Abstract
In contrast with the limited sequence divergence accumulated after separation of higher primate lineages, marked cytogenetic variation has been associated with the genome evolution in these species. Studying the impact of such structural variations on defined molecular processes can provide valuable insights on how genome structural organization contributes to organismal evolution. Here, we show that telomeres on chromosome arms carrying subtelomeric heterochromatic caps in the chimpanzee, which are completely absent in humans, replicate later than telomeres on chromosome arms without caps. In gorilla, on the other hand, a proportion of the subtelomeric heterochromatic caps present in most chromosome arms are associated with large blocks of telomere-like sequences that follow a replication program different from that of bona fide telomeres. Strikingly, telomere-containing RNA accumulates extrachromosomally in gorilla mitotic cells, suggesting that at least some aspects of telomere-containing RNA biogenesis have diverged in gorilla, perhaps in concert with the evolution of heterochromatic caps in this species.
INTRODUCTION
Chromosome evolution in eukaryotes has been largely shaped by rearrangements and localized chromosome instabilities that may have played a role in the emergence of lineage-specific characteristics (1,2). Because of their function, it is likely that telomeres have had a major influence on the way karyotypes have evolved. Indeed, telomeres, which are nucleoprotein structures found at the extremities of linear chromosomes, are absolutely essential for end protection and genome stability. On the other hand, the presence of large telomere-like repeats at interstitial locations (most of the time near centromeres) in different species suggests the participation of ancestral termini into chromosome rearrangements that contributed to shape lineage-specific karyotypes (3).
The repeated sequences that constitute the primary structure of telomeres have been extremely well conserved during evolution, being in all vertebrates the hexameric repeat T2AG3 (4). In contrast to their remarkable stability throughout evolution, the number of telomeric repeats at every chromosome end is inherently unstable during a cell’s lifespan. This instability is explained by shortening events because of the natural inability of the cell machinery to completely replicate a DNA end, to the processing reactions required to recreate a functional telomere after replication, to excision events because of length homeostasis mechanisms and to telomere loss because of damage (5). The shortening of telomeres can be counterbalanced by elongation mechanisms, such as the enzymatic activity of telomerase, a dedicated reverse transcriptase able to add telomere repeats to the 3′-end of chromosomes (6). In humans and other primates, however, telomerase is limiting in most tissues, and telomeres tend to progressively shorten with cell divisions and with age (7–10). In such a context, an efficient telomere replication becomes the most crucial mechanism for the preservation of telomere function.
Because of their repetitive nature and their lack of replication origins, telomeres have been considered difficult-to-replicate sequences. In fact, any perturbation in the progression of the replication fork will potentially lead to loss of distal sequences because of the impossibility of rescuing a fork blockage or collapse by a converging replication fork. Independently of the mechanisms that allow normal progression of the replication fork up to the tip of the chromosome, replication must be initiated by the firing of the most distal active origin. In a recent work, we have characterized the timing of replication of individual telomeres in primary human cells (11). We found that telomeres on specific chromosome arms replicate at specific moments during S-phase. This timing of replication was similar in different individuals and was not related to telomere length or influenced by the presence of telomerase. On the other hand, we showed that human telomeres that replicated late during S-phase were connected to subtelomeric regions that shared structural characteristics, such as the presence of potentially heterochromatinizing sequences. These results supported the contention that structural features in subtelomeric regions have a direct impact on telomere metabolism. Indeed, a more recent study confirmed the existence in humans of chromosome-specific telomere replication programs, which depended on replisomes found in subtelomeric regions (12). Importantly, that study also showed that these programs were conserved in different cell types, including both embryonic stem and primary cells, reinforcing the idea that telomere length variation or the presence of telomerase activity has little impact on the timing of telomere replication (12).
In humans, the subtelomeric regions contain a patchwork of more or less large subdomains that may be present at several extremities (13,14). These repeated domains, which may span several hundreds of kilobases, carry a high degree of homology, suggesting that mechanisms akin to gene conversion have been at work during evolution (15). Interestingly, some of the subtelomeric domains present in the human complement are also present in other high primates, particularly chimpanzee (Pan troglodytes) and gorilla (Gorilla gorilla), suggesting both had a common ancestor and a conserved function (16). In contrast to these conserved features, however, a simple cytogenetic comparison between metaphase chromosomes from these species reveals a striking structural difference affecting chromosome ends: at least half of the chromosome arms in chimpanzee and most of the chromosome arms in gorilla present strong heterochromatic terminal blocks (visible in a G-banding staining), a feature that is completely absent in human chromosomes (17,18). The origin and function of such blocks is not well understood, but recent data suggest that such terminal structural variation has been acquired independently and in parallel in both gorilla and chimpanzee through segmental duplication events, perhaps connected to lineage-specific genomic rearrangements that occurred early in evolution (19,20). Nevertheless, although the overall structure of these subterminal elements is different in both species, it has been shown that a 32-bp AT-rich minisatellite (hereafter referred to as Cht7) is associated with all heterochromatic caps in both chimpanzee and gorilla, whereas it is completely absent in the human genome (21), suggesting a close relationship between the presence of these minisatellite sequences and heterochromatin cap formation.
In this work, we have studied the way structural changes acquired during evolution have impacted basic aspects of telomere biology, in particular telomere replication and telomere-containing RNA (TERRA) expression, in higher primates.
MATERIALS AND METHODS
Cell lines
Gorilla and chimpanzee fibroblast cell lines were obtained as described previously (22). Chimpanzee fibroblasts were examined at early passages. Gorilla fibroblasts, which were already at late passages, were immortalized by the introduction of hTERT as previously described (23). Primary human fibroblasts WI-38 (ATCC CCL-75) were also maintained in culture for RNA preparation. Cells were maintained in Dulbecco’s modified Eagle’s medium + FCS (10%).
Telomeric DNA fluorescence in situ hybridization
Chromosome spreads were prepared by incubating cells with 0.01 μg/ml of colcemid for 1 h. Harvested cells were incubated in NaCitrate (8 g/l) for 20 min at 37°C, followed by ethanol/acid acetic fixation (3:1). Metaphase chromosomes were spread onto glass slides and air-dried overnight. Air-dried slides were rehydrated and fixed in 3.7% formaldehyde in 1× phosphate-buffered saline. A pepsin digestion (1 mg/ml) was performed for 10 min at 37°C, followed by fixation with 3.7% formaldehyde. Slides were dehydrated through an ethanol series (70, 85, 90 and 100%, 2 min each) and hybridized with telomeric Cy3-(CCCTAA)3 peptide nucleic acid (PNA) or locked nucleic acids (LNA) probes (Applied Biosystems, 100 nM in 70/50% formamide and 25 mM Tris, pH 7.4, respectively) or G-rich probes conjugated with either fluorescein isothiocyanate or Cy3. Co-denaturation was performed at 80°C for 5 min. Hybridization was carried out at room temperature for a minimum of 2 h in the dark. Slides were washed with 2× 15 min 70% formamide, 20 mM Tris–HCl, pH 7.4, with agitation, followed with 3× 5 min Tris–NaCl–Tween. DNA was stained with 100 ng/ml of DAPI in Vectashield (Vector). Images were obtained with a Zeiss UV microscope equipped with appropriate excitation/emission filters. Images were captured with an HQ-Coolsnap camera (Photometrics) using the IPlab software.
