Abstract
The clamp-loader complex plays a crucial role in DNA replication by loading the β-clamp onto primed DNA to be used by the replicative polymerase. Relatively little is known about the stoichiometry, structure and assembly pathway of this complex, and how it interacts with the replicative helicase, in Gram-positive organisms. Analysis of full and partial complexes by mass spectrometry revealed that a hetero-pentameric τ3-δ-δ′ Bacillus subtilis clamp-loader assembles via multiple pathways, which differ from those exhibited by the Gram-negative model Escherichia coli. Based on this information, a homology model of the B. subtilis τ3-δ-δ′ complex was constructed, which revealed the spatial positioning of the full C-terminal τ domain. The structure of the δ subunit was determined by X-ray crystallography and shown to differ from that of E. coli in the nature of the amino acids comprising the τ and δ′ binding regions. Most notably, the τ-δ interaction appears to be hydrophilic in nature compared with the hydrophobic interaction in E. coli. Finally, the interaction between τ3 and the replicative helicase DnaB was driven by ATP/Mg2+ conformational changes in DnaB, and evidence is provided that hydrolysis of one ATP molecule by the DnaB hexamer is sufficient to stabilize its interaction with τ3.
INTRODUCTION
The speed and processivity of DNA synthesis during replication is greatly increased by interaction of the core polymerase with a ring-shaped sliding clamp that encircles double-stranded DNA and slides along it (1). In bacteria, this protein is known as the β-clamp and is composed of two β subunits arranged as a head-to-tail ring with 6-fold symmetry (2,3). The β-clamp is loaded onto DNA by the clamp-loader complex, a hetero-multimeric assembly whose activity is fuelled by the binding and hydrolysis of ATP (4,5). In Escherichia coli, the minimal functional clamp-loader is composed of three different subunits (γ, δ and δ′), with a γ3-δ-δ′ stoichiometry (6). The DnaX gene coding for γ also codes for a longer version of this protein named τ, which contains a C-terminal domain (Cτ) that interacts with the α subunit of the core polymerase (7,8) and with the DnaB helicase, the activity of which significantly increases with this interaction (7,8). Both γ and τ can interact with δ and δ′ and form the trimeric central core of the E. coli clamp-loader (9). Structural studies have revealed a crescent-shaped complex with δ and δ′ subunits located at the ends, and a closed collar formed by the C-terminus of each single subunit (10–12). The assembly pathway of the complex in this organism has also been recently studied by electrospray ionization mass spectrometry (ESI-MS). In isolation, γ/τ exist predominantly as tetramers. Addition of δ′ is essential not only to break the γ/τ tetramer apart and trap the smaller oligomers formed, but also to promote subsequent binding of δ to the complex (13).
Although the E. coli clamp-loader structure and activity have been well characterized, limited information is available on the clamp-loader of Gram-positive bacteria. In these organisms, it is assumed that the premature translational termination resulting in the truncated γ protein does not occur, and only τ is produced, based on the expression of the DnaX gene from Streptococcus pyogenes. In the same study, it was observed that the minimal functional clamp-loader of this organism is composed of τ, δ and δ′ subunits, with densitometry analysis suggesting a τ4-δ-δ′ stoichiometry (14). Further studies on the Bacillus subtilis τ subunit have provided additional information on the oligomerization of this protein. Analytical ultracentrifugation data suggest the formation of a τ pentamer in the absence of δ and δ′ (15), while atomic force microscopy revealed that this oligomer adopts a crescent shape, similar to the shape of the E. coli clamp-loader complex (16). These results suggest that the composition of the Gram-positive clamp-loader differs somewhat from that of E. coli. However, there is no further structural information for the full complex. The assembly pathway of the Gram-positive clamp-loader is also unknown and the stoichiometry has not been confirmed by other techniques. Limited information concerning the interaction between the Gram-positive clamp-loader and other components of the replisome is available. Similar to the interaction between τ and DnaB helicase described in E. coli, evidence for the interaction between the τ subunit from B. subtilis and the replicative helicase from Bacillus stearothermophilus exists, showing that this interaction also occurs within the replisome of Gram-positive organisms (15,16). In fact, the last C-terminal residues of B. subtilis τ suppress the activity of the B. stearothermophilus primase in a primase-helicase–clamp-loader ternary complex in vitro, and may act as a functional gateway during DNA replication to regulate primer synthesis by τ-interacting components of the replisome (17).
Here we have applied ESI-MS to study the composition of the B. subtilis clamp-loader and its interaction with the B. stearothermophilus helicase DnaB. ESI is a gentle ionization technique that preserves non-covalent interactions between proteins (18,19), providing valuable information about composition, stoichiometry, solution dynamics and topology of protein assemblies (20–22). Our results show an oligomeric composition of the B. subtilis τ protein different from that reported previously. New information about the assembly pathway of the minimal clamp-loader complex from this organism is provided, which is distinctly different than that suggested for the E. coli clamp-loader complex. The correct stoichiometry of the clamp-loader complex from a Gram-positive organism is revealed for the first time and new insights into its structure are presented, including a homology model of the τ3-δ-δ′ core complex indicating the relative spatial position of the unknown Cτ domain. Additionally, the X-ray crystal structure of the δ subunit has been determined and used to assess the model. The binding regions of δ with δ′ and τ were found to be different than in E. coli. Finally, the interaction between the B. subtilis τ subunit and the replicative helicase DnaB has been studied, and a requirement for an ATP-induced conformational change in DnaB that strengthens the DnaB-τ complex is described. We provide evidence that the hydrolysis of one molecule of ATP to ADP may be sufficient to induce this conformational change.
MATERIALS AND METHODS
Cloning, expression and purification
Bacillus subtilis τ subunit and B. stearothermophilus DnaB helicase (82% identical and 92% similar to the B. subtilis DnaC helicase) were expressed and purified as described elsewhere (15,23,24). The yqeN gene (coding for δ) was amplified from B. subtilis genomic DNA by PCR, using the oligonucleotides yqeN-NcoI-F and yqeN-BamHI-R (Supplementary Table S1), and cloned into the NcoI and BamHI restriction sites of the vector pET-28a(+) (Merck Chemicals Ltd., UK) to generate pET28aYqeN, expressing native δ. The holB gene (coding for δ′) was amplified from B. subtilis genomic DNA by PCR, using the oligonucleotides holB-EcoRV-F and holB-XhoI-R (Supplementary Table S1) and cloned into the EcoRV and XhoI restriction sites of the vector pET-duet-1 in multiple cloning site 2. Expression plasmids pET28aYqeN and pETDuet1HolB were transformed into E. coli BL21-DE3 cells for the overexpression of B. subtilis δ and δ′ proteins, respectively. To increase the expression level of δ, it was co-expressed with δ′ by transformation with both expression vectors. Typically, cells were grown in 2 L lysogeny broth (LB) medium at 37°C in the presence of kanamycin (30 μg/ml) and ampicillin (100 μg/ml) for δ, and only ampicillin (100 μg/ml) for δ′, until the OD600 was 0.6. Expression was induced by addition of 1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG), and the temperature was changed to 20°C, with overnight expression for δ and 4 h expression for δ′. Pelleted cells containing overexpressed proteins were suspended in the appropriate buffer (δ–50 mM Tris, pH 7.5, 2 mM EDTA, 10% w/v sucrose, 100 mM NaCl; δ′–50 mM Tris, pH 8.0) and lysed by sonication in the presence of 1 mM phenylmethyl sulphonyl fluoride. The supernatants were clarified by centrifugation and used for purification. Supernatant containing δ was loaded onto a Resource Q equilibrated with 50 mM Tris, pH 7.5, 2 mM EDTA, 1 mM dithiothreitol (DTT). Protein was eluted with a gradient from 0 to 200 mM NaCl and loaded onto a Superdex 75 column equilibrated in 50 mM Tris, pH 7.5, 2 mM EDTA, 1 mM DTT, 100 mM NaCl. Purified δ protein was collected and stored. Supernatant containing δ′ was loaded onto a DEAE column equilibrated in 50 mM Tris, pH 8.0. Protein was eluted with a gradient from 0 to 500 mM NaCl and a 20% w/v ammonium sulphate cut was applied. The obtained supernatant was loaded onto a Phenyl HP column equilibrated in 50 mM Tris, pH 8.0, 800 mM ammonium sulphate. Protein was eluted with a reverse gradient from 800 to 0 mM ammonium sulphate and loaded onto a Superdex 75 column equilibrated in 50 mM Tris, pH 8.0, 150 mM NaCl. Purified δ′ protein was collected and stored. Proteins were quantified spectrophotometrically by measurement of the absorbance at 280 nm and using the specific extinction coefficients for each protein, calculated as described elsewhere (25).
