Summary
The QseC sensor kinase regulates virulence in multiple gram-negative pathogens, by controlling the activity of the QseB response regulator. We have previously shown that qseC deletion interferes with dephosphorylation of QseB thus unleashing what appears to be an uncontrolled positive feedback loop stimulating increased QseB levels. Deletion of QseC downregulates virulence gene expression and attenuates enterohemorrhagic and uropathogenic Escherichia coli (EHEC and UPEC), Salmonella typhimurium, and Francisella tularensis. Given that these pathogens employ different infection strategies and virulence factors, we used genome-wide approaches to better understand the role of the QseBC interplay in pathogenesis. We found that deletion of qseC results in misregulation of nucleotide, amino acid, and carbon metabolism. Comparable metabolic changes are seen in EHEC ΔqseC, suggesting that deletion of qseC confers similar pleiotropic effects in these two different pathogens. Disruption of representative metabolic enzymes phenocopied UPEC ΔqseC in vivo and resulted in virulence factor downregulation. We thus propose that in the absence of QseC, the constitutively active QseB leads to pleiotropic effects, impairing bacterial metabolism, and thereby attenuating virulence. These findings provide a basis for the development of anti-microbials targeting the phosphatase activity of QseC, as a means to attenuate a wide range of QseC-bearing pathogens.
Introduction
Bacterial survival and establishment of infection require a pathogen to sense and respond quickly and appropriately to environmental cues. Two-component regulatory systems are the major paradigm for this environmental adaptation in bacteria (Stock et al., 2000). Two-component systems recognize signaling molecules via a membrane sensor kinase and modulate gene expression accordingly through a cognate response regulator (Stock et al., 2000). QseBC is a two-component regulatory system that is ubiquitous among pathogens (Rasko et al., 2008). The QseC sensor kinase becomes activated in response to host and bacterial signals, and phosphorylates the QseB response regulator, a transcription factor that regulates virulence gene expression (Clarke et al., 2006, Sperandio et al., 2002). Studies in enterohemorrhagic Escherichia coli (EHEC) have shown that QseC is an α-adrenergic receptor, and becomes phosphorylated in the presence of epinephrine/norepinephrine (Clarke et al., 2006), however, adrenergic hormone-mediated virulence in Salmonella may involve additional or alternative pathways (Pullinger et al., 2010, Karavolos et al., 2011, Spencer et al., 2010). Even though this may suggest that QseC is tailored to respond to signals specific to the niche of each pathogen, all current studies converge to the fact that deletion of qseC attenuates important human pathogens, including EHEC, Salmonella typhimurium, Francisella tularensis, and uropathogenic E. coli (UPEC) (Bearson & Bearson, 2008, Rasko et al., 2008, Kostakioti et al., 2009).
We previously examined the role of QseBC in UPEC during urinary tract infection (UTI) (Kostakioti et al., 2009). UTIs are among the predominant bacterial infections in women, responsible for 3.5 billion dollars in annual health care costs in the US (Griebling, 2007). UPEC are the primary etiological agent, causing nearly 85% of community-acquired UTIs (Griebling, 2007). The pathogenesis of UPEC in a murine model of UTI has been extensively characterized (Justice et al., 2004, Hunstad & Justice, 2010) and presents an ideal system for exploring the function of virulence regulators such as QseBC. Upon UPEC introduction into the bladder, type 1 pili facilitate colonization of the luminal epithelium via the FimH adhesin, which recognizes surface mannosylated receptors in humans and mice (Zhou et al., 2001, Bouckaert et al., 2005, Wellens et al., 2008, Thumbikat et al., 2009, Hung et al., 2002, Eto et al., 2007). These interactions result in bacterial internalization into human bladder cells (Eto et al., 2007, Martinez et al., 2000) and in the murine epithelium (Thankavel et al., 1997, Bishop et al., 2007), which activates a TLR-4 dependent process expelling the bacteria in exocytic vesicles (Bishop et al., 2007). However, UPEC can escape into the host cell cytoplasm, where they are able to subvert expulsion and innate defenses by replicating into biofilm-like intracellular bacterial communities (IBCs) (Wright et al., 2007, Anderson et al., 2003, Justice et al., 2004, Rosen et al., 2007, Garofalo et al., 2007). Subsequently, UPEC disperse from the IBC, escape into the bladder lumen, and re-initiate the process by binding and invading naive epithelial cells (Justice et al., 2004). UPEC form IBCs in diverse mouse strain backgrounds as well as in humans (Garofalo et al., 2007, Rosen et al., 2007). The FimH-dependent IBC cycle that potentiates the establishment of infection was commonly observed in a 4 year clinical study in urines of women with recurrent UTI (rUTI) (Rosen et al., 2007). Virulence factors that increase the fitness of UPEC in the urinary tract are predicted to be under positive selection (Chen et al., 2006). For example, the fimH gene is under positive selection in clinical isolates of UPEC (Chen et al., 2009), consistent with an important role for FimH in human disease. These findings, argue for strong parallels between the murine model and human infection.
Type 1 pili belong to a class of extracellular fibers assembled by the chaperone-usher pathway (CUP) (Waksman & Hultgren, 2009). Multiple CUP pili within E. coli genomes are thought to be required for tuning adhesive properties specific for different environmental niches (Kline et al., 2010, Morschhauser et al., 1993, Mulvey et al., 1998, Uhlin et al., 1985). Other factors important for UPEC virulence include the salmochelin and yersiniabactin iron scavenging siderophores, which are expressed at higher levels in urinary strains than in the gut strains from the same patients suffering from UTI (Henderson et al., 2009), surface structures (such as flagella and curli) (Cegelski et al., 2009, Wright et al., 2005), stress response pathways (such as sulA, surA) (Justice et al., 2005, Justice et al., 2006), and core metabolic genes (Alteri et al., 2009).
Although not typically considered as virulence determinants, metabolic factors play a vital role during infection, as modulation of metabolism ensures survival and replication (Dalebroux et al., 2010). Studies in UPEC revealed that amino acids feeding into the TCA cycle are critical in vivo (Alteri et al., 2009) while studies in F. tularensis, identified the CarAB enzyme, involved in pyrimidine metabolism, to be critical for phagosome escape (Meibom & Charbit, 2010). Many other investigations have connected metabolism with bacterial virulence (Dalebroux et al., 2010, Wolfe, 2010) arguing that regulation of metabolic state is a general requirement during infection and suggesting that metabolic and virulence genes are co-regulated, regardless of infection site or pathogenic strategy.
We have previously shown deletion of qseC impairs IBC formation and attenuates UPEC (Kostakioti et al., 2009), while a single qseB or double qseBC deletion do not affect UPEC virulence (Kostakioti et al., 2009). We found that QseC dephosphorylates and deactivates QseB (Kostakioti et al., 2009), thus in the absence of QseC, QseB is constitutively phosphorylated (presumably by another kinase or phosphodonor molecule) and represses virulence-associated genes important for UTI, including type 1 pili, curli, and flagella. This phenomenon (where deletion of only qseC attenuates infection) is also seen in EHEC and S. typhimurium (Clarke et al., 2006, Bearson et al., 2010, Kostakioti et al., 2009).
Given that absence of QseC affects virulence of pathogens that engage in diverse host-pathogen interactions and cause different diseases, we asked whether attenuation stems from a misregulation of common pathways rather than specific virulence genes. We thus, performed genome-wide analyses of transcription, protein expression, and metabolite utilization patterns. We verified and extended observations that deletion of qseC dysregulates virulence factors. Interestingly, we discovered that the majority of misregulated targets in UTI89ΔqseC are devoted to metabolism; deletion of qseC leads to increased pyrimidine production and utilization, decreased synthesis and catabolism of amino acids important for coupling carbon and nitrogen metabolism (arginine, aspartate, glutamate/glutamine), and upregulation of the energetically less efficient glyoxylate shunt. We confirmed that many of these metabolic changes also occur in a qseC deletion mutant in EHEC, a pathogen which has >25% sequence divergence from UPEC (Darling et al., 2010, Brzuszkiewicz et al., 2006) and is an exclusively extracellular pathogen in the GI tract (Horne et al., 2002). The similarity in metabolic changes between UPEC and EHEC further suggests that these ΔqseC-mediated changes play an important role in pathogenesis. Indeed, deletion in UPEC of carbon metabolism genes that are dysregulated in the absence of QseC, phenocopied ΔqseC in vivo and affected virulence factor production. Therefore we propose that attenuation of E. coli and possibly other pathogens deleted for qseC stems from pleiotropic effects imparted by the uncontrolled activity of constitutively phosphorylated QseB, which compromises bacterial physiology resulting in downregulation of virulence gene expression and pathogen attenuation. Thus, blocking QseC phosphatase activity could impair metabolic processes in E. coli (and possibly other) QseC-bearing pathogens, thereby impeding their ability to express virulence factors, which opens avenues for the development of novel anti-virulence agents.