Telomeric Q-FISH and ReDFISH
Telomere quantitative-fluorescence in situ hybridization (Q-FISH) and replicative detargeting FISH (ReDFISH) were carried out as previously described (11,24). In brief, metaphase spreads, prepared as described earlier in the text, were fixed with 3.7% formaldehyde and digested with pepsin (1 mg/ml). Spreads were denatured at 80°C for 3 min in the presence of a Cy3-C-rich PNA or fluorescein isothiocyanate-G-rich LNA probes and incubated at room temperature for 2 h. After washes and ethanol dehydration, preparations were mounted in Vectashield (Vector) with DAPI (1 µg/ml). Images were acquired as described earlier in the text.
To ascertain chromosome assignment of the paracentromeric domain carrying telomere-like sequences in chimpanzee, metaphases were co-hybridized with the G-rich LNA probe and BAC RP11-54D5 (17q24.3).
ReDFISH analysis
The analysis was carried out exactly as described previously (11). For each pulse, 30–50 metaphases were captured and analyzed. For each metaphase, a karyotype was carried out, and chromosome ends with detargeted telomeres (defined as telomeres bearing non–co-localized red and green signals on different chromatids) were identified. For each BrdU/C pulse, the average percentage of detargeted telomeres in the population of metaphases was calculated for each pair of chromosome ends. The addition of these percentages from all pulses spanning the entire S-phase was adjusted to 100%. Pondered mean replication timings were calculated for every chromosome extremity by taking the sum of all percentages multiplied by the number of their corresponding pulse (percentage of telomeres replicating in pulse 1 × 1 + percentage of telomeres replicating in pulse 2 × 2 + etc …) and divided by 6 (the total number of pulses analyzed).
Telomere restriction and Bal31 digestion analyses
Five micrograms of gorilla genomic DNA was digested with Bal31 for 10 min, 30 min or 1 h at 37°C. The conditions for all the other enzymatic digestions, as well as specific enzymatic inactivation, were performed according to the manufacturer’s instructions (New England Biolabs). The restricted fragments were separated by electrophoresis in a 1% agarose gel and blotted. Hybridization and probe detection were performed as described later in the text.
Gorilla TERRA detection
Total RNA was extracted with RNeasy Mini Kit (Qiagen), according to the manufacturer’s instructions. Remaining traces of DNA were digested with DNase I (Qiagen). For northern blot experiments, 10 or 20 µg of total RNA from both gorilla and human fibroblasts (WI-38) was separated by alkaline electrophoresis in 1% agarose 1.3% formaldehyde gel and transferred to nylon membranes. Blots were blocked with Church buffer and hybridized overnight with Cht7, Tel-C-rich or Tel-G-rich 32P γ-ATP-labeled probes. The blots were then washed 2 × 5 min with 2× SSC 0.1% SDS and 5 min with 0.2× SSC, 0.1% SDS, neutralized in 2× SSC, exposed to PhosphorImager screens and visualized on a Typhoon PhosphorImager using ImageQuant software.
TERRA detection by FISH was performed similarly to the FISH protocol described earlier in the text, with the exception of an RNase A digestion (10 mg/ml) for 1 h 37°C and with no denaturation step, when mentioned as well as RNase-free conditions throughout the protocol. XpYp TERRA quantification by RT–qPCR was performed exactly as previously described (25) using a telomeric primer for reverse transcription (5′-CCC TAA CCC TAA CCC TAA CCC TAA CCC TAA-3′) and chromosome-specific primers 5′-GCA AAG AGT GAA AGA ACG AAG CTT-3′ and 5′-CCC TCT GAA AGT GGA CCA ATC A-3′ [described previously (26)] as forward and reverse primers, respectively. Level of expression was normalized to β-actin as previously described (25).
For analysis of potential TERRA-ChT7 hybrid molecules, 1 µg of total RNA was reverse transcribed with Superscript III reverse transcriptases (Invitrogen) using a telomeric C-rich oligonucleotide (5′-TGC TCC GTG CAT CTG GCA TCC CCT AAC-3′). PCR amplifications used the Teltail oligonucleotide (5′-TGC TCC GTG CAT CTG GCA TC-3′) in combination with Cht7-sequence-specific panF or panR (5′-GAT ATT TCC ATG TTT ATA CAG ATA GCG GTG AG-3′ and 5′-TCA CCG CTA TCT GTA TAA ACA TGG AAA TAT CTC-3′, respectively). The mixture was denatured at 95°C for 5 min and PCR amplified for 35 cycles of 20 s at 96°C, 30 s at 68°C and 5 min 72°C, followed by a final 10 min at 72°C step. The blotting was performed as aforementioned and hybridized overnight to a DIG–LNA-C-rich–DIG telomeric probe (Exiqon, modified bases are preceded by a + sign, 5′ CCCTA+AC+CCT+A+AC+CCTAAC+CCT+A+ACCCT+A+ACCCT+AA-3′).
Digoxigenin signals were detected with an anti-DIG–anti-phosphatase antibody (Roche) and revealed with the CDP-Star, ready-to use kit (Roche).
siRNA transfection and RT–PCR analysis
Cells were transfected with 40 nM of siRNA against either XRN2 (siXRN2A: 5′-GCCAGUAAACCUAAUCCAA-3′ and siXRN2B 5′-AAGAUGAAAUGAUGGUUGCA-3′) or luciferase (5′-UCGAAGUAUUCCGCGUACG) as a control, using Lipofectamine 2000 (Invitrogen) following manufacturer’s instructions. Total RNA was extracted with RNeasy Mini Kit (Qiagen), reverse transcribed with hexamers following the instructions of the kit ‘Cloned AMV first-strand’ (Invitrogen). The XRN2 and the normalizer β-actin mRNA levels were detected with the following primers: XRN2-forward 5′-TCC TTC GGC TTA ATG TTC TTC-3′, XRN2-reverse 5′-AGA TGT GAA ACT CGT ATT AGG-3′, actin-forward 5′-AAA GAC CTG TAC GCC AAC AC-3′, actin-reverse 5′-GTC ATA CTC CTG CTT GCT GAT-3′. PCR cycling conditions are as follow: 95°C for 5 min, 30 cycles of 96°C for 20 s, 50°C for 30 s, 68°C for 1 min, followed by a final incubation at 68°C for 5 min. Products were separated by electrophoresis in 1% agarose gel, and the quantification of the ethidium bromide images was performed with IMAGEJ software.
Statistical analyses
The R package was used for comparisons using Fisher exact tests and Spearman’s rank correlation coefficient calculations.
