Abstract
A major hurdle to evolutionary engineering approaches for multigenic phenotypes is the ability to simultaneously modify multiple genes rapidly and selectively. Here, we describe a method for in vivo-targeted mutagenesis in yeast, targeting glycosylases to embedded arrays for mutagenesis (TaGTEAM). By fusing the yeast 3-methyladenine DNA glycosylase MAG1 to a tetR DNA-binding domain, we are able to elevate mutation rates >800 fold in a specific ∼20-kb region of the genome or on a plasmid that contains an array of tetO sites. A wide spectrum of transitions, transversions and single base deletions are observed. We provide evidence that TaGTEAM generated point mutations occur through error-prone homologous recombination (HR) and depend on resectioning and the error-prone polymerase Pol ζ. We show that HR is error-prone in this context because of DNA damage checkpoint activation and base pair lesions and use this knowledge to shift the primary mutagenic outcome of targeted endonuclease breaks from HR-independent rearrangements to HR-dependent point mutations. The ability to switch repair in this way opens up the possibility of using targeted endonucleases in diverse organisms for in vivo-targeted mutagenesis.
INTRODUCTION
Directed evolution of proteins using in vitro-targeted mutagenesis of single genes has been an effective strategy for increasing enzyme stability, specificity and catalysis (1), enhancing viral vectors for gene therapy (2) and creating microbes that tolerate and/or overproduce desirable metabolites (3). Similar combinatorial strategies are now being applied to improve more complex multigenic cellular phenotypes, such as microbial fuel or chemical production. Targeted approaches (4–6) fine-tune expression levels of pathway components to optimize flux but have been restricted to manipulating a single gene, with one important exception (7). Global approaches involve mutations that combinatorially sample gene expression state, with little control over the number and extent to which different genes’ expression is altered (8–10). This makes it difficult to incorporate knowledge of pathways and fluxes fed-in from rational approaches.
New strategies are needed for the targeted evolution of complex, multigenic phenotypes. Scaling in vitro-targeted mutagenesis techniques to many genes is difficult and depends on the ease of introducing and modifying genetic material. High-efficiency automated transformation of Escherichia coli through multiplex automated genome engineering enabled targeted mutagenesis to 24 genes in E. coli, resulting in improved lycopene production in 3 days (7). However, the high transformation efficiency required is untenable in many organisms. Alternatively, in vivo mutagenesis techniques work by mutating the entire genome, but without specificity, the mutation rate is limited by organism viability. Phage-assisted continuous evolution sidesteps this issue by confining mutagenesis to a bacteriophage genome and has enabled evolution of variant T7 polymerases with altered promoter and initiation specificity (11). However, phage-assisted continuous evolution is also limited to evolving interactions within E. coli. Similar approaches using retroviruses and mammalian cell culture (12,13) do not sample as large a diversity of mutants.
An alternative strategy is to develop a technique for in vivo-targeted mutagenesis, ideally applicable in diverse organisms, that combines the specificity of in vitro-targeted mutagenesis with the absence of genetic manipulation in in vivo mutagenesis. A natural example of in vivo-targeted mutagenesis is somatic hypermutation (14), which has been used for the directed evolution of fluorescent proteins (15–17) but is restricted to a narrow range of cell types. A second in vivo-targeted mutagenesis strategy is the expression of a mutant, low fidelity DNA polymerase I in E. coli that elevates mutation rates in a 2-kb target region on a ColE1 plasmid (18). A third strategy uses zinc-finger nucleases to increase mutation rates within 1 kb from a targeted DNA-binding site and has been successful in Drosophila gametes (19) and Arabidopsis thaliana seedlings (20).
We describe TaGTEAM, a new strategy for in vivo-targeted mutagenesis in Saccharomyces cerevisiae that targets a chimeric fusion of a DNA glycosylase and DNA-binding domain to an array of cognate binding sites. TaGTEAM elevates mutation rates within a 20-kb region flanking the array >800-fold in a controllable manner. TaGTEAM generates both point mutations and rearrangements at the target locus through homologous recombination (HR). By elucidating how cellular context affects repair choice, we are able to shift the primary mutagenic outcome of endonuclease-induced double-strand breaks (DSBs) at the array from HR-independent rearrangements to HR-dependent base pair substitutions.
MATERIALS AND METHODS
Plasmid and yeast strain construction
Plasmids and yeast strains used in this study are listed in Supplementary Tables S1 and S2 and are available by request from the authors or through Addgene. A complete primer list is given in Supplementary Table S3.
Growth, fluorimetry, fluorescence microscopy and flow cytometry
Yeast strains containing plasmids were grown at 30°C in yeast nitrogen base with appropriate amino acids containing 2% dextrose (SD), except when induction by 2% galactose (SG) or a balance of galactose and raffinose (2% total sugar), was required. Experiments to measure growth rate and fluorescent protein expression were carried out by diluting cells from either a liquid starter culture or fresh plate in appropriate media at a density of 105 or 106 cells/ml. Growth was measured by optical density at 600 nm (OD) at various time points on a Varioskan Flash plate reader (Thermo Scientific). Fluorescence measurements were taken from exponentially growing cells at similar OD by fluorimetry (Varioskan Flash, Thermo Scientific), flow cytometry (LSR 2, Becton Dickinson), or fluorescence microscopy (Zeiss Axiovert 200 M).
Fluctuation analysis
Fluctuation analysis was carried out similar to methods in (21,22). Briefly, 12 parallel cultures were grown without agitation from low density (10 000 cells/ml) to saturation for 3–4 days in SG for induction. Small (20 μl) and large (0.5–1 ml) volume cultures were used to measure high and low mutation rates, respectively. To convert OD to cell density, a calibration factor was determined by growing 48 parallel cultures to saturation in the same conditions and plating dilutions on rich yeast peptone media with 2% glucose. After determining the OD, the entire culture was plated on 30-mm diameter plates to facilitate analysis of many cultures. Selection plates consisted of SD media and 1 g/l of 5-Fluoroorotic acid (5-FOA) (USBiological) for klura3 mutants or SD media without arginine and 600 mg/l of canavanine (Sigma) for can1 mutants. Selection on 5-FOA and canavanine plates required 2 and 4 days of growth, respectively. Plates were imaged at 4× magnification, and colony number was scored using custom image analysis software written in MATLAB (Mathworks). For most strains, a maximum likelihood estimate of mutation rate for the distribution of mutants in each culture was found using a MATLAB implementation of the Ma-Sarkar-Sandri equation (23). The 95% confidence intervals (CI) were calculated using Equation (3) in the study conducted by Stewart (24). For sgs1 exo1 strains expressing Mag1-sctetR or sctetR-FokI and sml1 ddc2 strains expressing sctetR-FokI, mutation rates were calculated using Drake’s method, Equation (10) in the study conducted by Foster (21). The 95% CI was found using Drake’s method with the 3rd and 10th most number of mutants (of 12 cultures) (21).
