Abstract
The HIV-1 nucleocapsid protein (NCp7) is a nucleic acid chaperone required during reverse transcription. During the first strand transfer, NCp7 is thought to destabilize cTAR, the (−)DNA copy of the TAR RNA hairpin, and subsequently direct the TAR/cTAR annealing through the zipping of their destabilized stem ends. To further characterize the destabilizing activity of NCp7, we locally probe the structure and dynamics of cTAR by steady-state and time resolved fluorescence spectroscopy. NC(11–55), a truncated NCp7 version corresponding to its zinc-finger domain, was found to bind all over the sequence and to preferentially destabilize the penultimate double-stranded segment in the lower part of the cTAR stem. This destabilization is achieved through zinc-finger–dependent binding of NC to the G10 and G50 residues. Sequence comparison further revealed that C•A mismatches close to the two G residues were critical for fine tuning the stability of the lower part of the cTAR stem and conferring to G10 and G50 the appropriate mobility and accessibility for specific recognition by NC. Our data also highlight the necessary plasticity of NCp7 to adapt to the sequence and structure variability of cTAR to chaperone its annealing with TAR through a specific pathway.
INTRODUCTION
The nucleocapsid protein (NCp7) of the human immunodeficiency virus type 1 (HIV-1) is a small basic protein resulting from the protease-mediated cleavage of the Gag polyprotein precursor. NCp7 is formed by two highly conserved CCHC zinc-finger motifs connected by a short flexible linker and flanked by poorly folded N- and C-terminal basic domains (1,2) (Figure 1). NCp7 is critical during the early and late steps of the HIV-1 viral life cycle (3–11). Most NCp7 functions rely on its RNA- and DNA-chaperone properties that direct the rearrangement of nucleic acids into their most thermodynamically stable structure (12–15). These chaperone properties are thought to play a critical role in reverse transcription (5,16,17). For instance, during the first strand-transfer of reverse transcription, NCp7 favours the transfer of the newly synthesized minus strong-stop DNA [(−) ssDNA] to the 3′-end of the viral RNA genome, by promoting the base pairing of the complementary repeat (R) sequences at the 3′-ends of the (−)ssDNA and genomic RNA, respectively (18). In vitro, the stable stem–loop (SL) of the transactivation response element (TAR) sequence within the R domain of the genomic RNA does not readily anneal with the complementary cTAR sequence of the (−)ssDNA in the absence of NCp7 (18–21). Addition of NCp7 strongly increases the rate and extent of the cTAR/TAR annealing reaction (18–23) and switches the reaction pathway to direct the hybridization of these sequences through the end of their double-stranded (ds) stems (20,21). The main chaperone properties involved in promoting the first strand transfer have been characterized as two interconnected components: a destabilization activity and a hybridization-promoting activity. Promotion of the annealing of complementary sequences requires an efficient aggregation of nucleic acids (24). This is achieved by the numerous basic residues of NCp7 that decrease the electrostatic repulsion between complementary oligonucleotides by neutralizing the negatively charged phosphate groups (5,21,25). The annealing facilitator property of NCp7 is shared with many other basic proteins that are also able to promote the cTAR/TAR DNA hybridization (26–28). On the contrary, the ability to destabilize the cTAR sequence appears to be a hallmark of NCp7. NCp7 activates the transient opening (fraying) of the lower half of the cTAR stem (29–34) constituted by a series of short ds-segments separated by a bulge and two mismatches. This activation follows a non–two-state mechanism, which results in complex spatial and temporal fluctuations of the cTAR ends that greatly facilitate cTAR/TAR hybridization. This activity has been assigned to the conserved hydrophobic platform formed by the Val13, Phe16, Thr24, Ala25, Trp37, Gln45 and Met46 residues at the surface of the folded zinc-finger (ZF) domains (1,2,35). More specifically, the Trp37 residue plays a critical role through stacking with G residues (17,36–43). This hydrophobic platform has also been found critical for binding (35,38). Nevertheless, the precise molecular mechanisms by which NCp7 destabilizes cTAR remain poorly described. Here, we selectively modified the 57-nt cTAR stem–loop sequence of the Lai strain with 2-aminopurine (2-Ap) at different positions to investigate by fluorescence spectroscopy the structural and dynamic modifications that occur within cTAR in response to NCp7 binding. In contrast to most techniques that monitor only global structural changes, the exquisite sensitivity of 2-Ap fluorescence to its local environment allows region-specific characterization of the structural modifications within the cTAR hairpin sequence. The 2-Ap has been previously used to site-specifically characterize the NCp7-induced changes in the dynamics of short single-stranded sequences (44,45), as well as in the loop of the primer-binding site sequence of HIV-1 (46). These studies showed that binding of the intact ZF domain was responsible for a decrease of the DNA base stacking and for a strong restriction of the base mobility on a picosecond–nanosecond timescale. By selectively replacing different adenine positions of the cTAR sequence by 2-Ap (Figure 1), we aimed to picture the preferential destabilization sites and to gain insight into the molecular mechanism by which the NCp7 ZFs destabilize the cTAR stem–loop. Although NC(11–55) was found to bind all over the sequence, it preferentially restricted the dynamics and destabilized the bases of the penultimate ds-segment sandwiched between two C•A mismatches in the lower part of the cTAR stem. These NC(11–55)-induced changes are highly specific, as they were not observed when NC(11–55) was substituted by the SSHS–NC(11–55) mutant where the Cys residues in the ZFs were substituted by Ser residues to prevent the binding of zinc and the folding of the ZFs. These structural changes in this part of cTAR play likely a critical role in the ability of NCp7 to promote cTAR/TAR annealing through a specific zipping pathway.
Figure 1.
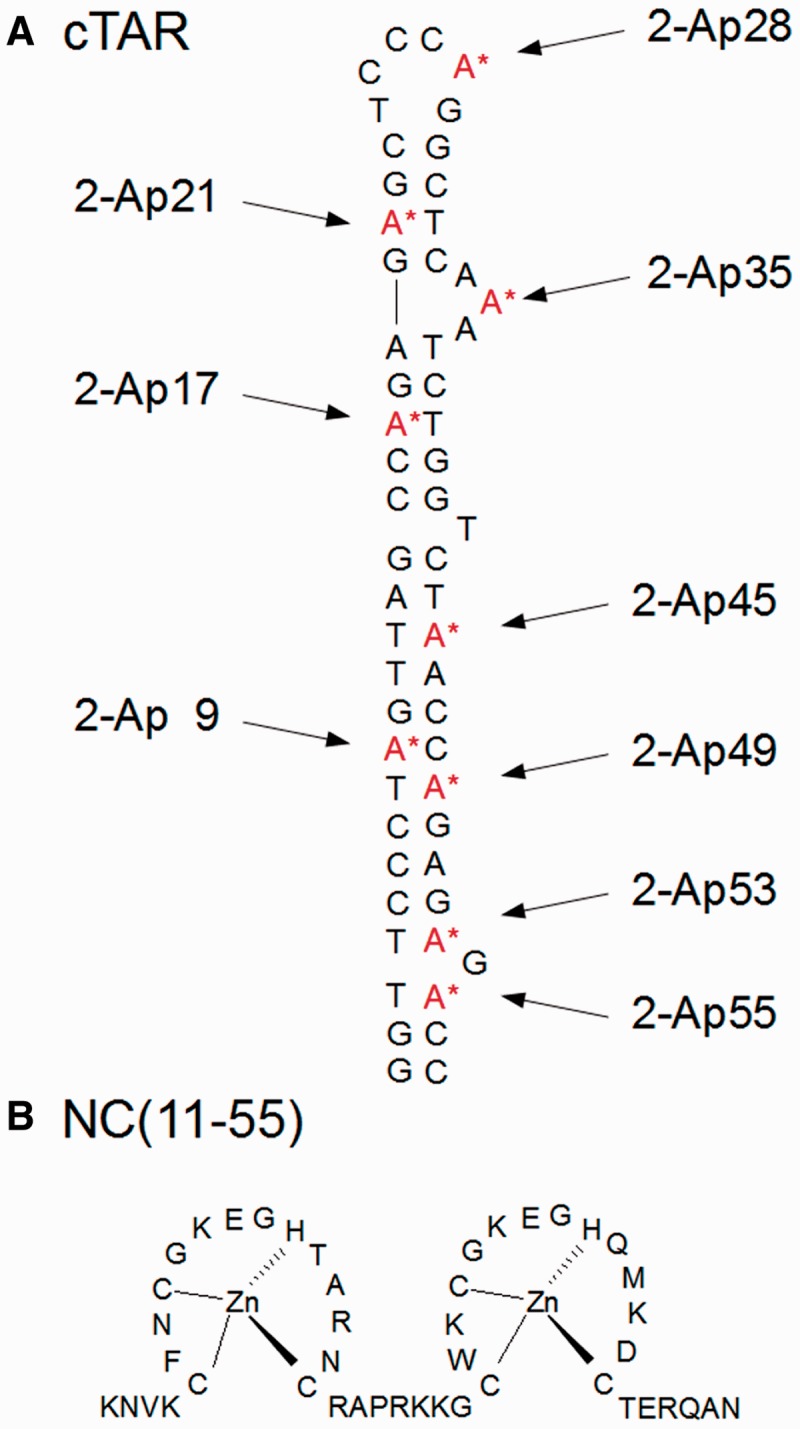
The 2-Ap substituted cTAR (A) and NC(11–55) (B) sequences. The cTAR sequence from the Lai strain of HIV-1 was selectively modified either at position 9, 17, 21, 28, 35, 45, 49, 53 or 55 with 2-Ap.
MATERIALS AND METHODS
NC(11–55) (Figure 1) was synthesized as previously described (47) and stored lyophilized in its zinc-bound form. An extinction coefficient of 5.700 M−1 cm−1 at 280 nm was used to determine its concentration. The oligonucleotides selectively labelled at different positions with 2′-deoxyribosyl-2-aminopurine (2-Ap) (Figure 1) were synthesized at a 1 µmol scale by IBA GmbH Nucleic Acids Product Supply (Göttingen, Germany) and purified by the manufacturer by double reverse-phase high-performance liquid chromatography.
Steady-state fluorescence
Experiments were performed in 25 mM Tris–HCl, pH 7.5, 30 mM NaCl and 0.2 mM MgCl2 at 20°C. Excitation and emission spectra were recorded with a Fluorolog or FluoroMax spectrofluorometer (Jobin Yvon Instruments, S.A. Inc.). All fluorescence intensities were corrected for buffer emission, lamp fluctuations and instrumental wavelength-dependent bias. Kinetics of cTAR/TAR annealing were monitored in real-time using fluorescent doubly labelled cTAR sequences and non-labelled TAR (20). The cTAR sequences were doubly labelled with 5/6 fluorescein (Fl) and 5/6 tetramethylrhodamine (TMR), at their 3′- and 5′-ends, respectively. Excitation and emission wavelengths were at 480 and 520 nm, respectively, to monitor the Fl fluorescence restoration resulting from the formation of the cTAR/TAR duplex. Concentrations of 5′TMR-cTAR-3′Fl and TAR were 10 and 100–600 nM, respectively, to ensure pseudo-first order conditions. Both reactants in identical volumes coated by NC peptides were mixed together to trigger the reaction. The apparent rate constants kobs were determined from kinetic traces, as previously described (20). All fitting procedures were carried out with the Microcal Origin 6.1 software based on non-linear least-squares methods, applying the Levenberg–Marquardt algorithm.
