Abstract
As a well-known phenomenon, total mRNAs poorly correlate to proteins in their abundances as reported. Recent findings calculated with bivariate models suggested even poorer such correlation, whereas focusing on the translating mRNAs (ribosome nascent-chain complex-bound mRNAs, RNC-mRNAs) subset. In this study, we analysed the relative abundances of mRNAs, RNC-mRNAs and proteins on genome-wide scale, comparing human lung cancer A549 and H1299 cells with normal human bronchial epithelial (HBE) cells, respectively. As discovered, a strong correlation between RNC-mRNAs and proteins in their relative abundances could be established through a multivariate linear model by integrating the mRNA length as a key factor. The R2 reached 0.94 and 0.97 in A549 versus HBE and H1299 versus HBE comparisons, respectively. This correlation highlighted that the mRNA length significantly contributes to the translational modulation, especially to the translational initiation, favoured by its correlation with the mRNA translation ratio (TR) as observed. We found TR is highly phenotype specific, which was substantiated by both pathway analysis and biased TRs of the splice variants of BDP1 gene, which is a key transcription factor of transfer RNAs. These findings revealed, for the first time, the intrinsic and genome-wide translation modulations at translatomic level in human cells at steady-state, which are tightly correlated to the protein abundance and functionally relevant to cellular phenotypes.
INTRODUCTION
As a major component of central dogma, the ribosome is a node in the flow of genetic information, adapting both the input of mRNA and the output of protein. With the recognition of poor correlations between the abundances of mRNAs and proteins in various species, with R2 ranging from ∼0.01 to 0.50 (1–8) [reviewed in (9)], it has been speculated for years that the amount of translating mRNAs (mRNAs bound to ribosome-nascent chain complex, RNC-mRNA) may better reflect protein abundances (10,11). However, this seemed to be uncertain at least in recent studies regarding yeasts (8), HEK293 cells (12) and tumour cells (13), with R2 < 0.37. These findings indicated a widespread and diversified translational modulation that may occur in all of the three stages of translation, namely, initiation, elongation and termination, as well as the spatial organization of mRNAs [reviewed in (14)]. However, studies on the translational kinetics revealed that the major influential factor of this modulation is translational initiation in general (15), which determines the fraction of mRNA molecules that are subjected to translation (16).
As a very upstream step of functional protein production, the translation offers a rapid and specific response to the environmental and physiological changes. Thus, transient alterations of genome-wide translational states were frequently observed in non-steady investigative systems, such as cell differentiation, T-cell activation and stress response [reviewed in (10,17)]. For example, the translation initiation of Saccharomyces cerevisiae is to be globally repressed in a few minutes after being shifted to a non-fermentable carbon source, and this is independent from the mRNA level (18). When exposed to other stresses including heat, acid, alkali, oxidation and salt, this yeast exhibits stress-specific patterns of translational control on gene expression for survival (19). Regarding systems at steady-state, such as drosophila, transcripts with the highest abundance are usually not those in ribosomes with the most abundances (20). This suggests that the translation initiation is an important factor to decouple the poor correlation of transcription and protein abundances, which is also relevant to other known factors, such as protein degradation [reviewed in (21)] and elongation rate [reviewed in (14)]. When comparing relative abundances between different cell populations at steady-state, the role of translation control can be more easily observed. For example, a cancer cell study, focusing on the ribosome-bound mRNA changes in a transforming growth factor beta-induced epithelial-mesenchymal transition (EMT) model, showed that differentially translated genes are phenotype relevant (22). These findings lead us to hypothesize that the relative abundances of translating mRNAs and proteins are strongly correlated in cells at steady states, when considering multivariate factors. Addressing this question is fundamentally significant for the characterization of various biosystems by quantitatively bridging the gap between transcriptome and proteome with translatome in the flow of genetic information.
Building this quantitative connection is an intriguing step forward to allow strategic advancement. In current human protein knowledge bases, there are numerous transcripts with no protein evidence, largely due to the extreme low abundances or biophysical properties of proteins for identification, incomplete or wrong annotations of genes, amino acid sequence variations, and the ubiquitous existence of non-translational mRNAs in the transcriptome (23–28). Other than these aspects, major obstacles exist in functional proteomics investigations regarding the data integration, relevant to both biological and technical variations in different laboratories (28,29). In contrast, deep sequencing on translating mRNAs is independent from these protein properties, conferring both translatomic annotation and potentially computational prediction on protein abundances to overcome these hindrances. To this end, a tight correlation of RNC-mRNAs versus proteins will emphasize the biological relevance of independently using nucleic acid information of translating mRNAs to investigate cellular functionalities and phenotypes.
Equally important is that a novel insight of global translation modulation can be achieved by addressing our hypothesis. The information of mRNA and RNC-mRNA abundances enables systematic evaluation of mRNA translation ratios (TR, defined as the abundance ratio of the translating mRNA to total mRNA regarding a certain gene), thus offering a new way to quantify the selection of mRNA molecules that are subjected to translation on genome-wide scale. Furthermore, the TR alterations of alternative spliced transcripts (ASTs) from mRNA to protein levels can be investigated precisely, demanded in current gene-centric studies (28,29).