Observation of Rad52-CFP foci
Cells expressing Rad52-CFP (cyan fluorescent protein) were grown as described earlier in the text, harvested at an OD between 1 and 2 and imaged on a Zeiss Axiovert 200M. Induction of the homothallic switching (HO) endonuclease was accomplished by overnight growth in 2% raffinose followed by 8 h induction in 2% galactose. Foci were counted by observing the change in brightness across a z-stack of images for the brightest 9 pixels in a cell. This change was used as a threshold, which was calibrated such that the HO-induced fractions of cells with dots matched those in the study conducted by Lisby et al. (25).
Determination of cell cycle distribution
Cells were grown overnight to an OD between 0.5 and 0.8 in SG media without leucine to induce mutators and select for plasmids. Cells were collected and fixed, and DNA was stained with SYTOX green (Invitrogen). Flow cytometry was performed on a BD Accuri C6 flow cytometer (Becton Dickinson).
RESULTS
Mag1-sctetR is a potent mutator of DNA and an avid binder to tetO sites
We chose to use a DNA glycosylase as our mutator enzyme and localize it by fusion with the tet repressor (tetR) that binds the 19-bp tet operator (tetO) sequence. DNA glycosylases normally function as the first step in base excision repair (BER) to remove chemically altered DNA bases. A build-up of unprocessed abasic sites leads to replication fork stalling and recruitment of error-prone trans-lesion polymerases (26). This faulty repair can lead to both point mutations and frameshifts (27). We tested the yeast 3-methyladenine glycosylase Mag1p, which is primarily responsible for excising alkylated bases, but has naturally broad substrate specificity (28) and is thought to excise normal base pairs when overexpressed (29). We fused Mag1p to single-chain (sc) tetR. Normal tetR forms a dimer; therefore, fusion to tetR may result in impaired enzymatic or binding activity because of a loss of conformational freedom caused by having two copies of Mag1p in such close proximity. Fusion of Mag1 to sctetR—a tandem repeat of tetR connected via a peptide linker (30)—results in one copy of Mag1 per tetR dimer and eliminates this problem (see Figure 1A). We then verified the mutator activity of Mag1-sctetR by overexpressing it in an apn1Δ background, as reduced AP (apurinic/apyrimidinic) endonuclease activity elevates mutation rates (29) (Supplementary Figure S1). Finally, we introduced a tetR-repressible promoter driving yeast fluorescent protein (YFP) (31) in cells expressing Mag1-sctetR to confirm it bound tetO sites. When doxycycline (dox) is added, it binds to and reduces the affinity of sctetR for tetO, relieving the repression and increasing YFP expression. Mag1-sctetR repressed YFP expression to nearly the same extent as sctetR (Supplementary Figure S1E). Therefore, Mag1-sctetR has both mutagenic and specific DNA-binding activity. Similar experiments using cytosine DNA glycosylase (CDG), a mutant of human uracil DNA glycosylase specific for excising cytosines (32), did not yield targeted mutagenesis, probably because of the low expression of the CDG–sctetR fusion (Supplementary Figures S1 and S2).
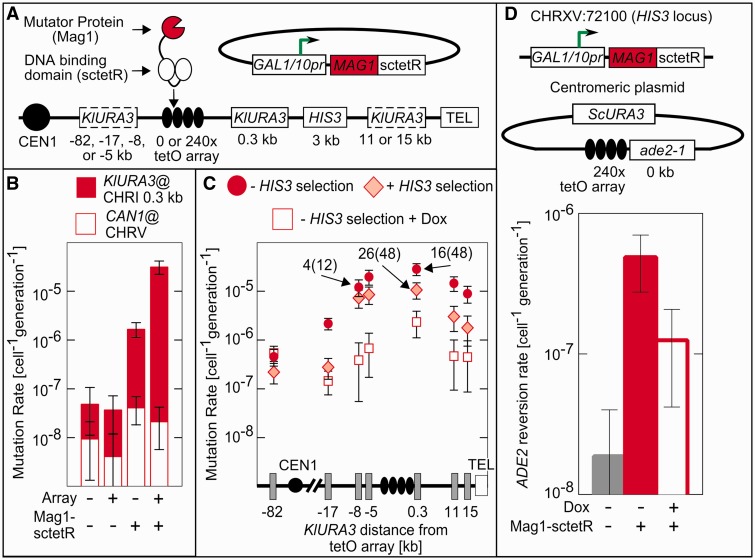
Figure 1.
TaGTEAM increases the mutation rate in a 20-kb region surrounding the tetO array. (A) The Mag1-sctetR fusion is expressed from a galactose-inducible promoter on a centromeric plasmid present in cells containing a 9-kb 240× tetO array integrated on the right arm of chromosome I. All distances are relative to the nearest edge of the tetO array, and KlURA3 markers indicted by dotted boxes show alternative integration sites used for measuring the distance dependence. (B) With an integrated tetO array and Mag1-sctetR expression, mutation rates at the 0.3 kb target are elevated 800-fold as measured by fluctuation assays, whereas rates at the CAN1 marker on chromosome V do not change. (C) This increase in mutation rate persists for at least 10 kb on either side of the array as measured in strains with KlURA3 integrated the specified distance from the tetO array (one instance per strain). Selection for HIS3 (diamonds) decreases the mutation rate slightly and addition of dox (squares) eliminates targeted mutagenesis completely. (D) TaGTEAM also functions when targeted to a plasmid containing the tetO array. Mutation rates were monitored using the gain of function marker ade2-1, which reverts through base pair substitutions at an internal stop codon. Labels on data points report the ability to PCR KlURA3 from a particular mutant, PCR+(total). Error bars are 95% CI.
TaGTEAM elevates the loss of function mutation rate in a 20-kb region surrounding an integrated 240× tetO array
To target Mag1-sctetR, we integrated a non-recombinogenic 240× tetO array (with each 19-bp tetO site separated by 10–30 bp of random sequence) (33) into the right arm of chromosome I (chrI:197000), adjacent to the SWH1 locus and ∼7-kb upstream of FLO1. No essential genes are present on the centromere-distal side of this site; on the centromere-proximal side the first essential gene is 21 kb away. To monitor targeted mutations, the Kluyveromyces lactis (Kl)URA3 marker was integrated at various distances surrounding the array (Figure 1A). To monitor or control the occurrence of chromosomal rearrangements that delete KlURA3 and a large portion of the nearby sequence, the HIS3 marker was placed centromere-distal to the array. Untargeted mutations were monitored at CAN1 on chromosome V. To control for locus and marker-specific effects, we also monitored mutation rates at KlURA3 in the presence of dox and in the absence of the tetO array.
Mutation rates at the targeted locus (Figure 1B) were 3.1 × 10−5 cell−1 gen−1 (generation−1), a >800-fold increase over the 3.9 × 10−8 cell−1 gen−1 mutation rate measured in the absence of Mag1-sctetR. Mag1-sctetR expression did not change the mutation rate at CAN1 significantly, but mutation rates at KlURA3 in the absence of the array were elevated 40-fold. This difference indicates Mag1-sctetR also causes a locus-dependent increase in the mutation rate that is unrelated to the tetR–tetO interaction. TaGTEAM creates a region of elevated mutagenesis that spans ∼10 kb on either side of the array (Figure 1C). On the centromeric side, mutation rates fall to background at between 17 and 82 kb away. We were unable to probe farther than 15 kb on the telomeric side because of difficulty integrating into the repetitive subtelomeric sequence.