Time-resolved fluorescence spectroscopy
Time-resolved fluorescence measurements were performed using the time-correlated single-photon counting technique described elsewhere (46). Briefly, excitation pulses at 315 nm were generated by a pulse-picked frequency-tripled titanium-sapphire laser (Tsunami, Spectra Physics) pumped by a diode-pumped solid-state Nd: YVO4 doubled laser (Millennia X, Spectra Physics). The emission was collected at 370 nm through a polarizer set at the magic angle and a 16-nm band-pass monochromator (H10, Jobin-Yvon). The single-photon events were detected with a micro-channel plate (R3809U, Hamamatsu) photomultiplier coupled to a pulse pre-amplifier (HFAC, Becker and Hickl GmbH) and recorded on a time correlated single photon counting board (SPC-130, Becker and Hickl GmbH). The full-width at half maximum of the instrumental response function was ∼50 ps. For time-resolved anisotropy measurements, the fluorescence decay curves were recorded at vertical and horizontal positions as detailed in the Supplementary Materials. Experimentally measured fluorescence decays were de-convoluted with the instrumental response function and fitted to retrieve the most probable lifetime distribution using the maximum entropy method (48,49). As the resulting lifetime distribution evidenced four well-individualized lifetime clusters, see Supplementary Figure S3 the decays were reported as the sum of discrete exponentials according to I(t) = Σαi × exp(−t/τi) where αi is the relative amplitude associated to the fluorescence lifetime τI representing a distribution of conformations in which 2-Ap experiences similar quenching rates. The mean lifetime 〈τ〉 was calculated according to 〈τ〉=Σαiτi. The population α0 of dark species was calculated by α0 = 1 − {τfree/(τsample × Rm)} where τfree is the lifetime of the free 2-Ap riboside, τsample is the measured lifetime of 2-Ap within the oligonucleotide (either free or bound to NCp7), and Rm is the ratio of their corresponding quantum yields. The remaining amplitudes of each fluorescent population were recalculated according to αιc = αi × (1−αo). All fitting procedures were carried out both with the maximum entropy method Pulse5 software and a software based on an iterative application of the Marquardt’s method in non-linear least-squares analysis (kindly provided by G. Krishnamoorthy).
Alignment of TAR sequences
TAR sequence alignments were performed using the HIV sequence database from the Los Alamos National Lab HIV (http://www.hiv.lanl.gov/content/sequence/HIV/mainpage.html). A position-weighted matrix was then computed using the open-source statistical software R from the Comprehensive R Archive Network (http://www.R-project.org/) (50). The sequence logo (51) was plotted using the seqLogo package (http://www.bioconductor.org/packages/devel/bioc/html/seqLogo.html).
RESULTS
Site-specific characterization of the 2Ap-labelled cTAR sequences in the absence of NCp7
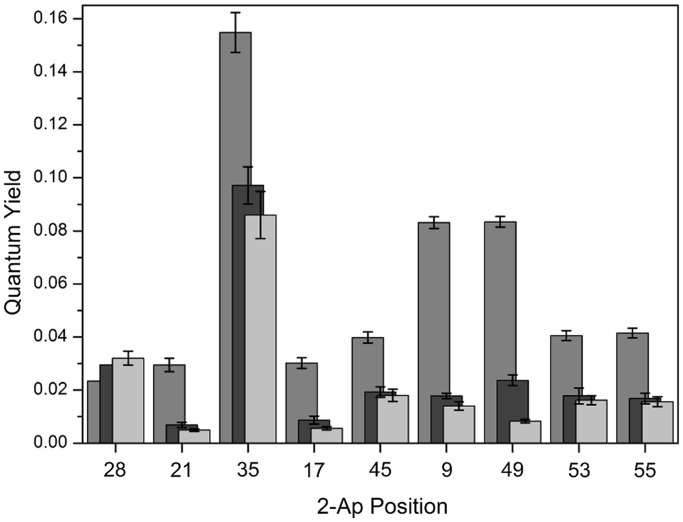
cTAR is an imperfect stem–loop characterized by a 3-nt internal loop, as well as by several bulged nucleotides and mismatches (Figure 1A). We selected labelling positions all along the cTAR Lai sequence. The 2-Ap in position 9 forms a mismatch with C within the lower part of the stem. Positions 28 and 35 are within the central and the internal loop, respectively, whereas positions 53 and 55 are flanking the G54 bulge, and positions 17, 21, 45 and 49 are distributed all over the stem within the different ds-segments. The fluorescence spectra of the different 2-Ap-labelled cTAR sequences exhibited the same maximum emission wavelength (at ∼370 nm) as the free 2-Ap nucleotide. In contrast, their quantum yields (QYs) were strongly reduced in comparison with the free probe (Figure 2), in line with static and dynamic quenching of 2-Ap by its close neighbours in the NAs (52–55). Although QYs were low in all labelled cTAR sequences, clear differences appeared as a function of the labelling positions. QYs were low when 2-Ap was inserted in ds-regions (positions 9, 17, 21, 45, 49, 53 and 55) that are known to favour base stacking, and thus the quenching of its fluorescence (53). The presence of guanine residues neighbouring 2-Ap was associated with a further decrease of the 2-Ap quantum yield, in full line with the most efficient quenching capabilities of Gs among bases (56). On the contrary, the presence of bulges or mismatches in the close vicinity of 2-Ap (positions 9, 53 and 55) was found to lead to somewhat higher QY values (57). By far, the highest QY value was observed when 2-Ap was inserted in the internal loop, suggesting a limited interaction of 2-Ap35 with its neighbours in this single-stranded segment of cTAR. In contrast to 2-Ap35, a significantly lower QY was observed for 2-Ap28, suggesting an efficient quenching of the probe by the G29 residue in the loop. In line with nuclear magnetic resonance (NMR) data showing that the loop of cTAR is highly flexible (58), fast conformational fluctuations may drive 2-Ap28 from unstacked to stacked conformations, resulting in a low QY value. Taken together, we observed a strong dependence of 2-Ap fluorescence signals on the sequence and/or the structure of the DNA segments in which the 2-Ap is inserted. The 2-Ap signals easily discriminated stable regular ds-segments from positions destabilized by bulges or mismatches and clearly showed heterogeneous 2-Ap environments along the cTAR sequence.
Figure 2.
Quantum yields of the different 2Ap-modified cTAR sequences in the absence and in the presence of NC(11–55). The QY values of the 2-Ap-labelled cTAR sequences were determined at 20°C in the absence (light grey bars) or in the presence of 1 (black bars) or 10 (dark grey bars) equivalents of NC(11–55). In the absence of NC, QY values revealed different base stacking extent within the different cTAR segments. Addition of NC(11–55) differently increased QYs. The strongest effects were observed for positions 9 and 49, suggesting a preferential destabilization in the proximity of these positions.
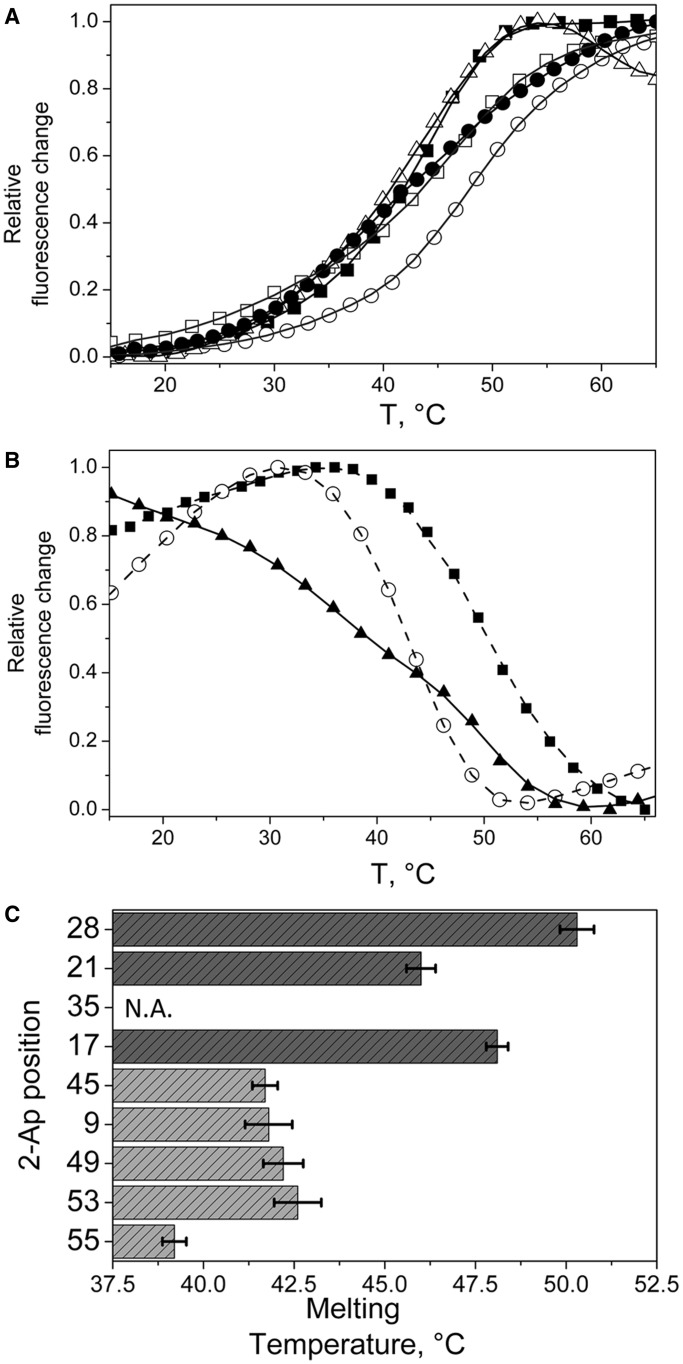
To further characterize the local 2-Ap environment in the various labelled cTAR sequences, thermal melting curves were recorded using the 2-Ap fluorescence signal. These curves allow evidencing local conformations and dynamics of 2-Ap and its close neighbours (59). On melting, two types of transitions were observed. When the 2-Ap residue was located in ds-segments (positions 17, 21, 45 and 49), its fluorescence emission strongly increased during duplex melting (Figure 3A). This was ascribed to the progressive loss of base stacking as the temperature increases. The 2-Ap9 presented a similar transition pattern, suggesting that 2-Ap9 is likely base paired with C48. On the contrary, downward transitions were observed for 2-Ap53 and 2-Ap55 (Figure 3B), evidencing an extra-helical conformation of G54 when the last two ds-segments are formed (60). Indeed, such an extra-helical conformation is thought to limit the dynamical quenching of G54 with both 2-Ap53 and 2-Ap55, explaining a higher QY for the folded state than for the melted form. Moreover, the low melting temperature associated with the 2-Ap55 residue indicated that the last ds-segment is the least stable one. In addition, the broad melting transition observed with this residue further suggested a high propensity of the last ds-segment to transiently melt at room temperature, in line with previous reports (29,31). More surprisingly, 2-Ap28 also underwent a downward melting transition (Figure 3B), probably as the consequence of the disappearance of the structural constraints within the apical loop when the upper ds-segment melted. Taken together, the range of melting temperatures of the different ds-segments of cTAR clearly supported a non–two-state melting transition (61,62) where ds-segments melted independently and where the upper part of the stem–loop seemed more stable than the lower part (Figure 3C).