Therefore, we analysed the mRNAs, RNC-mRNAs and proteome of lung cancer A549 and H1299 cells in comparison with normal human bronchial epithelial (HBE) cells, respectively. We discovered a novel multivariate linear correlation between translating mRNAs and proteins on genome-wide scale, by integrating the mRNA length as a key factor. We found the mRNA length is an important contributor in translation initiation and TR alterations, which are highly relevant to cancerous phenotypes.
MATERIALS AND METHODS
Cell lines
Human A549, H1299 and HBE cells were acquired from American Type Culture Collections (ATCC, Rockville, MD). Cells were maintained in Dulbecco’s modified Eagle’s medium (Invitrogen, Carlsbad, CA), supplemented with 10% fetal bovine serum (PAA Australia, Weike Biochemical Reagent, Shanghai, China), 1% penicillin/streptomycin and 10 μg/mL ciprofloxacin.
Ribosome-nascent chain complex extraction
The RNC extraction was performed as described by Esposito et al. (30) with certain modifications. In brief, cells were pre-treated with 100 μg/ml cycloheximide for 15 min, followed by pre-chilled phosphate buffered saline washes and addition of 2 ml cell lysis buffer [1% Triton X-100 in ribosome buffer (RB buffer) [20 mM HEPES-KOH (pH 7.4), 15 mM MgCl2, 200 mM KCl, 100 μg/ml cycloheximide and 2 mM dithiothreitol]. After 30-min ice-bath, cell lysates were scraped and transferred to pre-chilled 1.5 ml tubes. Cell debris was removed by centrifuging at 16 200 × g for 10 min at 4°C. Supernatants were transferred on the surface of 20 ml of sucrose buffer (30% sucrose in RB buffer). RNCs were pelleted after ultra-centrifugation at 185 000 × g for 5 h at 4°C.
RNA extraction
Total RNA and RNC-RNA were respectively isolated by using TRIzol® RNA extraction reagent (Ambion, Austin, TX), following the manufacturer’s instructions. Both total RNA and RNC-RNA samples were prepared from three independent experiments. Equal amount of total RNA or RNC-RNA from each preparation was pooled, respectively, for subsequent library construction and RNA-seq.
RNA-seq
The sequencing libraries were constructed following the TruSeq™ RNA Sample Preparation Guide (Illumina, San Diego, CA). Briefly, the polyA+ mRNA in the total mRNA or RNC-mRNA samples was isolated using the RNA Purification Beads (Illumina). The mRNA was fragmented by incubation in Elute-Prime-Fragment Mix at 94°C for 8 min to obtain 120–200 bp inserts. First-strand cDNA was synthesized with SuperScript II Reverse Transcriptase (Invitrogen) using random primer, and Ampure XP beads (Beckman Coulter, Beijing, China) were used to isolate double-stranded cDNA synthesized by Second Strand Master Mix. The adapters were ligated to the A-Tailing fragment, and 12 cycles of PCR were performed to enrich those DNA fragments that have adapter molecules on both ends and to amplify the amount of DNA in the library. Purified libraries were quantified by Qubit® 2.0 Fluorometer (Invitrogen) and validated by Agilent 2100 bioanalyzer (Agilent, Beijing, China). Clusters were generated by cBot with the library diluted to 10 pM and then were sequenced on the Illumina Genome Analyzer IIx for 75 cycles or HiSeq-2000 for 100 cycles (Illumina). Library construction and Illumina sequencing were performed at Shanghai Biotechnology Corporation. High quality reads that passed the Illumina quality filters were kept for the sequence analysis (Supplementary Table S1). The sequencing data sets are available at http://bioinformatics.jnu.edu.cn/software/sequencing_datasets/ and Gene Expression Omnibus database (access number GSE42006).
Sequence analysis
High quality reads were mapped to human mRNA reference sequence (RefSeq) for GRCh37/hg19 in UCSC genome browser (downloaded from http://hgdownload.cse.ucsc.edu/downloads, accessed on August 2nd, 2012) using FANSe v. 7.2 mapping algorithm (31) with the options –L78 –S8 –I0 –E9 –B1. The reads mapped to splice variants of one gene were summed. The mRNA abundance was normalized using both rpkM (reads per kilobase per million reads) (32) and edgeR package (33) methods. Genes with >10 mapped reads were considered as quantified genes (34). The TR of a gene g is calculated as:
BDP1 splice variants were detected and quantified from the deep sequencing data sets exactly using the method as we previously reported (35).
Stable isotope labelling with amino acids in cell culture
Cells were labeled with a stable isotope labelling with amino acids in cell culture (SILAC) quantitation kit (Pierce Biotechnology, Rockford, IL) as previously described (36). In brief, HBE cells were labelled by light lysine (12C6) containing media, whereas A549 and H1299 cells by heavy lysine (13C6) containing media. Cells were cultured in their respective media for at least six passages to allow maximum lysine incorporation. Equal amounts of protein from light and heavy lysine-labelled cell lines were mixed and separated by SDS–PAGE. Gel bands were excised and subjected to in-gel trypsin digestion as previously described (36).