RESULTS
Heterochromatic caps in chimpanzee are associated with a late telomere replication pattern
The ReDFISH approach to study telomere replication is based on a modification of the CO-FISH (chromosome-oriented FISH) approach in which G-rich and C-rich parental strands are differentially detected on sister chromatids (27). For practical reasons, including a greater insolubility of G-rich PNA probes and a much higher background associated with them, we use a C-rich PNA probe combined with a G-rich LNA probe to detect G-rich strands and C-rich strands, respectively. LNA probes have been proven, including by us, to be highly specific for telomere sequences in human cells and have been successfully used in quantitative approaches (11). When tested on chimpanzee metaphases, G-rich LNA and C-rich PNA probes yielded similar patterns at the end of all chromosomes (Supplementary Figure S1A). Interestingly, although, we noticed that the G-rich LNA probe revealed a strong paracentromeric signal on chromosome 17 (human nomenclature), which is not observed in the human complement, and it is not revealed by the C-rich PNA probe (Supplementary Figure S1). As single base mismatches deeply diminish the binding of PNA probes to their targets and two mismatches completely abolish it (28), the interstitial region revealed by the G-rich LNA probe must carry large blocks composed almost exclusively of somewhat degenerated telomere-like sequences.
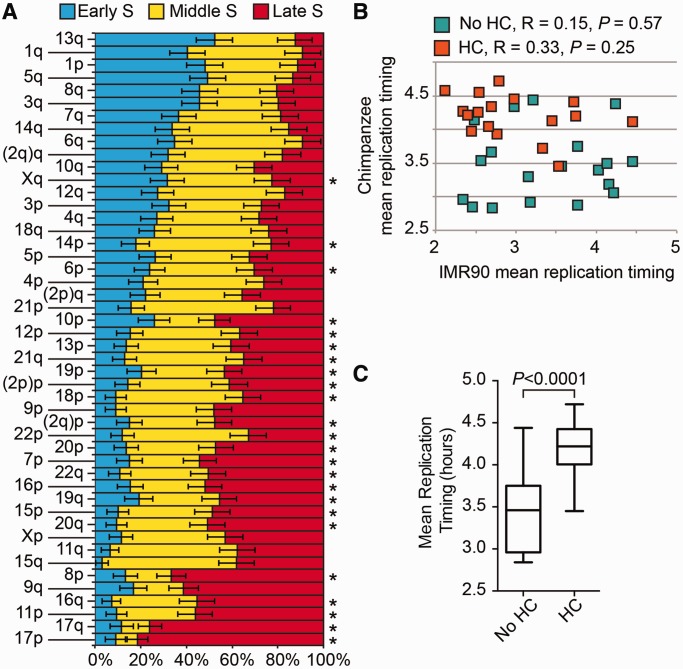
We next determined the timing of replication of individual telomeres in chimpanzee primary fibroblasts. We found that, similarly to humans (11), chimpanzee telomeres replicate all along the S-phase with a trend for specific chromosome extremities to replicate at particular moments (Figure 1A). To test whether the timing of replication for individual chromosomes was similar to the human replication pattern, we compared the mean replication scores calculated for each telomere in chimpanzee (Supplementary Table S1) to the mean replication scores calculated for the homologous extremities in the human fibroblast IMR90 [as determined in a previous study (11)]. As shown in Figure 1B, there is no significant correlation between the telomere-specific replication timing of these species, regardless of whether these extremities carried heterochromatic caps as in the chimpanzee. Interestingly, telomeres associated with chromosome extremities that carry heterochromatic caps seemed to replicate later than the others in the chimpanzee (Figure 1A and B). In fact, the mean replication timings of chimpanzee cap-associated telomeres are significantly higher than that of non-cap–associated telomeres in the same species (unpaired t-test: P < 0.0001) (Figure 1C). Therefore, although the order of replication of individual telomeres differs between human and chimpanzee, a late replication dynamics seems to be associated with the presence of heterochromatinization domains in the subtelomere in both species (11).
Figure 1.
Chimpanzee telomeres associated with heterochromatic caps replicate late. (A) Single telomeres in chimpanzee tend to replicate at particular moments of the S-phase. Single telomere replication was analyzed using ReDFISH as previously described (11). Briefly, synchronized cells were released in S-phase, and different batches were BrdU pulse-labeled for 1 h at different times. Metaphasic chromosomes were prepared and treated so that telomeres that had replicated during the pulse will be rendered single stranded and revealed by only one telomere probe (either G- or C-rich). The histogram represents the percentage of replicating telomeres for every chromosome arm detected during early S, middle S and late S pulses in horizontal bars. The total sum of partial percentages is normalized to 100%. An asterisk marks the extremities that bear a heterochromatic cap. Bar errors (α = 0.05) are indicated only for the mid-S percentages for simplification. (B) Total absence of correlation between the mean replication timing of chimpanzee extremities (as determined in this article), regardless of whether they bear a heterochromatic cap (orange and green squares, respectively), and that of the homologous human chromosome extremities [as determined previously (11)] (Supplementary Table S1). Correlation coefficients and P-values (Spearman correlation test) are indicated on top of the scatter plot. (C) The mean replication time of chimpanzee extremities bearing heterochromatic caps is significantly higher than that of extremities without caps.
Heterochromatic caps in gorilla contain telomere-like repeated domains that replicate independently of canonical telomere repeats
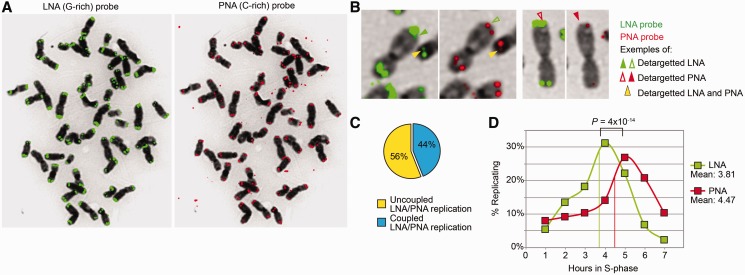
In preliminary experiments for the application of the ReDFISH technique to gorilla chromosomes, we noticed that, contrary to chimpanzee chromosomes, telomere G-rich LNA and C-rich PNA probes gave different patterns of telomere hybridization on the same metaphases (Figure 2A). Although the C-rich PNA probe showed the usual pattern (well-circumscribed round fluorescent signals at nearly all chromosome extremities), the G-rich LNA probe revealed much larger, stronger and less well-delimited signals closely associated with the heterochromatic caps, suggesting that sequences containing large amounts of telomere or telomere-like repeats were present at this level. CO-FISH experiments indicated that telomere sequences giving rise to LNA signals were always on different sister chromatids from telomere sequences revealed by the PNA probe, indicating that they had the same orientation than bona fide telomeres (Figure 2B and Supplementary Figure S2). Such large telomere signals associated with gorilla chromosome ends have already been detected by others using both oligonucleotide probes and the PRINS technique (29). However, our G-rich LNA probe did not detect the large paracentromeric signals detected by PRINS on gorilla chromosomes 17 and 18 (29). This observation suggests that, unlike in the case of chimpanzee chromosome 17, the interstitial sequences in the gorilla complement carry largely degenerated telomeric repeats not efficiently recognized by the LNA probe.