Figure 3.
Site-selective monitoring of the thermal melting of the 2-Ap-substituted cTAR sequences. (A and B) Melting curves are monitored through the 2-Ap fluorescence emission. Two types of transitions were observed. (A) Upward melting curves for 2-Ap9 (closed squares), 2-Ap17 (open circles), 2-Ap21 (open squares), 2-Ap45 (open triangles) and 2-Ap49 (closed circles). (B) Downward melting curves for 2-Ap28 (black squares), 2-Ap53 (open circles) and 2-Ap55 (closed triangles). (C) Local melting temperatures recorded for the various 2-Ap positions. The melting temperatures clearly show a lower stability of the lower part of cTAR stem (light grey bars) as compared with the upper cTAR part (dark grey bars). The last ds-segment probed by the 2-Ap55 residue seemed poorly stable. No melting transition was observed for 2-Ap35, (see Supplementary Figure S1) indicating that 2-Ap in the internal loop experienced interactions with its neighbours which are similar to those in single-stranded DNA.
Heterogeneity of the probe environment was further investigated using time-resolved experiments. In contrast to the mono-exponential fluorescence decay of free 2-Ap, the fluorescence decays of the 2-Ap-labelled cTAR sequences were multi-exponential. A satisfactory fit was obtained with 4 lifetime components, ranging from ∼100 ps to nearly 10 ns, demonstrating the large conformational heterogeneity explored by the 2-Ap residues at each position (Table 1). The longest-lived component τ4 that likely corresponded to conformations where 2-Ap was either unstacked or extrahelical (63,64) varied from 6.1 to 9.6 ns. The shortest τ4 values likely resulted from conformational dynamics of the 2-Ap residues in their excited state, which may drive them from an unstacked to a stacked conformation. The τ4 value is thus an indicator of the local dynamics of the labelled NA. The low-τ4 value retrieved for positions 9 and 49 revealed highly dynamic ns-fluctuations at these positions, whereas the high τ4 value for 2-Ap at positions 53 and 55 was fully consistent with the extra-helical G54 conformation that prevented stacking with its flanking residues. The three short-lived lifetime values were ∼0.1, 0.4–1 and 2.3–3.9 ns, respectively. These picoseconds–nanoseconds lifetimes could be ascribed to conformations in which collisions of the 2-Ap with its neighbours, lead to a lifetime shortening, mainly through a charge transfer mechanism (53,55). The mean lifetime of 2-Ap in cTAR (0.4–1.6 ns) was found to be 6- to 25-fold shorter than that of the free dye (Table 1). These differences in the mean lifetimes were far less than the ∼10–140 ratios of their corresponding QYs, indicating a significant population of ‘dark’ species (α0) (i.e. species with a lifetime shorter than the ∼40 ps detection limit of our equipment). Dark species were previously described as stacked conformations with ultra-fast dynamic quenching (55,65). Because of the close proximity of the τ1 value to the detection limit of our equipment, its corresponding population may be added to that of the dark species to provide the total amount of stacked conformations (Table 1). These conformations were largely dominant in all the positions, including the apical loop where they represented 90%. This confirmed that the cTAR loop exhibits high-conformational flexibility that allows efficient quenching of 2-Ap28 by the G29 residue. In sharp contrast, a much more limited population (56%) of strongly stacked conformations was observed in the internal loop, probably because of the combination of a less efficient quenching by the flanking A together with constraints in the internal loop that prevent efficient quenching by the neighbour bases.
Table 1.
Steady-state and time-resolved fluorescence parameters of 2-Ap-substituted cTARa
| NC | QY | α0 | τ1 (α1) | τ2 (α2) | τ3 (α3) | τ4 (α4) | 〈τ〉 | Ф1 (β1) | Ф2 (β2) | Ф3 (β3) | θ0 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Free 2-Ap | − | 0.69b | 10.2 (1.00) | 10.2 | 0.08 (1.0) | 90 | ||||||
| 2-Ap9 | − | 0.014 | 0.71 | 0.1 (0.21) | 0.6 (0.04) | 2.6 (0.03) | 6.7 (0.01) | 0.7 | 0.2 (0.42) | 1.1 (0.32) | 5 (0.26) | 34 |
| + | 0.085 | 0.48 | 0.1 (0.16) | 0.7 (0.15) | 3.1 (0.12) | 7.7 (0.11) | 2.3 | 0.2 (0.20) | 1.0 (0.13) | 32 (0.67) | 22 | |
| M | 0.043 | 0.56 | 0.2 (0.26) | 0.8 (0.08) | 3.0 (0.05) | 7.3 (0.05) | 1.5 | 0.2 (0.35) | 1.5 (0.25) | 21 (0.40) | 30 | |
| 2-Ap17 | − | 0.006 | 0.76 | 0.1 (0.18) | 0.5 (0.03) | 2.3 (0.01) | 7.9 (0.01) | 0.4 | 0.2 (0.22) | 1.2 (0.37) | 9 (0.41) | 23 |
| + | 0.031 | 0.67 | 0.1 (0.19) | 0.7 (0.06) | 3.1 (0.04) | 8.3 (0.03) | 1.4 | 0.2 (0.22) | 2.2 (0.19) | 31 (0.59) | 23 | |
| 2-Ap21 | − | 0.005 | 0.92 | 0.1 (0.06) | 0.5 (0.01) | 2.7 (0.01) | 8.5 (0.01) | 0.9 | 0.2 (0.33) | 1.5 (0.35) | 8 (0.32) | 29 |
| + | 0.031 | 0.68 | 0.1 (0.19) | 0.5 (0.05) | 2.9 (0.04) | 8.8 (0.04) | 1.5 | 0.1 (0.32) | 2.3 (0.19) | 31 (0.49) | 29 | |
| 2-Ap28 | − | 0.032 | 0.69 | 0.1 (0.21) | 0.8 (0.03) | 3.6 (0.02) | 8.3 (0.04) | 1.6 | 0.2 (0.48) | 1.2 (0.32) | 7 (0.20) | 37 |
| + | 0.022 | 0.79 | 0.1 (0.11) | 0.8 (0.04) | 3.8 (0.03) | 8.2 (0.01) | 1.6 | 0.2 (0.34) | 1.2 (0.16) | 34 (0.50) | 30 | |
| 2-Ap35 | − | 0.086 | 0.20 | 0.2 (0.34) | 1.0 (0.16) | 2.9 (0.26) | 8.2 (0.04) | 1.6 | 0.2 (0.20) | 1.4 (0.28) | 8 (0.52) | 22 |
| + | 0.156 | 0.14 | 0.2 (0.36) | 1.0 (0.11) | 3.5 (0.16) | 7.5 (0.22) | 2.8 | 0.2 (0.10) | 1.2 (0.16) | 33 (0.74) | 15 | |
| 2-Ap45 | − | 0.018 | 0.56 | 0.1 (0.34) | 0.8 (0.05) | 3.0 (0.03) | 7.1 (0.02) | 0.6 | 0.2 (0.36) | 0.6 (0.31) | 4 (0.33) | 31 |
| + | 0.039 | 0.71 | 0.1 (0.10) | 0.7 (0.08) | 3.1 (0.06) | 7.8 (0.04) | 2.0 | 0.2 (0.35) | 2.6 (0.30) | 31 (0.35) | 30 | |
| M | 0.026 | 0.42 | 0.1 (0.44) | 0.5 (0.06) | 2.1 (0.05) | 7.0 (0.03) | 0.7 | 0.2 (0.34) | 1.4 (0.32) | 11 (0.34) | 30 | |
| 2-Ap49 | − | 0.008 | 0.85 | 0.1 (0.09) | 0.8 (0.04) | 3.0 (0.02) | 6.1 (0.01) | 0.8 | 0.1 (0.41) | 1.1 (0.33) | 6 (0.26) | 33 |
| + | 0.085 | 0.26 | 0.1 (0.36) | 0.9 (0.18) | 3.6 (0.11) | 8.7 (0.08) | 1.7 | 0.1 (0.10) | 2.0 (0.33) | 32 (0.57) | 17 | |
| M | 0.029 | 0.52 | 0.1 (0.35) | 0.9 (0.06) | 2.7 (0.04) | 7.0 (0.03) | 0.9 | 0.1 (0.39) | 1.7 (0.21) | 15 (0.39) | 32 | |
| 2-Ap53 | − | 0.016 | 0.82 | 0.1 (0.14) | 0.6 (0.01) | 3.9 (0.01) | 9.6 (0.02) | 1.4 | 0.1 (0.44) | 0.8 (0.30) | 5 (0.26) | 35 |
| + | 0.042 | 0.73 | 0.1 (0.12) | 0.7 (0.06) | 3.5 (0.05) | 8.8 (0.05) | 2.3 | 0.1 (0.30) | 1.9 (0.23) | 24 (0.47) | 28 | |
| 2-Ap55 | − | 0.016 | 0.72 | 0.1 (0.16) | 0.4 (0.09) | 2.8 (0.02) | 8.9 (0.02) | 0.8 | 0.5 (0.70) | 4 (0.30) | 49 | |
| + | 0.043 | 0.62 | 0.1 (0.17) | 0.7 (0.08) | 3.0 (0.08) | 7.6 (0.04) | 1.7 | 0.2 (0.27) | 1.9 (0.28) | 27 (0.42) | 26 | |
| M | 0.021 | 0.69 | 0.2 (0.22) | 0.9 (0.05) | 2.7 (0.02) | 7.3 (0.02) | 1.0 | 0.5 (0.65) | 8 (0.35) | 46 |
aExperiments were performed with 1–5 µM of 2-Ap-labelled cTAR in 25 mM Tris, 30 mM NaCl and 0.2 mM MgCl2 at pH 7.5. Fluorescence parameters were determined in the absence (−) and in the presence of 10 equivalents of NC(11–55) (+) or SSHS2NC(11–55) (M).
bFrom Ward et al. (42).
τi (ns) are the fluorescence lifetimes and αi their amplitudes. The amplitude α0 of the dark species, as well as the amplitudes of the various lifetimes, was calculated as described in the ‘Materials and Methods’ section. 〈τ〉 is the mean fluorescence lifetime. SDs for the lifetimes and amplitudes are <15%. θ0, the cone semi-angle describing the conformational space explored by 2-Ap, is given in degrees.