Results and Discussion
UTI89ΔqseC is defective in cellular processes central for virulence and bacterial physiology
To investigate the hypothesis that deletion of qseC impacts circuits that extend beyond species-specific virulence factors, we used microarrays to compare the transcriptional profiles of the cystitis isolate UTI89, UTI89ΔqseC, UTI89ΔqseBC and UTI89ΔqseC/pQseC (which carries qseC under its native promoter (Kostakioti et al., 2009)), after static growth for 18 hours (h), conditions previously used to study UTI89ΔqseC (Kostakioti et al., 2009). We found that 443 genes were significantly altered in UTI89ΔqseC compared to wild type (wt) UTI89 (Fig. 1A and Table S1) and 99.3% of these were restored in UTI89ΔqseC/pQseC (Table S1). The qseB gene was the 3rd most highly affected target, with >500-fold elevated transcription in UTI89ΔqseC (Table S1). This observation corroborates previous qRT-PCR analyses showing that qseB is highly expressed in UTI89ΔqseC, an effect that is most likely a direct outcome of the increased QseB activity in the absence of QseC (Kostakioti et al., 2009). Thus, since deletion of qseC interferes with dephosphorylation of QseB (Kostakioti et al., 2009), the ΔqseC mutation appears to unleash an uncontrolled positive feedback loop stimulating increased levels of QseB, resulting in a massive pleiotropic alteration of gene expression. In addition, the expression patterns of UTI89ΔqseBC did not significantly deviate from those of wt UTI89 (Table S1), indicating that the UTI89ΔqseC transcriptional differences are primarily connected to the presence of QseB in the absence of QseC.
Fig. 1.
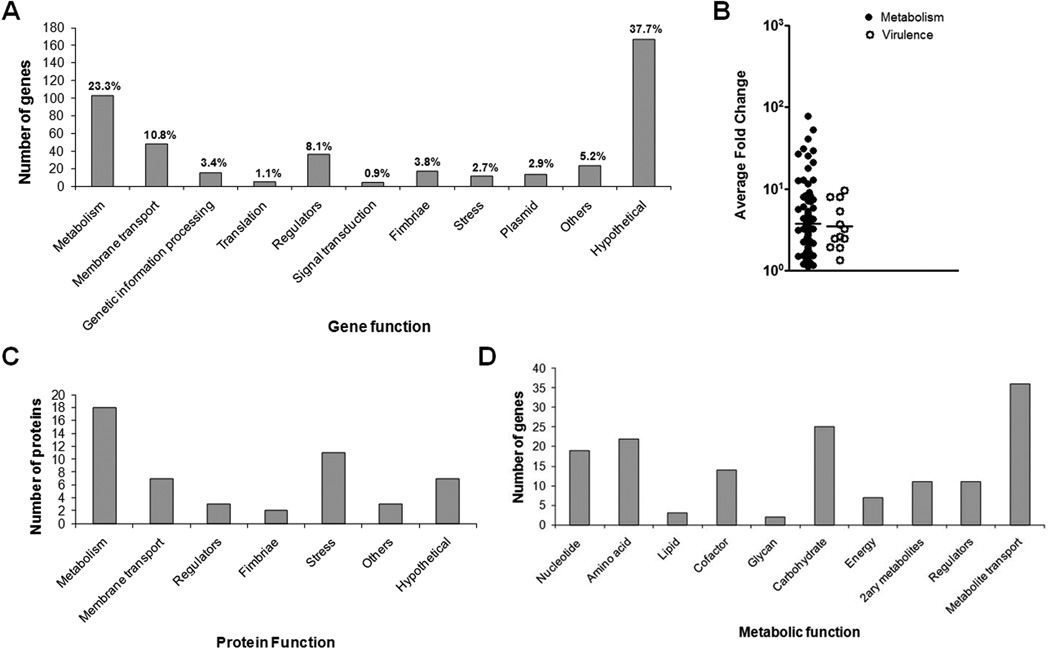
Deletion of qseC affects not only virulence factors, but primarily core metabolic processes. A) Classification of the 443 genes affected in UTI89ΔqseC, based on microarray analyses. Each gene is represented once, classified in the most relevant category. Percentage indicates the total number of affected factors in each category, relative to the total number of affected genes in the array. B) Average fold change of upregulated metabolic genes versus upregulated virulence genes. C) Graph showing the 53 altered proteins identified by GC/MS in UTI89ΔqseC. Functional classification was performed using KEGG and EcoCyc. D) The effects of qseC deletion on metabolism (Graph depicts the breakdown of all metabolic factors, including metabolite transporters, grouped in the “metabolism” and membrane transport” categories in 1A).
Among the 443 targets affected upon qseC deletion, genes encoding proteins with known functions (276 total) were classified into 11 broad categories (Fig. 1A) using KEGG and EcoCyc (Keseler et al., 2009, Kanehisa & Goto, 2000). Interestingly, the most upregulated gene in our array was ygiV, encoding a hypothetical transcriptional activator in an operon with ygiW, which was previously identified as the most abundant protein in UTI89ΔqseC whole cell lysates by N-terminal sequencing (unpublished data). QseBC and ygiVW are in close proximity and divergently transcribed. Thus, one possible mechanism for the increased ygiV expression could be that QseB binding to the qseBC promoter region might result in formation of open promoter complexes for both gene loci. Alternatively, ygiV could be upregulated due to a specific role it plays in the absence of QseC. We are currently investigating these hypotheses and the role of ygiV in virulence. Of note, although purified KdpE and QseF response regulators have been previously shown to be phosphorylated by QseC in vitro (Hughes et al., 2009), our analyses showed only a 1.8-fold increase in qseF expression and no altered expression of kdpE, suggesting a more minor role for these proteins in the observed ΔqseC-mediated defects.
Our microarray analyses confirmed that type 1 and S pili expression was altered in UTI89ΔqseC, in agreement with previous studies (Kostakioti et al., 2009), and identified other putative virulence-associated genes dysregulated in the absence of QseC (fimbriae, Fig. 1A). However, only 4% of the affected genes were dedicated to virulence factor production. The vast majority of known genes with altered expression in UTI89ΔqseC belonged to conserved processes, including metabolism, membrane transport, genetic information processing (DNA/RNA synthesis), translation, and stress responses (Fig. 1A). Furthermore, deletion of qseC altered the expression of 36 regulators, 52% of which modulate metabolism (Table S2). In addition to being numerically dominant, metabolic genes had higher fold-changes in expression compared to virulence factors (Fig. 1B). For example, the most highly upregulated virulence-related gene was sfaG with a 9.6-fold change, but 18 known or putative metabolic factors had >10-fold elevated transcription in UTI89ΔqseC (Table S1).
We also performed whole-cell proteome analysis (2D-DIGE) to capture changes between UTI89ΔqseC and wt UTI89 at the protein level (post-transcriptional and post-translational effects). UTI89ΔqseC had 536 protein spots with significantly different concentration/migration patterns. Fifty of these spots, selected to represent upregulated and downregulated proteins (cut-off P<0.0085), were identified by mass spectrometry (GC/MS). Our findings validated the expression patterns of 7 microarray targets (14% of identified proteins) and overall revealed effects in the same pathways as those identified by the transcriptional profiling (Fig. 1C). Considering the time-lapse between transcription and translation, the proteome is not expected to exactly reflect the mRNA pool present at that time. However, the same pathways were elucidated by both techniques, which support the argument that these pathways are compromised in UTI89ΔqseC. Consistent with the microarray data, most of the 53 identified proteins were dedicated to conserved bacterial processes, with 45.3% implicated in metabolism (Fig. 1C, Table S3). Therefore, in terms of the number of genes affected, the strength of transcriptional regulation and actual changes in steady-state protein expression, conserved metabolic genes are the predominant factors affected by the qseC deletion.