TaGTEAM generates both rearrangements and point mutations
The loss of function mutation rates measured at KlURA3 do not distinguish between point mutations and rearrangements. To assess the fraction of point mutations at the target locus, we used polymerase chain reaction (PCR) to probe for the KlURA3 cassette in the genome of mutants (Figure 1A). A third of mutants at both −8 and 0.3 kb were PCR+ (i.e. KlURA3 detectable) (Figure 1C; labels on data points indicate number of PCR+ mutants out of total assayed in parentheses). We sequenced KlURA3 in PCR+ mutants (Table 1). Similar to spontaneous mutagenesis, TaGTEAM generates a broad spectrum of both transitions and transversions. Roughly a quarter of mutants were single-base deletions, and one complex mutation was observed, containing three base substitutions within 10 bp.
Table 1.
Mutagenic spectra of various mutator/chemical treatments
| Point Mutation Type | MAG- sctetR (N = 49) | sctetR-FokI + Mag1 (N = 48) | sctetR-FokI + 0.003% MMS (N = 70) | Empty vector (N = 23) |
|---|---|---|---|---|
| Transitions | 16.3% | 18.8% | 31.4% | 21.7% |
| TA>CG | 6.1% | 14.6% | 1.4% | 13.0% |
| CG>TA | 10.2% | 4.2% | 30.0% | 8.7% |
| Transversions | 59.2% | 52.1% | 64.3% | 47.8% |
| TA>GC | 0.0% | 6.3% | 2.9% | 17.4% |
| GC>TA | 26.5% | 25.0% | 40.0% | 21.7% |
| TA>AT | 18.4% | 16.7% | 17.1% | 4.3% |
| GC>CG | 14.3% | 4.2% | 4.3% | 4.3% |
| Deletions | 24.5% | 29.2% | 4.3% | 30.4% |
We found all PCR+ mutants retained the ability to grow on media lacking histidine (His+), whereas >95% of PCR-mutants were His− and had lost the nearby HIS3 cassette. The correlated loss of KlURA3 and HIS3 suggests a rearrangement that results in a deletion spanning multiple kilo-base pairs. We could bias against deletions by selecting for HIS3 using media-lacking histidine. Here, mutation rates decreased by roughly one-third in the target region and the fraction of PCR+ mutants increased (Figure 1C), making point mutagenesis the dominant mutagenic event.
To assess TaGTEAM’s functionality at an alternative locus, the 240× tetO array was placed on a centromeric plasmid adjacent to an ade2-1 allele, which reverts to an Ade+ phenotype on mutation of an internal stop codon (Figure 1D). Because targeted mutations must lead to specific single base pair substitutions for growth on media lacking adenine, this gain of function marker better estimates the per base pair point mutation rate. Galactose-inducible Mag1-sctetR was integrated into the HIS3 locus on chromosome XV. The targeted mutation rate was 4.9 × 10−7 cell−1 gen−1 (Figure 1D). Accounting for the target size of KlURA3 [∼165 bp as calculated by the method of Lang and Murray (22)], this mutation rate was the same order of magnitude as the per base pair rate observed at the target locus on chromosome I. Surprisingly, addition of dox reduced the mutation rate only 4-fold, rather than 20-fold as compared with the rate measured in the absence of Mag1-sctetR expression. Because the mutation rate at CAN1 is unaffected by Mag1-sctetR expression, plasmids may also exhibit a specific propensity for non-specific Mag1-sctetR damage similar to the locus-specific effects seen at chromosome I.
Mutations are created during repair of targeted damage by homologous recombination
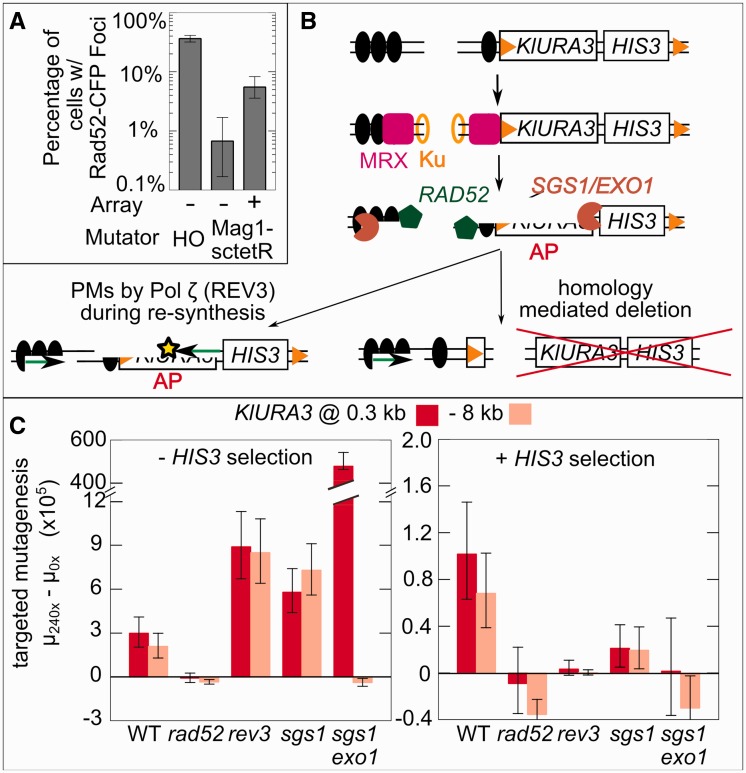
Two features of TaGTEAM are inconsistent with the model that Mag1-sctetR–mediated point mutations are generated by increased abasic site generation leading to mutagenesis via trans-lesion synthesis during replication. First is the long-range point mutagenesis; given Mag1 is tethered to sctetR by a short (∼20 nm) peptide linker, it is unclear how it acts to create abasic sites in a 20-kb region flanking the array with roughly equal frequency. Second is the combination of point mutations and rearrangements; trans-lesion synthesis of isolated abasic sites should not trigger the large deletions observed. The importance of promoting TaGTEAM’s desirable point mutations motivated us to explore an alternative model, whereby Mag1 damage generates intermediates for HR repair, which are then repaired in an error-prone manner. Indeed, when we expressed CFP-tagged Rad52 in exponentially growing Mag1-sctetR-expressing cells, we observed fluorescent foci indicative of HR repair (25) whose number increased in an array-dependent manner (Figure 2A). Mag1-generated abasic sites could lead to double-strand ends (DSEs) through fork collapse (26), or abasic sites clustered within a few helical turns could lead to DSBs directly (34).
Figure 2.