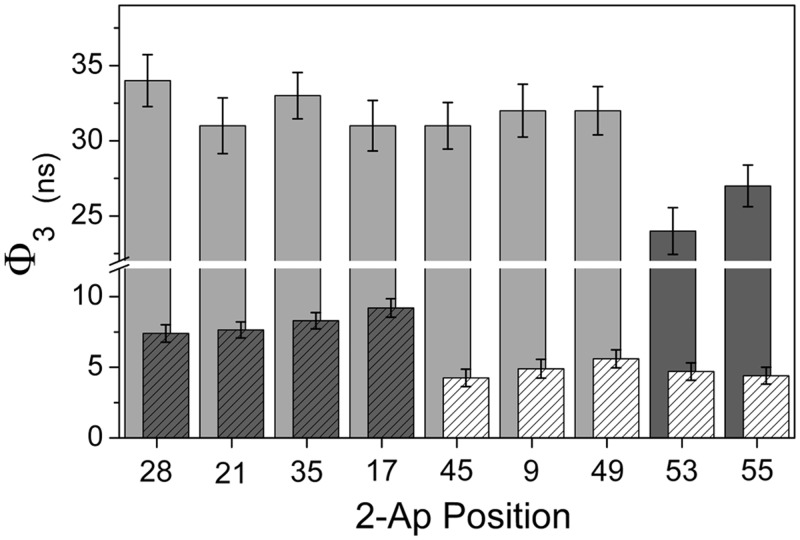
Beside the snapshot picture of the 2-Ap conformations distribution given by the time-resolved fluorescence decays, rotational dynamics of 2-Ap monitored through time-resolved fluorescence anisotropy provide information about the local and global dynamics of the cTAR sequences. Anisotropy decays were satisfactorily fitted using a three exponential model, with the exception of 2-Ap at position 55 for which only two components were sufficient. The short correlation time (Ф1), 0.1–0.5 ns, was assigned to the local motion of 2-Ap with respect to the strand backbone, whereas the longest rotational correlation time (Ф3) described the whole tumbling of the molecule. The presence of an intermediate rotational correlation time Ф2 (∼1 ns) with an amplitude almost constant of about ∼ 30% evidenced additional conformational fluctuations in cTAR. The amplitude of the short rotational correlation time reflected the conformational space explored by the 2-Ap residue during the excited time. The extent of the explored space was quantified by the cone semi-angle value θ0 (Table 1). The β1 and θ0 values were low for 2-Ap residues located in ds-segments, but large for 2-Ap9 and 2-Ap49, indicating a high rotational freedom for the latter, in line with the higher propensity of mismatched bases to explore geometries outside from the helix. The values of the long rotational correlation time (Ф3) ranged from 4 to 9 ns, depending on the labelled position. For 2-Ap residues in the upper half-part of the hairpin loop (at positions 17, 21, 28 or 35) (Figure 4), the Ф3 value was ∼8–9 ns, in full line with the theoretical correlation time calculated for the tumbling motion of a cylinder rod of length 95 Å and diameter 20 Å (66). In contrast, the Ф3 values were substantially lower (∼4–5 ns) for 2-Ap residues in the bottom of the cTAR sequence (positions 45, 49, 53 or 55) and were thus likely a combination of the overall tumbling of the cTAR sequence and of the segmental ‘fraying’ motion of the cTAR ends (30,34). Similarly to the Ф3 value, the Ф2 value was probably shortened in 2-Ap55 because of the larger segmental motions in the cTAR terminal ds-segment, making it impossible to resolve from Ф1. This hypothesis was further supported by the unexpectedly high β1 value observed for 2-Ap at position 55.
Figure 4.
Representation of the longest rotational correlation time Ф3 in the absence and in the presence of NC(11–55). Ф3 values for the 2-Ap residues inserted in the lower half of cTAR stem (hatched white bars) are significantly shorter than the values observed in the upper part of the sequence (hatched grey bars), indicating significant segmental motions in the lower part of the cTAR stem. In the presence of saturating concentrations of NC(11–55) (non hatched bars), NC(11–55) largely decreases the segmental mobility of the DNA segments containing 2-Ap residues at position 9, 45 and 49, whereas segmental motions persist in the last ds-segments of the cTAR stem (dark grey).
Effect of NC(11–55) on cTAR
Because of their low quantum yield, high concentrations of labelled NAs were required (typically 1–5 µM) to characterize the 2-Ap fluorescence parameters. This impeded us to use the full-length NCp7, which strongly aggregated nucleic acids, when added at these concentrations. The NC-induced destabilization of cTAR was thus investigated using NC(11–55), a mutant that lacks the major determinants of aggregation of the wild-type sequence but which is sufficient for specific binding and destabilization (24,35). Substitution of 2-Ap for an adenine in the cTAR sequence induced no detectable differences in the binding curves monitored through tryptophan quenching experiments, suggesting that 2-Ap did minimally affect NC(11–55) binding (see Supplementary Material and Supplementary Figure S4).
Fluorescence parameters of 2Ap-substituted cTAR in the presence of NC(11–55).
We investigated the effect of NC(11–55) with incremental addition of 1, 4, 7, 10 and 15 equivalents of peptide to the different 2-Ap-labelled cTAR sequences. NC(11–55) binding was confirmed by the progressive increase of the longest rotational correlation time Ф3, as NC(11–55) concentration increased. Ф3 reached values between 24 and 34 ns (Table 1 and Figure 4), confirming that ∼10 NC(11–55) molecules bound to cTAR. When the 2-Ap residue was not located at the bottom of the stem, the Ф3 values tended to be somewhat higher than the 30 ns value expected for a sphere with the same molecular mass (72 kDa) (66), suggesting that the global shape of the NC:cTAR complexes may differ from a sphere. With such a high-rotational correlation time, Ф3 likely described the overall tumbling motion of the whole complex, with marginal contribution of segmental motions. Shorter Ф3 values were only observed in the lower half part of the cTAR stem for the 2-Ap residues at positions 53 and 55 (Figure 4), suggesting that only the two terminal ds-segments kept a substantial mobility in the presence of NC(11–55). As a consequence, the binding of NC(11–55) to cTAR resulted in a significant decrease of the segmental mobility of the DNA segments containing 2-Ap9, 2-Ap45 and 2-Ap49, which were shown to undergo large segmental motions in the absence of NC(11–55).
Addition of NC(11–55) also increased the 2-Ap QY. In the presence of saturating concentrations of NC(11–55), the 2-Ap quantum yields varied from almost unchanged (2-Ap28) to an increase by a factor up to nearly 14 (2-Ap49) (Figure 2). The fluorescence changes were dependent on the amount of NC bound and on the position of the 2-Ap within the sequence. Time-resolved parameters further show that NC(11–55) did marginally change the lifetime values but induced a redistribution of the 2-Ap conformational states towards the longer-lived conformations (Table 1). The extent of this redistribution was dependent on the labelling position and on NC(11–55) concentration, being the most important at positions 9 and 49, showing that NC(11–55) strongly reduced base stacking at these two positions (Figure 5A). Stacked conformations were also decreased in positions 17, 21, 45, 53 and 55, albeit to a lesser extent, showing that NC(11–55) was able to bind and to decrease the base stacking all along the cTAR sequence (67). NCp7 seemed to be thus efficient to prevent the stacking of 2-Ap with its flanking bases, especially when 2-Ap is located close to a G. This likely resulted from the insertion of the neighbouring guanine into the hydrophobic platform at the top of the folded zinc-fingers of NC, which in turn prevents its collision with 2-Ap and thus, reduces its quenching efficiency (38,39,42,44). Moreover, the reduction of the amplitude associated with the shortest rotational correlation time Φ1, further indicates that NC(11–55) additionally restricts the base mobility on binding. Again, the Φ1 amplitude was the most decreased for 2-Ap at position 9 and 49, suggesting stable interactions of NC(11–55) with bases in the surrounding of 2-Ap in these two positions. The fluorescence parameters associated to 2-Ap9 and 2-Ap49 suggested that the collisions of the 2-Ap with their neighbouring guanines (G10 and G50, respectively) were strongly prevented by NC(11–55).
Figure 5.
Dependence of the populations of the stacked-conformations (A) and the local mobility expressed as a cone semi-angle (B) of the 2-Ap residues as a function of NC(11–55) concentrations. Points from dark to light grey correspond to 0, 1, 4, 7, 10 and 15 equivalents of NC(11–55), respectively. Error bars correspond to SD of at least three independent experiments. (A) NC(11–55) generally decreased the stacked conformations, albeit to different extents, being the most efficient for residues at positions 9 and 49. (B) The cone semi-angle of the conformational space explored by 2-Ap is largely decreased by NC(11–55), when the 2-Ap residue was located in the lower part of the cTAR stem (at positions 9, 49, 53 and 55).
Interestingly, in equimolar conditions where statistically one NC is bound per cTAR, a strong increase (∼2-fold) in quantum yield was observed for 2-Ap49, whereas only marginal changes were observed for all other positions (Figure 2). The populations of stacked conformations and the cone semi-angle θ0 value of 2-Ap at these two positions were also significantly reduced (Figure 5A and B). These large fluorescence changes at a 1:1 stoichiometry suggest a high affinity of NC(11–55) for these positions. Therefore, it is likely that G10 and G50 constitute preferentially targeted guanines along cTAR and thus, constitute initial destabilization sites within the cTAR stem.
Effect of SSHS2NC(11-55) mutant.
To confirm that the decrease in base stacking and the restriction in base and/or segmental mobility observed on NC(11–55) binding resulted from a specific activity of NC ZFs on cTAR, we investigated the 2-Ap fluorescence parameters in the presence of (SSHS)2NC(11–55), a NC(11–55) mutant in which all cysteine residues have been changed for serines to prevent zinc binding, and thus the folding of the ZFs (68). Addition of saturating levels of (SSHS)2NC(11–55) affected poorly the mobility of cTAR bases, as seen from the small decrease of the θ0 value induced by (SSHS)2NC(11–55) at the different 2-Ap positions tested (Table 1). Moreover, the longest rotational correlation time was increased in the presence of (SSHS)2NC(11–55) [showing that (SSHS)2NC(11–55) was binding to cTAR] but remained significantly shorter than that observed in the presence of NC(11–55). Thus, cTAR still underwent significant segmental motions when NC(SSHS)2 was bound. Consequently, (SSHS)2NC(11–55) was unable to freeze cTAR dynamics on the picosecond–nanosecond timescale. Moreover, (SSHS)2NC(11–55) was only able to moderately increase the 2-Ap QY (from 1.3- to 3.6-fold) as a result of the redistribution of the dark species (α0 conformations) towards the τ1 species. The most striking effect of (SSHS)2NC(11–55) was observed on 2-Ap9 where its mean lifetime was doubled. This was mainly because of the large increase of the population of τ4 conformations, which suggested that (SSHS)2NC(11–55) was able to modify the conformation of the mismatched 2-Ap9 residue, in line with the ability of basic peptides to tilt non-specifically the nucleobases (69). Nevertheless, the effect of (SSHS)2NC(11–55) on cTAR was considerably lower than that of NC(11–55), in full agreement with the severely impaired ability of (SSHS)2NC(11–55) to destabilize nucleic acids (35). Taken together, these observations indicate that the (SSHS)2NC(11–55) peptide marginally affects the cTAR structure and dynamics on binding, highlighting the unique role of the NC ZFs to remodel cTAR structure and dynamics.