Sample preparation for mass spectrometry
Purified protein samples were buffer exchanged into 1 M ammonium acetate using Vivaspin 500 centrifugal filters (Sartorius, Göttingen, Germany). This concentration of ammonium acetate was found to be the ideal for the analysis of the intact DnaB helicase hexamer, without affecting the analysis of the τ oligomers or the clamp-loader complex and subcomplexes. The clamp-loader complex was obtained by mixing equimolar amounts of τ, δ and δ′ before buffer exchange, while only δ or δ′ were added to τ to obtain partial complexes. The complex between τ and DnaB helicase was obtained by mixing both proteins in a τ3-DnaB6 stoichiometry before buffer exchange into 1 M ammonium acetate or into the same buffer containing 0.5 mM magnesium or 0.1 mM ATP or both. A final concentration between 3 and 5 µM of protein was used for ESI-MS analysis.
Electrospray ionization mass spectrometry
Samples were analysed using a Waters Synapt High Definition Mass Spectrometer (Manchester, UK)—a hybrid quadrupole/ion mobility/orthogonal acceleration time of flight instrument—equipped with a nanoelectrospray source. Samples were electrosprayed from thin wall Nanoflow Probe Tips (Waters, Manchester, UK), and experiments were typically conducted at a capillary voltage of 1.3–1.5 kV, nanoflow gas pressure of 0.1–0.3 Bar, source temperature of 50°C and sampling cone voltage of 50 V, with the source operating in positive ion mode. Backing pressure was maintained between 5.0 and 6.0 mBar to provide collisional cooling of ions in the intermediate vacuum region of the instrument. The collisional energy in the trap was 70 V for τ, 80 V for the full clamp-loader and its partial complexes and 160 V for the τ-DnaB complex. The collisional energy in the transfer was 50 V for τ, 50 V for the full clamp-loader and its partial complexes and 100 V for the τ-DnaB complex. Collisional dissociation of the clamp-loader complex was obtained with a trap collisional energy of 160 V. The trap gas (argon) flow was 8.0 ml.min−1 for τ and 9.0 ml.min−1 for the full/partial clamp-loader complexes and τ-DnaB complex. Spectra were acquired and processed using Masslynx 4.1 software (Waters, Manchester, UK).
Homology modelling
The structure of the B. subtilis clamp-loader was predicted using the iterative threading assembly refinement (I-TASSER) server, a platform for automated protein structure prediction (26). For the individual δ and δ′ subunits, each sequence was separately submitted and the highest confidence score models were chosen. To model the τ subunit, the Cτ domain sequence was submitted separately and the highest confidence score models were chosen as before. The full τ sequence was then submitted using the Cτ domain models as structural template for the folding of the C-terminal region, while the remaining domains were modelled by the automated structure prediction. The obtained full τ models were analysed according to the confidence score of each model and topological location of the Cτ domain. The model with the best confidence score and satisfying the spatial restraints on combination with adjacent subunits was selected. These τ, δ and δ′ models were aligned with their equivalents within the E. coli clamp-loader crystal structure to obtain an approximate model of the full B. subtilis clamp-loader.
Protein production for crystallization
δ was overexpressed in E. coli BL21(λDE3) cells grown in LB medium supplemented with 15 mg ml−1 kanamycin at 37°C. At an OD600 nm of 0.6, 1 mM IPTG was added, and the induced cells were then grown for a further 4 h at 20°C before harvesting by centrifugation (5000 g, 20 min, 4°C). To obtain selenomethionine incorporated (Se-Met) δ, cells were cultured in M9 medium supplemented with l-selenomethionine and other natural amino acids. Induction conditions were as for native protein. Cell pellets were suspended in 50 mM Tris–HCl, pH 8.0, disrupted by sonication (Soniprep 150) at 15 micron amplitude using 3–4 cycles for 30 s and the supernatant clarified by centrifugation at 24 500g for 15 min. Both native and l-selenomethionine labelled δ proteins were purified using the same protocol. The protein was applied onto a 25 ml-DEAE sepharose column (GE healthcare) equilibrated in 50 mM Tris–HCl buffer, pH 8.0 and then eluted with a linear gradient of 0–1 M NaCl in the same buffer. Fractions containing δ were pooled, the protein was precipitated with 0.8 M ammonium sulphate, centrifuged at 24 500 g for 15 min and the pellet was dissolved in 50 mM Tris–HCl pH 8.0. It was then applied onto a Resource Q column (GE healthcare) equilibrated in 50 mM Tris–HCl buffer, pH 8.0, and then eluted with a linear gradient of 0–1 M NaCl in the same buffer. The δ-containing fractions were pooled and applied onto a Superdex 200 gel filtration column equilibrated in 50 mM Tris–HCl, pH 8.0, 0.5 M NaCl. The δ-containing fractions were pooled once more and concentrated to 5 mg/ml using a Vivaspin column (MWCO 10 000, Sartorius Stedim Biotech). Purity of δ was assessed by SDS-PAGE and the identities of native and Se-Met δ were confirmed by mass spectrometry (data not shown). The protein was tested for crystallization with a variety of commercial screens, and crystals of both native and Se-Met δ were obtained in a condition constituting 0.1 M Bis-Tris, pH 5.5, 1 M (NH4)2SO4 and 6% w/v PEG 3350. The crystals reached an average size of 0.1 × 0.2 × 0.05 mm3 within 12 h at 17°C by the sitting drop vapour diffusion method.
Data collection and structure determination
Before data collection at 100 K, crystals were mounted in LithoLoops (Molecular Dimensions Ltd) and soaked briefly in a solution of 30% (v/v) glycerol, 1 M (NH4)2SO4, 0.1 M Bis-Tris, pH 5.5, and 6% (w/v) PEG 3350. Diffraction from native and Se-Met δ crystals was measured with synchrotron radiation at the Diamond Light Source (DLS) and extended to 2.1 and 3.3 Å, respectively. The native datasets were collected on beamline I-02, while the multiwavelength anomalous dispersion (MAD) data collection strategy was used with the Se-Met crystals, and data at three wavelengths were collected on beamline I-04.
The data from native and Se-Met crystals were processed using iMOSFLM (27) and analysed, scaled and merged using POINTLESS and SCALA in the CCP4 software suite (28) and shown to belong to space group P43212. The asymmetric unit was predicted to contain one copy of δ with a solvent content of 62%. An initial set of phases were obtained from the Se-Met data using SOLVE (29) followed by density modification using RESOLVE (30) in the PHENIX suite (31). The model building was carried out using BUCANEER (32). Further iterations of manual building using the COOT program (33) were followed by maximum likelihood refinement against the native data using phenix.refine in the PHENIX suite. Water molecules were added in the final refinement cycles. Model validity and quality were assessed using MOLPROBITY (34).
A summary of the relevant data statistics is shown in Table 1. Structure factors and coordinates have been deposited at the protein data bank (PDB) with the accession code 3zh9. All structure figures were generated using PyMOL (Schrödinger, LLC).
Table 1.