Mutations caused by TaGTEAM are created during HR repair of targeted damage. (A) TaGTEAM in a strain carrying a CFP-tagged version of Rad52p shows that damage at the array is repaired through HR. Error bars are bootstrapped 95% CI. (B) A model for mutagenesis through HR that generates rearrangements because of short repetitive sequences (orange triangles) or point mutations (PMs) through resection, DNA damage and pol ζ recruitment during resynthesis. MRX refers to the MRE11-RAD50-XRS2 complex that initially binds to DSBs and helps initiate resectioning. Ku is the YKU70-YKU80 dimer that initiates NHEJ. All other genes are described in the text. (C) Knockout mutants of pathway components in (B) demonstrate that targeted mutagenesis depends on HR (RAD52), and that point mutations (dominant under HIS3 selection) depend on REV3 and SGS1 +EXO1. Error bars are 95% CI.
Although the repair of such intermediates by HR is generally error-free, it can be error-prone in certain circumstances. In the presence of repetitive sequence, incorrect homology choice can lead to rearrangements. Repair of DSBs generated by the HO endonuclease has been shown to generate point mutations that are dependent on the error-prone polymerase ζ (REV3-REV7 in S. cerevisiae) (35,36), as has repair of I-SceI-induced DSBs in the presence of DNA base pair-damaging agents (37,38). We hypothesized that targeted point mutations were occurring through the HR-dependent localized hyper-mutagenesis (LHM) process (37,38), where resectioning of broken ends by EXO1 or the SGS1-TOP1-RMI3 complex (39) exposes ssDNA, which is used to search for homology in a RAD52-directed process. Any damage of ssDNA requires lesion bypass by Pol ζ during re-synthesis generating point mutations (Figure 2B). As resectioning can precede many kilo-base pairs from a break (40), this can explain long-range point mutations. Although a DSB is pictured, similar resectioning can occur with a DSE intermediate (26).
The LHM model predicts that RAD52, REV3 and the exonuclease activity of either SGS1 or EXO1 are necessary for targeted mutagenesis (Figure 2B). We measured the mutation rate at −8 and 0.3 kb in deletion backgrounds of each repair enzyme with and without selection for HIS3, and subtracted it from the mutation rate in the same deletion background lacking the array (Figure 2C). This ‘targeted mutation rate’ accounts for global changes because of the deletion. Regardless of selection for HIS3, all targeted mutagenesis requires RAD52, confirming HR as the key repair process. Under selection for HIS3, the targeted point mutants that predominate depend absolutely on REV3 (Pol ζ) and SGS1 + EXO1 (resectioning activity).
In the absence of selection, the targeted mutation rate in an sgs1 exo1 double mutant increased two orders of magnitude at 0.3 kb and was eliminated at −8 kb. De novo telomere addition is known to be a dominant repair pathway in this background (41), and this is consistent with the asymmetric results observed. The mutation rate increase is likely because breaks that could be repaired error-free at KlURA3 using HR now must be repaired using de novo telomere addition, which always results in a mutation on the telomeric side of the array. Although it is not known that de novo telomere addition is upregulated in a rev3 mutant, it is possible that HR of the damaged DNA around the array is less effective without Pol ζ, and more breaks are repaired by de novo telomere addition as a consequence.
The RAD52 dependence even without HIS3 selection implies that large deletions spanning both HIS3 and KlURA3 are HR-dependent. When integrating the array and mutator, short repetitive sequence elements were introduced that could explain these correlated deletions (see Supplementary Figure S3 for a description of how these elements could combine to delete sections of the targeted region). To confirm their involvement, we integrated an 85× tetO array with KlURA3 that lacked the repeated sequences. We found a similar targeted mutation rate (2.4 × 10−5 cell−1 gen−1), but almost all mutants (11/12) were PCR+, suggesting the repeated sequences are responsible for almost all rearrangements. The tetO sites within the array could also cause aberrant recombination, leading to changes in array size or deletion in mutants. Still, PCR+, His+ mutants always contain an array as probed by fluorescent foci formed by localized tetR-YFP (Supplementary Figure S4).
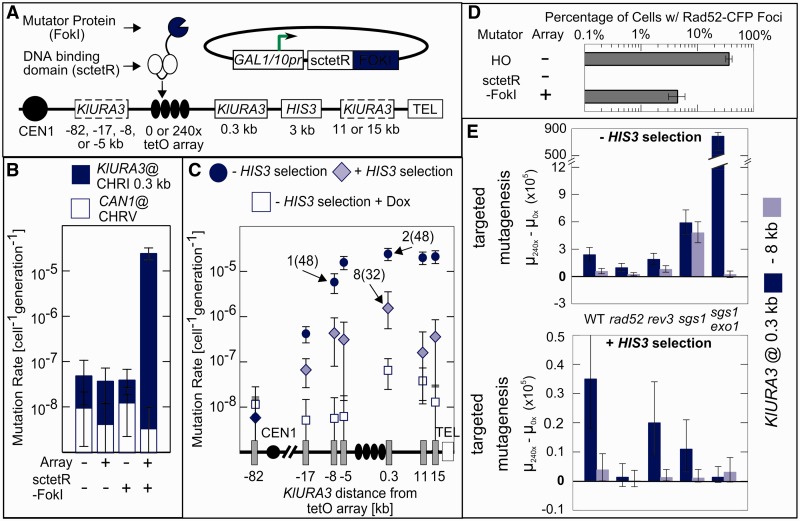
Targeted FokI leads to rearrangements but not point mutations
If the sole role of Mag1-sctetR were to create substrates with DNA ends to be repaired by HR, then creating DSBs in the array using an endonuclease might be sufficient for targeted mutagenesis. Although site-specific endonucleases have been associated with neighboring damage (20), such enzymes repeatedly cleave the DNA until mutagenic repair of the recognition site prevents further cleavage. Mag1-sctetR generates significantly fewer Rad52-CFP foci-containing cells than the site-specific HO endonuclease (Figure 2A). To better mimic this infrequent damage at the array, we created a C-terminal fusion of the nuclease domain of FokI to sctetR and expressed it in a strain containing the 240× array and KlURA3 marker at various positions (Figure 3A). We expected lower efficiency cleavage because the monomeric sctetR-FokI must dimerize to be active, and potential partners bound to the array may not be optimally spaced. Similar to Mag1-sctetR, sctetR-FokI elevated the mutation rate at the target 620-fold (Figure 3B). However, sctetR-FokI had no effect on the background mutation rate, either at CAN1 or at KlURA3 in the absence of the array. In addition, sctetR-FokI exhibited an asymmetric distance dependence profile, and few mutants (2/48) were PCR+ (Figure 3C). Although the fraction of cells with Rad52-CFP foci in cells experiencing sctetR-FokI damage at the array was similar to Mag1-sctetR (Figure 3D), RAD52 deletion did not completely eliminate targeted mutagenesis in the absence of HIS3 selection (Figure 3E). Therefore, a large fraction of mutations created by sctetR-FokI are RAD52-independent rearrangements. As expected, this rearrangement did not require the short repetitive sequences present near the array because their elimination using the aforementioned 85× array construct did not decrease the mutation rate (2.7 × 10−5 cell−1 gen−1) and most mutants remained rearrangements (2/12 were PCR+). Under HIS3 selection, mutation rates throughout the target region were further decreased as compared with Mag1-sctetR (Figure 3C). The remaining mutagenesis was independent of Pol ζ (Figure 3E) and still predominantly rearrangements (8/32 were PCR+). Therefore, processing of FokI-generated damage—presumably DSBs—results in loss of KlURA3 function largely through RAD52-independent rearrangements rather than the LHM process in Figure 2B.