Role of the ZF domain in the cTAR–TAR annealing reaction.
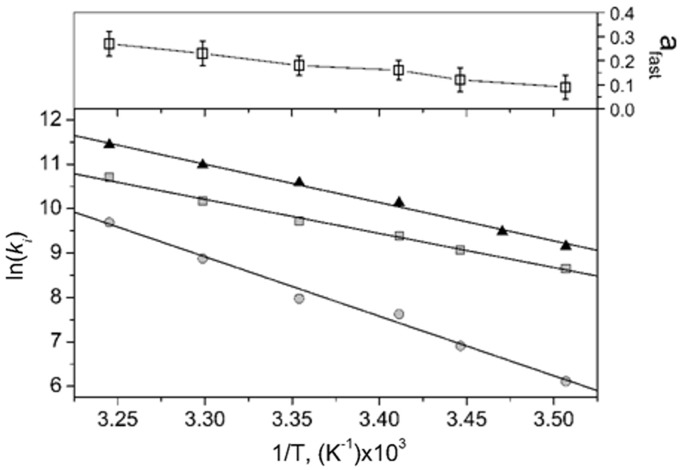
To evaluate whether the NC(11–55)-induced remodelling of cTAR could facilitate the cTAR/TAR annealing reaction, we compared the cTAR/TAR annealing kinetics in the presence of NC(11–55) and (SSHS)2NC(11–55). We reacted 10 nM of doubly labelled 5′-TMR-cTAR-3′Fl together with TAR in pseudo-first order conditions, in the presence of either of the two peptides at a ratio of 5 nt per peptide and monitored the hybridization reaction in real-time through the Fl fluorescence restoration occurring as the ds-DNA/RNA duplex forms. As their initial part was too fast to be monitored, the kinetic curves in the presence of NC(11–55) could be fitted with a single kinetic rate constant of 2.5 ± 0.2 × 104 M−1s−1, close to the 1.8 ± 0.2 × 104 M−1s−1 value found for the NC(12–55)-promoted annealing reaction of the cTAR and TAR Mal strain sequences (20). In contrast, the kinetic curves observed in the presence of (SSHS)2NC(11–55) were clearly bi-exponential with rate constants of 1.9 ± 0.2 ×1 04 M−1s−1 and 1.5 ± 0.4 × 103 M−1s−1 for the fast and slow component, respectively (Table 2). Thus, the slow component with (SSHS)2NC(11–55) was at least one order of magnitude slower than that observed with NC(11–55). Thermodynamic parameters of the reactions associated with the two NC mutants were determined (Table 2). The transition state enthalpy was estimated from the temperature dependence of the annealing reaction (Figure 6) according to:
| (1) |
Table 2.
Thermodynamic parameters of the cTAR/TAR annealing reaction in the presence of NC(11–55) or SSHS2 NC(11–55)
Figure 6.
Arrhenius analysis of the cTAR/TAR annealing reaction in the presence of NC(11–55) or (SSHS)2NC(11–55). The temperature dependence of the cTAR/TAR annealing reaction was determined by reacting 10 nM of the doubly labelled TMR-5′-cTAR-3′-Fl species together with 200 nM of TAR in the presence of NC(11–55) (closed triangle) or (SSHS)2NC(11–55) (squares correspond to the fast component and circles to the slow one) added at a ratio of one peptide per 5 nt. The retrieved thermodynamic parameters are reported in Table 2. In the presence of (SSHS)2NC(11–55), the fraction afast of the fast component (upper panel open square) increased with temperature.
Compared with NC(11–55), the slow component of (SSHS)2NC(11–55) presented a largely increased ΔH‡ value, indicating that more base pairs need to be melted for the reaction to proceed (Table 2). In contrast, the thermodynamic parameters observed with NC(11–55) were close to those associated with the fast component of (SSHS)2NC(11–55). Together with the increase of the amplitude of this fast component with temperature (Figure 6), this suggests that this component may correspond to the (SSHS)2NC(11–55)-promoted annealing of a cTAR population, which was partly melted by temperature before it was reacted with TAR. Thus, in full line with related studies describing the effect of NC on nucleic acid thermodynamics (71,72), NC(11–55) likely decreases the transition free energy of base pair melting of cTAR.
More than destabilizing the cTAR sequence to generate the cTAR reactive species, NC(11–55) also likely increases the efficiency of the nucleation-limited reaction by promoting the competent reactive species. This could be a consequence of the ability of NC(11–55) to freeze the bottom of cTAR stem at the picosecond–nanosecond timescale into specific conformations (Figure 5B), notably through direct specific interactions with the G10 and G50 residues.
Conservation of the cTAR sequence and structure
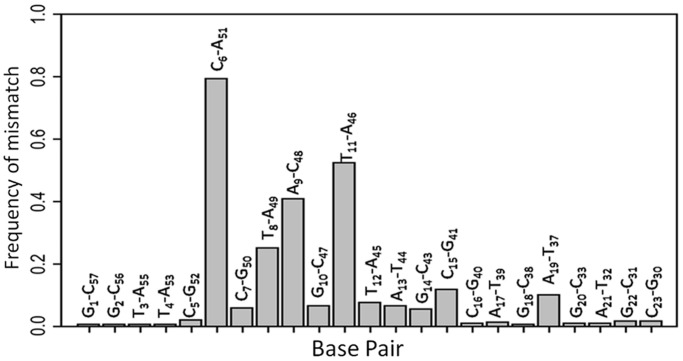
If G10 and G50 residues are critically involved in NCp7 recognition sites needed for efficient strand transfer, it is likely that these positions must be conserved. To check whether these positions corresponded to conserved motifs within the HIV subtypes, we aligned the cTAR sequence presented in Figure 1 to the LANL HIV subtype reference database (http://www.hiv.lanl.gov). The 587 sequences retrieved from the QuickAlign procedure were used to calculate a position weight matrix from which a sequence logo was derived (Figure 7). This sequence logo is drawn such that the overall height of the stack indicates the information content at each position (derived from the Shannon entropy), whereas the height of the base symbols within the stack indicates their relative frequency (51,73,74). The information content, which can reach a maximal theoretical value of 2 bits for nucleic acids, is usually interpreted as an indicator of evolutionary conservation. The cTAR sequence seems to be well conserved, including the G10 and G50 residues. In contrast, residues A9 and T11 flanking G10 were associated to low information content as compared with the other residues of the cTAR ends, suggesting that the nature of the bases at these positions is of limited importance. As a result of this low base conservation, A9 was replaced by a T in one-third of the sequences. In addition, the C48 residue was frequently replaced by an A or a T. Therefore, the A9–C48 mismatch is barely as probable as the canonical A–T base pair at this position (Figure 8B). However, a C11–A46 mismatch was also frequent, indicating that the position of the distal CA mismatch seems to be exchangeable on both sides of the G10–C47 base pair (data not shown). Similarly, a C8–A49 mismatch was also frequently found when the C6–A51 mismatch was absent. It resulted that albeit in different positions, CA mismatches were largely conserved, in close proximity of the conserved C7–G50 and G10–C47 base pairs, highlighting that their presence is required to finely tune the thermodynamic stability of the two terminal ds-segments of cTAR (75).
Figure 7.

Sequence logo of cTAR. Sequence logo resulting from 587 aligned cTAR sequences showed the globally strong conservation of the cTAR residues. Residues flanking the G10–C47 base pairs seemed to be among the least conserved.
Figure 8.
Histogram of the mismatch frequencies for the different base pairs within cTAR. Mismatch base pairs are preferentially flanking the two well-conserved G10 and G50 residues identified as directly interacting with NC(11–55). These mismatch base pairs are conserved, suggesting a critical role of the mismatches for NC recognition.
DISCUSSION
In this work, we investigated the steady-state and time-resolved fluorescence properties of cTAR labelled by 2-Ap residues at different positions. Fluorescence decay kinetics enabled the visualization of various 2-Ap conformers as snapshot pictures, whereas time-resolved fluorescence anisotropy provided complementary information on the associated rotational dynamics. This approach was thus well suited to monitor the structure and dynamics of the cTAR stem–loop and to probe the structural and dynamical perturbations induced on NC(11–55) binding. The base stacking patterns of 2-Ap seemed to be highly heterogeneous, adopting at least five conformations where the most stacked ones were the most represented. cTAR seemed to be highly dynamic at the picosecond–nanosecond timescale, especially in its lower half part that underwent segmental motions as the consequence of the fraying process that governs the dynamics of the cTAR stem ends (30,32,34). The terminal loop was found highly flexible, whereas the internal loop seemed to be highly constrained, in agreement with the slow micro-to-millisecond motions of the residues of the internal loop reported by NMR (58). Taken together, our data fit well with the NMR data of the top-half of cTAR (58) and the chemical probing experiments (67), as well as with the m-fold structure prediction of cTAR (76), except for the last ds-segment of cTAR that we found transiently annealed at 20°C in our conditions. Our data constitute thus a refined structural and dynamical model on the environment and the available conformational space for the base within the cTAR sequence.
Binding of NC(11–55)-induced local fluorescence changes that depended on the location of the 2-Ap residue within the sequence and on the amount of protein molecules bound to cTAR. Although at a 1:1 NC(11–55)/cTAR ratio, the peptide shows a preferential binding to the ds-segments encompassing residues 9 and 49, NC(11–55) molecules were shown to bind all over the cTAR sequence at saturating concentrations, suggesting that they can adjust to the heterogeneous structures and dynamics present all along cTAR (Figure 9). Nevertheless, major NC(11–55)-induced changes in the cTAR structure and dynamics were observed in the lower half of the stem and notably at positions 9 and 49. The dramatic changes in the 2-Ap fluorescence signals at these two positions were similar to those attributed to the stacking interaction of the aromatic Trp37 residue of NC with a Guanine flanking a 2-Ap residue on the interaction of NC(11–55) with small 2-Ap-labelled oligonucleotides or (−)primer-binding site (44,45,77). Thus, 2-Ap9 and 2-Ap49 fluorescence parameters strongly suggest that G10 and G50 residues directly interact with NC(11–55), likely through a stacking interaction with the Trp37 residue of NC. The resulting ‘freezing’ of cTAR at these two positions is thought to constitute a prerequisite for the NC-induced destabilization of cTAR stem. Indeed, this NC-induced restriction of the local and segmental flexibility of the NA likely prevents fast transitions back to the stably stacked conformations, and thus allows the formation of locally destabilized domains with longer lifetime and probably with exposed bases that are competent for annealing with the complementary TAR sequence. The specific restriction of the local DNA mobility by NC ZFs likely constitutes a key component of the NC destabilization mechanism that is required to further facilitate the cTAR/TAR hybridization. This hypothesis is confirmed by the fact that the (SSHS)2NC(11–55) mutant, which is unable to constrain the cTAR structure and dynamics, seems to be much less efficient than NC(11–55) to promote the cTAR/TAR annealing reaction.