Mass spectrometry
Peptides were analysed by a Finnigan Surveyor HPLC system coupled with LTQ-Orbitrap mass spectrometer (Thermo Electron, Beijing, China) as previously described with minor modifications (36). In brief, the peptides were loaded in a C18 reverse-phase column, followed by a 0–40% gradient wash with acetonitrile buffer over 90 min. The eluent was real-time analysed by the LTQ-Orbitrap under data-dependent mode with capillary temperature of 200°C and spray voltage of 1.80 kV. Mass range of 400–1800 m/z was scanned in the Orbitrap at resolution r = 60 000 at m/z 400, followed by 10 mass spectrometry (MS)2 scans for each MS in the LTQ with Dynamic Exclusion setting: a repeat count of 2, a repeat duration of 30 s and an exclusion duration of 90 s. Database searching and protein quantification were performed by employing the MaxQuant software (37,38).
Reverse transcription and PCR
RNC-RNA, isolated from both A549 and HBE cells, were reverse transcribed to cDNA as templates with poly-dT primer using RevertAid™ Premium Reverse Transcriptase (Fermentas, Hunover, MD), respectively, by following the manufacturer’s instructions. The quantitative real-time PCR (qPCR) was then performed with gene-specific primers and the iTaq™ universal SYBR® Green Supermix (Bio-rad, Hercules, CA) on a Bio-rad MiniOpticon real-time PCR system (Bio-rad) by following the manufacturer’s instructions. The primers were listed in the Supplementary Table S2. The specificity of the primers was verified by both in silico computation (NCBI Primer-BLAST) and melting curve measurement after the qPCR amplification. To double check the existence of the PCR product, another conventional PCR was performed by using the DreamTaq™ polymerase (Fermentas) with cDNA template and gene-specific primers. The PCR program was set identical to the quantitative PCR. The reaction mixture was resolved using a 2.5% agarose gel electrophoresis for in-gel visualization confirmation.
Ingenuity pathway analysis
Ingenuity pathway analysis (IPA) was performed as described previously with minor modifications (39,40). Briefly, the differentially expressed proteins (DEPs) and/or genes with different TRs were uploaded into www.ingenuity.com (Ingenuity Systems, Inc., Redwood City, CA). Core analyses were performed to identify top canonical pathways, biological networks, bioprocesses and effects on functions.
Statistics
The Spearman correlation coefficients were calculated to determine bivariate relationships. The regressions, data distribution and standard deviations were calculated by using MATLAB R2012a software package (MathWorks, Natick, MA). Data are shown as mean ± standard deviation. Statistical difference was accepted when P < 0.001.
RESULTS
Translatome sequencing and quantitative proteome profilling
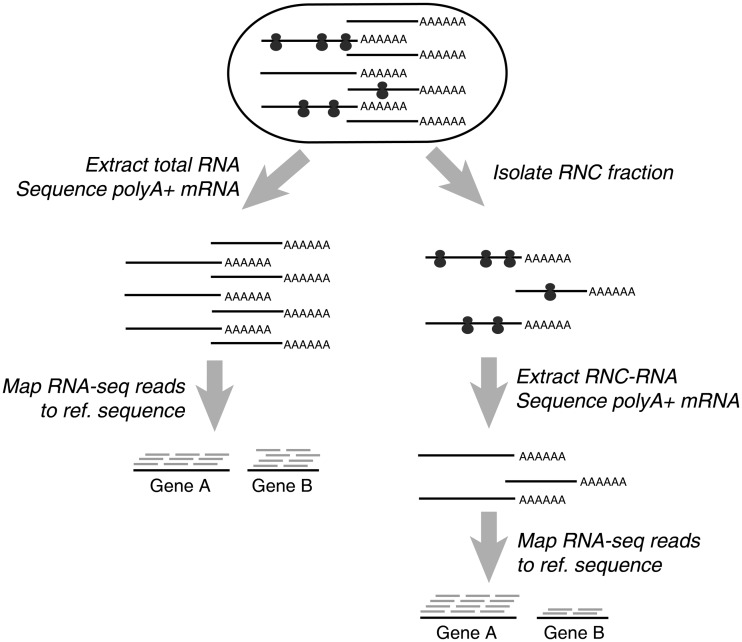
We performed RNA-seq on both mRNAs (transcriptome sequencing) and RNC-mRNAs (translatome sequencing) of A549 and HBE cells (Figure 1), and compared the proteomic difference between the two cell lines by using SILAC-based MS. To ensure the complete detection of mappable reads and the unbiased quantification of low-abundance mRNAs, we used our published FANSe algorithm to map the sequencing reads (31).
Figure 1.
Schematic procedure for translatome and transcriptome sequencing.