Figure 3.
Targeted DSBs generated by FokI lead to HR-independent rearrangements and not point mutations. (A) Expression of sctetR-FokI in the same strain background as Mag1-sctetR (B) leads to a similar (620-fold) increase in targeted mutation rates without any increase in background mutation rates. (C) The sctetR-FokI distance dependence is asymmetric, selection for HIS3 leads to a more severe drop in mutation rate, and few PCR+ mutants are generated as compared with Mag1-sctetR. (D) Rad52-CFP repair foci show that sctetR-FokI damage is repaired by HR in roughly the same fraction of cells as Mag1-sctetR, but (E) targeted mutagenesis is not RAD52-dependent, and even under selection for HIS3, there is no REV3-dependence on. Error bars as in Figure 2.
Checkpoint activation and genome-wide DNA damage are sufficient to bias repair towards error-prone HR that generates point mutations
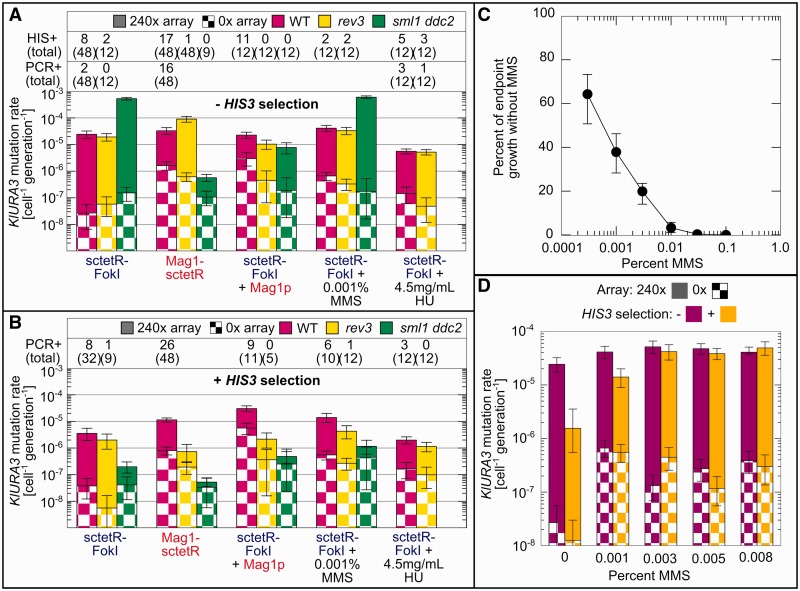
As sctetR-FokI damage increases Rad52 foci (Figure 3D), much of it must be repaired via HR without mutating KlURA3. Understanding why these HR repair events do not lead to point mutations and why the dominant mutagenic event is RAD52-independent rearrangements could allow us to increase point mutations and potentially use any DSB to generate them. We hypothesized that differences between sctetR-FokI and Mag1-sctetR were either because of the nature of the break intermediate or the cellular context in which the break was repaired. In support of the second hypothesis, Mag1-sctetR, but not sctetR-FokI, has a non-specific DNA damaging activity that increases background mutation rates (Figure 1B and C) and increases the fraction of cells with Rad52-CFP foci in the absence of the array (Figures 2B and 3D). To test whether the non-specific DNA damage activity of Mag1-sctetR explains the difference in types of mutations generated by each mutator, we co-expressed untargeted Mag1p with sctetR-FokI (Figure 4). Mag1p co-expression was sufficient to switch mutations generated by sctetR-FokI to predominantly point mutations (11/12 were PCR+). HIS3 selection caused no drop in the observed mutation rate, and like Mag1-sctetR, targeted mutagenesis was REV3-dependent. The mutation spectrum was also similar to Mag1-sctetR (Table 1), consistent with mutations occurring at bases damaged by Mag1.
Figure 4.
Global DNA damage redirects mutagenic repair of sctetR-FokI–induced breaks towards HR-dependent point mutations via checkpoint activation and DNA lesions. Mutation rates generated by sctetR-FokI expression in WT, Pol ζ-deficient (rev3) and checkpoint-deficient (sml1 ddc2) strains were measured and compared with those in the presence of co-expressed Mag1p, MMS or HU. (A) In the absence of selection for HIS3, co-expression of Mag1p with sctetR-FokI makes checkpoint- and Pol ζ-dependent point mutagenesis the dominant mutagenic outcome, as indicated by scoring of mutants for a His+ and/or PCR+ phenotype (listed above bars). HU, on the other hand, decreases HR-independent rearrangements without creating point mutations. In the absence of Mag1 activity, loss of checkpoint activation leads to very high (>10−4 cell−1 gen−1) mutation rates that correspond to rearrangements. (B) HIS3 selection reveals Pol ζ-dependent point mutations generated by the addition of MMS. (C) Overnight growth of cells in various levels of MMS compared with growth without MMS. (D) Mutation rates in cells expressing sctetR-FokI reach a maximum at 0.003% MMS. Selection for HIS3 reveals that the majority of mutations at this level of MMS are point mutations. In every case observed, His− mutants were never PCR+. Addition of HU to sml1 ddc2 strains eliminates growth, preventing measurement of the mutation rate. Error bars are 95% CI.
We hypothesized the non-specific DNA damage activity of Mag1p promotes HR-mediated point mutations in at least two ways: (i) by activating the DNA damage checkpoint, biasing repair towards HR and (ii) by creating DNA lesions that must be bypassed after resection using Pol ζ. Repair pathway choice is affected by checkpoint activation (39), and LHM surrounding a I-SceI–generated DSB occurs only with the addition of the DNA methylating agent methyl methanesulfonate (MMS) (38).
SYTOX green staining of DNA in growing cells (Supplementary Figure S5) showed more cells with 2C DNA content when Mag1-sctetR or Mag1 (and sctetR-FokI) was expressed versus sctetR-FokI alone, consistent with increased checkpoint activation. We then measured mutation rates in a sml1 ddc2 background deficient in the Mec1p-dependent DNA damage checkpoint (42). Targeted mutagenesis by Mag1-sctetR or sctetR-FokI and Mag1p was completely eliminated under selection for HIS3 (Figure 4B), confirming that point mutagenesis depends on Mec1p/ATR checkpoint activation. In the absence of HIS3 selection (Figure 4A), mutation rates in strains expressing sctetR-FokI increased significantly to 5.79 × 10−4 cell−1 gen−1 (−MMS) and 6.51 × 10−4 cell−1 gen−1 (+MMS). Therefore, without checkpoint activation His− rearrangements increase, likely through an HR-independent pathway. Strains co-expressing sctetR-FokI and Mag1p did not show an increase in mutation rate, but mutations switched from point mutants (11/12 PCR+) to rearrangements (0/12 PCR+).