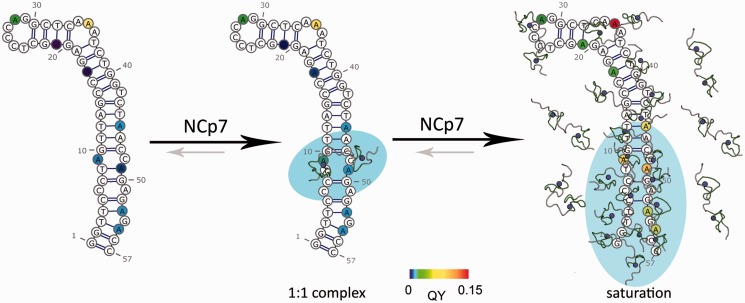
Figure 9.
Proposed mechanism for NC-induced destabilization of cTAR. NC(11–55) preferentially recognizes G10 and G50 and initiate the destabilization of cTAR. In the presence of higher concentrations, NC(11–55) can adjust to the heterogeneous structures and dynamics to bind all along cTAR and generate the reactive species needed to anneal the complementary TAR sequence.
Moreover, our data further suggest that the cTAR destabilization process is the result of the progressive coating of cTAR by NC(11–55). In contrast to 2-Ap9 and 2-Ap49 residues, which show subsequent changes in at least one of their fluorescence parameters at already low NC(11–55)/cTAR ratios, the 2-Ap45 residue exhibited fluorescence signal changes only for higher concentrations of NC(11–55), suggesting that the upper ds-DNA segment probed by 2-Ap45 can be destabilized only when its lower part is destabilized by NC(11–55). Moreover, as position 45 is only a few nucleotides apart from positions 9 or 49, the effects of NC on cTAR seem to be highly localized around its initial binding site. The latter is a prerequisite for transient interaction (78,79). Another consequence of this local NA remodelling is that effective chaperone activity requires a sufficient amount of bound NC, a situation which is achieved in the viral nucleoprotein complex context. The requirement for a sufficient level of coating to induce NA destabilization distant from its initiation site is in line with the fact that NC shows optimal chaperone activity when the target oligonucleotides are fully coated (21,29). This requirement also suggests that the initial and limited destabilization of cTAR can be extended by the binding of additional NC molecules, resulting in larger destabilized domains that likely persist long enough to efficiently react with the complementary TAR sequence. A progressive destabilization of cTAR stem as NC concentration increases was previously described (29) and likely explains the NC-coating dependent progressive switch in the cTAR/TAR annealing mechanism from a loop/loop mechanism to a zipper mechanism through the ends of their ds-stems (21).
The preferential binding of NC(11–55) to G10 and G50 residues showed that NC can discriminate and specifically target these two G residues within cTAR. Surprisingly, these guanines were located within motifs with low information content, suggesting that the nature of the bases flanking the targeted G seemed to be of limited importance for NC(11–55) recognition. However, the CA mismatches on the side of the G10–C47 and C7–G50 base pairs likely destabilize these base pairs and confer to the G10 and G50 residues the high degree of mobility (80), which was recently shown to be essential for recognition by NC (81). Thus, these intrinsic destabilizing CA motifs within the target DNA may facilitate the initial interaction of NC with the NA. The presence of mismatch base pairs seemed to play a more critical role in G recognition and discrimination than the nature of the NA sequence per se. Similar conclusions have been previously made on MuLV Gag and NC showing that local context can considerably increase NC affinity for particular nucleic acid motifs (82). The conservation of the low stability of cTAR stem, probably co-evoluting with NC chaperone properties, was shown to be critical for cTAR destabilization (31). This was confirmed by the early restoration of CA motifs within revertant viruses, which further highlights the importance of these destabilizing motifs in cTAR stem for the virus infectivity (83). Thus, this substantiates the hypothesis that NC destabilizes preferentially motifs with low stability, in line with the weak destabilizing activity of the NC ZFs (71,84). This finely tuned match between NC activity and the stability of the target NA sequence appears as an elegant way for NC to selectively chaperone a limited set of sequences within the HIV RNA genome and its DNA copies.
Moreover, the variability of the CA mismatch positions around G10 and G50 within the HIV-1 subtypes further highlights the necessary plasticity of NC to efficiently chaperone the first strand transfer. The flexible nature of NC must ensure sufficient plasticity to facilitate the correct and specific intercalation of the ZF aromatic residues to initiate the destabilization of cTAR, irrespective of the relative positions of the target G residues in respect with the CA mismatches. The poor sequence specificity and the high flexibility of NC likely ensure that NC can chaperone its various target nucleic acid sequences in the context of the high mutation rate of the virus. In addition, the high plasticity of NC and its unique ability to destabilize NA must provide NC a preferential and specific role in respect to other viral or cellular proteins (5,26,85,86) to timely and specifically enhance the cTAR/TAR annealing and to prevent self-priming (19). As HIV-1 has evolved to select such a complex interplay between NC and TAR structures and properties, the ability of NC to melt cTAR and to induce a specific annealing pathway through the bottom of these sequences is probably of major importance for the production of stable vDNA. As a result, therapeutic agents disturbing the fine-tuned equilibrium between cTAR stability and NC activity could probably significantly alter the specificity of the annealing pathway during the first strand transfer, and thus compromise the faithful and timely production of viral DNA (87–89).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–5 and Supplementary Methods.
FUNDING
French agency against AIDS (ANRS); Agence Nationale de la Recherche [ANR-10-BLAN-1529]; University of Strasbourg. Funding for open access charge: Centre national de la recherche scientifique (CNRS).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors are grateful to Hugues de Rocquigny for the peptide synthesis and purification. This article is dedicated to the memory of Dr Valérie Goldschmidt.
REFERENCES
- 1.Morellet N, Jullian N, De Rocquigny H, Maigret B, Darlix JL, Roques BP. Determination of the structure of the nucleocapsid protein NCp7 from the human immunodeficiency virus type 1 by 1H NMR. EMBO J. 1992;11:3059–3065. doi: 10.1002/j.1460-2075.1992.tb05377.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Summers MF, Henderson LE, Chance MR, Bess JW, South TL, Blake PR, Sagi I, Perez-Alvarado G, Sowder RC, Hare DR. Nucleocapsid zinc fingers detected in retroviruses: EXAFS studies of intact viruses and the solution-state structure of the nucleocapsid protein from HIV-1. Protein Sci. 1992;1:563–574. doi: 10.1002/pro.5560010502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Darlix JL, Cristofari G, Rau M, Péchoux C, Berthoux L, Roques B. Nucleocapsid protein of human immunodeficiency virus as a model protein with chaperoning functions and as a target for antiviral drugs. Adv. Pharmacol. 2000;48:345–372. doi: 10.1016/s1054-3589(00)48011-7. [DOI] [PubMed] [Google Scholar]
- 4.Darlix J-L, Garrido JL, Morellet N, Mély Y, De Rocquigny H. Properties, functions, and drug targeting of the multifunctional nucleocapsid protein of the human immunodeficiency virus. Adv. Pharmacol. 2007;55:299–346. doi: 10.1016/S1054-3589(07)55009-X. [DOI] [PubMed] [Google Scholar]
- 5.Levin JG, Guo J, Rouzina I, Musier-Forsyth K. Nucleic acid chaperone activity of HIV-1 nucleocapsid protein: critical role in reverse transcription and molecular mechanism. Prog. Nucleic Acid Res. Mol. Biol. 2005;80:217–286. doi: 10.1016/S0079-6603(05)80006-6. [DOI] [PubMed] [Google Scholar]
- 6.Thomas JA, Gorelick RJ. Nucleocapsid protein function in early infection processes. Virus Res. 2008;134:39–63. doi: 10.1016/j.virusres.2007.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mirambeau G, Lyonnais S, Gorelick RJ. Features, processing states, and heterologous protein interactions in the modulation of the retroviral nucleocapsid protein function. RNA Biol. 2010;7:85–95. doi: 10.4161/rna.7.6.13777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Muriaux D, Darlix J-L. Properties and functions of the nucleocapsid protein in virus assembly. RNA Biol. 2010;7:744–753. doi: 10.4161/rna.7.6.14065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kafaie J, Song R, Abrahamyan L, Mouland AJ, Laughrea M. Mapping of nucleocapsid residues important for HIV-1 genomic RNA dimerization and packaging. Virology. 2008;375:592–610. doi: 10.1016/j.virol.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 10.Lee S-K, Harris J, Swanstrom R. A strongly transdominant mutation in the human immunodeficiency virus type 1 gag gene defines an Achilles heel in the virus life cycle. J. Virol. 2009;83:8536–8543. doi: 10.1128/JVI.00317-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ohishi M, Nakano T, Sakuragi S, Shioda T, Sano K, Sakuragi J. The relationship between HIV-1 genome RNA dimerization, virion maturation and infectivity. Nucleic Acids Res. 2011;39:3404–3417. doi: 10.1093/nar/gkq1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Herschlag D. RNA chaperones and the RNA folding problem. J. Biol. Chem. 1995;270:20871–20874. doi: 10.1074/jbc.270.36.20871. [DOI] [PubMed] [Google Scholar]
- 13.Rein A, Henderson LE, Levin JG. Nucleic-acid-chaperone activity of retroviral nucleocapsid proteins: significance for viral replication. Trends Biochem. Sci. 1998;23:297–301. doi: 10.1016/s0968-0004(98)01256-0. [DOI] [PubMed] [Google Scholar]
- 14.Cristofari G, Darlix J-L. The ubiquitous nature of RNA chaperone proteins. Prog. Nucleic Acid Res. Mol. Biol. 2002;72:223–268. doi: 10.1016/s0079-6603(02)72071-0. [DOI] [PubMed] [Google Scholar]