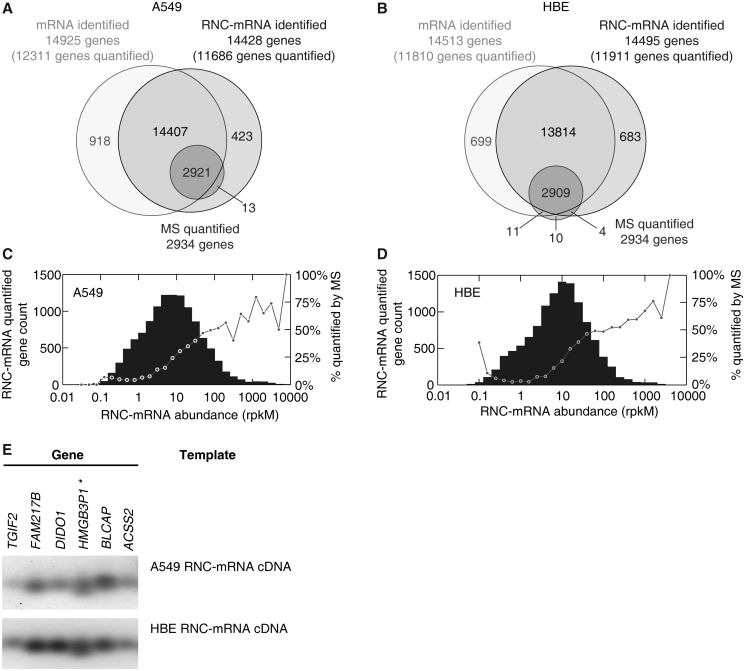
Genes detected by mRNA and RNC-mRNA sequencing in both cell lines showed a remarkable overlap, indicating that the majority of transcribed mRNAs were translated (Figure 2A and B, Supplementary Table S3). Regarding RNC-mRNA data set, a total of 11 686 genes in A549 cells (Figure 2A) and 11 911 genes in HBE cells (Figure 2B) were mapped with ≥10 reads, which is considered as the threshold of quantifiable genes in RNA-seq data (34). In the SILAC experiments (A549 versus HBE cells), a total of 4803 proteins were identified, among which 3045 proteins were identified with at least two unique peptides, and 2934 proteins contained quantifiable information (Ratio H/L Normalized) (Figure 2A and B). The detailed protein list is included in the Supplementary Table S4. We observed that all of the MS-quantified proteins were also detected in RNC-mRNAs in A549 cells (Figure 2A). Similar detection pattern was also observed in experiments performed with HBE cells, although minor proportion of outliers was noted (Figure 2B).
Figure 2.
Gene identification in translatome and transcriptome sequencing, in comparison with SILAC-based mass spectrometry. (A and B) Number of genes and proteins identified with RNA-seq (lighter circles) and MS (dark circle), respectively, in A549 cells (A) and HBE cells (B). (C and D) RNC-mRNA abundance distribution in A549 cells (C) and HBE cells (D). Genes were step-wise classified, based on abundances of quantified RNC-mRNA. Each bar indicates gene number of detection in its respective category. In each category, the percentage of the number of MS quantifiable protein to the number of genes that are detected by RNC-mRNA sequencing is shown with a dot. (E) Validation of gene detection in RNC-mRNA, extracted from A549 and HBE cells, respectively. Six randomly selected genes were subjected to RT-PCR assays and indicated by HGNC gene names.
In general, lognormal-like distributions of RNC-mRNA abundances were observed in both A549 (Figure 2C) and HBE cells (Figure 2D). In addition, proteins with higher RNC-mRNA abundances tended to be more detectable by MS, in comparison of those with lower such abundances (≤50 rpkM) (Figure 2C and D). It is known that protein abundance is a critical factor for successful MS identification. This observation implies a potential correlation between the abundances of RNC-mRNAs and proteins on genome-wide scale.
To validate RNC-mRNA identification, we performed reverse transcription PCR (RT-PCR) on six randomly selected genes that were not detected by MS but quantified by RNC-mRNA deep sequencing with abundance ranging from 4 to 300 rpkM, low to medium range. All of these genes were detected in the RNC-mRNA fraction by both real-time quantitative RT-PCR and conventional RT-PCR, evidencing the reliability of this high-throughput method (Figure 2E). The gene HMGB3P1 (RefSeq: NR_002165) has not been included in either the NCBI reference sequence (RefSeq) protein sequence database or the UniProtKB/Swiss-Prot protein knowledgebase, indicating that its protein product has never been detected.
