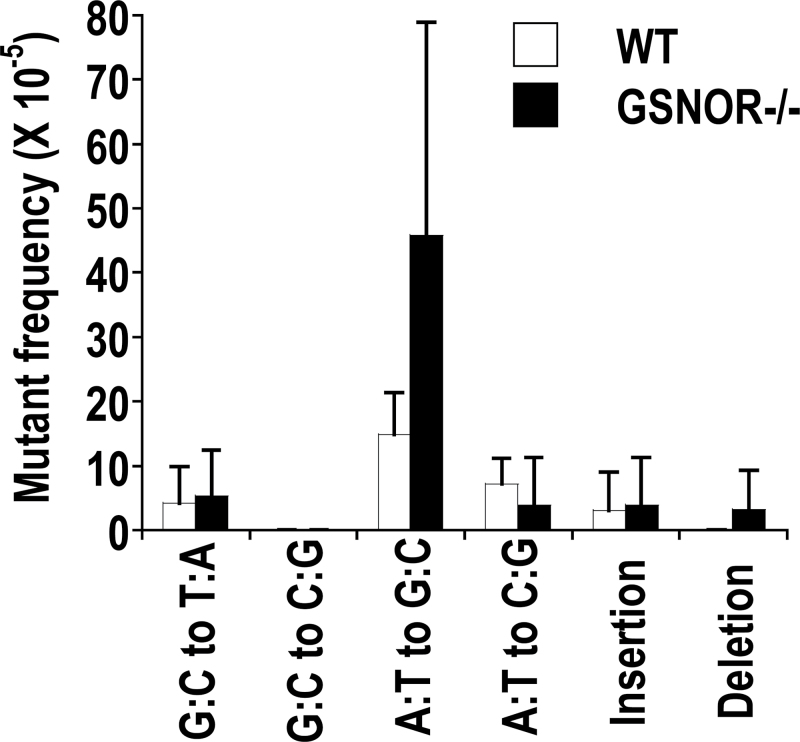
DEN challenge increased the frequencies of A:T to G:C transitions and A:T to C:G transversions in wild-type mice, which nevertheless were not significantly different from those in GSNOR−/− mice (Figure 5). In addition, DEN challenge did not significantly affect the frequency of G:C to T:A and G:C to C:G transversions, nucleotide insertions or nucleotide deletions in either wild-type or GSNOR−/− mice (Figure 5). GSNOR deficiency thus does not appear to increase frequencies of mutations indiscriminately.
Fig. 5.
Mutations not significantly affected by GSNOR deficiency. The data (mean + SD) are from DEN-treated GSNOR−/− and wild-type mice (n = 4 in each group).
Because the conversion of DNA lesions into mutations after DEN treatment depends on DNA replication in affected cells, the amount of DEN-derived mutations can be affected by the rates of both DNA repair and cell proliferation. Our immunohistochemical analysis of Ki67, a marker of proliferating cells, showed that the number of Ki67-positive hepatocytes in DEN-treated GSNOR−/− mice was comparable with that in wild-type control (Supplementary Figure S3, available at Carcinogenesis Online). Thus, in this animal model using infant mice with high rates of hepatocyte proliferation (41), GSNOR deficiency did not appear to increase hepatocyte proliferation significantly. Our findings, therefore, suggest that GSNOR deficiency likely increases DNA mutations through affecting DNA lesions in the liver, not hepatocyte proliferation.
Discussion
Employing Big Blue transgenic reporter mice, we examined in this study the impact of GSNOR deficiency on DNA mutations in vivo. We found that while spontaneous mutations occurred comparably in the liver of young GSNOR−/− and wild-type mice, the rate of single-nucleotide substitutions following DEN treatment was significantly increased in the liver of GSNOR−/− mice compared with wild-type control. The mutations occurring more frequently in DEN-challenged GSNOR−/− mice included both G:C to A:T transitions and A:T to T:A transversions. GSNOR deficiency did not appear to significantly affect the frequencies of other single-nucleotide changes or increase hepatocyte proliferation. Our findings thus demonstrate that GSNOR deficiency can significantly raise the rate of select DNA mutations, underscoring the importance of GSNOR to genomic integrity.