We further asked whether the changes in metabolic gene transcription and protein expression resulted in measurable differences in metabolism, using Biolog metabolic phenotype microarrays. We found that UTI89ΔqseC did not grow well in the presence of metabolites consumed in pathways which are downregulated upon qseC deletion (Table 1, discussed further in later sections). Conversely, UTI89ΔqseC grew better than wt UTI89 on metabolites utilized in pathways which are upregulated in the absence of QseC (Table 1). The most striking effects on UTI89ΔqseC growth were observed in the presence of nucleotide, amino acid, and TCA cycle intermediates, corresponding to the metabolic pathways most affected upon qseC deletion (Fig. 1D, Table 1). Moreover, administration of substrates imported by metabolite transporters that were downregulated in UTI89ΔqseC (Fig. S1) did not support efficient growth of UTI89ΔqseC (Table 1). Thus, global analyses of transcription, protein expression, and metabolic potential showed that deletion of qseC primarily alters multiple metabolic pathways besides specific virulence factors.
Table 1.
| a. Phenotypes lost by the qseC mutant as shown by metabolome analysis. | ||
|---|---|---|
| A. Phenotypes lost by UTI89ΔqseC | ||
| Test Metabolite | Growth difference1 | Mode of action/Metabolic Pathway |
| L-Pyroglutamic acid | −20.53 | C-Source/AA2, TCA |
| L-Ornithine | −20.86 | C-Source/AA, Urea |
| Fumaric acid | −21.95 | C-source/TCA |
| Ala-Asp | −29.23 | N-source/AA, TCA, Pyruvate |
| Ala-Glu | −28.68 | N-source/AA, TCA, Pyruvate |
| Spermine | −24.89 | Nutritional supplement |
| D,L-Thioctic acid | −23.4 | Nutritional supplement |
| Choline | −21.65 | Nutritional supplement |
| D,L-Mevalonic acid lactone | −21.31 | Nutritional supplement |
| a-Ketobutyric acid | −47.11 | Nutritional supplement/AA |
| a-Hydroxybutyric acid | −28.31 | Nutritional supplement/TCA, Pyruvate |
| Butyric acid | −29.06 | Nutritional supplement/TCA, Butanoate |
| D,L-Carnitine | −20.58 | Nutritional supplement/TCA, AA |
| Cysteamine-S-phosphate | −30.3 | P-source |
| Phosphocreatine | −47.99 | P-source/AA, TCA |
| Tripolyphosphate | −45.33 | P-source |
| D,L-Lipoamide | −43.84 | S-source |
| Glutathione | −29.45 | S-source |
| Thiourea | −27.99 | S-source |
| Sulfate | −37.67 | S-source |
| Taurocholic acid | −29.04 | S-source |
| Taurine | −27.16 | S-source |
| Hypotaurine | −26.12 | S-source |
| N-Acetyl-L-cysteine | −22.14 | S-source/AA, TCA, Pyruvate |
| L-Methionine | −21.47 | S-source/AA, TCA, Propanoate |
| L-Djenkolic acid | −37.87 | S-source/AA, TCA, Pyruvate |
| 1-Thio-b-D-glucose | −34.42 | S-source/Glycolysis, Glucose |
| b. Phenotypes gained by the qseC mutant as shown by metabolome analysis. | ||
|---|---|---|
| B.Phenotypes gained by UTI89ΔqseC | ||
| Uridine 5'- monophosphate | 60.54 | P-source/Pyrimidine |
| D-Glucose-6-phosphate | 61.24 | P-source/Glycolysis, Glucose |
| b-Glycerol phosphate | 60.63 | P-source/Glycolysis |
| Adenosine 5'-monophosphate | 52.1 | P-source/Purine |
| Glyoxylic acid | 51.34 | C-source/Glyoxylate |
| Inositol hexaphosphate | 45.41 | P-source |
| O-Phospho-D-serine | 45.1 | P-source/AA, TCA, Pyruvate |
| Maltose | 41.88 | C-source/Glycolysis, Glucose |
| Methylene diphosphonic acid | 41.83 | P-source |
| L-Asparagine | 37.11 | C-source/AA, TCA |
| Uridine 2'-monophosphate | 34.44 | P-source/Pyrimidine |
| b-D-Allose | 31.93 | C-source/Pentose |
| L-Glutamine | 31.84 | Nutritional supplement/AA, TCA |
| D-Trehalose | 28.35 | C-source/ Glycolysis, Glucose |
| Quinolinic acid | 26.83 | Nutritional supplement/NAD |
| D-Galactose | 26.72 | C-source/Galactose |
| D-Mannose | 25.35 | C-source/Glycolysis |
| (−)Shikimic acid | 23.59 | Nutritional supplement |
| b-Cyclodextrin | 22.75 | C-source |
| g-Cyclodextrin | 22.65 | C-source |
| D-Saccharic acid | 22.47 | C-source/Glycolysis, Glucose, Ascorbate, Aldarate |
| Dextrin | 22.34 | C-source |
| Nitrate | 21.87 | N-source/Urea |
| Gelatin | 21.73 | C-source |
| N-Acetyl-D-glucosamine | 21.21 | C-source/Glycolysis, Glucose |
Growth difference, Differences in growth rate between parent and mutant strains are determined by measuring the difference in average height of the kinetic plots in arbitrary units;
AA, Amino acid
Pathogen-specific virulence networks are dysregulated in UTI89 in the absence of QseC
CUP pili have been extensively implicated in uropathogenesis (Uhlin et al., 1985, Mulvey et al., 1998, Waksman & Hultgren, 2009, Kline et al., 2010). UTI89 harbors 10 gene clusters encoding known or putative CUP pili; fim, pap, sfa, yeh, yqi, fml, F17-like, auf, yad, and yfc (Chen et al., 2006). We have previously shown that deletion of qseC in UTI89 leads to reduced fim and increased sfa transcription (Kostakioti et al., 2009). Microarray analysis verified these observations (Fig. 2A), and revealed that sfaB and iscR were both upregulated in UTI89ΔqseC (Table S2). SfaB is a transcriptional activator of the sfa operon (Morschhauser et al., 1993), and IscR is a repressor of type 1 pili (Wu & Outten, 2009). Thus, increased expression of sfaB and iscR correlate with sfa upregulation and fim downregulation, respectively, suggesting a mechanism for how QseBC coordinates the expression of multiple CUP pili. Besides fim and sfa, 5 additional CUP pili systems were significantly affected in UTI89ΔqseC compared to wt UTI89; yeh, yqi, auf were downregulated and the fml and F17-like systems were upregulated (Fig. 2A). This is the first study to show that, yqi, auf, and F17-like gene clusters are expressed in UTI89 and that yeh and fml are expressed in E. coli. Given that our previous studies confirm the fim and sfa transcriptional effects (Kostakioti et al., 2009), we selected 2 more CUP systems (yeh and F17-like) and validated their expression changes by qRT-PCR (Fig. 2B).
Fig. 2.
QseC is implicated in the fimbrial regulatory networks. A) Expression of 7 CUP systems in UTI89ΔqseC, relative to UTI89, determined by microarray analysis. B) Relative fold change of yeh, F17-like, csgD and rstA in UTI89ΔqseC (red bar) compared to UTI89 (blue bar) by qRT-PCR. Values are normalized to the 16s rrsH gene.
In addition to CUP systems, we have previously shown that qseC deletion abolishes expression of curli fibers, by affecting the transcription of the csgD curli positive regulator (Kostakioti et al., 2009). Our microarray data revealed a 2.7-fold increase in the expression of rstA (Table S2), encoding a transcriptional repressor of csgD (Barnhart & Chapman, 2006), which could be responsible for the observed curli downregulation in the absence of QseC. We validated the increased rstA transcription and the corresponding reduction in csgD transcript in UTI89ΔqseC by qRT-PCR (Fig. 2B).
Given the prominent role of extracellular adhesive fibers in host cell colonization, invasion, and IBC formation (Anderson et al., 2003, Cegelski et al., 2009, Waksman & Hultgren, 2009), deregulation of these organelles in UTI89ΔqseC could confer a disadvantage at the early and acute infection stages.
Conserved metabolic pathways are affected upon qseC deletion
Since metabolic factors are predominantly dysregulated in UTI89ΔqseC (Fig. 1A, Table S4), we examined the effects on nucleotide, amino acid, and carbohydrate metabolism and iron homeostasis (Fig. S2).