Strong multivariate linear correlation exists among relative abundances of RNC-mRNAs and proteins, together with mRNA lengths
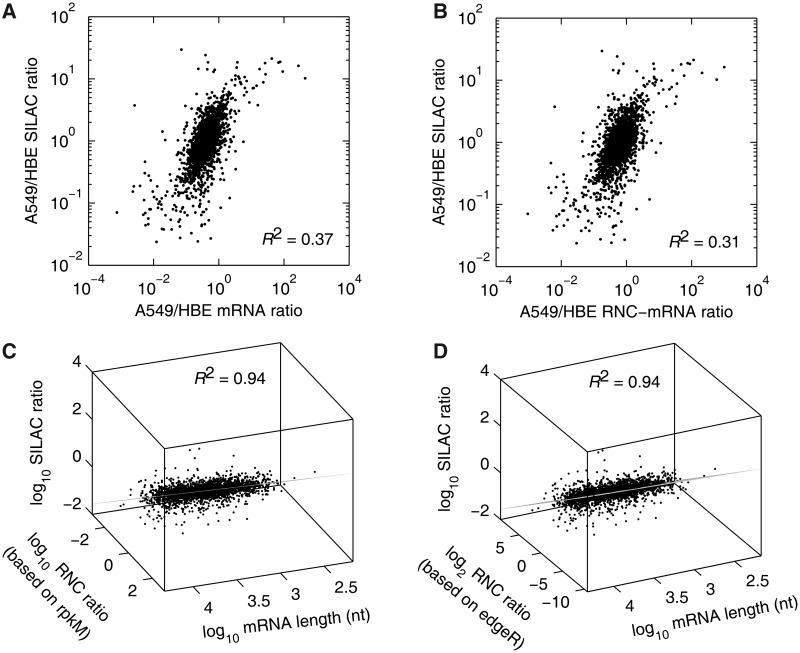
We observed that the mRNA ratio of A549 to HBE correlated poorly with the SILAC ratio (R2 = 0.37, Figure 3A), and that the correlation between the RNC-mRNA ratio and the SILAC ratio showed no increase (R2 = 0.31, Figure 3B). These results suggest that an extra factor must exist if our hypothesis on the tight correlation of mRNA/RNC-mRNA with protein abundance is true. We previously found that translation efficiency is affected by mRNA length (41), leading us to hypothesize that the mRNA length may serve as this additional factor. Therefore, we tested two multivariate linear models including mRNA length as candidates:
| (1) |
| (2) |
where SR = SILAC ratio, MR = mRNA ratio, RR = RNC-mRNA ratio and L = mRNA length.
Figure 3.
Multivariate linear correlation among the relative abundances of mRNA, RNC-mRNA and protein. (A and B) Bivariate correlation comparing mRNA (A) and RNC-mRNA ratios (A549/HBE) (B) with SILAC ratio (A549/HBE), respectively. (C and D) Multivariate linear model, fitting SILAC ratio (A549/HBE), mRNA length and RNC-mRNA ratio (A549/HBE), calculated based on rpkM (C) and edgeR (D) normalizations regarding RNA-seq data. The viewpoints were on the fitted planes.
First, we used the stepwise regression to examine whether the mRNA length contributes to the linear fitting. When analysing Equation (1), the mRNA length does not contribute to the regression (P = 0.208), thus being excluded from the analysis, whereas in Equation (2), regarding RNC-mRNA ratio (based on rpkM normalization), the mRNA length contributes significantly to the regression (P = 6.69 × 10−12) (Table 1). Next, we applied the least absolute residual robust iterative method to refine the regression. The regression converged within 500 iterations and resulted in a = 0.5998 (95% CI 0.5917–0.6079), b = 0.1509 (95% CI 0.1401–0.1616) and c = −0.4004 (95% CI −0.4370–−0.3638), with the correlation coefficient reaching R2 = 0.94 (Figure 3C and Supplementary Figure S1A). The data points distributed evenly along with the fitted plane determined by the Equation (2), showing a tight multivariate linear correlation of the data set (Figure 3C). When excluding SILAC value as a factor in Equation (2), the data points were sparsely scattered on the top-view plane with no correlation (R2 = 0.085), implicating that mRNA length and RNC-mRNA ratio are not correlated (Supplementary Figure S2). To address whether the different normalization method can affect the correlation, we calculated the RNC-mRNA ratio with edgeR package (33). EdgeR uses the trimmed mean of M-values method based on the negative binomial distribution, which is able to reliably detect the differential expression (42). In our case, the multivariate correlation remained exactly the same when using the RNC-mRNA ratio calculated using edgeR (Figure 3D, Supplementary Figure S1B), showing that this tight correlation is not dependent on the normalization method.
Table 1.
Stepwise regression with the multivariate linear model
| log10SR = a·log10MR + b·log10L + c |
log10SR = a·log10RR + b·log10L + c |
||||||
|---|---|---|---|---|---|---|---|
| Coefficient | Value | 95% CI | P-value | Coefficient | Value | 95% CI | P-value |
| a | 0.6004 | 0.5743–0.6264 | <10−308 | a | 0.5795 | 0.5534–0.6055 | 1.89 × 10−317 |
| b | −0.0210 | −0.0537–0.0117 | 0.2084 | b | 0.1213 | 0.0868–0.1558 | 6.69 × 10−12 |
| c | 0.2892 | c | −0.3167 | ||||
SR, SILAC ratio; MR, mRNA ratio; RR, RNC-mRNA ratio; L, mRNA length; CI, confidence interval.