Figure 2.
Large telomere-like sequences detected by LNA telomeric probes replicate independently of bona fide gorilla telomeres. (A) G-rich LNA (left, in green) and C-rich PNA (right, in red) probes yield different telomere FISH profiles on the same gorilla metaphase. The C-rich PNA probe detects round and well-defined fluorescent signals, typical of telomeres. The G-rich LNA probe detects a much larger and not so well-defined area, which is associated with the heterochromatic caps. Contrary to the PRINS technique (29), the LNA probe does not detect paracentromeric telomere-like sequences present on particular gorilla chromosomes (29). Of note, extrachromosomal telomere signals are uniquely recognized by the C-rich probe (right). (B) The ReDFISH approach allows to examine the dynamics of replication of sequences revealed by PNA alone, LNA alone or both. The figure shows examples of extremities for which the detargeting occurred either simultaneously (yellow arrowhead), specifically for the LNA probe (green arrowhead) or specifically for the PNA probe (red arrowhead). (C) The percentage of LNA-revealed and PNA-revealed telomere sequences that replicated at different moments, thus suggesting distinct replication timings, is higher than that of sequences replicating together. (D) The replication of LNA-revealed (presumably subtelomeric) telomere sequences peaks significantly earlier than that of PNA-revealed (bona fide) telomere sequences (Fisher exact test), suggesting that the former follow a different replication program. Mean replication timings are indicated for both structures.
Given the heterogeneity of telomere lengths, the poor resolution of subtelomeric domains on metaphase chromosomes and the fact that LNA probes also recognize canonical telomere sequences, the fluorescence intensity criterion alone is insufficient to formally distinguish between subtelomeric domains carrying repeated telomere sequences and bona fide telomeres. Nevertheless, we took advantage of the analytic power of the ReDFISH approach to determine whether they form distinct domains and their respective replication timing. We assumed that if the telomere repeats revealed by the LNA probes belonged to functionally separated chromosome domains, their replication timing might be different from that of true telomeres. Therefore, we registered the moment at which every probe revealed only one signal in one sister chromatid (i.e. the other chromatid had been ‘detargeted’ for these sequences), and we counted the number of events (extremities) showing either unique (LNA or PNA) or simultaneous (both LNA and PNA) detargetings. First of all, the percentage of extremities that showed chromatid detargeting for both probes (presumably corresponding to ends with unique telomere domains) at any time during S-phase was lower than that of extremities showing a differential detargeting (and presumably bearing distinct domains) (Figure 2B and C). This result suggested that in many instances, the LNA-revealed telomere blocks and the PNA-revealed (bona fide) telomeres replicated separately. Indeed, a comparison of the dynamics during S-phase of the replication timings of these two domains indicated that bona fide telomeres replicate significantly later than more interstitial telomere-like blocks (Fisher exact test: P < 10−14) (Figure 2D). In conclusion, the experiments demonstrate that at least some of the heterochromatic caps found in gorilla chromosomes are associated with large blocks of telomere-like repeats, and that these blocks probably constitute functionally distinct chromosome domains that replicate independently of bona fide telomeres.
Telomere repeat-containing RNA molecules accumulate extrachromosomally in gorilla mitotic cells
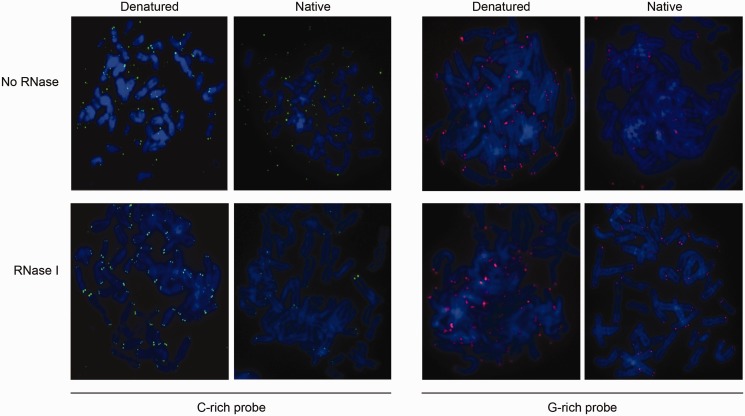
In the course of telomeric FISH analyses carried out with gorilla cells, we noticed the presence of abundant extrachromosomic telomeric signals, a feature normally absent in human cells and not observed in chimpanzee. These signals were detected exclusively with C-rich (both LNA and PNA) telomeric probes, suggesting that they contained G-rich telomeric repeats of single-stranded composition (Figure 3). That these molecules were indeed single stranded was further indicated by the fact that their detection did not require a denaturation step (Figure 3).
Figure 3.
TERRA accumulates extrachromosomally in gorilla mitotic cells. Telomeric FISH on metaphasic spreads revealed the presence of extrachromosomal telomeric-sequences. These extrachromosomal sequences are G-rich, as they are exclusively detected with C-rich probes (upper panel, left) and not with G-rich probes (upper panel, right). These molecules correspond to single-stranded telomeric-repeats containing RNA (TERRA), as they are RNase I sensitive (bottom panel, left), and their detection does not require a denaturation step (upper panel, second from left).
As telomeric repeats have been shown to be expressed in most organisms (30), we investigated whether the extrachromosomal molecules detected in gorilla metaphases were in fact telomere repeats containing RNA (TERRA). First, an RNase treatment completely prevented the detection of the extrachromosomal signals, strongly suggesting that these were indeed TERRA molecules (Figure 3). Second, a northern blot analysis of chimpanzee and gorilla RNA revealed G-rich signals with large size distribution, similar to that described in humans (31), indicating that TERRA is present in both chimpanzee and gorilla cells (Supplementary Figure S3A). The amount of G-rich RNA signals detected was apparently higher in gorilla cells (Supplementary Figure S3A). On the other hand, gorilla cells that express telomerase seem to accumulate less TERRA, as detected both by northern blot and RT–qPCR (Supplementary Figure S3A and B). This result suggests a telomere position effect, as already shown for TERT-expressing human cells (25).