To see whether checkpoint activation was sufficient to shift the mutagenic outcome of a sctetR-FokI break towards HR-mediated point mutagenesis, we added hydroxyurea (HU) to activate the DNA damage checkpoint without creating lesions. The addition of HU, which depletes nucleotide pools leading to fork stalling and collapse, to cells expressing sctetR-FokI (Figure 4) decreased the mutation rate in the absence of selection 10-fold, such that HIS3 selection no longer had any effect on the mutation rate. Unlike Mag1p or Mag1-sctetR, checkpoint activation via HU addition decreases HR-independent rearrangements without adding REV3-dependent point mutations.
Finally, we added MMS to generate DNA lesions in cells experiencing sctetR-FokI–induced breaks at a concentration (0.001%) we found has minimal impact on growth (Figure 4C). Mutation rates under HIS3 selection increased in a REV3-dependent manner and 6/10 mutants were PCR+, consistent with an increase in the rate of point mutagenesis. Increasing the MMS concentration to 0.003% further increased the mutation rate (Figure 4D) to 5.14 × 10−5 cell−1 gen−1, with 9/12 His+ mutants even without HIS3 selection. Further increases in MMS did not affect the mutation rate, and growth was impaired. The mutation spectrum generated by MMS (Table 1) was different from Mag1 or Mag1-sctetR and consistent with MMS damage occurring at cytosine residues (>70% of base pair substitutions were CG>TA or CG>AT), as previously reported (38).
DISCUSSION
By fusing a DNA glycosylase to a DNA-binding domain and localizing it to an array of binding sites in S. cerevisiae, we have created a 20-kb region of elevated point mutagenesis. Given a 165-bp target size for KlURA3 (22), we estimate point mutations are created by TaGTEAM at a rate of ∼10−7 bp−1 gen−1. In applications, the targeted region will encompass a set of genes to be evolved; therefore, in a population of 3 × 107 cells, every single base pair change in the region will be represented with >95% probability assuming a uniform mutation rate. This population size is easily achieved for yeast in bench scale shake flask or chemostat culture. As a comparison, under the same assumptions, the WT mutation rate of ∼10−10 bp−1 gen−1 yields single base pair coverage with a population size of 3 × 1010 cells, which could also be achieved in 1 l culture but without the 1000× coverage of single mutants realized by TaGTEAM that may be necessary for rapid selection of desired mutants instead of their loss to drift.
TaGTEAM represents a novel method for targeted mutagenesis in yeast, and the first method to our knowledge where mutagenesis occurs continuously in vivo without the need for rounds of genetic transformation. Because of yeast’s industrial relevance, TaGTEAM is an important step towards the in vivo-directed evolution of multigenic cellular phenotypes, including metabolic pathways, synthetic regulatory networks and tolerance to chemicals of interest present in industrial fermentation. In fact, TaGTEAM can generate point mutations in industrially relevant prototrophic strains with the mutator under control of a constitutive promoter (Supplementary Figure S6). The stability of the array may limit TaGTEAM for long-term continuous evolution, but we find point mutants retain the ability to localize tetR-YFP (Supplementary Figure S4), and targeted mutagenesis should continue in subsequent generations, provided the mutator is expressed stably. Another limitation is that the mutation rate currently achieved by TaGTEAM requires the use of a selection scheme to enrich for mutations of interest.
Combining our analysis of Mag1-sctetR– and sctetR-FokI–generated damage suggests that genome-wide DNA damage and DNA damage checkpoint activation serve as two distinct control points that switch the primary mutagenic outcome resulting from clustered DNA damage (Figure 5). When applied to endonuclease-derived breaks, these control points allow for the downregulation of HR-independent rearrangements (checkpoint activation with HU) and the upregulation of point mutagenesis (genome-wide DNA damage with MMS). It remains unclear which control point is rate limiting in generating point mutations. Even though the targeted mutation rate plateaus with increasing levels of MMS (Figure 4D), this may be due to growth defects rather than saturating ssDNA lesions. Moreover, the use of HU leads to activation of an intra-S checkpoint rather than a G2/M checkpoint. Because both these checkpoints are Mec1p-dependent and require DDC2 (43), we have shown that lack of checkpoint activation decreases HR-dependent point mutations. Whether greater checkpoint activation or arrest in S versus G2/M affects the number of breaks undergo extensive resectioning, or if ssDNA-specific damaging agents can lead to multiple point mutations per lesion event will be the subject of future work.
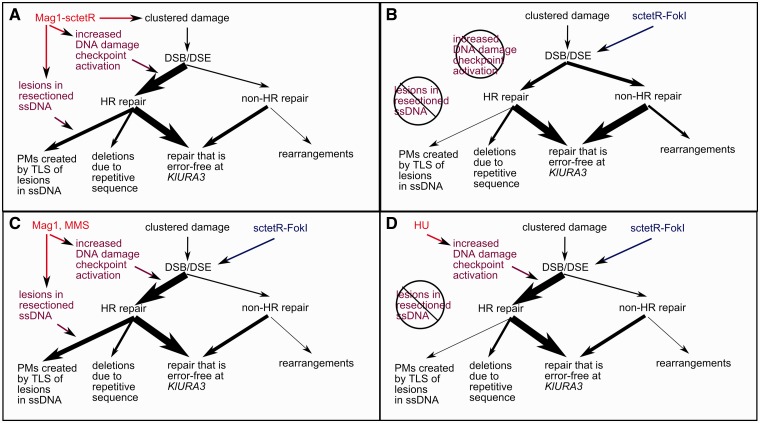
Figure 5.
A model for the mutagenic outcome at KlURA3 because of targeted damage generated by Mag1-sctetR or sctetR-FokI. Mag1-sctetR and sctetR-FokI both generate lesions that lead to DSBs, but the mutagenic repair outcome at KlURA3 depends on two conditions: (i) checkpoint activation and (ii) base pair damage. Repair that is error-free at KlURA3 and cell death are other possible outcomes of targeted damage. (A) In cells expressing Mag1-sctetR, both conditions are present, leading to high rate point mutagenesis and minimal HR-independent rearrangement. (B) sctetR-FokI expressing cells do not activate the DNA damage checkpoint to the same extent or experience base pair damage and the primary mutagenic event is HR-independent rearrangement. Co-expression of untargeted Mag1p with sctetR-FokI or addition of MMS (C) mimics Mag1-sctetR mutagenesis. Addition of 4.5 mg/ml HU (D) demonstrates that the transition in primary mutagenic outcome from HR-independent rearrangements to HR- and REV3-dependent point mutations occurs only when both conditions are met.