Nucleic acid metabolism
Compared to UTI89, UTI89ΔqseC upregulated genes involved in de novo pyrimidine biosynthesis and downregulated genes devoted to purine synthesis (Fig. 3A–B). Specifically, CarAB, catalyzing the first step in pyrimidine biosynthesis, was significantly upregulated compared to wt UTI89 (Fig. 3A, Tables S1, S3), suggesting increased carbamoyl-phosphate production. In addition, pyrIB, encoding the enzyme that breaks down carbamoyl-phosphate (Fig. 3B), were the most highly upregulated (29- and 31-fold respectively) metabolic genes (Fig. 3A, Table S4). In addition, pyrH, important for UMP-UDP interconversions was also upregulated (Fig. 3A), pointing towards increased levels of PyrH, which could explain the efficient growth of UTI89ΔqseC on 5´- and 2´-UMP, compared to wt UTI89 (Table 1). In contrast, the purine biosynthetic genes purK, purM, and purT, the ribonucleoside reductase nrdF, and the transcriptional activator xapR (involved in purine interconversions) were all downregulated in UTI89ΔqseC (Fig. 3A, Table S2), suggesting reduced purine synthesis and defects in some of the purine utilization pathways. Administration of GMP or AMP bypassed the corresponding purine synthesis defects in UTI89ΔqseC, resulting in growth that resembled or surpassed that of the parent strain respectively (data not shown and Table 1b). Given that deletion of qseC did not result in detectable effects in the pathways converting AMP to ATP and GMP to GTP, provision of AMP or GMP could be used for generation of ATP or GTP, respectively, and result in efficient UTI89ΔqseC growth possibly by accounting for the energy shortcomings of this mutant. The ability of UTI89ΔqseC to utilize administered AMP and GMP may suggest a defect that arises prior to the branch in the biosynthetic pathway where AMP and GMP synthesis diverge, most likely prior to IMP formation during de novo purine biosynthesis.
Fig. 3.
Effects of qseC deletion on nucleotide and amino acid metabolism. A) Nucleotide metabolism genes differentially expressed in UTI89ΔqseC, as determined by microarray analysis. B) Schematic of nucleotide metabolism. Upregulated genes, based on microarray analysis, are shown in green; downregulated genes in red; solid arrows indicate single steps; dashed arrows indicate more than one step. C) Differentially expressed amino acid metabolism genes. D) Schematic of the amino acid metabolic pathways affected upon qseC deletion. Color coding is same as in (B).
Collectively, our data suggest an imbalanced pyrimidine:purine ratio in ΔqseC, which could increase nucleotide mismatches during DNA replication and transcription. Interestingly, genes involved in DNA replication and repair (recG, recB, recD, recN, dinG, the dinG-like UTI89_C2002, ycaJ, rmuC, mfd and hepA) were upregulated in UTI89ΔqseC (Fig. S3, Table S1), indicating a stress response that may occur upon accumulation of nucleotide mismatches.
Amino acid metabolism
Previous studies reported that the arginine pathway is induced in UPEC during growth in urine, supporting a role for this metabolite in bacterial survival inside the host (Darling et al., 2010, Alteri et al., 2009). We observed that argC (involved in ornithine production from glutamate), and argF, argG and argR (implicated in utilization of ornithine for arginine synthesis) were downregulatred in UTI89ΔqseC (Fig. 3C–D). In agreement with downregulation of ornithine utilization genes, UTI89ΔqseC displayed a growth defect on L-ornithine (Table 1), supporting the hypothesis that this mutant cannot efficiently produce arginine (Fig. 3C–D). Notably, arginine- and de novo pyrimidine biosynthesis are coupled, as both pathways utilize carbamoyl-phosphate (Fig. 3B). It is thus possible that downregulation of arginine biosynthetic genes leads to accumulation of carbamoyl-phosphate and its subsequent shunt towards pyrimidine production, accounting for the upregulation of pyrimidine synthesis genes. Alternatively, upregulation of pyrimidine synthesis may be titrating carbamoyl-phosphate away from the arginine pathway, resulting in downregulation of arginine biosynthetic genes.
Arginine biosynthesis is tied to glutamate metabolism since ornithine is produced from glutamate conversions (Fig. 3D). Thus, downregulation of arginine biosynthetic genes may be due to reduced glutamate availability, which could influence glutamine and aspartate production (Fig. 3D). Indeed, we observed a 2-fold downregulation of ybaS involved in glutamate/glutamine interconversions (Fig. 3C–D), and a reduction in the abundance of glutamine (UTI89_C0814, 3.6-fold) and glutamate/aspartate (UTI89_C0651, 7-fold) transporters in UTI89ΔqseC (Fig. S1). Consistent with a defect in transport/metabolism of these amino acids, UTI89ΔqseC could not grow well on L-glutamate, L-pyroglutamate and L-aspartate (Table 1).
L-aspartate can be used for the production of L-asparagine, L-methionine, and L-cysteine (Fig. 3D) (Phillips & Stockley, 1996). Studies have shown that reduction of L-asparagine upregulates asnA and asnB, implicated in aspartate/asparagine interconversions (Thaw et al., 2006). Expression of asnA was higher in UTI89ΔqseC suggesting lower asparagine levels, which further supports a decrease in aspartate (Fig. 3C). In parallel, we observed downregulation of asnC (Table S2), an asnA repressor expressed in high asparagine concentrations (Thaw et al., 2006). Taken together, these observations imply a reduction in asparagine upon QseC deletion. In addition, the methionine biosynthesis genes metL, metA, and metF and the regulators metJ and metR were downregulated (2 to 3.6-fold) in UTI89ΔqseC (Fig. 3C, Table S2), suggestive of lower production of methionine, which could lead to defects in protein synthesis and DNA replication, eliciting stress responses (Fig. S1A, S3). Downregulation of met genes, along with downregulation of gadB important for cysteine utilization (Fig. 3C–D), indicate reduced availability of cysteine and its derivatives. This was supported by a 2- and 3-fold reduction of the transporters FliY (cysteine) and TauB (taurine) (Fig. S1), and defective growth of UTI89ΔqseC on N-acetyl-L-cysteine, cysteamine-S-phosphate, glutathione, taurine, hypotaurine, and taurocholic acid (Table 1). Although, no annotated genes implicated in methionine utilization were downregulated in our analyses, UTI89ΔqseC could not grow well on methionine (Table 1a). However, given that many of the UTI89 methionine utilization genes are not annotated, the inability of the qseC mutant to utilize methionine for growth could be attributed to some of the effects involving hypothetical proteins. Overall, our analyses indicate that deletion of qseC leads to dysregulation of genes and proteins involved in pathways that are inter-connected and could thus lead to pleiotropic effects on metabolism and bacterial physiology.