G:C to A:T transitions are a major type of DNA mutation significantly increased in the liver of DEN-treated GSNOR−/− mice. In unchallenged GSNOR−/− and wild-type mice, the transition occurs at comparable frequencies and mostly at CpG sites, suggesting that the mutation results largely from spontaneous deamination of 5-methylcytosine (42) and is not affected by GSNOR deficiency. The substantial increase of G:C to A:T transitions in GSNOR−/− mice after DEN challenge is consistent with our previous findings of persistent elevation of O 6-ethylguanines (18), the major lesion from DEN treatment that causes this mutation (30). As we reported previously, the repair of O 6-ethylguanines is impaired in the liver of DEN-treated GSNOR−/− mice, probably because of nitrosative inactivation of AGT (18). It is unclear if repair of O 6-ethylguanines might be affected by other DNA repair systems (25), or if the G:C to A:T transition might additionally result from N3-ethylcytosine, a minor product from DEN exposure (30). NO and related molecules originated from endogenous NOS activity might directly cause DNA mutation by damaging DNA and in cell culture and animal models of severe nitrosative stress, the predominantly increased mutation is G:C to T:A transversions (43,44). However, G:C to T:A transversion is not significantly increased by DEN treatment and GSNOR deficiency, suggesting that direct contribution by NO to mutagenesis may be minimal or limited in this study. Whether protection against G:C to A:T transition by GSNOR results solely from its effect on AGT or additional mechanisms might be addressed by comparison of DEN-induced mutations in GSNOR−/− and GSNOR-competent mice in AGT-null background. Our results obtained so far have provided strong evidence for the mechanism that GSNOR deficiency in our animal model results in nitrosative inactivation of AGT, impaired DNA repair and persistent elevation of O 6-ethylguanines, leading to increased G:C to A:T transitions. Interestingly, G:C to A:T transition is genome-wide one of the most common mutations in human HCC (2). In human HCC unassociated with aflatoxin B1 exposure, G:C to A:T transition occurs also frequently in the tumor suppressor gene TP53 (3).
GSNOR appears to also protect against DNA mutation through AGT-independent mechanisms. In the liver of DEN-challenged mice, GSNOR deficiency significantly increases the frequency of A:T to T:A transversions. This transversion is the predominant mutation in wild-type cells treated with the ethylating agents DEN and ethylnitrosourea (38–40). The frequency of the transversion induced either by ethylating agents in cell culture (45,46) or by an alkylating agent in a mouse model (47) is unaffected by deficiency of AGT, indicating little protection from the transversion by AGT. The causative DNA lesion(s) of the mutation has not been established and repair of the DNA lesion in cells remains unknown. The predominant thymine lesion from DEN and ethylnitrosourea is O 2-ethylthymine (48), which in DNA replication assays in vitro can cause the A:T to T:A transversion (49). In contrast to rapid repair of O 6-ethylguanine by AGT, O 2-ethylthymine is persistent in cells long after DEN challenge (30). The half-life of O 2-ethylthymine intriguingly is much longer than that of O 2-methylthymine (48). Among the other ethylthymidines from DEN, N3-ethylthymidine is a minor product, whereas O 4-ethylthymine may cause T:A to C:G transitions but not A:T to T:A transversions (30). In addition, ethylation of adenines by DEN is believed to contribute little to DNA point mutation (30). O 2-ethylthymine, therefore, is hypothesized to be the major ethylation product that causes the A:T to T:A transversion. O 2-alkylthymine, which can be repaired by E.coli AlkA glycosylase, is not repaired by mammalian methylpurine DNA glycosylase (50). It has been suggested that proliferating cells, in contrast to quiescent cells, effectively repair ethylnitrosourea-induced DNA lesions, presumably including the major O 2-ethylthymine lesion, through an unknown mechanism (51). This proliferation-dependent DNA repair mechanism would have minor effects on the total amount of O 2-ethylthymines in DEN-treated livers consisting of mostly quiescent cells, but it could have a significant impact on O 2-ethylthymine levels in proliferating hepatocytes that determine the amount of related mutations in the liver. We have not detected significant increases in the total amount of O 2-ethylthymines in livers of DEN-treated GSNOR−/− mice compared with wild-type control (18). It remains to be determined if GSNOR deficiency may increase the amount of O 2-ethylthymines in proliferating hepatocytes.