Data collection and refinement statistics
| Item | Native data | Se-Met derivative (peak) | Se-Met derivative (inflection) | Se-Met derivative (low-remote) |
|---|---|---|---|---|
| Beam line (DLS) | I0-2 | I0-4 | I0-4 | I0-4 |
| Wavelength (Å) | 0.97950 | 0.98020 | 0.98040 | 0.97630 |
| Space group | P43212 | P43212 | P43212 | P43212 |
| Unit cell (Å) | a = 81.6 Å, | a = 82.2 Å, | a = 82.5 Å, | a = 82.4 Å, |
| b = 81.6 Å, | b = 82.2 Å, | b = 82.5 Å, | b = 82.4 Å, | |
| c = 157.7 Å, | c = 157.099 Å, | c = 158.5 Å, | c = 158.2 Å, | |
| α = β = γ = 90° | α = β = γ = 90° | α = β = γ = 90° | α = β = γ = 90° | |
| Resolution range (Å) | 56.70–2.10 | 58.12–3.30 | 58.31–3.30 | 58.23–3.50 |
| No of measured reflections | 212 733 | 111 867 | 115 009 | 9686 |
| No of unique reflections | 31 753 | 8661 | 8796 | 7397 |
| Completeness (%)a | 99.6 (99.9) | 100 (100) | 100 (100) | 100 (100) |
| Rpima,b | 0.038 (0.294) | 0.069 (0.195) | 0.045 (0.209) | 0.1000 (0.210) |
| Mn (I/sd)a | 11.5 (2.7) | 8.0 (3.9) | 12.3 (4) | 6.3 (3.4) |
| Refinement | ||||
| R/Rfreec,d | 0.191/0.239 | |||
| Overall B-factor (Åc) | 52 | |||
| Composition of AU | 1 polypeptide chain (residues 1–339), 178 waters, missing residues 340–347 | |||
| Geometry | ||||
| RMSD in bond distances (Å) | 0.007 | |||
| RMSD in bond angles (°) | 0.99 | |||
| Ramachandan | ||||
| % most favored | 99 | |||
| % additionally allowed | 1 | |||
aData in parentheses correspond to the highest resolution shell.
bRpim = Σ[1/(N − 1)]1/2 Σ|Ii(hkl) − I(hkl) Σ Σ Ii(hkl).
cR-factor = Σ | Fobs − Fcalc | / Σ Fobs.
dRfree calculated as in c but using 5% of experimental data excluded from refinement for validation.
RESULTS
Assembly pathways and stoichiometry of the clamp-loader complex
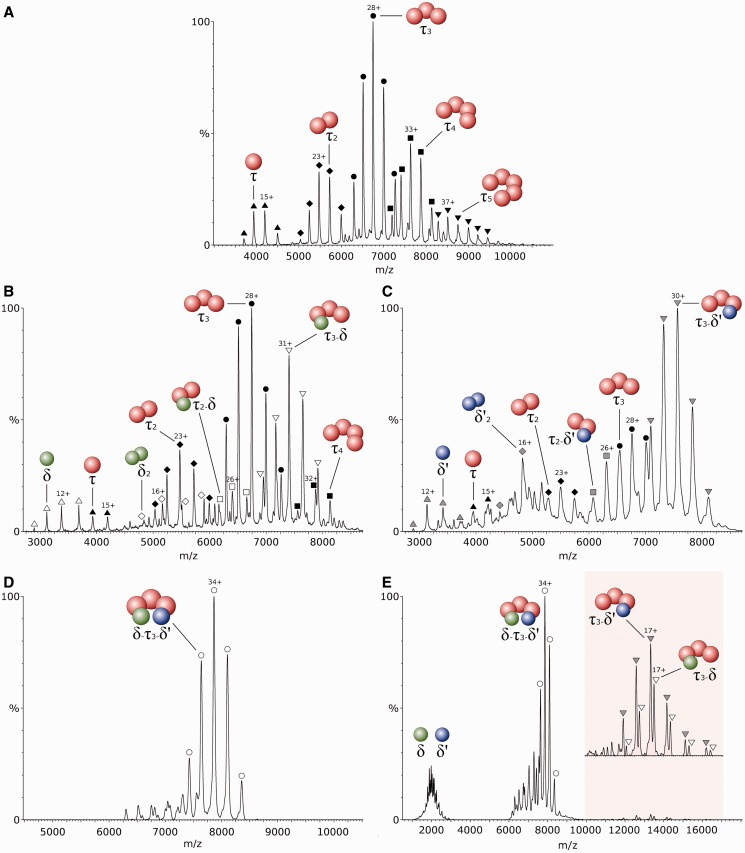
To identify the oligomeric state adopted by the τ subunit from B. subtilis in isolation, a mass spectrum of the purified protein was recorded. Five charge-state distributions were observed corresponding to five different oligomers (Figure 1A), with the most abundant having a measured mass of 188 712 ± 12 Da. This value is in close agreement with the calculated mass of the τ trimer, 188 288 Da. The measured mass of the other four charge state distributions were found to correspond to the monomeric, dimeric, tetrameric and pentameric forms of τ. This result suggests a solution equilibrium between these five species.
Figure 1.
Electrospray ionization-mass spectra of different combinations of the clamp-loader subunits from B. subtilis sprayed from 1 M ammonium acetate, pH7.5. The τ subunits are represented by red spheres, while the δ and δ′ subunits are represented by green and blue spheres, respectively. A, τ subunit in isolation; B, equimolar mixture of τ and δ subunits; C, equimolar mixture of τ and δ′ subunits; D, equimolar mixture of τ3, δ and δ′ subunits, resulting in the formation of the clamp-loader complex. E, The clamp-loader complex analysed under high collisional energy, resulting in a partial dissociation of δ and δ′ subunits (peaks corresponding to the low charged τ3-δ and τ3-δ′ after dissociation are magnified in the shaded region of the spectrum).
To obtain information on the assembly pathway of the clamp-loader in B. subtilis, δ and δ′ were cloned and expressed, as described in the Experimental Procedures, and separately mixed with τ to analyse the interactions between them. Several different complexes were observed on addition of equimolar δ to τ (Figure 1B). The masses of the two predominant species correspond to the τ trimer and a τ3-δ complex. In the low m/z region of the spectrum, another species was observed with a mass corresponding to the isolated δ. This result shows an incomplete binding of δ to τ. In the region around m/z 5700, two different species were resolved, corresponding to τ and δ dimers. Close examination of the spectrum also revealed a small population of τ2-δ ions, suggesting that δ may not bind exclusively to the τ trimer. Other small populations detected included a τ monomer and tetramer. When equimolar δ′ was added to τ, the spectrum showed a major species corresponding to τ3-δ′ (Figure 1C). A charge distribution for the τ trimer was also observed but its relative amount was considerably lower than that in Figure 1B. This result suggests a stronger interaction between δ′ and τ3 than was observed with δ. A small amount of δ′ monomer was also identified, and some other minor species present include a τ monomer and trimer, a δ′ dimer and a τ2-δ′ complex (Figure 1C). In both cases, complete saturation of τ was never achieved, suggesting an eventual equilibrium in solution between bound and unbound species.
The addition of equimolar concentrations of δ and δ′ to τ resulted in the formation a single major species with a molecular mass of 267 405 ± 53 Da, in close agreement with the calculated mass of 266 413 Da for the full τ3-δ-δ′ complex. This result allows us to propose with confidence the stoichiometry for the B. subtilis clamp-loader, as no other stoichiometries were observed (Figure 1D). It was clear that all the different subcomplexes obtained in both spectra from Figure 1A and B were converted to τ3-δ-δ′ and this is not consistent with the existence of one single mechanism for the assembly of this complex. Therefore, multiple pathways are likely responsible for the assembly of the B. subtilis clamp-loader, including the initial binding of δ to τ oligomers. This result clearly differs from the more specific mechanism for E. coli, where the assembly of this complex always involves the initial binding of δ′ to τ, promoting the later binding of δ, which is not capable of binding on its own (13). Under collisional activation, δ and δ′ were ejected from the complex as seen by two charge state distributions shown in the shaded region of the spectrum in Figure 1E, corresponding to the low-charged complex stripped of δ or δ′ subunits. The corresponding highly charged δ and δ′ subunits were observed in the low m/z region of the spectrum, around m/z 2000. In this technique, a protein complex ion is accelerated against neutral gas particles and its kinetic energy is partially converted into internal energy by the multiple collisions experienced. The accumulation of internal energy results in the disruption of the interactions between subunits and consequent dissociation, with peripheral subunits being more easily ejected. The distribution of charges on dissociation is usually asymmetric, with leaving subunits carrying a high number of charges (35). This result suggests an external location for these two subunits, making them more susceptible to collisional unfolding and dissociation. No τ subunits were dissociated from the complex.