- 15.Woodson SA. Taming free energy landscapes with RNA chaperones. RNA Biol. 2010;7:677–686. doi: 10.4161/rna.7.6.13615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Levin JG, Mitra M, Mascarenhas A, Musier-Forsyth K. Role of HIV-1 nucleocapsid protein in HIV-1 reverse transcription. RNA Biol. 2010;7:754–774. doi: 10.4161/rna.7.6.14115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Darlix J-L, Godet J, Ivanyi-Nagy R, Fossé P, Mauffret O, Mély Y. Flexible nature and specific functions of the HIV-1 nucleocapsid protein. J. Mol. Biol. 2011;410:565–581. doi: 10.1016/j.jmb.2011.03.037. [DOI] [PubMed] [Google Scholar]
- 18.You JC, McHenry CS. Human immunodeficiency virus nucleocapsid protein accelerates strand transfer of the terminally redundant sequences involved in reverse transcription. J. Biol. Chem. 1994;269:31491–31495. [PubMed] [Google Scholar]
- 19.Guo J, Henderson LE, Bess J, Kane B, Levin JG. Human immunodeficiency virus type 1 nucleocapsid protein promotes efficient strand transfer and specific viral DNA synthesis by inhibiting TAR-dependent self-priming from minus-strand strong-stop DNA. J. Virol. 1997;71:5178–5188. doi: 10.1128/jvi.71.7.5178-5188.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Godet J, De Rocquigny H, Raja C, Glasser N, Ficheux D, Darlix J-L, Mély Y. During the early phase of HIV-1 DNA synthesis, nucleocapsid protein directs hybridization of the TAR complementary sequences via the ends of their double-stranded stem. J. Mol. Biol. 2006;356:1180–1192. doi: 10.1016/j.jmb.2005.12.038. [DOI] [PubMed] [Google Scholar]
- 21.Vo M-N, Barany G, Rouzina I, Musier-Forsyth K. HIV-1 nucleocapsid protein switches the pathway of transactivation response element RNA/DNA annealing from loop-loop ‘kissing’ to ‘zipper’. J. Mol. Biol. 2009;386:789–801. doi: 10.1016/j.jmb.2008.12.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Darlix JL, Vincent A, Gabus C, De Rocquigny H, Roques B. Trans-activation of the 5’ to 3’ viral DNA strand transfer by nucleocapsid protein during reverse transcription of HIV1 RNA. C. R. Acad. Sci. III. 1993;316:763–771. [PubMed] [Google Scholar]
- 23.Lapadat-Tapolsky M, Pernelle C, Borie C, Darlix JL. Analysis of the nucleic acid annealing activities of nucleocapsid protein from HIV-1. Nucleic Acids Res. 1995;23:2434–2441. doi: 10.1093/nar/23.13.2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stoylov SP, Vuilleumier C, Stoylova E, De Rocquigny H, Roques BP, Gérard D, Mély Y. Ordered aggregation of ribonucleic acids by the human immunodeficiency virus type 1 nucleocapsid protein. Biopolymers. 1997;41:301–312. doi: 10.1002/(SICI)1097-0282(199703)41:3<301::AID-BIP5>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 25.Williams MC, Rouzina I, Wenner JR, Gorelick RJ, Musier-Forsyth K, Bloomfield VA. Mechanism for nucleic acid chaperone activity of HIV-1 nucleocapsid protein revealed by single molecule stretching. Proc. Natl Acad. Sci. USA. 2001;98:6121–6126. doi: 10.1073/pnas.101033198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boudier C, Storchak R, Sharma KK, Didier P, Follenius-Wund A, Muller S, Darlix J-L, Mély Y. The mechanism of HIV-1 Tat-directed nucleic acid annealing supports its role in reverse transcription. J. Mol. Biol. 2010;400:487–501. doi: 10.1016/j.jmb.2010.05.033. [DOI] [PubMed] [Google Scholar]
- 27.Sharma K, kant, Didier P, Darlix JL, De Rocquigny H, Bensikaddour H, Lavergne J-P, Pénin F, Lessinger J-M, Mély Y. Kinetic analysis of the nucleic acid chaperone activity of the hepatitis C virus core protein. Nucleic Acids Res. 2010;38:3632–3642. doi: 10.1093/nar/gkq094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guichard C, Ivanyi-Nagy R, Sharma KK, Gabus C, Marc D, Mély Y, Darlix J-L. Analysis of nucleic acid chaperoning by the prion protein and its inhibition by oligonucleotides. Nucleic Acids Res. 2011;39:8544–8558. doi: 10.1093/nar/gkr554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bernacchi S, Stoylov S, Piémont E, Ficheux D, Roques BP, Darlix JL, Mély Y. HIV-1 nucleocapsid protein activates transient melting of least stable parts of the secondary structure of TAR and its complementary sequence. J. Mol. Biol. 2002;317:385–399. doi: 10.1006/jmbi.2002.5429. [DOI] [PubMed] [Google Scholar]
- 30.Azoulay J, Clamme JP, Darlix JL, Roques BP, Mély Y. Destabilization of the HIV-1 complementary sequence of TAR by the nucleocapsid protein through activation of conformational fluctuations. J. Mol. Biol. 2003;326:691–700. doi: 10.1016/s0022-2836(02)01430-4. [DOI] [PubMed] [Google Scholar]
- 31.Beltz H, Azoulay J, Bernacchi S, Clamme J-P, Ficheux D, Roques B, Darlix J-L, Mély Y. Impact of the terminal bulges of HIV-1 cTAR DNA on its stability and the destabilizing activity of the nucleocapsid protein NCp7. J. Mol. Biol. 2003;328:95–108. doi: 10.1016/s0022-2836(03)00244-4. [DOI] [PubMed] [Google Scholar]
- 32.Cosa G, Harbron EJ, Zeng Y, Liu H-W, O’Connor DB, Eta-Hosokawa C, Musier-Forsyth K, Barbara PF. Secondary structure and secondary structure dynamics of DNA hairpins complexed with HIV-1 NC protein. Biophys. J. 2004;87:2759–2767. doi: 10.1529/biophysj.104.043083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu H-W, Cosa G, Landes CF, Zeng Y, Kovaleski BJ, Mullen DG, Barany G, Musier-Forsyth K, Barbara PF. Single-molecule FRET studies of important intermediates in the nucleocapsid-protein-chaperoned minus-strand transfer step in HIV-1 reverse transcription. Biophys. J. 2005;89:3470–3479. doi: 10.1529/biophysj.105.065326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cosa G, Zeng Y, Liu H-W, Landes CF, Makarov DE, Musier-Forsyth K, Barbara PF. Evidence for non-two-state kinetics in the nucleocapsid protein chaperoned opening of DNA hairpins. J. Phys. Chem. B. 2006;110:2419–2426. doi: 10.1021/jp054189i. [DOI] [PubMed] [Google Scholar]
- 35.Beltz H, Clauss C, Piémont E, Ficheux D, Gorelick RJ, Roques B, Gabus C, Darlix J-L, De Rocquigny H, Mély Y. Structural determinants of HIV-1 nucleocapsid protein for cTAR DNA binding and destabilization, and correlation with inhibition of self-primed DNA synthesis. J. Mol. Biol. 2005;348:1113–1126. doi: 10.1016/j.jmb.2005.02.042. [DOI] [PubMed] [Google Scholar]
- 36.De Guzman RN, Wu ZR, Stalling CC, Pappalardo L, Borer PN, Summers MF. Structure of the HIV-1 nucleocapsid protein bound to the SL3 psi-RNA recognition element. Science. 1998;279:384–388. doi: 10.1126/science.279.5349.384. [DOI] [PubMed] [Google Scholar]
- 37.Fisher RJ, Rein A, Fivash M, Urbaneja MA, Casas-Finet JR, Medaglia M, Henderson LE. Sequence-specific binding of human immunodeficiency virus type 1 nucleocapsid protein to short oligonucleotides. J. Virol. 1998;72:1902–1909. doi: 10.1128/jvi.72.3.1902-1909.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morellet N, Déméné H, Teilleux V, Huynh-Dinh T, De Rocquigny H, Fournié-Zaluski MC, Roques BP. Structure of the complex between the HIV-1 nucleocapsid protein NCp7 and the single-stranded pentanucleotide d(ACGCC) J. Mol. Biol. 1998;283:419–434. doi: 10.1006/jmbi.1998.2098. [DOI] [PubMed] [Google Scholar]
- 39.Vuilleumier C, Bombarda E, Morellet N, Gérard D, Roques BP, Mély Y. Nucleic acid sequence discrimination by the HIV-1 nucleocapsid protein NCp7: a fluorescence study. Biochemistry. 1999;38:16816–16825. doi: 10.1021/bi991145p. [DOI] [PubMed] [Google Scholar]
- 40.Amarasinghe GK, Zhou J, Miskimon M, Chancellor KJ, McDonald JA, Matthews AG, Miller RR, Rouse MD, Summers MF. Stem-loop SL4 of the HIV-1 psi RNA packaging signal exhibits weak affinity for the nucleocapsid protein. structural studies and implications for genome recognition. J. Mol. Biol. 2001;314:961–970. doi: 10.1006/jmbi.2000.5182. [DOI] [PubMed] [Google Scholar]
- 41.Bourbigot S, Ramalanjaona N, Boudier C, Salgado GFJ, Roques BP, Mély Y, Bouaziz S, Morellet N. How the HIV-1 nucleocapsid protein binds and destabilises the (−)primer binding site during reverse transcription. J. Mol. Biol. 2008;383:1112–1128. doi: 10.1016/j.jmb.2008.08.046. [DOI] [PubMed] [Google Scholar]
- 42.Mori M, Dietrich U, Manetti F, Botta M. Molecular dynamics and DFT study on HIV-1 nucleocapsid protein-7 in complex with viral genome. J. Chem. Inf. Model. 2010;50:638–650. doi: 10.1021/ci100070m. [DOI] [PubMed] [Google Scholar]
- 43.Bazzi A, Zargarian L, Chaminade F, Boudier C, De Rocquigny H, René B, Mély Y, Fossé P, Mauffret O. Structural insights into the cTAR DNA recognition by the HIV-1 nucleocapsid protein: role of sugar deoxyriboses in the binding polarity of NC. Nucleic Acids Res. 2011;39:3903–3916. doi: 10.1093/nar/gkq1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Avilov SV, Piemont E, Shvadchak V, De Rocquigny H, Mély Y. Probing dynamics of HIV-1 nucleocapsid protein/target hexanucleotide complexes by 2-aminopurine. Nucleic Acids Res. 2008;36:885–896. doi: 10.1093/nar/gkm1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Avilov SV, Godet J, Piémont E, Mély Y. Site-specific characterization of HIV-1 nucleocapsid protein binding to oligonucleotides with two binding sites. Biochemistry. 2009;48:2422–2430. doi: 10.1021/bi8022366. [DOI] [PubMed] [Google Scholar]
- 46.Godet J, Ramalanjaona N, Sharma KK, Richert L, De Rocquigny H, Darlix J-L, Duportail G, Mély Y. Specific implications of the HIV-1 nucleocapsid zinc fingers in the annealing of the primer binding site complementary sequences during the obligatory plus strand transfer. Nucleic Acids Res. 2011;39:6633–6645. doi: 10.1093/nar/gkr274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.De Rocquigny H, Ficheux D, Gabus C, Fournié-Zaluski MC, Darlix JL, Roques BP. First large scale chemical synthesis of the 72 amino acid HIV-1 nucleocapsid protein NCp7 in an active form. Biochem. Biophys. Res. Commun. 1991;180:1010–1018. doi: 10.1016/s0006-291x(05)81166-0. [DOI] [PubMed] [Google Scholar]