To validate whether this strong correlation is a random phenomenon, we performed a biological validation analysing another pair of human cells at steady-state, H1299 and HBE cells, with the same strategy as described in the Figure 1. We quantified >11 868 genes from the RNC-mRNA data of H1299 cells (Supplementary Table S5). A total of 2353 quantifiable proteins that were identified with at least two unique peptides were obtained by the SILAC experiment, comparing the relative protein abundance between H1299 and HBE cells (Supplementary Table S6). Interestingly, the strong multivariate linear correlation among the mRNA length and the relative abundances of RNC-mRNAs and proteins was affirmatively observed, with R2 = 0.97 (Supplementary Figure S3). Even distribution of data points along with the fitted plane was also observed.
Genome-wide upregulation of mRNA TRs in cancer cells and correlation analysis with the mRNA length
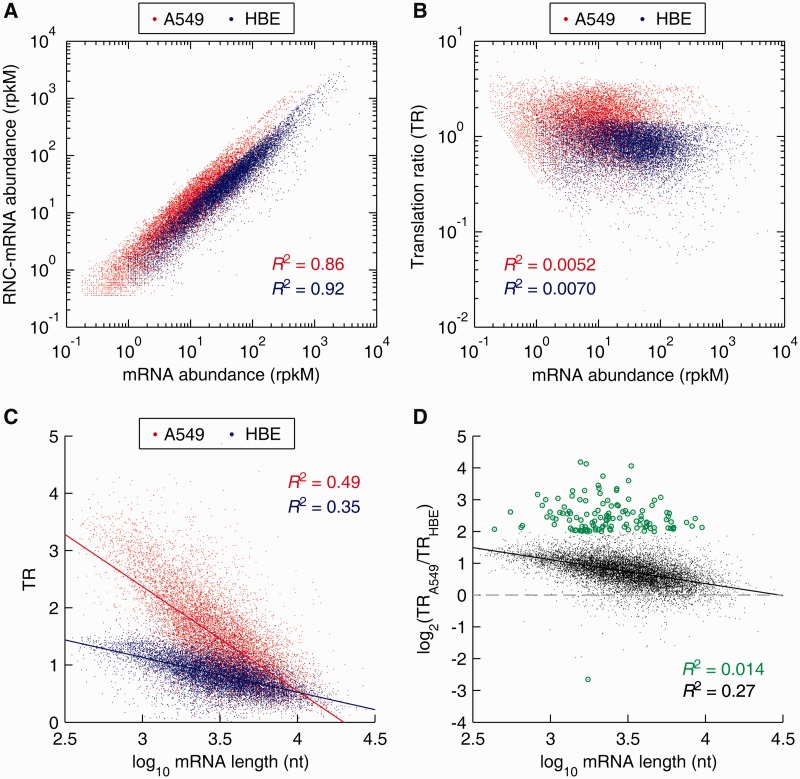
This tight multivariate correlation between translating mRNAs and proteins suggests a close relationship between translational modulation and phenotypes. In this regard, gene-specific transfer of mRNA to ribosomes is an essential step in protein biogenesis. We then proceeded to analyse the TR changes of a total of 10 626 genes that were detected in A549 and HBE cells (Figure 4). Indeed, the mRNA abundances showed good correlation with RNC-mRNA abundances in both A549 and HBE cells (R2 were both greater than 0.85) (Figure 4A), whereas TR values did not correlate with mRNA abundances at all (R2 were approaching to 0) (Figure 4B).
Figure 4.
Distribution and correlation analysis of mRNA TRs, comparing A549 cells with HBE cells. (A) Correlation of mRNA and RNC-mRNA abundances in A549 and HBE cells, respectively. (B) Correlation of mRNA ratio (A549/HBE) and RNC-mRNA ratio (A549/HBE). (C) Correlation of TRs and mRNA lengths. (D) Correlation of TR fold changes (A549/HBE) and mRNA lengths. The genes with TR ratio changes greater than 4 folds are indicated by green dots.
We next performed correlation analysis on the TR and the mRNA length to address the contribution of the mRNA length to translational modulation. The two parameters were significantly and negatively correlated with R2 of 0.49 and 0.35, in A549 and HBE cells, respectively (both P < 10−16) (Figure 4C). The slope of the regression line in A549 cells was −1.82, approximately three times sharper than that of the HBE cells (−0.61) (Figure 4C). Especially regarding genes with mRNA lengths of, 1000 nt, the TR distribution in A549 cells was largely between 2 and 5, whereas between 1 and 2 in HBE cells, suggesting a remarkable upregulation of TR in genes with shorter mRNA lengths (Figure 4C).
Genome-wide upregulation of TRs was observed in A549 cells, with detection of 10 160 upregulations and 446 downregulations (Figure 4D). By examining TR fold change differences of all genes in a chromosome-by-chromosome manner, a widespread distribution of TR upregulated genes across chromosomes was observed, comparing A549 with HBE cells; however, chromosome 19 was noted to contain ∼10% of all TR downregulated genes (44 of the 446), indicating chromosomal enrichment and uneven distribution of such genes (Supplementary Figure S4). The TR fold change (A549/HBE) and the mRNA length exhibited significant negative correlation as well (R2 = 0.27, P < 10−16) (Figure 4D). Interestingly, this correlation was non-significant regarding genes with considerable TR fold changes, which were greater than 4 folds (R2 = 0.0141, P = 0.1913) (Figure 4D).