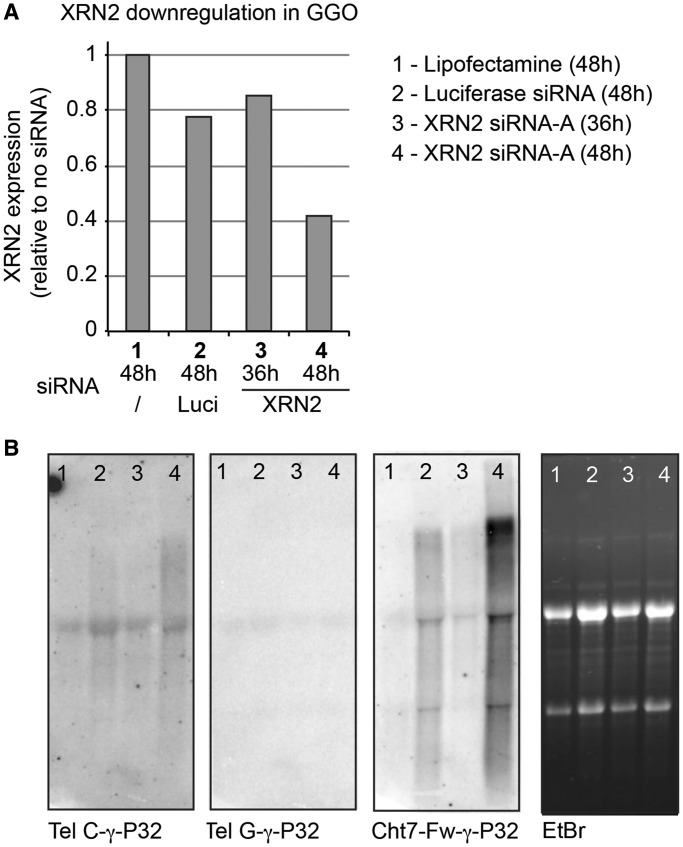
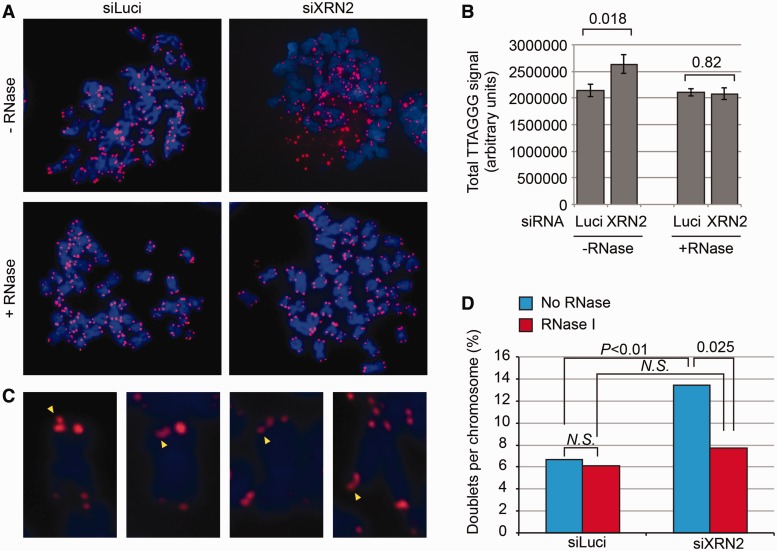
It has been shown in the budding yeast that the RNA exonuclease Rat1P controls the levels of yeast TERRA by inducing its degradation (32). Although the role of the Rat1P-homologue XRN2 (5′–3′ exoribonuclease 2) in controlling TERRA levels remains to be demonstrated in higher primates, we tested whether XRN2 was involved in TERRA regulation in gorilla cells. Using an siRNA approach, we achieved a partial expression knockdown of the gorilla XRN2 gene (60% as determined by RT–PCR, Figure 4A) after 48 h of treatment. Strikingly, this decrease in the expression of XRN2 was associated with an increase in the detection of TERRA molecules by northern blot (Figure 4B). Remarkably, an increase in extrachromosomal TERRA molecules was also detected during mitosis, leading to a significant increase in the total fluorescence intensity associated with metaphases from cells treated with siXRN2 when compared with cells treated with an irrelevant siRNA (Figure 5A and B). This increase completely disappeared when metaphase preparations were treated with RNase (Figure 5A and B). Although most TERRA seemed to accumulate extrachromosomally, it is possible that TERRA also accumulated in association with chromosome ends, as suggested both by the fact that the proportion of sister chromatids bearing telomeric doublets was increased in siXRN2-treated cells and by the fact that this increase was lost on RNase treatment (Figure 5C and D). At the same time, as there was not detectable difference in the overall intensity of telomeres after RNase treatment (Figure 5D), telomere replication was likely not affected.
Figure 4.
XRN2 downregulation leads to TERRA and Cht7 RNA accumulation. (A) XRN2 expression was downregulated in gorilla cells using a specific siRNA. Levels of downregulation were estimated using RT–qPCR to quantify XRN2 mRNA levels 36 and 48 h after transfection. (B) After siXRN2 treatment, total RNA was analyzed by northern blot, and TERRA molecules were revealed with a telomeric C-rich (Tel C) probe (γ-P32-labeled) (left panel). Relative to the amount of RNA loaded (rightmost panel), levels of TERRA increased in cells after 48 h of treatment with siXRN2. The same membrane was stripped and re-hybridized with a telomeric G-rich probe (γ-P32-labeled) and with a probe (γ-P32-labeled) against the Cht7 minisatellite repeat (second to right panel). RNAs containing this repeat also became more abundant in siXRN2-treated cells.
Figure 5.
TERRA accumulates in metaphases of siXRN2-treated gorilla cells. Q-FISH analyses show an accumulation of extrachromosomal telomeric signals (A) and an increase in the mean intensity of total telomere signals (B) revealed by the C-rich probe in siXRN2-treated cells. This increase is eliminated by an RNase treatment of the slides, indicating that this increase is due to TERRA accumulation. Numbers above bars indicate P-values (student t-test). On the other hand, the Q-FISH analyses also revealed the presence of telomeric doublets (sister chromatids bearing two telomeric signals, yellow arrows) in control cells, as well as in siXRN2-treated cells (C). In the latter, however, doublets were significantly more abundant, suggesting that many of these doublets were the consequence of TERRA accumulation (D). This conclusion is also supported by the fact that an RNase I treatment of the slides leads to a significant reduction of the doublets (D). At least 600 chromosomes were considered per condition. Numbers above bars indicate P-values (student t-test). N.S., no significant difference.