Homology model of the B. subtilis clamp-loader complex
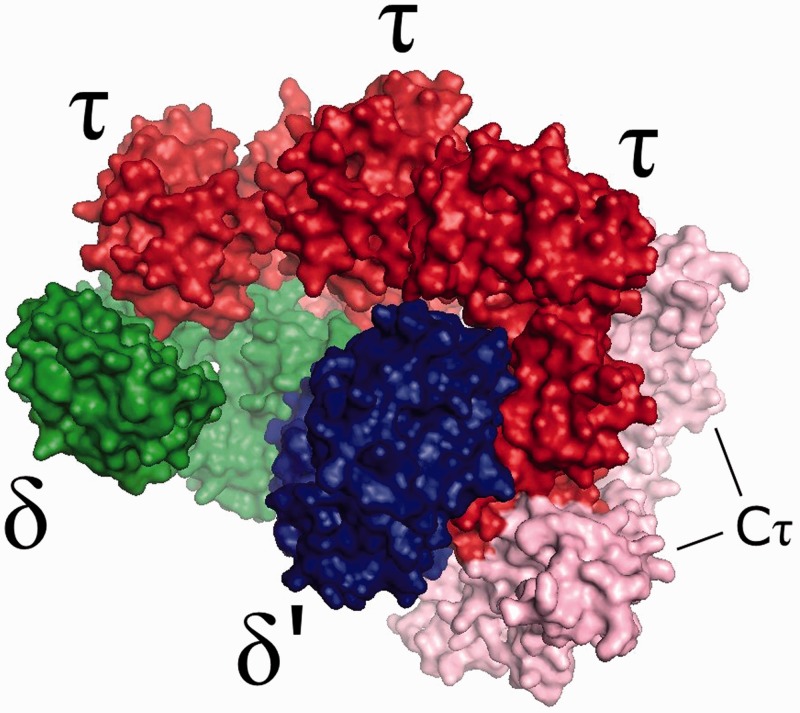
Using the structural information described above, together with homology modelling techniques, a model for the B. subtilis clamp-loader complex was produced. Homology models of the individual B. subtilis clamp-loader subunits were generated using the I-TASSER automated protein structure prediction server. The structural templates used to model each individual protein were automatically selected by the server through sequence alignment against a database of proteins of known structure, with the higher homology sequences being chosen. The obtained δ and δ′ models were principally based on their E. coli equivalents, resulting in similar tertiary structures. The τ region corresponding to E. coli γ was modelled using this protein as the main template, resulting in a similar structure. The Cτ domain of τ was modelled separately, and the obtained structure resulted from the contribution of several different protein structures with considerable sequence homology, including a partial solution structure of the E. coli Cτ domain and the structures of DnaA (a protein involved in the initiation of DNA replication) from three different organisms. The γ equivalent region and Cτ were also combined using I-TASSER. The assembly of the individual modelled subunits was made by structural alignment with their equivalents within the E. coli clamp-loader crystal structure, assuming a similar topology and shape for both complexes. The obtained model is depicted in Figure 2. ESI-ion mobility-MS measurements were conducted on the clamp-loader complex. However, the high degree of structural collapse seen in the gas phase rendered these results of limited structural value (see Supplementary Figure S3). Such behaviour of protein complexes with relatively open structures has been described previously (36).
Figure 2.
A homology model of the B. subtilis clamp-loader complex, with δ, δ′ and τ subunits represented in green, blue and red, respectively. The Cτ domain of the τ subunit is represented in pink.
Crystal structure of the B. subtilis δ subunit
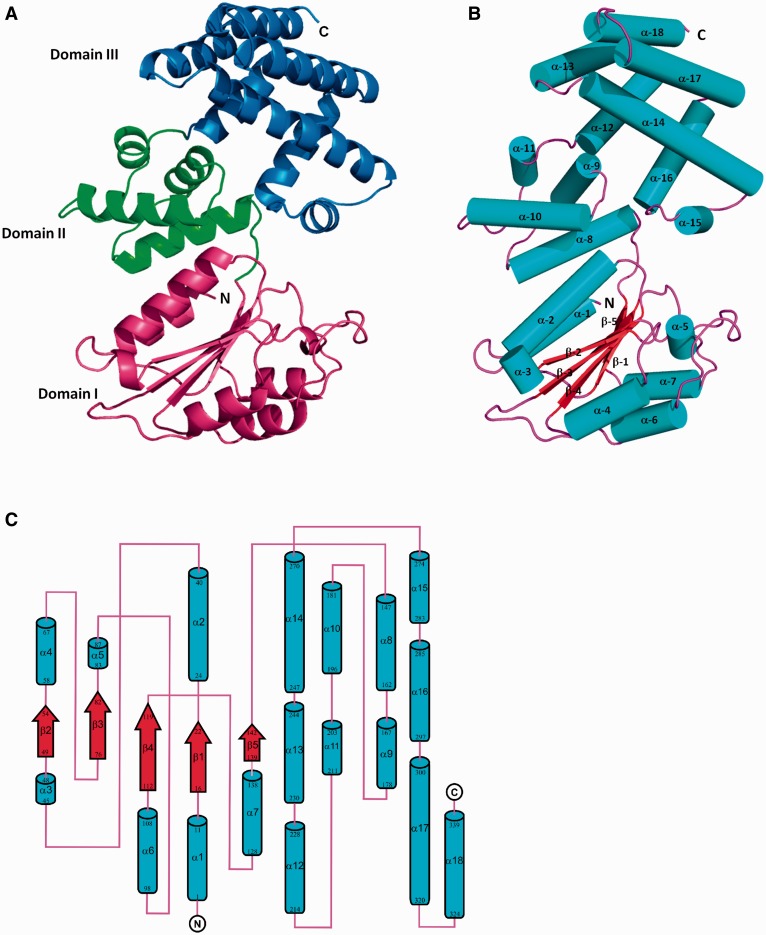
To assess the accuracy of the clamp-loader model described above, the crystal structure of δ (coded by the B. subtilis yqeN gene) was determined at 2.1 Å resolution. It revealed a protein composed of three domains: domain I (residues 1–143), domain II (residues 144–214) and domain III (residues 215–339). The overall structure and the topology diagram of δ are shown in Figure 3. The N-terminal domain I consists of a central five-stranded parallel β sheet (β1–β5) surrounded by seven α-helices (α1–α7). This domain has a RecA-like fold that resembles the core of the nucleotide binding domain of RecA (37). Domain II is composed of just four α-helices (α8–α11). Finally, the C-terminal domain III is also predominantly helical and consists of seven α-helices (α12–α18). Examination of the crystal packing of the protein confirms the monomeric state observed from gel filtration during protein purification, and the overall surface area of δ calculated by the PISA server (38) was ∼17 000 Å2.
Figure 3.
The crystal structure of B. subtilis δ (yqeN). (A) Cartoon representation of the overall structure of δ. Domain I (residues 1–143), Domain II (residues 144–214) and Domain III (residues 215–339) are coloured magenta, green and blue, respectively. (B) Cartoon representation of δ, with α-helices, β-strands and loops shown as cyan cylinders, red arrows and magenta lines, respectively. (C) Topology diagram of δ with individual elements and residues labelled for α-helices (cyan cylinders) and β-sheets (red arrows) with connecting loops (magenta lines). Produced using TOPDRAW from the CCP4 suite.
The structure of δ was submitted to the DALI server, and the best match was obtained to DNA polymerase III delta subunit (δ) from the clamp-loader complex of E. coli (PDB; 3glh chain F) (5). Sequence identity between the B. subtilis δ and E. coli δ is only ∼16%, and the main structural difference between them is that in E. coli δ there is the addition of one β-strand in domain I and an additional two-stranded β-sheet in domain II. Superimposition of B. subtilis δ onto E. coli δ gave an overall root mean square deviation (RMSD) of 4.6 Å for 312 Cα positions.
The crystal structure and homology model (vide infra) of the B. subtilis δ were found to be similar. Superimposition of the two structures resulted in an overall RMSD of 4.6 Å for 329 Cα positions (see Supplementary Figure S4 for an overlay).