Translational modulation is highly phenotype relevant
We previously reported that A549 cells exhibit EMT-polarized phenotypes in contrast to HBE cells, based on functional proteomic evidence (40). With the detection of TR difference in this study, we posit that these outliers with considerable TR fold changes, as shown in the Figure 4, is relevant to the malignant phenotypes of A549 cells. We therefore performed IPA on 123 genes with considerable TR changes (TR fold change ≥ 4.0) (Supplementary Table S7) as well as 1505 differentially expressed proteins [DEPs, fold change ≥ 1.5 (43)] identified by SILAC MS (Supplementary Table S4), respectively. IPA analysis on the DEPs pointed towards cancer cell phenotypes in the general reports (Supplementary Figure S5A); however, TR analysis specifically revealed airway pathology as one of the top canonical pathways (Supplementary Figure S5B).
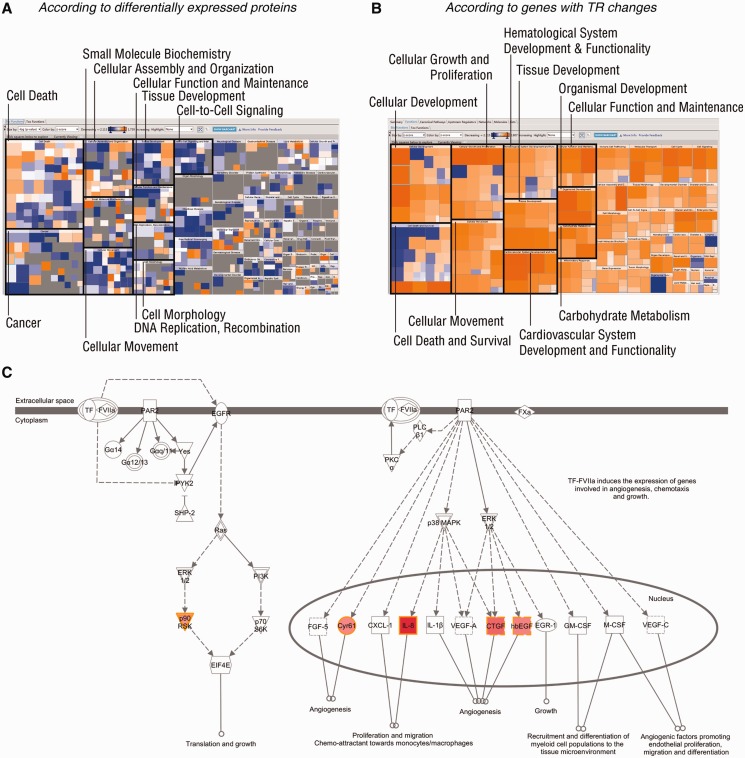
This increase of computational specificity in TR analysis was also confirmed by effect-on-function assays in IPA (Figure 5). The effects of DEPs on biological processes were largely mixed with contradictory predictions, showing both promotion and inhibition on the same processes (Figure 5A). However, the specificity was improved when analysing TR-changed genes, indicating homogenous regulation on functions (Figure 5B). These TR-predicted and promoted bioprocesses included cell growth and proliferation, cell movement and development, specifically reflecting the features of EMT phenotype of A549 cells (40) (Figure 5B). The top canonical pathway regulated by these TR-changed genes fell into the role of tissue factor in cancer (P = 2.96 × 10−4, Fisher’s Exact test provided by IPA) (Supplementary Figure S5B). The endpoint biological functions regulated by this pathway included cell growth and proliferation, angiogenesis and migration (Figure 5C). With the results shown earlier in the text, TR-based pathway analysis exhibited unique advantages in focusing investigative bioprocesses.
Figure 5.
IPA. DEPs and genes with considerable TR fold changes (A549/HBE) were subjected to IPA. (A and B) Heat maps of effects on biological processes, regulated by DEPs (A) and TR-changed genes (B). Top 10 Category Level I bioprocesses are indicated by black blocks. An orange square represents an enhanced Category Level II bioprocess with a positive z-score, provided by IPA, whereas suppressed such bioprocesses, with negative z-scores, are shown in blue squares. Insignificant bioprocesses are indicated by grey squares. (C) The top canonical pathway regulated by TR-changed genes (A549/HBE). Experimentally detected genes are indicated in red shapes, and the colour intensity represents the grade of regulation. Shapes of inversed triangles, circles and squares represent kinases, complexes and cytokines, respectively. Solid and dashed lines with arrows represent direct and indirect promotion, respectively. Full names of the genes (HGNC nomenclature) in red shapes are protease-activated kinase II (p90RSK), cysteine-rich angiogenic inducer 61 (Cyr61), interleulin-8 (IL-8), connective tissue growth factor (CTGF) and heparin-binding EGF-like growth factor (hbEGF).