There is recent evidence that the same mechanism responsible for point mutagenesis in TaGTEAM (Figure 2B) may be the source of point mutations in multiple sequenced human cancers (44), including 21 sequenced breast cancer genomes (45). In these cases, the location of point mutations suggests native APOBEC proteins are responsible for ssDNA lesions in resected DNA. If our results apply to higher eukaryotes, the risk of such mutations may be heightened by other stresses that promote robust checkpoint activation and/or HR as opposed to other repair pathways. The ability to switch the mutagenic outcome of a DSB opens up the possibility for carrying out in vivo-targeted mutagenesis in a variety of organisms. FokI-based targeted nucleases (like zinc-finger nucleases and transcription activator-like effector nucleases) have been functionally expressed in mammalian cell lines, plants, mice and yeast (20,46,47). They are primarily used to promote recombination of artificial DNA at a particular site or to create knockouts through imprecise non-homologous end-joining (NHEJ). Using insights about checkpoint activation and low-level DNA damage gained from this study and knowledge of DSB repair pathway choice (39), it is possible that repair of these DSBs could be switched from NHEJ to error-prone HR that generates point mutations in the nearby region.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–3, Supplementary Figures 1–6 and Supplementary References [48–51].
FUNDING
MIT Reed Research Fund, National Institute of Environmental and Health Sciences Pilot [P30-ES002109]; MIT startup funds (to N.M.); National Science Foundation graduate fellowship (to S.F.-M.). Funding for open access charge: St. Laurent Career Development Chair in Chemical Engineering.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank L. Samson, B. Engelward and K. Prather for discussions.
REFERENCES
- 1.Farinas ET, Bulter T, Arnold FH. Directed enzyme evolution. Curr. Opin. Biotechnol. 2001;12:545–551. doi: 10.1016/s0958-1669(01)00261-0. [DOI] [PubMed] [Google Scholar]
- 2.Maheshri N, Koerber JT, Kaspar BK, Schaffer DV. Directed evolution of adeno-associated virus yields enhanced gene delivery vectors. Nat. Biotechnol. 2006;24:198–204. doi: 10.1038/nbt1182. [DOI] [PubMed] [Google Scholar]
- 3.Tyo KE, Alper HS, Stephanopoulos GN. Expanding the metabolic engineering toolbox: more options to engineer cells. Trends Biotechnol. 2007;25:132–137. doi: 10.1016/j.tibtech.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 4.Pfleger BF, Pitera DJ, Smolke CD, Keasling JD. Combinatorial engineering of intergenic regions in operons tunes expression of multiple genes. Nat. Biotechnol. 2006;24:1027–1032. doi: 10.1038/nbt1226. [DOI] [PubMed] [Google Scholar]
- 5.Alper H, Fischer C, Nevoigt E, Stephanopoulos G. Tuning genetic control through promoter engineering. Proc. Natl Acad. Sci. USA. 2005;102:12678–12683. doi: 10.1073/pnas.0504604102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jensen PR, Hammer K. The sequence of spacers between the consensus sequences modulates the strength of prokaryotic promoters. Appl. Environ. Microbiol. 1998;64:82–87. doi: 10.1128/aem.64.1.82-87.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang HH, Isaacs FJ, Carr PA, Sun ZZ, Xu G, Forest CR, Church GM. Programming cells by multiplex genome engineering and accelerated evolution. Nature. 2009;460:894–898. doi: 10.1038/nature08187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Park KS, Seol W, Yang HY, Lee SI, Kim SK, Kwon RJ, Kim EJ, Roh YH, Seong BL, Kim JS. Identification and use of zinc finger transcription factors that increase production of recombinant proteins in yeast and mammalian cells. Biotechnol. Prog. 2005;21:664–670. doi: 10.1021/bp049658x. [DOI] [PubMed] [Google Scholar]
- 9.Alper H, Moxley J, Nevoigt E, Fink GR, Stephanopoulos G. Engineering yeast transcription machinery for improved ethanol tolerance and production. Science. 2006;314:1565–1568. doi: 10.1126/science.1131969. [DOI] [PubMed] [Google Scholar]
- 10.Santos CN, Stephanopoulos G. Combinatorial engineering of microbes for optimizing cellular phenotype. Curr. Opin. Chem. Biol. 2008;12:168–176. doi: 10.1016/j.cbpa.2008.01.017. [DOI] [PubMed] [Google Scholar]
- 11.Esvelt KM, Carlson JC, Liu DR. A system for the continuous directed evolution of biomolecules. Nature. 2011;472:499–503. doi: 10.1038/nature09929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Das AT, Zhou X, Vink M, Klaver B, Verhoef K, Marzio G, Berkhout B. Viral evolution as a tool to improve the tetracycline-regulated gene expression system. J. Biol. Chem. 2004;279:18776–18782. doi: 10.1074/jbc.M313895200. [DOI] [PubMed] [Google Scholar]
- 13.Davis JN, van den Pol AN. Viral mutagenesis as a means for generating novel proteins. J. Virol. 2010;84:1625–1630. doi: 10.1128/JVI.01747-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barreto VM, Ramiro AR, Nussenzweig MC. Activation-induced deaminase: controversies and open questions. Trends Immunol. 2005;26:90–96. doi: 10.1016/j.it.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 15.Wang CL, Yang DC, Wabl M. Directed molecular evolution by somatic hypermutation. Protein Eng. Des. Sel. 2004;17:659–664. doi: 10.1093/protein/gzh080. [DOI] [PubMed] [Google Scholar]
- 16.Wang L, Jackson WC, Steinbach PA, Tsien RY. Evolution of new nonantibody proteins via iterative somatic hypermutation. Proc. Natl Acad. Sci. USA. 2004;101:16745–16749. doi: 10.1073/pnas.0407752101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arakawa H, Kudo H, Batrak V, Caldwell RB, Rieger MA, Ellwart JW, Buerstedde JM. Protein evolution by hypermutation and selection in the B cell line DT40. Nucleic Acids Res. 2008;36:e1. doi: 10.1093/nar/gkm616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Camps M, Naukkarinen J, Johnson BP, Loeb LA. Targeted gene evolution in escherichia coli using a highly error-prone DNA polymerase I. Proc. Natl Acad. Sci. USA. 2003;100:9727–9732. doi: 10.1073/pnas.1333928100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bibikova M, Golic M, Golic KG, Carroll D. Targeted chromosomal cleavage and mutagenesis in drosophila using zinc-finger nucleases. Genetics. 2002;161:1169–1175. doi: 10.1093/genetics/161.3.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lloyd A, Plaisier CL, Carroll D, Drews GN. Targeted mutagenesis using zinc-finger nucleases in arabidopsis. Proc. Natl Acad. Sci. USA. 2005;102:2232–2237. doi: 10.1073/pnas.0409339102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Foster PL. Methods for determining spontaneous mutation rates. Methods Enzymol. 2006;409:195–213. doi: 10.1016/S0076-6879(05)09012-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lang GI, Murray AW. Estimating the per-base-pair mutation rate in the yeast saccharomyces cerevisiae. Genetics. 2008;178:67–82. doi: 10.1534/genetics.107.071506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sarkar S, Ma WT, Sandri GH. On fluctuation analysis: a new, simple and efficient method for computing the expected number of mutants. Genetica. 1992;85:173–179. doi: 10.1007/BF00120324. [DOI] [PubMed] [Google Scholar]