ATP and Mg2+ drive the τ3-DnaB helicase interaction
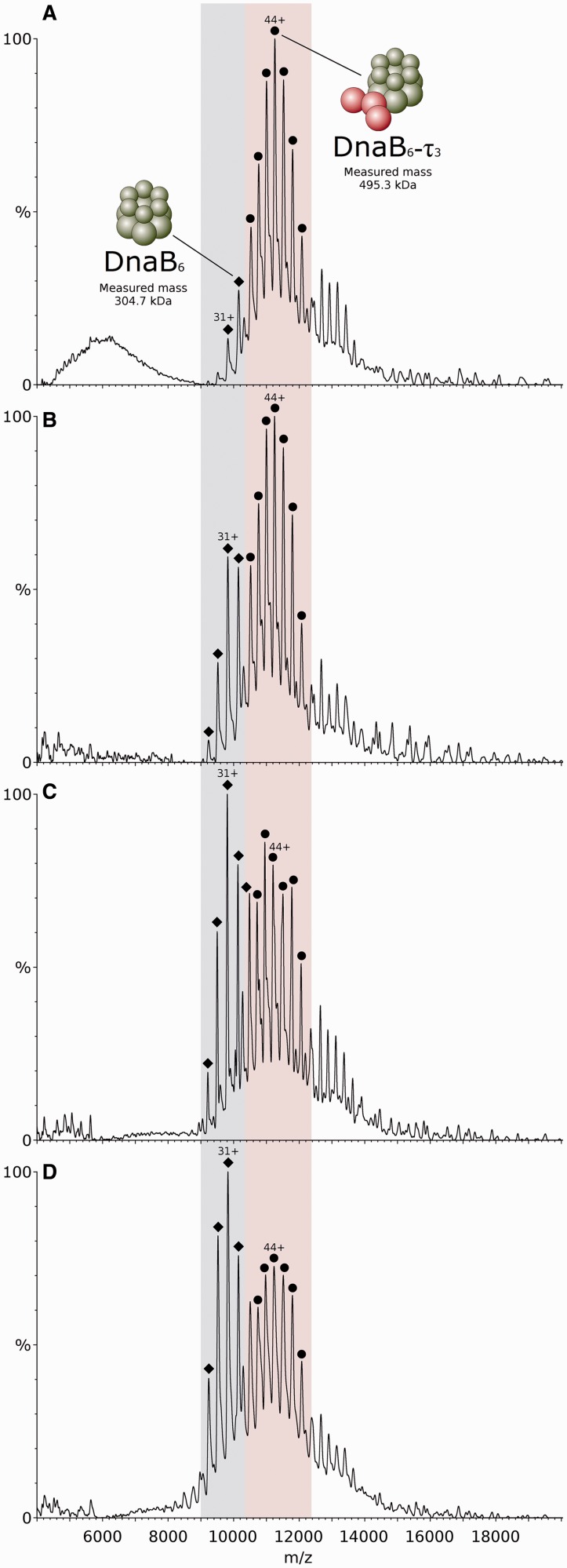
To study the interaction between τ and DnaB helicase, proteins were mixed in a τ3-DnaB6 stoichiometry before buffer exchange and analysis by mass spectrometry. When proteins were sprayed in the ESI-MS experiment using 1 M ammonium acetate buffer, pH 7.5, containing 0.5 mM Mg2+ and 0.1 mM ATP, one major species was observed, corresponding to the τ3-DnaB6 complex, with little unbound DnaB hexamer (Figure 4A). When only ATP was added to the buffer, the relative amount of unbound DnaB hexamer increased considerably, but was still less abundant than the τ3-DnaB6 complex (Figure 4B). Adding only Mg2+ (and no ATP) to the buffer resulted in a larger increase in the unbound DnaB signal, with it now becoming the major species, as seen in Figure 4C. The highest relative amount of unbound DnaB was observed when the proteins were sprayed from 1 M ammonium acetate only, as seen in Figure 4D, suggesting a stabilization of the interaction between τ3 and DnaB6 by both ATP and Mg2+.
Figure 4.
Mass spectra of the B. subtilis τ3 subunit interaction with the B. stearothermophilus DnaB helicase. Proteins were mixed in a τ3-DnaB6 stoichiometry and sprayed from different buffer compositions. Signal for the τ3-DnaB6 complex is located in the pink region of the spectra, while unbound DnaB6 is located in the blue region. (A) Proteins sprayed from 1 M ammonium acetate containing 0.5 mM Mg2+ and 0.1 mM ATP. The τ subunit is represented by red spheres, while the DnaB hexameric ring is represented in green. (B) Proteins sprayed from 1 M ammonium acetate containing 0.1 mM ATP. (C) Proteins sprayed from 1 M ammonium acetate containing 0.5 mM Mg2+. (D) Proteins sprayed from 1 M ammonium acetate only.
Both DnaB and the τ subunit of the clamp-loader complex possess ATPase activity. To establish whether ATPase activity of DnaB is required to promote its interaction with τ3, a catalytically inactive Thr217Ala mutant of the former was used. This mutant cannot coordinate the Mg2+ ion in the active site and hence lacks ATPase and helicase activities because it is unable to undergo functional conformational changes (39). Performing the binding study described above using Thr217Ala-DnaB and τ3 revealed that little or no complex formed, even on addition of ATP and Mg2+ (see Supplementary Figure S8). This result strongly suggests that ATP-driven conformational change of DnaB is required for an effective interaction with the τ3 core of the clamp-loader complex. In a separate experiment, designed to examine the ATP binding stoichiometry of the DnaB hexamer, this protein was exposed to 0.5 mM Mg2+ and 0.1 mM ATP, concentrations that would favour the formation of the τ3-DnaB6 complex. At this concentration of ATP, each DnaB hexamer peak was split in two (Supplementary Figure S9). A mass difference of 446 ± 20 Da was calculated after measurement of the distance between the two peaks. This difference is close to the mass of the mono-ammonium salt of ADP (444.20 Da), although the mass of ADP alone (427.20 Da) or the mass of ADP + Mg2+ (451.50 Da) also lie within the margin of error. The mass of ATP (507.18 Da), in contrast, is significantly different from the measured value, suggesting that this is not the species bound to the DnaB hexamer. This result is an indication that a single ATP→ADP binding and hydrolysis event is sufficient to promote the formation of the τ3-DnaB6 assembly.
DISCUSSION
The native mass spectrum of the τ subunit from B. subtilis in isolation revealed the existence of five different oligomers (monomer to pentamer), with the most abundant corresponding to the trimer. In a previous sedimentation equilibrium ultracentrifugation study, a mass of 309 kDa was obtained for τ (15). This value is higher than the tetramer (251 kDa) and lower than the pentamer (314 kDa), but is closer to the latter. This result appears inconsistent with the predominant trimer observed by ESI-MS. However, this type of discrepancy has been observed before, a good example being the small heat-shock protein Acr1 from Mycobacterium tuberculosis. This assembly has been determined to be composed of nine subunits by sedimentation equilibrium analytical ultracentrifugation and dynamic light scattering (40), while by ESI-MS, the only complex detected was composed of 12 subunits, in agreement with three-dimensional data from negative stain electron microscopy images (41). The reason for this difference may be related to a dependence of some techniques on the shape of the complex, resulting in apparently different stoichiometries. ESI-MS can assign the mass of a complex without any interference from its spatial arrangement (21). The ionization and MS detection occurs on a millisecond scale time frame, which is fast enough to detect all the existing oligomeric species in solution equilibrium. This is not possible to detect by most of the other available biophysical techniques. Here we report for the first time the presence of several different oligomeric species in solution for the τ subunit of a Gram-positive organism. To guard against any artefacts associated with MS measurement, care was taken to optimize instrumental conditions for survival of the τ complexes. It is unlikely, but not impossible, that weakly associated peripheral subunits may dissociate from a τ5 species to yield the τ3 core; however, no increase in the relative proportion of τ-pentamer to τ-trimer signals was observed on further reduction of MS voltages. The energetics associated with the desolvation events may also cause dissociation of weakly bound complexes, but, once again, no variation in the relative amount of the different oligomers was observed by changing the ESI parameters to minimize this possibility. These observations suggest that the obtained signal should be a close representation of the oligomeric states present in dilute solution.
Previous studies on the E. coli τ protein have also revealed heterogeneity in solution. Equilibrium sedimentation analysis showed an equilibrium between the τ tetramer and a free τ monomer (6), while a recent ESI-MS study revealed an equilibrium composed of four oligomeric species ranging from the monomer to the tetramer, with a higher relative amount of the latter (13). The biological significance of this heterogeneity of the τ subunit in isolation is not known. However, the difference observed in the predominant species for the two organisms (trimer for B. subtilis and tetramer for E. coli) may be an indication of different assembly pathways of the clamp-loader complex in different organisms as discussed below.