Our findings of protection by GSNOR against alkylation-induced mutagenesis may have important implications in human cancer. GSNOR deficiency has been implicated in the development of human HCC (18,19). Alkylating N-nitroso compounds can be formed endogenously and are widely present in the environment, including in various food products (21–23). Exposure of GSNOR-deficient cells to alkylating agents may cause increase in mutagenesis, providing the primary drive for carcinogenesis. Pharmacological inhibition of nitrosative stress in patients with GSNOR deficiency and concurrent iNOS overexpression may provide a therapeutic strategy to prevent HCC or its reoccurrence after tumor resection.
Supplementary material
Supplementary Figures 1–3 can be found at http://carcin.oxfordjournals.org/
Funding
Sandler Family Supporting Foundation and the National Institutes of Health (R01CA122359 to L.L.; P30 DK026743 to UCSF Liver Center).
Supplementary Material
Acknowledgements
We thank Dr Chi-Hui Tang for reading the article and helpful comments.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations:
- AGT
O 6-alkylguanine-DNA alkyltransferase
- DEN
diethylnitrosamine
- GSNOR
S-nitrosoglutathione reductase
- HCC
hepatocellular carcinoma
- iNOS
inducible nitric oxide synthase
- LPS
lipopolysaccharide
- NO
nitric oxide.
References
- 1. Loeb L.A. (2011). Human cancers express mutator phenotypes: origin, consequences and targeting. Nat. Rev. Cancer, 11, 450–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Totoki Y., et al. (2011). High-resolution characterization of a hepatocellular carcinoma genome. Nat. Genet., 43, 464–469 [DOI] [PubMed] [Google Scholar]
- 3. Li M., et al. (2011). Inactivating mutations of the chromatin remodeling gene ARID2 in hepatocellular carcinoma. Nat. Genet, 43, 828–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sjöblom T., et al. (2006). The consensus coding sequences of human breast and colorectal cancers. Science, 314, 268–274 [DOI] [PubMed] [Google Scholar]
- 5. Perz J.F., et al. (2006). The contributions of hepatitis B virus and hepatitis C virus infections to cirrhosis and primary liver cancer worldwide. J. Hepatol., 45, 529–538 [DOI] [PubMed] [Google Scholar]
- 6. Majano P.L., et al. (1998). Inducible nitric oxide synthase expression in chronic viral hepatitis. Evidence for a virus-induced gene upregulation. J. Clin. Invest., 101, 1343–1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kane J.M., 3rd, et al. (1997). Chronic hepatitis C virus infection in humans: induction of hepatic nitric oxide synthase and proposed mechanisms for carcinogenesis. J. Surg. Res., 69, 321–324 [DOI] [PubMed] [Google Scholar]
- 8. Rahman M.A., et al. (2001). Coexpression of inducible nitric oxide synthase and COX-2 in hepatocellular carcinoma and surrounding liver: possible involvement of COX-2 in the angiogenesis of hepatitis C virus-positive cases. Clin. Cancer Res., 7, 1325–1332 [PubMed] [Google Scholar]
- 9. McNaughton L., et al. (2002). Distribution of nitric oxide synthase in normal and cirrhotic human liver. Proc. Natl Acad. Sci. USA, 99, 17161–17166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hess D.T., et al. (2005). Protein S-nitrosylation: purview and parameters. Nat. Rev. Mol. Cell Biol., 6, 150–166 [DOI] [PubMed] [Google Scholar]
- 11. Liu L., et al. (2001). A metabolic enzyme for S-nitrosothiol conserved from bacteria to humans. Nature, 410, 490–494 [DOI] [PubMed] [Google Scholar]
- 12. Liu L., et al. (2004). Essential roles of S-nitrosothiols in vascular homeostasis and endotoxic shock. Cell, 116, 617–628 [DOI] [PubMed] [Google Scholar]
- 13. Yang Z., et al. (2010). Lymphocyte development requires S-nitrosoglutathione reductase. J. Immunol., 185, 6664–6669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nagai H., et al. (1997). Comprehensive allelotyping of human hepatocellular carcinoma. Oncogene, 14, 2927–2933 [DOI] [PubMed] [Google Scholar]