Subtelomeric Cht7 repeats are connected to, and co-transcribed with, telomeric repeats
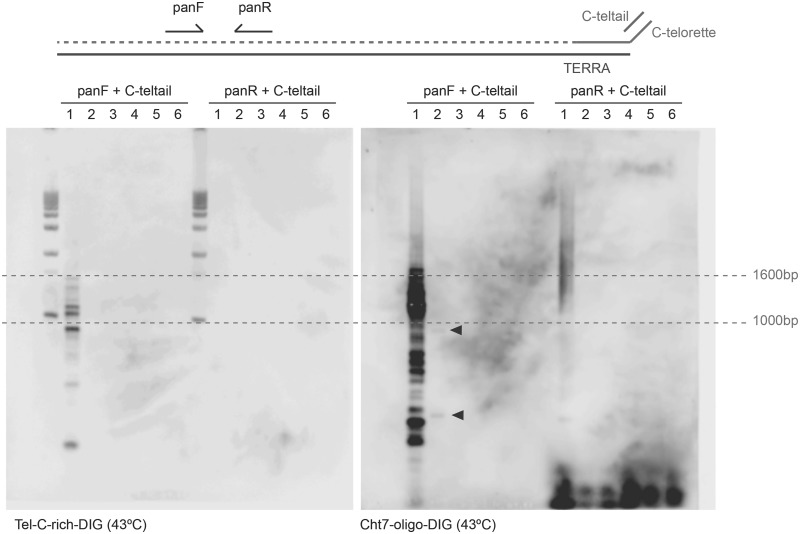
Previously published data demonstrate that Cht7 minisatellite sequences are associated with the heterochromatic caps exclusive to chimpanzees and gorillas, and suggest that Cht7 may be physically connected to telomere repeats (21). A restriction fragment analysis by Southern blotting revealed that both telomere and Cht7 repeats are sensitive to Bal31 digestion, thus confirming their position close to a chromosome end (Supplementary Figure S4). Furthermore, PCR reactions using a C-telorette (partly complementary to telomere repeats) and one oligonucleotide representing the Cht7 repeat unit (here called panF) allowed the amplification, from genomic DNA, of molecules containing both telomeric and Cht7 repeats, further supporting the idea that these sequences are physically connected (Figure 6, left panel, lane 1). This physical proximity raised the possibility that both sequences may be co-transcribed.
Figure 6.
Telomere repeats and the Cht7 minisatellite are physically linked on gorilla chromosomes and are co-transcribed. A PCR approach was adapted to determine whether Cht7 and telomeric sequences were physically linked and co-transcribed. A telomeric C-rich primer, ending with a unique 3′-end (C-telorette) was used to reverse transcribe total gorilla RNA (top). The unique sequence (C-teltail) was used to prime PCR reactions in combination with primers specific to Cht7-sequence, either in the sense (panF) or in the antisense (panR) orientation. PCR products were separated in a 1% agarose gel and hybridized with either digoxigenin-labeled telomeric C-rich probes (bottom left) or Cht7-oligo (bottom right). PCR templates included: lane 1: genomic DNA; lane 2: cDNA transcribed with C-telorette3; lane 3: same RNA sample treated as in 2 but leaving out the reverse transcriptase (RT); lane 4: same as in 3 but treated with DNAse I; lane 5: same as in 3 but treated with RNase I; and lane 6: RT reaction with no RNA. Both the telomeric and the Cht7 probes revealed amplified products from genomic DNA exclusively obtained with the combination C-teltail/panF, indicating proximity of both sequences and allowing orienting the minisatellite repeats with regard to the telomeric sequences. On the other hand, the Cht7 probe reveals rare molecules (arrows) amplified using the same couple of primers and the C-telorette cDNA, suggesting the presence of RNA molecules carrying both telomere and Cht7 repeats.
Although Cht7-containing RNA molecules were barely detectable in unperturbed cells, we detected an increase in cells treated with siXRN2, thus indicating that this minisatellite is indeed transcribed, and that Cht7 transcripts are also under the control of the XRN2 exonuclease (Figure 4B). To test whether Cht7 RNA molecules also contain telomere repeats, we reverse transcribed total RNA preparations using the C-telorette. Then, we carried out PCR reactions using an oligonucleotide complementary to the non-telomeric part of the C-telorette and the panF oligonucleotide. Using this combination, we detected a few discrete bands, suggesting the presence of hybrid RNA molecules. These molecules seemed to be specific, as they were only detected in reverse transcribed RNA samples and not in non-reverse transcribed or RNase/DNase treated samples (Figure 6, right panel). Our experiments suggest that at least some portion of the TERRA molecules detected in gorilla cells include sequences associated with the heterochromatic caps.
DISCUSSION
Despite the high sequence conservation between genomes of higher primates at the nucleotide level, significant variations at the chromosomal structural level exist (33–35). One striking example is constituted by the heterochromatic caps found at the end of chimpanzee and gorilla chromosomes and absolutely absent from the orangutan and human chromosome ends. Recent in-depth analyses of sequence composition of duplicated domains linked to the heterochromatic caps indicated that these chromosome regions have evolved independently in the gorilla and chimpanzee lineages (19). Interestingly, although, the subterminal satellite Cht7 is associated with these heterochromatic caps both in gorilla and chimpanzee (21), supporting the notion that this association precedes the segmental expansion events that independently took place in both species. In addition, the Cht7 minisatellite sequences also co-localize with a heterochromatic region found in the middle of the long arm of chromosome 7 in chimpanzee (21), suggesting an association between minisatellite sequences and heterochromatic potential.
Our studies on telomere replication in chimpanzee have indicated that the presence of heterochromatic caps has a genuine impact on telomere metabolism by delaying the timing of replication of associated telomeres. This is in agreement with previous works showing that the subtelomeric domain composition of particular human chromosome arms directly affects the timing of telomere replication (11). Here, the effect is exerted on about half of the chimpanzee chromosome arms, thus impacting the overall pattern of telomere replication timing. On the other hand, the replication timing of chromosome ends that do not carry heterochromatic caps was also different from the replication timing of the homologous arms in humans. This observation suggests that in spite of high sequence conservation between both species, other, still undetermined, factors have intervened during evolution to influence the timing of telomere replication.
Interestingly, the use of a telomeric LNA probe allowed the detection of a large paracentromeric domain on chromosome 17 in the chimpanzee that contains slightly degenerated telomere repeats, not detected by PNA probes. The origin of this domain is not known, but it may be associated with the pericentromeric inversion that affected that chromosome in chimpanzees (36). In any case, the presence of paracentromeric domains carrying large blocks of telomere-like repeats are not uncommon in other primates (29) and may represent scars of ancient points of breakage associated with fragile sites (37).
Our analysis of telomere replication in gorilla was complicated by the fact that part of the heterochromatic caps contained a distinct domain composed of telomere-like sequences. The precise nucleotide composition of these domains is not known, but they most likely contain slightly degenerated telomere repeats, so that they are efficiently recognized by LNA (but not by PNA) probes. As we only detected cap-associated telomere-like blocks in gorilla, these sequences probably emerged after separation of this lineage from the common ancestor. Recent studies using molecular cytogenetic approaches and sequence analyses (19,20) have revealed in detail the composition of the heterochromatic caps in chimpanzee and gorilla. The caps are composed of segmental duplications, present at high number of copies towards the tip of the chromosome, interspersed with Cht7 satellite sequences, which in turn may have played a central role in local segmental expansion and spread among extremities (19,20). These studies point to a completely independent evolution of caps in chimpanzee and gorilla and may explain the emergence of large blocks of telomere-like repeated sequences only in the latter. Intriguingly, no interspersed blocks of telomere repeats were reported in sequencing analyses of gorilla BAC clones (19,20). Of note, however, the authors do report that 30% of the BACs completely failed to assemble or else assembled into sequence contigs substantially shorter than the expected length, indicating particular difficulties in sub-cloning and in in silico sequence assembly procedures of these regions (19,20), a difficulty perhaps enhanced by the presence of long tracks of telomere-like repeats.