The δ and δ′ subunits were individually mixed with τ, resulting in the observation of several different bound and unbound species. In both cases, the binding was incomplete with free δ/δ′ and τ being observed, showing that saturation is not achieved. The δ and δ′ subunits were found to bind mostly the τ trimer, with binding to the dimer being observed in smaller amounts. In E. coli, it is now accepted that the major oligomeric state of the τ subunit in isolation is the tetramer. This tetramer must then be converted into a trimer on binding of δ and δ′. It has been demonstrated by ESI-MS that δ′ acts as an oligomer breaker, with smaller oligomers of τ being formed on δ′ binding, resulting in τ-δ′, τ2-δ′ and τ3-δ′ complexes, with no τ4-δ′ observed. In contrast, when δ is added to τ, no interaction is observed, not even with the τ trimer present in solution to form the subcomplex τ3-δ. These findings suggest that a conformational change promoted by δ′ may be essential for δ binding. Another possible explanation is the cooperative interaction between δ′ and τ, allowing δ to bind the complex (13). Here we report a different behaviour with the equivalent subunits from the B. subtilis clamp-loader. The most prominent difference is the binding of δ to τ without the presence of δ′. When δ is added to τ, several different species were observed, including τ2-δ and τ3-δ complexes, four different oligomers of τ ranging from monomer to tetramer and free δ in the monomeric and dimeric form, as seen in Figure 1B. In the analogous experiment, with δ′ instead of δ, a similar result was obtained. However, the relative amount of τ3-δ′ compared with τ3 was significantly higher than that observed for τ3-δ, and no τ4 was present (Figure 1C). The larger amount of τ3-δ′ suggests a higher affinity of δ′ for τ3 than that of δ, while the absence of τ4 is consistent with the oligomer breaker function of δ′ suggested in E. coli, which does not seem to occur with δ. These facts suggest that the clamp-loader complex can form by multiple assembly pathways in B. subtilis, in addition to the sequential mechanism described for E. coli, where δ′ breaks τ4 into smaller oligomers and binds them before δ. The addition of both δ and δ′ to τ resulted exclusively in the formation of the τ3-δ-δ′ complex. It was clear that all the different subcomplexes obtained in both spectra from Figure 1A and B were converted to τ3-δ-δ′. This is not consistent with the existence of one single mechanism for the assembly of this complex and provides further indirect evidence that multiple pathways are likely responsible for the assembly of the B. subtilis clamp-loader. Under collisional activation, only the δ and δ′ subunits were ejected from the complex, suggesting an external location of these subunits. Interestingly, the intensity of the signal for the low charge τ3-δ′ assembly resulting from dissociation is higher than the intensity for τ3-δ, supporting the previous observation that δ′ interacts more strongly with τ3 than δ, with this difference being maintained on transfer to the gas phase.
The τ3-δ-δ′ stoichiometry is reported here for a Gram-positive organism for the first time, after a previous study suggesting a τ4-δ-δ′ stoichiometry based on densitometry analysis (14). This new result is in agreement with the described stoichiometry in E. coli (6), but the assembly pathways of the two complexes are likely different as suggested by our data. The fact that the clamp-loader in both organisms is composed of the same subunits assembled with the same stoichiometry, albeit with different pathways, suggests a similar topology and shape. The E. coli clamp-loader is crescent-shaped with an internal core formed by the τ trimer. The other two subunits, δ and δ′, occupy an external location, only interacting with each other through the C-terminal domains (10,11). This peripheral location should make these subunits more susceptible to dissociation than the internal τ subunits, which would be consistent with our collisional activation data for the B. subtilis clamp-loader.
Using the structural information obtained by ESI-MS, together with homology modelling techniques, a model for the B. subtilis clamp-loader was produced. To assess the accuracy of this model, the crystal structure of δ was determined, revealing a protein composed of three domains, as shown in Figure 3. This structure was submitted to the DALI server, and the best match was obtained to its E. coli equivalent. Superimposition of B. subtilis δ onto E. coli δ gave an overall RMSD of 4.6 Å for 312 Cα positions. However, this RMSD value masks closer similarities at the individual domain level and reflects the relative movement of domain III to the other two domains in the two proteins. In the family of structures of the E. coli protein in various complexes, domain III can be seen to adopt a range of conformations including rotations of up to ∼90° relative to domains I and II, which are critical for biological function. The crystal structure and homology model of the B. subtilis δ were also found to be similar. Superimposition of the two structures resulted in an overall RMSD of 4.6 Å for 329 Cα positions.
The structure of δ in the δ:β-sliding clamp complex in E. coli (42) suggests that only domain I of δ is involved in the interactions with the β-sliding clamp. The β-interaction elements on δ are located in helix 4 and the loop that flanks it, where the key hydrophobic residues involved in the β-sliding clamp interaction in E. coli are Met-71, Leu-73 and Phe-74. The Leu-73 and Phe-74 protrude out to form a hydrophobic plug, which sits into a hydrophobic pocket of the β-sliding clamp. This region is equivalent to residues 57–72 in B. subtilis δ, where the corresponding residues are Phe-69, Phe-71 and Met-72 (Supplementary Figure S5).
Studies on the structure of the processivity clamp-loader γ complex of E. coli DNA polymerase III (10) revealed that δ interacts with the γ subunit 3 (γ3) via residues in α-8 (residues 177–192) of domain II with some contribution from domain I (Leu-29). Hydrophobic interactions at the interface are located in two places. The first region consists of Leu-190 and Leu-191 from domain II of δ protein and Phe-173 and Ala-27 from γ3, and the second region involves Leu-29 and Leu-179 from δ protein and Val-164 and Leu-167 from γ3. The equivalent residues in B. subtilis δ are Tyr-26, Ser-183, Thr-194 and Phe-195 and are strikingly dissimilar, being largely hydrophilic. This implies that the interface could have distinctly different properties. Indeed, examination of our homology model reveals that the τ subunit (equivalent to γ3) of the B. subtilis clamp-loader may use the hydrophilic amino acid residue Ser-168 to interact with Tyr-26 and Ser-183 of δ, and Thr-27 to interact with Thr-194 of δ (Supplementary Figure S6). In addition, a number of hydrophobic residues remain conserved. The absence of a high-resolution structure for the complex in B. subtilis means that the possibility of an altered local conformation in δ on complex formation cannot be discounted.
The E. coli clamp-loader complex (10) also revealed that domain III of δ is important for the interactions between δ and δ′ as well as between δ and γ3. The C-terminal domain of δ is bound between the corresponding domains of γ3 and δ′, closing the circle formed by the clamp-loader subunits. The interface formed by δ with δ′ protein in E. coli involves helix α-12 and helix α-14 of δ and is composed of a high number of hydrophobic residues. It also contains six hydrogen bonds. Of the 14 residues in E. coli δ that interact with δ′, only Arg-299, Leu-305 and Lys-313 are completely conserved between it and B. subtilis δ (Arg-305, Leu-311 and Lys-319) (Supplementary Figure S5). Despite this low level of sequence identity, there is some conservation of residue type in this region. Thus, it is possible that this region of δ from B. subtilis might also interact with δ′ in B. subtilis in a similar manner to δ with δ′ in E. coli. Examination of the homology model for the interface between δ and δ′ in B. subtilis reveals reasonable coincidence of hydrophobic residues on the two subunits, as well as alignment of pairs of potential hydrogen bond-forming side chains (Supplementary Figure S7). The interface formed by domain III of δ with γ3 protein in E. coli involves Glu-325, His-333, Lys-334 and Asp-338 from helix α-15 of δ and helix α-12 of γ3. Of these residues, only the glutamic acid is identical in B. subtilis δ (Glu-330) but the others are conserved in type (Supplementary Figure S6). More recent investigation of E. coli δ suggests that Trp-279 facilitates clamp-loader complex interaction with primer-template DNA via base stacking at the junction point of single- and double-strand DNA. However, in B. subtilis δ, the equivalent residue is His-285 and is unlikely to form equivalent interactions (Supplementary Figure S5). Thus B. subtilis δ might have a somewhat different mode of DNA binding.