TCA cycle
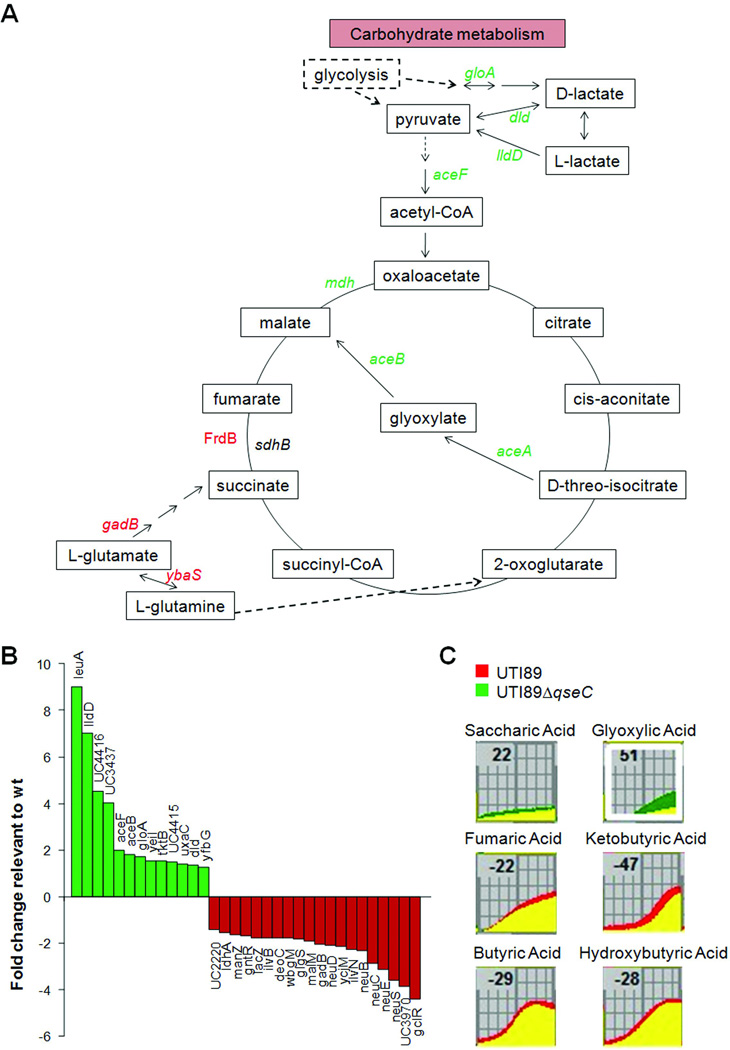
Many of the amino acid pathways affected in UTI89ΔqseC are used for the replenishment of TCA intermediates (Alteri et al., 2009, Hanson & Cox, 1967). L-glutamate gives rise to succinate and glutamine, which can be used for production of 2-oxoglutarate (Fig. 4A). Thus, the downregulation of the corresponding amino acid pathways in UTI89ΔqseC may impact TCA cycle progression. In further support of a disruption in the TCA cycle, we observed a 7-fold reduction in the levels of FrdB (Fig. 4A, Table S3), the enzyme implicated in fumarate to succinate conversion. Moreover, reduction in the formation of fumarate in UTI89ΔqseC could arise from the downregulation of the purine pathway, since fumarate is a byproduct of AMP biosynthesis. In parallel, we observed a 2–7 fold upregulation of pyruvate metabolism genes (aceF, lldD, dld, gloA, and aceB) and proteins (Mdh) in UTI89ΔqseC (Fig. 4A–B, Tables S1, S3), supporting that part of acetyl-CoA is preferentially consumed in the glyoxylate shunt (Fig. 4A). In agreement with a shift towards the glyoxylate shunt, UTI89ΔqseC grows better than UTI89 on saccharic and glyoxylic acids (Table 1), which can be used for the production of malate and oxaloacetate (Fig. 4A). In contrast, neither butyric, ketobutyric, or a-hydorxybutyric acids (Table 1), which are all used to generate oxaloglutarate, nor fumarate (Fig. 4B) sustain efficient growth of UTI89ΔqseC (Table 1). Given that TCA cycle intermediates are in turn consumed in numerous pathways, TCA cycle depression could play a key role in the severity of the qseC deletion defects; particularly, defects in 2-oxoglutarate production could impair formation of glutamate, which is central to various processes, including nitrogen metabolism. In further support of a defect in TCA cycle progression, is the upregulation of the iscRSUA, hscA, and fdx genes, suggestive of low [Fe-S] cluster assembly (further discussed in SI), which could impact the activity of several iron-sulfur TCA cycle enzymes. In addition, the UTI89ΔqseC proteome revealed a 3-fold upregulation of Pta and AckA, which utilize acetyl-CoA and acetate for the production of acetyl phosphate. Since acetyl-phosphate is a major phosphodonor molecule in the cell, a potential increase in its levels could account for, or be the outcome of the increased phosphorylation of QseB.
Fig. 4.
Deletion of qseC impedes TCA cycle completion. A) Schematic of carbohydrate metabolism. Upregulated genes, based on microarray analysis, are shown in green; downregulated genes in red; solid arrows indicate single steps; dashed arrows indicate more than one step. B) Carbohydrate metabolism genes differentially expressed in the absence of QseC. C) Growth curves of UTI89ΔqseC (green) and wt UTI89 (red) in the presence of metabolites implicated in the glyoxylate shunt and the TCA cycle, generated by the Omnilog PM software. UTI89ΔqseC has a growth defect on metabolites that require completion of the TCA cycle (bottom 4 panels) and shows a preference for metabolites of the glyoxylate shunt (top 2 panels).
Collectively, our data support that deletion of qseC tilts the nucleotide balance towards pyrimidine synthesis, a defect that is reflected in interconnected amino acid pathways feeding into the TCA cycle. Moreover, the reduced expression of genes involved in succinate and 2-oxoglutarate production, combined with upregulation of genes and proteins important for the glyoxylate shunt show a defect in the ability of UTI89ΔqseC to efficiently complete the TCA cycle and generate wt energy levels. The effects conferred upon qseC deletion are not specific to the LB growth conditions as similar pathways were shown to be affected in both LB (used for microarray and proteome), and minimal media (metabolome profiling). In addition, qPCR analyses probing for qseB and aceB expression suggest similar expression patterns during growth in human urine (Fig. S4). Given that energy production is crucial during infection, and biochemical intermediates, especially those from central pathways, are formed and utilized in a variety of anabolic and catabolic processes, the misregulation of these central metabolic pathways in the absence of QseC leads to the hypothesis that a controlled QseBC interplay is an important fitness determinant of pathogenic bacteria.
The metabolic dysregulation in the absence of QseC is not a UPEC-specific phenomenon
To examine whether the metabolic defects caused by qseC deletion are conserved among QseC-bearing pathogens, we performed metabolic profiling of EHEC strain 86-24 and its isogenic qseC deletion mutant. EHEC and UPEC display significant genomic divergence of at least 25%, and occupy distinct pathogenic niches (Brzuszkiewicz et al., 2006, Darling et al., 2010, Horne et al., 2002). The EHEC qseC mutant displayed defects in the same metabolic pathways as UPEC, and in many cases exhibited more severe phenotypes (Table S5). In particular, similar to UPEC, 86-24ΔqseC had defects growing on L-aspartate, L-glutamine, cysteine and its derivatives (N-acetyl-L-cysteine and cysteamine-S-phosphate), D-methionine, taurine, hypotaurine, and taurocholic acid (Table S5). Moreover, the TCA intermediates fumaric, a-ketobutyric, oxaloacetic, a-hydroxybutyric and butyric acids did not support efficient growth of 86-24ΔqseC, verifying that the TCA cycle is compromised in both pathotypes (Table S5). Notably, 86-24ΔqseC exhibited more severe defects, being unable to utilize glycolysis and pyruvate substrates, indicating that qseC deletion has a higher impact on carbohydrate metabolism in EHEC. More pronounced defects were also observed in nucleotide metabolism, as 86-24ΔqseC did not grow well on purine or pyrimidine substrates, indicative of a defect in both metabolic branches (Table S5). The differences in the extent of metabolic perturbation may reflect the distinct lifestyle and energy requirements of EHEC inside the host.
Deletion of qseC from two different bacterial pathogens affects similar core metabolic processes, resulting in altered nucleotide metabolism, downregulation of amino acid pathways that feed into the TCA cycle, and impeding TCA cycle progression. Previous transcriptional profiling studies with an EHEC qseC mutant, focusing on the virulence gene dysregulation of this mutant also reported defects on metabolism gene expression (Hughes et al., 2009), however, the significance and extent of this differential gene expression was not addressed. Our metabolome analyses extend the observations of Hughes et al., and demonstrate that disruption of QseC interferes with metabolic processes. The similarity in metabolic changes in the two pathotypes is even more striking given the differences in specific virulence factors affected and the distinct niches of these pathogens. In addition, transcriptional analyses of Salmonella qse mutants also revealed effects on metabolic gene expression (Merighi et al., 2009). Thus, we propose that the attenuation of qseC mutants is due, at least in part, to these metabolic effects in the absence of QseC.
Disruption of metabolic pathways in UTI89 results in a ΔqseC-like phenotype in vivo
We examined whether perturbation of metabolic pathways interferes with establishment of infection, using the TCA cycle as a proxy. The results described above revealed that deletion of qseC impedes TCA cycle progression, and in the case of UPEC, results in engagement of the glyoxylate shunt. We investigated whether the inability of UTI89ΔqseC to complete the TCA cycle is associated with its attenuation in vivo. We created non-polar deletions of aceA, sdhB or mdh in UTI89. AceA converts isocitrate to glyoxylate, and its deletion disrupts the glyoxylate shunt (Fig. 4A). The sdhB gene encodes the succinate dehydogenase iron-sulfur subunit, required for the conversion of succinate to fumarate (Fig. 4A), and its deletion interferes with TCA cycle completion. The malate dehydrogenase, Mdh, oxidizes malate to oxaloacetate, participating in both the TCA cycle and the glyoxylate shunt.