Independently of the exact organization of these large telomere-like repeated domains in gorilla, our work shows that these sequences replicate at different moments than bona fide telomeres (only detected by PNA probes), demonstrating that they both constitute physically separated domains and likely belong to different genome replication domains. The possibility that both domains were replicated by the same fork cannot be entirely excluded, as the mean replication timing for subtelomeric blocks peaked a little earlier than that of bona fide telomeres, i.e. the most distal sequences in the chromosome. Experiments using single molecule analysis will be required to address this question.
We also showed here that gorilla telomeres are transcribed, like in many other species (26,38). A striking difference, although, is that gorilla TERRA accumulated extrachromosomally during mitosis, although the mechanisms leading to this accumulation remain to be explored. Interestingly, the accumulation of TERRA seemed to be under the control of XRN2, a putative 5′ > 3′ RNA exonuclease involved in the control of RNA synthesis and trafficking (39). This is indeed the first study to show that, as predicted based on findings in the budding yeast (32), XRN2 impacts TERRA biogenesis in higher primates. The fact that, in cells deficient in XRN2, extrachromosomal TERRA molecules accumulated even more in mitotic cells without perturbing telomere maintenance suggests that TERRA is efficiently displaced from telomeres, perhaps at the passage of the replication fork. We have also found that XRN2 controls the levels of RNA molecules transcribed from the Cht7 sequences. Intriguingly, we detected RNA molecules carrying both telomeric and Cht7 repeats, suggesting that both sequences are co-transcribed. This observation predicts a site for transcription initiation either within Cht7 or on proximal (related to centromere) flanking regions. Whether this is the same transcription starting point for all gorilla TERRA molecules remains to be explored.
Independently of the many questions elicited by our observations, our finding clearly illustrates the impact exerted by species-specific chromosome structural variations on telomere metabolism.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Table 1 and Supplementary Figures 1–4.
FUNDING
Association pour la Recherche contre le Cancer (ARC); Agence National pour la Recherche (ANR); Curie Institute (to C.N.); Telomere and Cancer laboratory is ‘labellisé Ligue’. Funding for open access charge: Institutional funds.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Eichler EE, Sankoff D. Structural dynamics of eukaryotic chromosome evolution. Science. 2003;301:793–797. doi: 10.1126/science.1086132. [DOI] [PubMed] [Google Scholar]
- 2.Larkin DM, Pape G, Donthu R, Auvil L, Welge M, Lewin HA. Breakpoint regions and homologous synteny blocks in chromosomes have different evolutionary histories. Genome Res. 2009;19:770–777. doi: 10.1101/gr.086546.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weber B, Allen L, Magenis RE, Goodfellow PJ, Smith L, Hayden MR. Intrachromosomal location of the telomeric repeat (TTAGGG)n. Mamm. Genome. 1991;1:211–216. doi: 10.1007/BF00352327. [DOI] [PubMed] [Google Scholar]
- 4.Meyne J, Ratliff RL, Moyzis RK. Conservation of the human telomere sequence (TTAGGG)n among vertebrates. Proc. Natl Acad. Sci. USA. 1989;86:7049–7053. doi: 10.1073/pnas.86.18.7049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hug N, Lingner J. Telomere length homeostasis. Chromosoma. 2006;115:413–425. doi: 10.1007/s00412-006-0067-3. [DOI] [PubMed] [Google Scholar]
- 6.Blackburn EH. The telomere and telomerase: nucleic acid-protein complexes acting in a telomere homeostasis system. A review. Biochemistry. 1997;62:1196–1201. [PubMed] [Google Scholar]
- 7.Allsopp RC, Chang E, Kashefi-Aazam M, Rogaev EI, Piatyszek MA, Shay JW, Harley CB. Telomere shortening is associated with cell division in vitro and in vivo. Exp. Cell Res. 1995;220:194–200. doi: 10.1006/excr.1995.1306. [DOI] [PubMed] [Google Scholar]
- 8.Gomes NM, Ryder OA, Houck ML, Charter SJ, Walker W, Forsyth NR, Austad SN, Venditti C, Pagel M, Shay JW, et al. Comparative biology of mammalian telomeres: hypotheses on ancestral states and the roles of telomeres in longevity determination. Aging Cell. 2011;10:761–768. doi: 10.1111/j.1474-9726.2011.00718.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Herbig U, Ferreira M, Condel L, Carey D, Sedivy JM. Cellular senescence in aging primates. Science. 2006;311:1257. doi: 10.1126/science.1122446. [DOI] [PubMed] [Google Scholar]
- 10.Jeyapalan JC, Ferreira M, Sedivy JM, Herbig U. Accumulation of senescent cells in mitotic tissue of aging primates. Mech. Ageing Dev. 2007;128:36–44. doi: 10.1016/j.mad.2006.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arnoult N, Schluth-Bolard C, Letessier A, Drascovic I, Bouarich-Bourimi R, Campisi J, Kim SH, Boussouar A, Ottaviani A, Magdinier F, et al. Replication timing of human telomeres is chromosome arm-specific, influenced by subtelomeric structures and connected to nuclear localization. PLoS Genet. 2010;6:e1000920. doi: 10.1371/journal.pgen.1000920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Drosopoulos WC, Kosiyatrakul ST, Yan Z, Calderano SG, Schildkraut CL. Human telomeres replicate using chromosome-specific, rather than universal, replication programs. J. Cell Biol. 2012;197:253–266. doi: 10.1083/jcb.201112083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Flint J, Bates GP, Clark K, Dorman A, Willingham D, Roe BA, Micklem G, Higgs DR, Louis EJ. Sequence comparison of human and yeast telomeres identifies structurally distinct subtelomeric domains. Hum. Mol. Genet. 1997;6:1305–1313. doi: 10.1093/hmg/6.8.1305. [DOI] [PubMed] [Google Scholar]
- 14.Der-Sarkissian H, Vergnaud G, Borde YM, Thomas G, Londono-Vallejo JA. Segmental polymorphisms in the proterminal regions of a subset of human chromosomes. Genome Res. 2002;12:1673–1678. doi: 10.1101/gr.322802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mefford HC, Trask BJ. The complex structure and dynamic evolution of human subtelomeres. Nat. Rev. Genet. 2002;3:91–102. doi: 10.1038/nrg727. [DOI] [PubMed] [Google Scholar]