- 24.Stewart FM. Fluctuation tests: how reliable are the estimates of mutation rates? Genetics. 1994;137:1139–1146. doi: 10.1093/genetics/137.4.1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lisby M, Rothstein R, Mortensen UH. Rad52 forms DNA repair and recombination centers during S phase. Proc. Natl Acad. Sci. USA. 2001;98:8276–8282. doi: 10.1073/pnas.121006298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boiteux S, Guillet M. Abasic sites in DNA: repair and biological consequences in saccharomyces cerevisiae. DNA Repair (Amst) 2004;3:1–12. doi: 10.1016/j.dnarep.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 27.Gibbs PE, Lawrence CW. Novel mutagenic properties of abasic sites in saccharomyces cerevisiae. J. Mol. Biol. 1995;251:229–236. doi: 10.1006/jmbi.1995.0430. [DOI] [PubMed] [Google Scholar]
- 28.O'Brien PJ, Ellenberger T. Dissecting the broad substrate specificity of human 3-methyladenine-DNA glycosylase. J. Biol. Chem. 2004;279:9750–9757. doi: 10.1074/jbc.M312232200. [DOI] [PubMed] [Google Scholar]
- 29.Glassner BJ, Rasmussen LJ, Najarian MT, Posnick LM, Samson LD. Generation of a strong mutator phenotype in yeast by imbalanced base excision repair. Proc. Natl Acad. Sci. USA. 1998;95:9997–10002. doi: 10.1073/pnas.95.17.9997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krueger C, Berens C, Schmidt A, Schnappinger D, Hillen W. Single-chain tet transregulators. Nucleic Acids Res. 2003;31:3050–3056. doi: 10.1093/nar/gkg421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Murphy KF, Balazsi G, Collins JJ. Combinatorial promoter design for engineering noisy gene expression. Proc. Natl Acad. Sci. USA. 2007;104:12726–12731. doi: 10.1073/pnas.0608451104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kavli B, Slupphaug G, Mol CD, Arvai AS, Peterson SB, Tainer JA, Krokan HE. Excision of cytosine and thymine from DNA by mutants of human uracil-DNA glycosylase. EMBO J. 1996;15:3442–3447. [PMC free article] [PubMed] [Google Scholar]
- 33.Charvin G, Cross FR, Siggia ED. A microfluidic device for temporally controlled gene expression and long-term fluorescent imaging in unperturbed dividing yeast cells. PLoS One. 2008;3:e1468. doi: 10.1371/journal.pone.0001468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eccles LJ, O’Neill P, Lomax ME. Delayed repair of radiation induced clustered DNA damage: Friend or foe? Mutat. Res. 2011;11:134–141. doi: 10.1016/j.mrfmmm.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Holbeck SL, Strathern JN. A role for REV3 in mutagenesis during double-strand break repair in saccharomyces cerevisiae. Genetics. 1997;147:1017–1024. doi: 10.1093/genetics/147.3.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rattray AJ, Shafer BK, McGill CB, Strathern JN. The roles of REV3 and RAD57 in double-strand-break-repair-induced mutagenesis of Saccharomyces cerevisiae. Genetics. 2002;162:1063–1077. doi: 10.1093/genetics/162.3.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang Y, Sterling J, Storici F, Resnick MA, Gordenin DA. Hypermutability of damaged single-strand DNA formed at double-strand breaks and uncapped telomeres in yeast Saccharomyces cerevisiae. PLoS Genet. 2008;4:e1000264. doi: 10.1371/journal.pgen.1000264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang Y, Gordenin DA, Resnick MA. A single-strand specific lesion drives MMS-induced hyper-mutability at a double-strand break in yeast. DNA Repair. 2010;9:914–921. doi: 10.1016/j.dnarep.2010.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Symington LS, Gautier J. Double-strand break end resection and repair pathway choice. Annu. Rev. Genet. 2011;45:247–271. doi: 10.1146/annurev-genet-110410-132435. [DOI] [PubMed] [Google Scholar]
- 40.Chung WH, Zhu Z, Papusha A, Malkova A, Ira G. Defective resection at DNA double-strand breaks leads to de novo telomere formation and enhances gene targeting. PLoS Genet. 2010;6:e1000948. doi: 10.1371/journal.pgen.1000948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lydeard JR, Lipkin-Moore Z, Jain S, Eapen VV, Haber JE. Sgs1 and exo1 redundantly inhibit break-induced replication and de novo telomere addition at broken chromosome ends. PLoS Genet. 2010;6:e1000973. doi: 10.1371/journal.pgen.1000973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Paciotti V, Clerici M, Lucchini G, Longhese MP. The checkpoint protein Ddc2, functionally related to S. pombe Rad26, interacts with Mec1 and is regulated by Mec1-dependent phosphorylation in budding yeast. Genes Dev. 2000;14:2046–2059. [PMC free article] [PubMed] [Google Scholar]
- 43.Garber PM, Rine J. Overlapping roles of the spindle assembly and DNA damage checkpoints in the cell-cycle response to altered chromosomes in Saccharomyces cerevisiae. Genetics. 2002;161:521–534. doi: 10.1093/genetics/161.2.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roberts S, Sterling J, Thompson C, Harris S, Mav D, Shah R, Klimczak L, Kryukov G, Malc E, Mieczkowski P, et al. Clustered mutations in yeast and in human cancers can arise from damaged long single-strand DNA regions. Mol. Cell. 2012;46:424–435. doi: 10.1016/j.molcel.2012.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nik-Zainal S, Alexandrov LB, Wedge DC, Van Loo P, Greenman CD, Raine K, Jones D, Hinton J, Marshall J, Stebbings LA, et al. Mutational processes molding the genomes of 21 breast cancers. Cell. 2012;149:979–993. doi: 10.1016/j.cell.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Durai S, Mani M, Kandavelou K, Wu J, Porteus MH, Chandrasegaran S. Zinc finger nucleases: custom-designed molecular scissors for genome engineering of plant and mammalian cells. Nucleic Acids Res. 2005;33:5978–5990. doi: 10.1093/nar/gki912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li T, Huang S, Zhao X, Wright DA, Carpenter S, Spalding MH, Weeks DP, Yang B. Modularly assembled designer TAL effector nucleases for targeted gene knockout and gene replacement in eukaryotes. Nucleic Acids Res. 2011;39:6315–6325. doi: 10.1093/nar/gkr188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gietz RD, Woods RA. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 2002;350:87–96. doi: 10.1016/s0076-6879(02)50957-5. [DOI] [PubMed] [Google Scholar]
- 49.Liti G, Carter DM, Moses AM, Warringer J, Parts L, James SA, Davey RP, Roberts IN, Burt A, Koufopanou V, et al. Population genomics of domestic and wild yeasts. Nature. 2009;458:337–341. doi: 10.1038/nature07743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lau IF, Filipe SR, Soballe B, Okstad OA, Barre FX, Sherratt DJ. Spatial and temporal organization of replicating escherichia coli chromosomes. Mol. Microbiol. 2003;49:731–743. doi: 10.1046/j.1365-2958.2003.03640.x. [DOI] [PubMed] [Google Scholar]
- 51.Blake WJ, KAern M, Cantor CR, Collins JJ. Noise in eukaryotic gene expression. Nature. 2003;422:633–637. doi: 10.1038/nature01546. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.