Female C3H/HeN mice were transurethrally inoculated with wt UTI89, UTI89ΔsdhB, UTI89Δmdh, UTI89ΔaceA, or UTI89ΔqseC and the ability of each mutant to survive in vivo was assessed at 6h and 16h post infection (p.i.) by cfu enumeration and confocal microscopy, to capture the mid- and late-stages of IBC formation (Justice et al., 2004). Our data showed that disruption of the glyoxylate shunt alone had minor effects on urinary tract colonization and IBC formation as indicated by the UTI89ΔaceA phenotypes (Fig. 5), suggesting that this pathway is not essential for UPEC in vivo survival. However, UTI89ΔsdhB and UTI89Δmdh, exhibited a severe survival defect within the bladder, indicated by a significant reduction in the recovered cfu compared to wt UTI89 (Fig. 5A). Reduced UTI89ΔsdhB and UTI89Δmdh bladder titers correlated with fewer IBCs formed by these strains (Fig. 5B), as was the case for UTI89ΔqseC (Fig. 5B and (Kostakioti et al., 2009)). Given that mdh deletion impacts both the TCA cycle and glyoxylate shunt, and since disruption of the glyoxylate shunt does not significantly affect UPEC virulence as evidenced by the UTI89ΔaceA phenotypes, attenuation of UTI89Δmdh is due to the disruption of the TCA cycle. These data argue that the ΔqseC in vivo defects are connected to its inability to complete the TCA cycle and are in agreement with previous studies demonstrating that the TCA cycle is important for uropathogenesis (Alteri et al., 2009). Deletion of sdhB or aceA in a qseC deletion background resulted in a qseC-like phenotype (data not shown), indicating that QseC is indeed upstream of these metabolic factors.
Fig. 5.
TCA cycle mutants are attenuated exhibiting ΔqseC-like phenotypes. A) Bladder titers of UTI89ΔsdhB, UTI89Δmdh and UTI89ΔaceA compared to UTI89 and UTI89ΔqseC. Experiment was repeated 3 times. B) IBC production by UTI89, UTI89ΔsdhB, UTI89Δmdh, UTI89ΔaceA and UTI89ΔqseC. C) HA titers of UTI89ΔsdhB, UTI89Δmdh and UTI89ΔaceA compared to UTI89 and UTI89ΔqseC. Mannose addition inhibits type 1-dependent HA. D) Western Blot analysis probing for the CsgG curli subunit in UTI89ΔsdhB, UTI89Δmdh and UTI89ΔaceA compared to UTI89 and UTI89ΔqseC. UTI89ΔsdhB and UTI89Δmdh are defective in curli expression. E) Motility assays showing that UTI89ΔsdhB and UTI89Δmdh exhibit defective swimming motility similar to UTI89ΔqseC and UTI89ΔflhDC (negative for flagella). ***, P<0.0007, **, P<0.0099 and *, P<0.05
Deletion of qseC impacts virulence gene expression by dysregulating core metabolism
We asked whether disruption of metabolic processes that are dysregulated in the qseC mutant affects virulence gene expression. We screened UTI89ΔsdhB, UTI89Δmdh and UTI89ΔaceA for production of type 1 pili in vitro by hemagglutination (HA). Our findings showed that, while UTI89ΔaceA displayed wt HA titers, UTI89ΔsdhB and UTI89Δmdh exhibited decreased mannose-sensitive HA similar to UTI89ΔqseC (Fig. 5C), indicative of reduced type 1 pili expression, further verified by Western blot analyses (data not shown). Similarly, UTI89ΔsdhB and UTI89Δmdh but not UTI89ΔaceA, displayed defects on other virulence-associated factors affected in UTI89ΔqseC, such as flagella and curli (Fig. 5D–E). Thus the reduced type 1 pili, curli and flagella expression in UTI89ΔqseC likely stems from its metabolic defects, supporting that the basis of attenuation of qseC mutants in vivo involves a complex metabolic circuitry that affects virulence gene expression.
Concluding remarks
In summary, the absence of QseC perturbs critical cellular processes that are common among various bacterial pathogens, compromising bacterial physiology and virulence, due to the aberrant activity of QseB. Thus, null mutations in qseC appear to unleash a potent QseB positive feedback loop that now operates uncontrolled. Increased levels of QseB result in more QseB-P by mass action, which then stimulates production of more QseB, which may explain the cause of the pleiotropy. We thus propose that, under normal conditions QseC tightly controls the phosphorylation of QseB so as to optimize expression patterns (metabolic and virulence genes). In the case of UPEC, this QseBC-controlled interplay may function to maximize fitness during infection by adjusting the internal (carbon and nitrogen metabolism) and the external (surface expressed pili) state of the bacteria during transition from the intestine to the urinary tract (enhancing bladder colonization), as well as during transition from extracellular to intracellular niches (enabling invasion and IBC formation). Disruption of QseC function interferes with QseB phosphorylation and results in an over-active regulator, which dysregulates targets that most likely are not part of its canonical regulon, leading to pronounced pleiotropic effects. Given that several of the affected pathways, like metabolic pathways, are highly conserved across bacteria, this could explain the attenuation conferred by deletion of QseC in diverse pathogens, with distinct virulence factors and disparate host-pathogen interactions. Indeed we have shown that this is true for UPEC and EHEC, whereas studies in Salmonella qse mutants showing metabolic perturbations support that our findings may pertain to non-E. coli QseC-bearing pathogens; however further studies are needed to verify whether attenuation of other pathogens deleted for qseC is related to metabolism. Thus, a proper QseBC interplay is critical for optimal bacterial expression patterns and establishment of infection in pathogenic E. coli and possibly other pathogens. Therefore, interfering with QseC and impeding its ability to dephosphorylate QseB, opens new avenues for targeting the virulence of QseC-bearing gram-negative pathogens and makes QseC an excellent candidate for the development of anti-virulence therapeutics.
Materials and Methods
Strains, constructs and growth conditions
UTI89ΔqseC, UTI89ΔqseC/pQseC and EHEC 86-24ΔqseC were created previously (Kostakioti et al., 2009). UTI89ΔsdhB, UTI89Δmdh and UTI89ΔaceA were created using λ Red Recombinase (Murphy & Campellone, 2003). Bacteria were incubated in Luria Bertani (LB) media at 37°C for 4h with shaking, subcultured (1:1000) in fresh LB media and incubated statically for 18h.
RNA extraction, microarray analyses and qRT-PCR
RNA was extracted using the RNeasy kit (Qiagen), DNase-treated, and reverse transcribed. Exogenous RNA spikes were added as internal controls for RT and labeling reactions. Resulting cDNA samples were fragmented, biotinylated and hybridized to GeneChip custom-made genome arrays (Affymetrix UTI89-01a520299F). Data were analyzed based on recommendations from the Golden Spike data set analysis (PPLR test threshold cutoff, 0.95) (Pearson, 2008). qRT-PCR was performed as previously described (Kostakioti et al., 2009) using primers listed in Table S6.
Proteomics
2-Dimensional difference gel electrophoresis (2D-DIGE), gel image generation and analysis were performed as previously described (Alban et al., 2003). Proteins were identified by mass spectrometry according to King et al. (King et al., 2007). Detailed description is provided in SI.
HA, motility and curli Western blots
HA was performed on normalized cells (OD600=1) as previously described (Kostakioti et al., 2009). Two-tailed Student’s t test was used for statistical analyses (P<0.05). Motility and curli expression were assessed as previously described (Kostakioti et al., 2009).
Metabolic phenotype microarrays
Metabolic profiling was performed according to the Biolog guidelines (www.biolog.com), using plates PM1–5. Detailed description is provided in SI. An average of 3 independent experiments is being reported.
Mouse infections
Female C3H/HeN mice (Harlan), were transurethrally infected with 107 bacteria carrying plasmid pCom-GFP as previously described (Kostakioti et al., 2009). Confocal microscopy was used for IBC enumeration (Kostakioti et al., 2009). Experiments were repeated 3 times. Statistical analyses were performed using two-tailed Mann Whitney (P <0.05). The Washington University animal studies committee has approved these studies.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Douglas E. Berg for helpful discussions and provision of the Biolog incubator (fund 1R41GM073965). We are grateful to Joe Palermo, Karen Dodson and Thomas Hannan for critical review of the manuscript. This work was supported by the NIH grants P50 DK64540-06, R01 AI048689-08 and R37 AI02549-18 (to S.J.H.). Mass spectrometry for enterobactin studies was supported by RR00954, DK20579 and DK56341.