- 48.Livesey AK, Brochon JC. Analyzing the distribution of decay constants in pulse-fluorimetry using the maximum entropy method. Biophys. J. 1987;52:693–706. doi: 10.1016/S0006-3495(87)83264-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brochon JC. Maximum entropy method of data analysis in time-resolved spectroscopy. Methods Enzymol. 1994;240:262–311. doi: 10.1016/s0076-6879(94)40052-0. [DOI] [PubMed] [Google Scholar]
- 50.Development Core Team R. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Development Core Team; 2009. [Google Scholar]
- 51.Schneider TD, Stephens RM. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 1990;18:6097–6100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ward DC, Reich E, Stryer L. Fluorescence studies of nucleotides and polynucleotides. I. Formycin, 2-aminopurine riboside, 2,6-diaminopurine riboside, and their derivatives. J. Biol. Chem. 1969;244:1228–1237. [PubMed] [Google Scholar]
- 53.Jean JM, Hall KB. 2-Aminopurine fluorescence quenching and lifetimes: role of base stacking. Proc. Natl Acad. Sci. USA. 2001;98:37–41. doi: 10.1073/pnas.011442198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jean JM, Hall KB. 2-Aminopurine electronic structure and fluorescence properties in DNA. Biochemistry. 2002;41:13152–13161. doi: 10.1021/bi020308y. [DOI] [PubMed] [Google Scholar]
- 55.Wan C, Xia T, Becker H-C, Zewail AH. Ultrafast unequilibrated charge transfer: A new channel in the quenching of fluorescent biological probes. Chem. Phys. Lett. 2005;412:158–163. [Google Scholar]
- 56.Jean JM, Hall KB. Stacking-unstacking dynamics of oligodeoxynucleotide trimers. Biochemistry. 2004;43:10277–10284. doi: 10.1021/bi049701p. [DOI] [PubMed] [Google Scholar]
- 57.Guest CR, Hochstrasser RA, Sowers LC, Millar DP. Dynamics of mismatched base pairs in DNA. Biochemistry. 1991;30:3271–3279. doi: 10.1021/bi00227a015. [DOI] [PubMed] [Google Scholar]
- 58.Zargarian L, Kanevsky I, Bazzi A, Boynard J, Chaminade F, Fossé P, Mauffret O. Structural and dynamic characterization of the upper part of the HIV-1 cTAR DNA hairpin. Nucleic Acids Res. 2009;37:4043–4054. doi: 10.1093/nar/gkp297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xu D, Evans KO, Nordlund TM. Melting and premelting transitions of an oligomer measured by DNA base fluorescence and absorption. Biochemistry. 1994;33:9592–9599. doi: 10.1021/bi00198a027. [DOI] [PubMed] [Google Scholar]
- 60.Jiao Y, Stringfellow S, Yu H. Distinguishing ‘looped-out’ and ‘stacked-in’ DNA bulge conformation using fluorescent 2-aminopurine replacing a purine base. J. Biomol. Struct. Dyn. 2002;19:929–934. doi: 10.1080/07391102.2002.10506795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jung J, Van Orden A. A three-state mechanism for DNA hairpin folding characterized by multiparameter fluorescence fluctuation spectroscopy. J. Am. Chem. Soc. 2006;128:1240–1249. doi: 10.1021/ja0560736. [DOI] [PubMed] [Google Scholar]
- 62.Lin MM, Meinhold L, Shorokhov D, Zewail AH. Unfolding and melting of DNA (RNA) hairpins: the concept of structure-specific 2D dynamic landscapes. Phys. Chem. Chem. Phys. 2008;10:4227–4239. doi: 10.1039/b804675c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Neely RK, Daujotyte D, Grazulis S, Magennis SW, Dryden DTF, Klimasauskas S, Jones AC. Time-resolved fluorescence of 2-aminopurine as a probe of base flipping in M.HhaI-DNA complexes. Nucleic Acids Res. 2005;33:6953–6960. doi: 10.1093/nar/gki995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Neely RK, Tamulaitis G, Chen K, Kubala M, Siksnys V, Jones AC. Time-resolved fluorescence studies of nucleotide flipping by restriction enzymes. Nucleic Acids Res. 2009;37:6859–6870. doi: 10.1093/nar/gkp688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Larsen OF, Van Stokkum IH, Gobets B, Van Grondelle R, Van Amerongen H. Probing the structure and dynamics of a DNA hairpin by ultrafast quenching and fluorescence depolarization. Biophys. J. 2001;81:1115–1126. doi: 10.1016/S0006-3495(01)75768-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lakowicz JR. Principles of Fluorescence Spectroscopy. New York, USA: Springer; 2006. [Google Scholar]
- 67.Kanevsky I, Chaminade F, Chen Y, Godet J, René B, Darlix J-L, Mély Y, Mauffret O, Fossé P. Structural determinants of TAR RNA-DNA annealing in the absence and presence of HIV-1 nucleocapsid protein. Nucleic Acids Res. 2011;39:8148–8162. doi: 10.1093/nar/gkr526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Guo J, Wu T, Anderson J, Kane BF, Johnson DG, Gorelick RJ, Henderson LE, Levin JG. Zinc finger structures in the human immunodeficiency virus type 1 nucleocapsid protein facilitate efficient minus- and plus-strand transfer. J. Virol. 2000;74:8980–8988. doi: 10.1128/jvi.74.19.8980-8988.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Helene C, Maurizot JC. Interactions of oligopeptides with nucleic acids. CRC Crit. Rev. Biochem. 1981;10:213–258. doi: 10.3109/10409238109113600. [DOI] [PubMed] [Google Scholar]
- 70.Dill KA, Bromberg S. Molecular Driving Forces: Statistical Thermodynamics in Chemistry and Biology. 1st edn. London, UK: Garland Science; 2002. [Google Scholar]
- 71.Stewart-Maynard KM, Cruceanu M, Wang F, Vo M-N, Gorelick RJ, Williams MC, Rouzina I, Musier-Forsyth K. Retroviral nucleocapsid proteins display nonequivalent levels of nucleic acid chaperone activity. J. Virol. 2008;82:10129–10142. doi: 10.1128/JVI.01169-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Vo M-N, Barany G, Rouzina I, Musier-Forsyth K. Mechanistic studies of mini-TAR RNA/DNA annealing in the absence and presence of HIV-1 nucleocapsid protein. J. Mol. Biol. 2006;363:244–261. doi: 10.1016/j.jmb.2006.08.039. [DOI] [PubMed] [Google Scholar]
- 73.Schneider TD, Stormo GD, Gold L, Ehrenfeucht A. Information content of binding sites on nucleotide sequences. J. Mol. Biol. 1986;188:415–431. doi: 10.1016/0022-2836(86)90165-8. [DOI] [PubMed] [Google Scholar]
- 74.Schneider TD. Consensus sequence Zen. Appl. Bioinformatics. 2002;1:111–119. [PMC free article] [PubMed] [Google Scholar]
- 75.Berkhout B, Klaver B, Das AT. Forced evolution of a regulatory RNA helix in the HIV-1 genome. Nucleic Acids Res. 1997;25:940–947. doi: 10.1093/nar/25.5.940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Godet J, Mély Y. Biophysical studies of the nucleic acid chaperone properties of the HIV-1 nucleocapsid protein. RNA Biol. 2010;7:687–699. doi: 10.4161/rna.7.6.13616. [DOI] [PubMed] [Google Scholar]
- 78.Cruceanu M, Gorelick RJ, Musier-Forsyth K, Rouzina I, Williams MC. Rapid kinetics of protein-nucleic acid interaction is a major component of HIV-1 nucleocapsid protein’s nucleic acid chaperone function. J. Mol. Biol. 2006;363:867–877. doi: 10.1016/j.jmb.2006.08.070. [DOI] [PubMed] [Google Scholar]
- 79.Doetsch M, Schroeder R, Fürtig B. Transient RNA-protein interactions in RNA folding. FEBS J. 2011;278:1634–1642. doi: 10.1111/j.1742-4658.2011.08094.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nag N, Rao BJ, Krishnamoorthy G. Altered dynamics of DNA bases adjacent to a mismatch: a cue for mismatch recognition by MutS. J. Mol. Biol. 2007;374:39–53. doi: 10.1016/j.jmb.2007.08.065. [DOI] [PubMed] [Google Scholar]
- 81.Bazzi A, Zargarian L, Chaminade F, De Rocquigny H, René B, Mély Y, Fossé P, Mauffret O. Intrinsic nucleic acid dynamics modulates HIV-1 nucleocapsid protein binding to its targets. PLoS One. 2012;7:e38905. doi: 10.1371/journal.pone.0038905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gherghe C, Lombo T, Leonard CW, Datta SAK, Bess JW, Jr, Gorelick RJ, Rein A, Weeks KM. Definition of a high-affinity Gag recognition structure mediating packaging of a retroviral RNA genome. Proc. Natl Acad. Sci. USA. 2010;107:19248–19253. doi: 10.1073/pnas.1006897107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Klaver B, Berkhout B. Evolution of a disrupted TAR RNA hairpin structure in the HIV-1 virus. EMBO J. 1994;13:2650–2659. doi: 10.1002/j.1460-2075.1994.tb06555.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cruceanu M, Urbaneja MA, Hixson CV, Johnson DG, Datta SA, Fivash MJ, Stephen AG, Fisher RJ, Gorelick RJ, Casas-Finet JR, et al. Nucleic acid binding and chaperone properties of HIV-1 Gag and nucleocapsid proteins. Nucleic Acids Res. 2006;34:593–605. doi: 10.1093/nar/gkj458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Henriet S, Sinck L, Bec G, Gorelick RJ, Marquet R, Paillart J-C. Vif is a RNA chaperone that could temporally regulate RNA dimerization and the early steps of HIV-1 reverse transcription. Nucleic Acids Res. 2007;35:5141–5153. doi: 10.1093/nar/gkm542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Derrington E, Gabus C, Leblanc P, Chnaidermann J, Grave L, Dormont D, Swietnicki W, Morillas M, Marck D, Nandi P, et al. PrPC has nucleic acid chaperoning properties similar to the nucleocapsid protein of HIV-1. C. R. Biol. 2002;325:17–23. doi: 10.1016/s1631-0691(02)01388-4. [DOI] [PubMed] [Google Scholar]
- 87.De Rocquigny H, Shvadchak V, Avilov S, Dong CZ, Dietrich U, Darlix J-L, Mély Y. Targeting the viral nucleocapsid protein in anti-HIV-1 therapy. Mini Rev. Med. Chem. 2008;8:24–35. doi: 10.2174/138955708783331603. [DOI] [PubMed] [Google Scholar]
- 88.Goldschmidt V, Miller Jenkins LM, De Rocquigny H, Darlix J-L, Mély Y. The nucleocapsid protein of HIV-1 as a promising therapeutic target for antiviral drugs. HIV Ther. 2010;4:179–198. [Google Scholar]
- 89.Grigorov B, Bocquin A, Gabus C, Avilov S, Mély Y, Agopian A, Divita G, Gottikh M, Witvrouw M, Darlix J-L. Identification of a methylated oligoribonucleotide as a potent inhibitor of HIV-1 reverse transcription complex. Nucleic Acids Res. 2011;39:5586–5596. doi: 10.1093/nar/gkr117. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.