To study the interaction between τ and DnaB helicase, proteins were mixed in a τ3-DnaB6 stoichiometry. The presence of 0.5 mM Mg2+ and 0.1 mM ATP in the spraying buffer was essential for the effective formation of the τ3-DnaB6 complex, showing that ATP binding to either or both proteins in the presence of Mg2+ promotes the interaction. Both DnaB and the τ subunit of the clamp-loader complex possess ATPase activity. In the case of the DnaB helicase, this fuels conformational changes in the hexameric ring, which drive unidirectional translocation along the lagging strand and separation of the parental DNA strands during DNA replication. In contrast, ATP binding and hydrolysis by the τ subunit is thought to drive conformational changes essential for loading of the β-clamp onto a primer-template junction (12). To establish whether ATPase activity of DnaB is required to promote its interaction with τ3, a catalytically inactive Thr217Ala mutant of the former was used, resulting in little or no complex formed even in the presence of ATP and Mg2+. This result strongly suggests that ATP-driven conformational change of DnaB is necessary for an effective interaction with the τ3 core of the clamp-loader. To analyse the binding of ATP to the DnaB ring, this protein was sprayed from a buffer containing Mg2+ and ATP at the same concentrations that led to the near complete formation of the τ3-DnaB6 assembly. Each peak corresponding to the DnaB hexamer was split in two similar peaks corresponding to the unbound and singly bound forms of the complex. The measured distance between the peaks corresponds to a mass difference of 446 ± 20 Da, suggesting a single ADP binding event. These findings indicate that ATP is immediately hydrolysed on binding to the DnaB ring and that binding and hydrolysis of one single ATP molecule in one of the six available ATP-binding sites is enough to promote the conformational change in the ring, necessary for the more effective binding to τ3. Although different ATP binding and hydrolysis models have been proposed based on different crystal structures of ring helicases (simple sequential for T7gp4, blocked sequential for E1 and Rho and concerted for SV40 LTag), in all of the proposed mechanisms, at least one of the six available active sites around the ring will be bound by an ATP at any time during the catalytic cycle, hence, promoting a stable interaction with the clamp-loader. Quantitative complex formation between τ3 and DnaB6 is only achieved when Mg2+ is present in the buffer. DnaB helicase belongs to the RecA-like family of ATPases whose highly conserved core contains the ATP-binding site. When ATP binds to this site, a network of hydrogen bonds/salt bridges is formed around ATP, with a Mg2+ ion also being involved (43). The stability of ATP binding and consequent increase in the conformationally altered DnaB population should then be enhanced by the presence of Mg2+ in solution. This fact together with our data suggests that ATP binding to DnaB in the presence of Mg2+ is an important event in the formation of an effective and stable interaction between the clamp-loader and the primosome, essential for the performance of the whole replisome during DNA replication. The primosome is especially prominent during lagging strand replication synthesizing repeated RNA primers, which are extended by DNA polymerase to form the Okazaki fragments. The ATP/Mg2+ driven helicase-clamp-loader interaction may facilitate the repeated loading of the β-clamp by the clamp-loader on the primed lagging strand template during DNA synthesis. We note that the mechanism for the helicase-clamp-loader interaction may differ in its requirements for ATP binding stoichiometry and hydrolysis from that of helicase action.
ACCESSION NUMBERS
3zh9.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Table 1, Supplementary Figures 1–9, Supplementary Methods and Supplementary References [44–49].
FUNDING
Research in the PS lab is supported by the Wellcome Trust [091968/Z/10/Z]. Research in the JBR lab was supported by the BBSRC [BB/E017576/1]; a studentship to CS from the Royal Thai Government. Funding for open access charge: University of Nottingham.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors are grateful to Dr Avinash Kale for helpful comments, and to the Wellcome Trust, the Biotechnology and Biological Sciences Research Council (BBSRC), and the University of Nottingham for funding.
REFERENCES
- 1.Johnson A, O’Donnell M. Cellular DNA replicases: components and dynamics at the replication fork. Annu. Rev. Biochem. 2005;74:283–315. doi: 10.1146/annurev.biochem.73.011303.073859. [DOI] [PubMed] [Google Scholar]
- 2.Kong XP, Onrust R, O’Donnell M, Kuriyan J. Three-dimensional structure of the beta subunit of E. coli DNA polymerase III holoenzyme: a sliding DNA clamp. Cell. 1992;69:425–437. doi: 10.1016/0092-8674(92)90445-i. [DOI] [PubMed] [Google Scholar]
- 3.Argiriadi MA, Goedken ER, Bruck I, O’Donnell M, Kuriyan J. Crystal structure of a DNA polymerase sliding clamp from a Gram-positive bacterium. BMC Struct. Biol. 2006;6:2. doi: 10.1186/1472-6807-6-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Indiani C, O’Donnell M. The replication clamp-loading machine at work in the three domains of life. Nat. Rev. Mol. Cell Biol. 2006;7:751–761. doi: 10.1038/nrm2022. [DOI] [PubMed] [Google Scholar]
- 5.Kelch B, Makino D, O’Donnell M, Kuriyan J. Clamp loader ATPases and the evolution of DNA replication machinery. BMC Biol. 2012;10:34. doi: 10.1186/1741-7007-10-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pritchard AE, Dallmann HG, Glover BP, McHenry CS. A novel assembly mechanism for the DNA polymerase III holoenzyme DnaX complex: association of deltadelta’ with DnaX(4) forms DnaX(3)deltadelta’. EMBO J. 2000;19:6536–6545. doi: 10.1093/emboj/19.23.6536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim S, Dallmann HG, McHenry CS, Marians KJ. Coupling of a replicative polymerase and helicase: a tau-DnaB interaction mediates rapid replication fork movement. Cell. 1996;84:643–650. doi: 10.1016/s0092-8674(00)81039-9. [DOI] [PubMed] [Google Scholar]
- 8.Dallmann HG, Kim S, Pritchard AE, Marians KJ, McHenry CS. Characterization of the unique C terminus of the Escherichia coli tau DnaX protein. Monomeric C-tau binds alpha and DnaB and can partially replace tau in reconstituted replication forks. J. Biol. Chem. 2000;275:15512–15519. doi: 10.1074/jbc.M909257199. [DOI] [PubMed] [Google Scholar]
- 9.Gao D, McHenry CS. tau binds and organizes Escherichia coli replication through distinct domains. Partial proteolysis of terminally tagged tau to determine candidate domains and to assign domain V as the alpha binding domain. J. Biol. Chem. 2001;276:4433–4440. doi: 10.1074/jbc.M009828200. [DOI] [PubMed] [Google Scholar]
- 10.Jeruzalmi D, O’Donnell M, Kuriyan J. Crystal structure of the processivity clamp loader gamma (gamma) complex of E. coli DNA polymerase III. Cell. 2001;106:429–441. doi: 10.1016/s0092-8674(01)00463-9. [DOI] [PubMed] [Google Scholar]
- 11.Kazmirski SL, Podobnik M, Weitze TF, O’Donnell M, Kuriyan J. Structural analysis of the inactive state of the Escherichia coli DNA polymerase clamp-loader complex. Proc. Natl Acad. Sci. USA. 2004;101:16750–16755. doi: 10.1073/pnas.0407904101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Simonetta KR, Kazmirski SL, Goedken ER, Cantor AJ, Kelch BA, McNally R, Seyedin SN, Makino DL, O’Donnell M, Kuriyan J. The mechanism of ATP-dependent primer-template recognition by a clamp loader complex. Cell. 2009;137:659–671. doi: 10.1016/j.cell.2009.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Park AY, Jergic S, Politis A, Ruotolo BT, Hirshberg D, Jessop LL, Beck JL, Barsky D, O’Donnell M, Dixon NE, et al. A single subunit directs the assembly of the Escherichia coli DNA sliding clamp loader. Structure. 2010;18:285–292. doi: 10.1016/j.str.2010.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bruck I, O’Donnell M. The DNA replication machine of a gram-positive organism. J. Biol. Chem. 2000;275:28971–28983. doi: 10.1074/jbc.M003565200. [DOI] [PubMed] [Google Scholar]