References
- Alban A, David SO, Bjorkesten L, Andersson C, Sloge E, Lewis S, Currie I. A novel experimental design for comparative two-dimensional gel analysis: two-dimensional difference gel electrophoresis incorporating a pooled internal standard. Proteomics. 2003;3:36–44. doi: 10.1002/pmic.200390006. [DOI] [PubMed] [Google Scholar]
- Alteri CJ, Smith SN, Mobley HL. Fitness of Escherichia coli during urinary tract infection requires gluconeogenesis and the TCA cycle. PLoS Pathog. 2009;5 doi: 10.1371/journal.ppat.1000448. e1000448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson GG, Palermo JJ, Schilling JD, Roth R, Heuser J, Hultgren SJ. Intracellular bacterial biofilm-like pods in urinary tract infections. Science. 2003;301:105–107. doi: 10.1126/science.1084550. [DOI] [PubMed] [Google Scholar]
- Barnhart MM, Chapman MR. Curli biogenesis and function. Annu Rev Microbiol. 2006;60:131–147. doi: 10.1146/annurev.micro.60.080805.142106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bearson BL, Bearson SM. The role of the QseC quorum-sensing sensor kinase in colonization and norepinephrine-enhanced motility of Salmonella enterica serovar Typhimurium. Microb Pathog. 2008;44:271–278. doi: 10.1016/j.micpath.2007.10.001. [DOI] [PubMed] [Google Scholar]
- Bearson BL, Bearson SM, Lee IS, Brunelle BW. The Salmonella enterica serovar Typhimurium QseB response regulator negatively regulates bacterial motility and swine colonization in the absence of the QseC sensor kinase. Microb Pathog. 2010;48:214–219. doi: 10.1016/j.micpath.2010.03.005. [DOI] [PubMed] [Google Scholar]
- Bishop BL, Duncan MJ, Song J, Li G, Zaas D, Abraham SN. Cyclic AMP-regulated exocytosis of Escherichia coli from infected bladder epithelial cells. Nat Med. 2007;13:625–630. doi: 10.1038/nm1572. [DOI] [PubMed] [Google Scholar]
- Bouckaert J, Berglund J, Schembri M, De Genst E, Cools L, Wuhrer M, Hung CS, Pinkner J, Slattegard R, Zavialov A, Choudhury D, Langermann S, Hultgren SJ, Wyns L, Klemm P, Oscarson S, Knight SD, De Greve H. Receptor binding studies disclose a novel class of high-affinity inhibitors of the Escherichia coli FimH adhesin. Mol Microbiol. 2005;55:441–455. doi: 10.1111/j.1365-2958.2004.04415.x. [DOI] [PubMed] [Google Scholar]
- Brzuszkiewicz E, Bruggemann H, Liesegang H, Emmerth M, Olschlager T, Nagy G, Albermann K, Wagner C, Buchrieser C, Emody L, Gottschalk G, Hacker J, Dobrindt U. How to become a uropathogen: comparative genomic analysis of extraintestinal pathogenic Escherichia coli strains. Proc Natl Acad Sci U S A. 2006;103:12879–12884. doi: 10.1073/pnas.0603038103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cegelski L, Pinkner JS, Hammer ND, Cusumano CK, Hung CS, Chorell E, Aberg V, Walker JN, Seed PC, Almqvist F, Chapman MR, Hultgren SJ. Small-molecule inhibitors target Escherichia coli amyloid biogenesis and biofilm formation. Nat Chem Biol. 2009;5:913–919. doi: 10.1038/nchembio.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen SL, Hung CS, Pinkner JS, Walker JN, Cusumano CK, Li Z, Bouckaert J, Gordon JI, Hultgren SJ. Positive selection identifies an in vivo role for FimH during urinary tract infection in addition to mannose binding. Proc Natl Acad Sci U S A. 2009;106:22439–22444. doi: 10.1073/pnas.0902179106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen SL, Hung CS, Xu J, Reigstad CS, Magrini V, Sabo A, Blasiar D, Bieri T, Meyer RR, Ozersky P, Armstrong JR, Fulton RS, Latreille JP, Spieth J, Hooton TM, Mardis ER, Hultgren SJ, Gordon JI. Identification of genes subject to positive selection in uropathogenic strains of Escherichia coli: a comparative genomics approach. Proc Natl Acad Sci U S A. 2006;103:5977–5982. doi: 10.1073/pnas.0600938103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke MB, Hughes DT, Zhu C, Boedeker EC, Sperandio V. The QseC sensor kinase: a bacterial adrenergic receptor. Proc Natl Acad Sci U S A. 2006;103:10420–10425. doi: 10.1073/pnas.0604343103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalebroux ZD, Svensson SL, Gaynor EC, Swanson MS. ppGpp conjures bacterial virulence. Microbiol Mol Biol Rev. 2010;74:171–199. doi: 10.1128/MMBR.00046-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darling AE, Mau B, Perna NT. progressiveMauve: Multiple Genome Alignment with Gene Gain, Loss and Rearrangement. PLoS One. 2010;5:e11147. doi: 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eto DS, Jones TA, Sundsbak JL, Mulvey MA. Integrin-mediated host cell invasion by type 1-piliated uropathogenic Escherichia coli. PLoS Pathog. 2007;3:e100. doi: 10.1371/journal.ppat.0030100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garofalo CK, Hooton TM, Martin SM, Stamm WE, Palermo JJ, Gordon JI, Hultgren SJ. Escherichia coli from urine of female patients with urinary tract infections is competent for intracellular bacterial community formation. Infect Immun. 2007;75:52–60. doi: 10.1128/IAI.01123-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griebling TL. Urologic Diseases in America. Washington, DC: NIH; 2007. pp. 587–645. [Google Scholar]
- Hanson RS, Cox DP. Effect of different nutritional conditions on the synthesis of tricarboxylic acid cycle enzymes. J Bacteriol. 1967;93:1777–1787. doi: 10.1128/jb.93.6.1777-1787.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson JP, Crowley JR, Pinkner JS, Walker JN, Tsukayama P, Stamm WE, Hooton TM, Hultgren SJ. Quantitative metabolomics reveals an epigenetic blueprint for iron acquisition in uropathogenic Escherichia coli. PLoS Pathog. 2009;5 doi: 10.1371/journal.ppat.1000305. e1000305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horne C, Vallance BA, Deng W, Finlay BB. Current progress in enteropathogenic and enterohemorrhagic Escherichia coli vaccines. Expert Rev Vaccines. 2002;1:483–493. doi: 10.1586/14760584.1.4.483. [DOI] [PubMed] [Google Scholar]
- Hughes DT, Clarke MB, Yamamoto K, Rasko DA, Sperandio V. The QseC adrenergic signaling cascade in Enterohemorrhagic E. coli (EHEC) PLoS Pathog. 2009;5 doi: 10.1371/journal.ppat.1000553. e1000553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung CS, Bouckaert J, Hung D, Pinkner J, Widberg C, DeFusco A, Auguste CG, Strouse R, Langermann S, Waksman G, Hultgren SJ. Structural basis of tropism of Escherichia coli to the bladder during urinary tract infection. Mol Microbiol. 2002;44:903–915. doi: 10.1046/j.1365-2958.2002.02915.x. [DOI] [PubMed] [Google Scholar]
- Hunstad DA, Justice SS. Intracellular lifestyles and immune evasion strategies of uropathogenic Escherichia coli. Annu Rev Microbiol. 2010;64:203–221. doi: 10.1146/annurev.micro.112408.134258. [DOI] [PubMed] [Google Scholar]
- Justice SS, Hung C, Theriot JA, Fletcher DA, Anderson GG, Footer MJ, Hultgren SJ. Differentiation and developmental pathways of uropathogenic Escherichia coli in urinary tract pathogenesis. Proc Natl Acad Sci U S A. 2004;101:1333–1338. doi: 10.1073/pnas.0308125100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Justice SS, Hunstad DA, Harper JR, Duguay AR, Pinkner JS, Bann J, Frieden C, Silhavy TJ, Hultgren SJ. Periplasmic peptidyl prolyl cis-trans isomerases are not essential for viability, but SurA is required for pilus biogenesis in Escherichia coli. J Bacteriol. 2005;187:7680–7686. doi: 10.1128/JB.187.22.7680-7686.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Justice SS, Hunstad DA, Seed PC, Hultgren SJ. Filamentation by Escherichia coli subverts innate defenses during urinary tract infection. Proc Natl Acad Sci U S A. 2006;103:19884–19889. doi: 10.1073/pnas.0606329104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karavolos MH, Bulmer DM, Spencer H, Rampioni G, Schmalen I, Baker S, Pickard D, Gray J, Fookes M, Winzer K, Ivens A, Dougan G, Williams P, Khan CM. Salmonella Typhi sense host neuroendocrine stress hormones and release the toxin haemolysin E. EMBO Rep. 2011;12:252–258. doi: 10.1038/embor.2011.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keseler IM, Bonavides-Martinez C, Collado-Vides J, Gama-Castro S, Gunsalus RP, Johnson DA, Krummenacker M, Nolan LM, Paley S, Paulsen IT, Peralta-Gil M, Santos-Zavaleta A, Shearer AG, Karp PD. EcoCyc: a comprehensive view of Escherichia coli biology. Nucleic Acids Res. 2009;37:D464–D470. doi: 10.1093/nar/gkn751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King JB, Gross J, Lovly CM, Piwnica-Worms H, Townsend RR. Identification of protein phosphorylation sites within Ser/Thr-rich cluster domains using site-directed mutagenesis and hybrid linear quadrupole ion trap Fourier transform ion cyclotron resonance mass spectrometry. Rapid Commun Mass Spectrom. 2007;21:3443–3451. doi: 10.1002/rcm.3223. [DOI] [PubMed] [Google Scholar]