- 15. Patil M.A., et al. (2005). Array-based comparative genomic hybridization reveals recurrent chromosomal aberrations and Jab1 as a potential target for 8q gain in hepatocellular carcinoma. Carcinogenesis, 26, 2050–2057 [DOI] [PubMed] [Google Scholar]
- 16. Wong N., et al. (1999). Assessment of genetic changes in hepatocellular carcinoma by comparative genomic hybridization analysis: relationship to disease stage, tumor size, and cirrhosis. Am. J. Pathol., 154, 37–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yeh S.H., et al. (2001). Chromosomal allelic imbalance evolving from liver cirrhosis to hepatocellular carcinoma. Gastroenterology, 121, 699–709 [DOI] [PubMed] [Google Scholar]
- 18. Wei W., et al. (2010). S-nitrosylation from GSNOR deficiency impairs DNA repair and promotes hepatocarcinogenesis. Sci. Transl. Med., 2, 19ra13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hoshida Y., et al. (2008). Gene expression in fixed tissues and outcome in hepatocellular carcinoma. N. Engl. J. Med., 359, 1995–2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wei W., et al. (2011). Targeted deletion of GSNOR in hepatocytes of mice causes nitrosative inactivation of O6-alkylguanine-DNA alkyltransferase and increased sensitivity to genotoxic diethylnitrosamine. Carcinogenesis, 32, 973–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Liu R.H., et al. (1991). Elevated formation of nitrate and N-nitrosodimethylamine in woodchucks (Marmota monax) associated with chronic woodchuck hepatitis virus infection. Cancer Res., 51, 3925–3929 [PubMed] [Google Scholar]
- 22. Gerson S.L. (2004). MGMT: its role in cancer aetiology and cancer therapeutics. Nat. Rev. Cancer, 4, 296–307 [DOI] [PubMed] [Google Scholar]
- 23. Lundberg J.O., et al. (2004). Nitrate, bacteria and human health. Nat. Rev. Microbiol., 2, 593–602 [DOI] [PubMed] [Google Scholar]
- 24. Pegg A.E. (2000). Repair of O(6)-alkylguanine by alkyltransferases. Mutat. Res., 462, 83–100 [DOI] [PubMed] [Google Scholar]
- 25. Bronstein S.M., et al. (1992). Efficient repair of O6-ethylguanine, but not O4-ethylthymine or O2-ethylthymine, is dependent upon O6-alkylguanine-DNA alkyltransferase and nucleotide excision repair activities in human cells. Cancer Res., 52, 2008–2011 [PubMed] [Google Scholar]
- 26. Iwakuma T., et al. (1997). High incidence of nitrosamine-induced tumorigenesis in mice lacking DNA repair methyltransferase. Carcinogenesis, 18, 1631–1635 [DOI] [PubMed] [Google Scholar]
- 27. Nakatsuru Y., et al. (1993). O6-methylguanine-DNA methyltransferase protects against nitrosamine-induced hepatocarcinogenesis. Proc. Natl Acad. Sci. USA, 90, 6468–6472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhou Z.Q., et al. (2001). Spontaneous hepatocellular carcinoma is reduced in transgenic mice overexpressing human O6- methylguanine-DNA methyltransferase. Proc. Natl Acad. Sci. USA, 98, 12566–12571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tang C.H., et al. (2012). Regulation of DNA repair by S-nitrosylation. Biochim. Biophys. Acta, 1820, 730–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Verna L., et al. (1996). N-nitrosodiethylamine mechanistic data and risk assessment: bioactivation, DNA-adduct formation, mutagenicity, and tumor initiation. Pharmacol. Ther., 71, 57–81 [DOI] [PubMed] [Google Scholar]
- 31. Burney S., et al. (1999). The chemistry of DNA damage from nitric oxide and peroxynitrite. Mutat. Res., 424, 37–49 [DOI] [PubMed] [Google Scholar]
- 32. Lambert I.B., et al. (2005). Detailed review of transgenic rodent mutation assays. Mutat. Res., 590, 1–280 [DOI] [PubMed] [Google Scholar]
- 33. Piegorsch W.W., et al. (1995). Study design and sample sizes for a lacI transgenic mouse mutation assay. Environ. Mol. Mutagen., 25, 231–245 [DOI] [PubMed] [Google Scholar]