- 16.Monfouilloux S, Avet-Loiseau H, Amarger V, Balazs I, Pourcel C, Vergnaud G. Recent human-specific spreading of a subtelomeric domain. Genomics. 1998;51:165–176. doi: 10.1006/geno.1998.5358. [DOI] [PubMed] [Google Scholar]
- 17.Yunis JJ, Sawyer JR, Dunham K. The striking resemblance of high-resolution G-banded chromosomes of man and chimpanzee. Science. 1980;208:1145–1148. doi: 10.1126/science.7375922. [DOI] [PubMed] [Google Scholar]
- 18.Haaf T, Schmid M. Chromosome heteromorphisms in the gorilla karyotype. Analyses with distamycin A/DAPI, quinacrine and 5-azacytidine. J. Hered. 1987;78:287–292. doi: 10.1093/oxfordjournals.jhered.a110389. [DOI] [PubMed] [Google Scholar]
- 19.Ventura M, Catacchio CR, Alkan C, Marques-Bonet T, Sajjadian S, Graves TA, Hormozdiari F, Navarro A, Malig M, Baker C, et al. Gorilla genome structural variation reveals evolutionary parallelisms with chimpanzee. Genome Res. 2011;21:1640–1649. doi: 10.1101/gr.124461.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ventura M, Catacchio CR, Sajjadian S, Vives L, Sudmant PH, Marques-Bonet T, Graves TA, Wilson RK, Eichler EE. The evolution of African great ape subtelomeric heterochromatin and the fusion of human chromosome 2. Genome Res. 2012;22:1036–1049. doi: 10.1101/gr.136556.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Royle NJ, Baird DM, Jeffreys AJ. A subterminal satellite located adjacent to telomeres in chimpanzees is absent from the human genome. Nat. Genet. 1994;6:52–56. doi: 10.1038/ng0194-52. [DOI] [PubMed] [Google Scholar]
- 22.Dutrillaux B. Chromosomal evolution in primates: tentative phylogeny from Microcebus murinus (Prosimian) to man. Hum. Genet. 1979;48:251–314. doi: 10.1007/BF00272830. [DOI] [PubMed] [Google Scholar]
- 23.Wen J, Cong YS, Bacchetti S. Reconstitution of wild-type or mutant telomerase activity in telomerase- negative immortal human cells. Hum. Mol. Genet. 1998;7:1137–1141. doi: 10.1093/hmg/7.7.1137. [DOI] [PubMed] [Google Scholar]
- 24.Londono-Vallejo JA, DerSarkissian H, Cazes L, Thomas G. Differences in telomere length between homologous chromosomes in humans. Nucleic Acids Res. 2001;29:3164–3171. doi: 10.1093/nar/29.15.3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arnoult N, Van Beneden A, Decottignies A. Telomere length regulates TERRA levels through increased trimethylation of telomeric H3K9 and HP1alpha. Nat. Struct. Mol. Biol. 2012;19:948–956. doi: 10.1038/nsmb.2364. [DOI] [PubMed] [Google Scholar]
- 26.Porro A, Feuerhahn S, Reichenbach P, Lingner J. Molecular dissection of telomeric repeat-containing RNA biogenesis unveils the presence of distinct and multiple regulatory pathways. Mol. Cell Biol. 2010;30:4808–4817. doi: 10.1128/MCB.00460-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zou Y, Gryaznov SM, Shay JW, Wright WE, Cornforth MN. Asynchronous replication timing of telomeres at opposite arms of mammalian chromosomes. Proc. Natl Acad. Sci. USA. 2004;101:12928–12933. doi: 10.1073/pnas.0404106101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Egholm M, Buchardt O, Christensen L, Behrens C, Freier SM, Driver DA, Berg RH, Kim SK, Norden B, Nielsen PE. PNA hybridizes to complementary oligonucleotides obeying the Watson-Crick hydrogen-bonding rules. Nature. 1993;365:566–568. doi: 10.1038/365566a0. [DOI] [PubMed] [Google Scholar]
- 29.Hirai H. Relationship of telomere sequence and constitutive heterochromatin in the human and apes as detected by PRINS. Methods Cell Sci. 2001;23:29–35. [PubMed] [Google Scholar]
- 30.Azzalin CM, Lingner J. Telomeres: the silence is broken. Cell Cycle. 2008;7:1161–1165. doi: 10.4161/cc.7.9.5836. [DOI] [PubMed] [Google Scholar]
- 31.Azzalin CM, Reichenbach P, Khoriauli L, Giulotto E, Lingner J. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science. 2007;318:798–801. doi: 10.1126/science.1147182. [DOI] [PubMed] [Google Scholar]
- 32.Luke B, Panza A, Redon S, Iglesias N, Li Z, Lingner J. The Rat1p 5′ to 3′ exonuclease degrades telomeric repeat-containing RNA and promotes telomere elongation in Saccharomyces cerevisiae. Mol. Cell. 2008;32:465–477. doi: 10.1016/j.molcel.2008.10.019. [DOI] [PubMed] [Google Scholar]
- 33.Fortna A, Kim Y, MacLaren E, Marshall K, Hahn G, Meltesen L, Brenton M, Hink R, Burgers S, Hernandez-Boussard T, et al. Lineage-specific gene duplication and loss in human and great ape evolution. PLoS Biol. 2004;2:E207. doi: 10.1371/journal.pbio.0020207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cheng Z, Ventura M, She X, Khaitovich P, Graves T, Osoegawa K, Church D, DeJong P, Wilson RK, Paabo S, et al. A genome-wide comparison of recent chimpanzee and human segmental duplications. Nature. 2005;437:88–93. doi: 10.1038/nature04000. [DOI] [PubMed] [Google Scholar]
- 35.Marques-Bonet T, Kidd JM, Ventura M, Graves TA, Cheng Z, Hillier LW, Jiang Z, Baker C, Malfavon-Borja R, Fulton LA, et al. A burst of segmental duplications in the genome of the African great ape ancestor. Nature. 2009;457:877–881. doi: 10.1038/nature07744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kehrer-Sawatzki H, Schreiner B, Tanzer S, Platzer M, Muller S, Hameister H. Molecular characterization of the pericentric inversion that causes differences between chimpanzee chromosome 19 and human chromosome 17. Am. J. Hum. Genet. 2002;71:375–388. doi: 10.1086/341963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ruiz-Herrera A, Nergadze SG, Santagostino M, Giulotto E. Telomeric repeats far from the ends: mechanisms of origin and role in evolution. Cytogenet. Genome Res. 2008;122:219–228. doi: 10.1159/000167807. [DOI] [PubMed] [Google Scholar]
- 38.Flynn RL, Centore RC, O’Sullivan RJ, Rai R, Tse A, Songyang Z, Chang S, Karlseder J, Zou L. TERRA and hnRNPA1 orchestrate an RPA-to-POT1 switch on telomeric single-stranded DNA. Nature. 2011;471:532–536. doi: 10.1038/nature09772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang M, Pestov DG. 5′-end surveillance by XRN2 acts as a shared mechanism for mammalian pre-rRNA maturation and decay. Nucleic Acids Res. 2011;39:1811–1822. doi: 10.1093/nar/gkq1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.