- Kline KA, Dodson KW, Caparon MG, Hultgren SJ. A tale of two pili: assembly and function of pili in bacteria. Trends Microbiol. 2010;18:224–232. doi: 10.1016/j.tim.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostakioti M, Hadjifrangiskou M, Pinkner JS, Hultgren SJ. QseC-mediated dephosphorylation of QseB is required for expression of genes associated with virulence in uropathogenic Escherichia coli. Mol Microbiol. 2009;73:1020–1031. doi: 10.1111/j.1365-2958.2009.06826.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez JJ, Mulvey MA, Schilling JD, Pinkner JS, Hultgren SJ. Type 1 pilus-mediated bacterial invasion of bladder epithelial cells. Embo J. 2000;19:2803–2812. doi: 10.1093/emboj/19.12.2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meibom KL, Charbit A. The unraveling panoply of Francisella tularensis virulence attributes. Curr Opin Microbiol. 2010;13:11–17. doi: 10.1016/j.mib.2009.11.007. [DOI] [PubMed] [Google Scholar]
- Merighi M, Septer AN, Carroll-Portillo A, Bhatiya A, Porwollik S, McClelland M, Gunn JS. Genome-wide analysis of the PreA/PreB (QseB/QseC) regulon of Salmonella enterica serovar Typhimurium. BMC Microbiol. 2009;9:42. doi: 10.1186/1471-2180-9-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morschhauser J, Vetter V, Korhonen T, Uhlin BE, Hacker J. Regulation and binding properties of S fimbriae cloned from E. coli strains causing urinary tract infection and meningitis. Zentralbl Bakteriol. 1993;278:165–176. doi: 10.1016/s0934-8840(11)80834-0. [DOI] [PubMed] [Google Scholar]
- Mulvey MA, Lopez-Boado YS, Wilson CL, Roth R, Parks WC, Heuser J, Hultgren SJ. Induction and evasion of host defenses by type 1-piliated uropathogenic Escherichia coli. Science. 1998;282:1494–1497. doi: 10.1126/science.282.5393.1494. [DOI] [PubMed] [Google Scholar]
- Murphy KC, Campellone KG. Lambda Red-mediated recombinogenic engineering of enterohemorrhagic and enteropathogenic E. coli. BMC Mol Biol. 2003;4:11. doi: 10.1186/1471-2199-4-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson RD. A comprehensive re-analysis of the Golden Spike data: towards a benchmark for differential expression methods. BMC Bioinformatics. 2008;9:164. doi: 10.1186/1471-2105-9-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips SE, Stockley PG. Structure and function of Escherichia coli met repressor: similarities and contrasts with trp repressor. Philos Trans R Soc Lond B Biol Sci. 1996;351:527–535. doi: 10.1098/rstb.1996.0051. [DOI] [PubMed] [Google Scholar]
- Pullinger GD, Carnell SC, Sharaff FF, van Diemen PM, Dziva F, Morgan E, Lyte M, Freestone PP, Stevens MP. Norepinephrine augments Salmonella enterica-induced enteritis in a manner associated with increased net replication but independent of the putative adrenergic sensor kinases QseC and QseE. Infect Immun. 2010;78:372–380. doi: 10.1128/IAI.01203-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasko DA, Moreira CG, Li de R, Reading NC, Ritchie JM, Waldor MK, Williams N, Taussig R, Wei S, Roth M, Hughes DT, Huntley JF, Fina MW, Falck JR, Sperandio V. Targeting QseC signaling and virulence for antibiotic development. Science. 2008;321:1078–1080. doi: 10.1126/science.1160354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen DA, Hooton TM, Stamm WE, Humphrey PA, Hultgren SJ. Detection of intracellular bacterial communities in human urinary tract infection. PLoS Med. 2007;4:e329. doi: 10.1371/journal.pmed.0040329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer H, Karavolos MH, Bulmer DM, Aldridge P, Chhabra SR, Winzer K, Williams P, Khan CM. Genome-wide transposon mutagenesis identifies a role for host neuroendocrine stress hormones in regulating the expression of virulence genes in Salmonella. J Bacteriol. 2010;192:714–724. doi: 10.1128/JB.01329-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperandio V, Torres AG, Kaper JB. Quorum sensing Escherichia coli regulators B and C (QseBC): a novel two-component regulatory system involved in the regulation of flagella and motility by quorum sensing in E. coli. Mol Microbiol. 2002;43:809–821. doi: 10.1046/j.1365-2958.2002.02803.x. [DOI] [PubMed] [Google Scholar]
- Stock AM, Robinson VL, Goudreau PN. Two-component signal transduction. Annu Rev Biochem. 2000;69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- Thankavel K, Madison B, Ikeda T, Malaviya R, Shah AH, Arumugam PM, Abraham SN. Localization of a domain in the FimH adhesin of Escherichia coli type 1 fimbriae capable of receptor recognition and use of a domain-specific antibody to confer protection against experimental urinary tract infection. J Clin Invest. 1997;100:1123–1136. doi: 10.1172/JCI119623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaw P, Sedelnikova SE, Muranova T, Wiese S, Ayora S, Alonso JC, Brinkman AB, Akerboom J, van der Oost J, Rafferty JB. Structural insight into gene transcriptional regulation and effector binding by the Lrp/AsnC family. Nucleic Acids Res. 2006;34:1439–1449. doi: 10.1093/nar/gkl009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thumbikat P, Berry RE, Zhou G, Billips BK, Yaggie RE, Zaichuk T, Sun TT, Schaeffer AJ, Klumpp DJ. Bacteria-induced uroplakin signaling mediates bladder response to infection. PLoS Pathog. 2009;5 doi: 10.1371/journal.ppat.1000415. e1000415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlin BE, Norgren M, Baga M, Normark S. Adhesion to human cells by Escherichia coli lacking the major subunit of a digalactoside-specific pilus-adhesin. Proc Natl Acad Sci U S A. 1985;82:1800–1804. doi: 10.1073/pnas.82.6.1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waksman G, Hultgren SJ. Structural biology of the chaperone-usher pathway of pilus biogenesis. Nat Rev Microbiol. 2009;7:765–774. doi: 10.1038/nrmicro2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellens A, Garofalo C, Nguyen H, Van Gerven N, Slattegard R, Hernalsteens JP, Wyns L, Oscarson S, De Greve H, Hultgren S, Bouckaert J. Intervening with urinary tract infections using anti-adhesives based on the crystal structure of the FimH-oligomannose-3 complex. PLoS One. 2008;3:e2040. doi: 10.1371/journal.pone.0002040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe AJ. Physiologically relevant small phosphodonors link metabolism to signal transduction. Curr Opin Microbiol. 2010;13:204–209. doi: 10.1016/j.mib.2010.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright KJ, Seed PC, Hultgren SJ. Uropathogenic Escherichia coli flagella aid in efficient urinary tract colonization. Infect Immun. 2005;73:7657–7668. doi: 10.1128/IAI.73.11.7657-7668.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright KJ, Seed PC, Hultgren SJ. Development of intracellular bacterial communities of uropathogenic Escherichia coli depends on type 1 pili. Cell Microbiol. 2007;9:2230–2241. doi: 10.1111/j.1462-5822.2007.00952.x. [DOI] [PubMed] [Google Scholar]
- Wu Y, Outten FW. IscR controls iron-dependent biofilm formation in Escherichia coli by regulating type I fimbria expression. J Bacteriol. 2009;191:1248–1257. doi: 10.1128/JB.01086-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou G, Mo WJ, Sebbel P, Min G, Neubert TA, Glockshuber R, Wu XR, Sun TT, Kong XP. Uroplakin Ia is the urothelial receptor for uropathogenic Escherichia coli: evidence from in vitro FimH binding. J Cell Sci. 2001;114:4095–4103. doi: 10.1242/jcs.114.22.4095. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.