- 15.Haroniti A, Till R, Smith MCM, Soultanas P. Clamp-loader-helicase interaction in Bacillus. Leucine 381 is critical for pentamerization and helicase binding of the Bacillus tau protein. Biochemistry. 2003;42:10955–10964. doi: 10.1021/bi034955g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haroniti A, Anderson C, Doddridge Z, Gardiner L, Roberts CJ, Allen S, Soultanas P. The clamp-loader-helicase interaction in Bacillus. Atomic force microscopy reveals the structural organisation of the DnaB-tau complex in Bacillus. J. Mol. Biol. 2004;336:381–393. doi: 10.1016/j.jmb.2003.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chintakayala K, Machón C, Haroniti A, Larson MA, Hinrichs SH, Griep MA, Soultanas P. Allosteric regulation of the primase (DnaG) activity by the clamp-loader (tau) in vitro. Mol. Microbiol. 2009;72:537–549. doi: 10.1111/j.1365-2958.2009.06668.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Loo JA. Studying noncovalent protein complexes by electrospray ionization mass spectrometry. Mass Spectrom. Rev. 1997;16:1–23. doi: 10.1002/(SICI)1098-2787(1997)16:1<1::AID-MAS1>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 19.Benesch JLP, Ruotolo BT, Simmons DA, Robinson CV. Protein complexes in the gas phase: technology for structural genomics and proteomics. Chem. Rev. 2007;107:3544–3567. doi: 10.1021/cr068289b. [DOI] [PubMed] [Google Scholar]
- 20.van den Heuvel RH, Heck AJR. Native protein mass spectrometry: from intact oligomers to functional machineries. Curr. Opin. Chem. Biol. 2004;8:519–526. doi: 10.1016/j.cbpa.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 21.Sharon M, Robinson CV. The role of mass Spectrometry in structure elucidation of dynamic protein complexes. Annu. Rev. Biochem. 2007;76:167–193. doi: 10.1146/annurev.biochem.76.061005.090816. [DOI] [PubMed] [Google Scholar]
- 22.Hernandez H, Robinson CV. Determining the stoichiometry and interactions of macromolecular assemblies from mass spectrometry. Nat. Protoc. 2007;2:715–726. doi: 10.1038/nprot.2007.73. [DOI] [PubMed] [Google Scholar]
- 23.Bird LE, Pan H, Soultanas P, Wigley DB. Mapping protein-protein interactions within a stable complex of DNA primase and DnaB helicase from Bacillus stearothermophilus. Biochemistry. 2000;39:171–182. doi: 10.1021/bi9918801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bird LE, Wigley DB. The Bacillus stearothermophilus replicative helicase: cloning, overexpression and activity. Biochim. Biophys. Acta. 1999;1444:424–428. doi: 10.1016/s0167-4781(99)00024-x. [DOI] [PubMed] [Google Scholar]
- 25.Gill S, Vonhippel P. Calculation of protein extinction coefficients from amino-acid sequence data. Anal. Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- 26.Roy A, Kucukural A, Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat. Protoc. 2010;5:725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leslie AGW, Powell HR. Processing diffraction data with Mosflm. In: Reid RJ, Sussman JL, editors. Evolving Methods for Macromolecular Crystallography. The Netherlands: Springer, Dordrecht; 2007. pp. 41–51. [Google Scholar]
- 28.Potterton E, Briggs P, Turkenburg M, Dodson E. A graphical user interface to the CCP4 program suite. Acta Crystallogr. D Biol. Crystallogr. 2003;59:1131–1137. doi: 10.1107/s0907444903008126. [DOI] [PubMed] [Google Scholar]
- 29.Terwilliger TC, Berendzen J. Automated MAD and MIR structure solution. Acta Crystallogr. D Biol. Crystallogr. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Terwilliger TC. Maximum-likelihood density modification. Acta Crystallogr. D Biol. Crystallogr. 2000;56:965–972. doi: 10.1107/S0907444900005072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung L-W, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cowtan K. The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr. D Biol. Crystallogr. 2006;62:1002–1011. doi: 10.1107/S0907444906022116. [DOI] [PubMed] [Google Scholar]
- 33.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 34.Chen VB, Arendall WB, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 2009;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Benesch JLP, Aquilina JA, Ruotolo BT, Sobott F, Robinson CV. Tandem mass spectrometry reveals the quaternary organization of macromolecular assemblies. Chem. Biol. 2006;13:597–605. doi: 10.1016/j.chembiol.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 36.Hogan CJ, Ruotolo BT, Robinson CV, de la Mora JF. Tandem differential mobility analysis-mass spectrometry reveals partial gas-phase collapse of the GroEL complex. J. Phys. Chem. B. 2011;115:3614–3621. doi: 10.1021/jp109172k. [DOI] [PubMed] [Google Scholar]
- 37.Story RM, Weber IT, Steitz TA. The structure of the E. coli recA protein monomer and polymer. Nature. 1992;355:318–325. doi: 10.1038/355318a0. [DOI] [PubMed] [Google Scholar]
- 38.Doublié S. Preparation of selenomethionyl proteins for phase determination. Methods Enzymol. 1997;276:523–530. [PubMed] [Google Scholar]
- 39.Soultanas P, Wigley DB. Site-directed mutagenesis reveals roles for conserved amino acid residues in the hexameric DNA helicase DnaB from Bacillus stearothermophilus. Nucleic Acids Res. 2002;30:4051–4060. doi: 10.1093/nar/gkf527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chang ZY, Primm TP, Jakana J, Lee IH, Serysheva I, Chiu W, Gilbert HF, Quiocho FA. Mycobacterium tuberculosis 16-kDa antigen (Hsp16.3) functions as an oligomeric structure in vitro to suppress thermal aggregation. J. Biol. Chem. 1996;271:7218–7223. [PubMed] [Google Scholar]
- 41.Kennaway CK, Benesch JLP, Gohlke U, Wang L, Robinson CV, Orlova EV, Saibi HR, Keep NH. Dodecameric structure of the small heat shock protein Acr1 from Mycobacterium tuberculosis. J. Biol. Chem. 2005;280:33419–33425. doi: 10.1074/jbc.M504263200. [DOI] [PubMed] [Google Scholar]
- 42.Jeruzalmi D, Yurieva O, Zhao Y, Young M, Stewart J, Hingorani M, O’Donnell M, Kuriyan J. Mechanism of processivity clamp opening by the delta subunit wrench of the clamp loader complex of E. coli DNA polymerase III. Cell. 2001;106:417–428. [PubMed] [Google Scholar]
- 43.Liao J-C. Mechanical transduction mechanisms of RecA-Like molecular motors. J. Biomol. Struct. Dyn. 2011;29:497–507. doi: 10.1080/07391102.2011.10507401. [DOI] [PubMed] [Google Scholar]
- 44.Bush MF, Hall Z, Giles K, Hoyes J, Robinson CV, Ruotolo BT. Collision cross sections of proteins and their complexes: a calibration framework and database for gas-phase structural biology. Anal. Chem. 2010;82:9557–9565. doi: 10.1021/ac1022953. [DOI] [PubMed] [Google Scholar]
- 45.Ruotolo BT, Benesch JLP, Sandercock AM, Hyung S-J, Robinson CV. Ion mobility-mass spectrometry analysis of large protein complexes. Nat. Protoc. 2008;3:1139–1152. doi: 10.1038/nprot.2008.78. [DOI] [PubMed] [Google Scholar]
- 46.Bush MF, Hall Z, Politis A, Barsky D, Robinson CV. 59th Annual Conference American Society Mass Spectrometry. 2011;Vol. 2 Denver, CO, WOB, p. 50. [Google Scholar]
- 47.Bertini I, Case DA, Ferella L, Giachetti A, Rosato A. A Grid-enabled web portal for NMR structure refinement with AMBER. Bioinformatics. 2011;27:2384–2390. doi: 10.1093/bioinformatics/btr415. [DOI] [PubMed] [Google Scholar]
- 48.Ritchie DW, Venkatraman V. Ultra-fast FFT protein docking on graphics processors. Bioinformatics. 2010;26:2398–2405. doi: 10.1093/bioinformatics/btq444. [DOI] [PubMed] [Google Scholar]
- 49.Hall Z, Politis A, Bush MF, Smith LJ, Robinson CV. Charge-state dependent compaction and dissociation of protein complexes – insights from ion mobility and molecular dynamics. J. Am. Chem. Soc. 2012;134:3429–3438. doi: 10.1021/ja2096859. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.