- 34. Kohler S.W., et al. (1991). Spectra of spontaneous and mutagen-induced mutations in the lacI gene in transgenic mice. Proc. Natl Acad. Sci. USA, 88, 7958–7962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Jakubczak J.L., et al. (1996). Analysis of genetic instability during mammary tumor progression using a novel selection-based assay for in vivo mutations in a bacteriophage lambda transgene target. Proc. Natl Acad. Sci. USA, 93, 9073–9078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Harbach P.R., et al. (1999). Spontaneous mutation spectrum at the lambda cII locus in liver, lung, and spleen tissue of Big Blue transgenic mice. Environ. Mol. Mutagen., 33, 132–143 [DOI] [PubMed] [Google Scholar]
- 37. Morgan C., et al. (2006). iMARS–mutation analysis reporting software: an analysis of spontaneous cII mutation spectra. Mutat. Res., 603, 15–26 [DOI] [PubMed] [Google Scholar]
- 38. O’Neill J.P. (2000). DNA damage, DNA repair, cell proliferation, and DNA replication: how do gene mutations result? Proc. Natl Acad. Sci. USA, 97, 11137–11139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mientjes E.J., et al. (1998). DNA adducts, mutant frequencies, and mutation spectra in various organs of lambda lacZ mice exposed to ethylating agents. Environ. Mol. Mutagen., 31, 18–31 [PubMed] [Google Scholar]
- 40. Madden C.R., et al. (2001). Hepatitis B virus X protein acts as a tumor promoter in development of diethylnitrosamine-induced preneoplastic lesions. J. Virol., 75, 3851–3858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Vesselinovitch S.D., et al. (1983). Kinetics of diethylnitrosamine hepatocarcinogenesis in the infant mouse. Cancer Res., 43, 4253–4259 [PubMed] [Google Scholar]
- 42. Barnes D.E., et al. (2004). Repair and genetic consequences of endogenous DNA base damage in mammalian cells. Annu. Rev. Genet., 38, 445–476 [DOI] [PubMed] [Google Scholar]
- 43. Kim M.Y., et al. (2012). Delivery method, target gene structure, and growth properties of target cells impact mutagenic responses to reactive nitrogen and oxygen species. Chem. Res. Toxicol., 25, 873–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sheh A., et al. (2010). Mutagenic potency of Helicobacter pylori in the gastric mucosa of mice is determined by sex and duration of infection. Proc. Natl Acad. Sci. USA, 107, 15217–15222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bronstein S.M., et al. (1991). Toxicity, mutagenicity, and mutational spectra of N-ethyl-N-nitrosourea in human cell lines with different DNA repair phenotypes. Cancer Res., 51, 5188–5197 [PubMed] [Google Scholar]
- 46. Yang J.L., et al. (1994). Comparison of mutation spectra induced by N-ethyl-N-nitrosourea in the hprt gene of Mer+ and Mer− diploid human fibroblasts. Carcinogenesis, 15, 939–945 [DOI] [PubMed] [Google Scholar]
- 47. Sandercock L.E., et al. (2008). Mgmt deficiency alters the in vivo mutational spectrum of tissues exposed to the tobacco carcinogen 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK). Carcinogenesis, 29, 866–874 [DOI] [PubMed] [Google Scholar]
- 48. Beranek D.T. (1990). Distribution of methyl and ethyl adducts following alkylation with monofunctional alkylating agents. Mutat. Res., 231, 11–30 [DOI] [PubMed] [Google Scholar]
- 49. Bhanot O.S., et al. (1992). In vitro DNA replication implicates O2-ethyldeoxythymidine in transversion mutagenesis by ethylating agents. Nucleic Acids Res., 20, 587–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. O’Brien P.J., et al. (2003). Human alkyladenine DNA glycosylase uses acid-base catalysis for selective excision of damaged purines. Biochemistry, 42, 12418–12429 [DOI] [PubMed] [Google Scholar]
- 51. Bielas J.H., et al. (2000). Proliferation is necessary for both repair and mutation in transgenic mouse cells. Proc. Natl Acad. Sci. USA, 97, 11391–11396 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.