Splice variants of the BDP1 gene are not equally translated
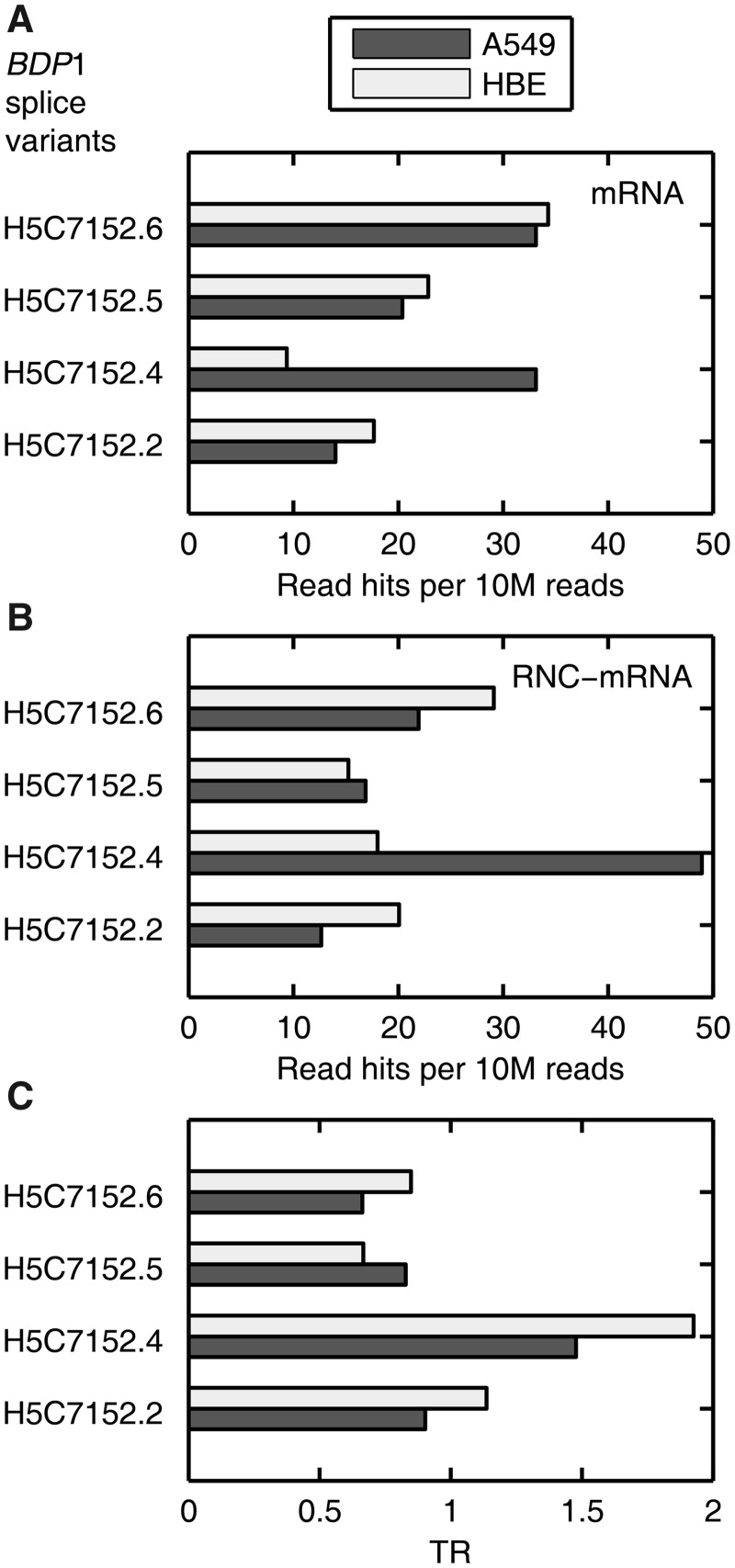
Proteomics and/or transcriptomics cannot address whether the genetic information of ASTs can be proportionally transmitted to translational level (44). However, comparison of the ASTs in mRNA and RNC-mRNA can reveal variant-specific TR information. We previously reported that ASTs of the BDP1 gene, coding one of the transcription factor IIIB subunits, exist with different abundances in various tissues (35). Therefore, we added RNC-mRNA information and analysed this gene again to serve as an example in this study (Figure 6). Consistent with our previous findings (35), we observed different and independent expression patterns of the BDP1 ASTs in both A549 and HBE cells at mRNA level (Figure 6A). These patterns were shifted at RNC-mRNA level in both cell lines (Figure 6B). Compared with other ASTs, the splice variant H5C7152.4 exhibited remarkably higher reads in the RNC-mRNA fraction than the mRNA fraction in A549 cells (Figure 6B). Furthermore, the splice variants H5C7152.5 and H5C7152.6 were less likely to be translated as their TRs ranged from 0.67 to 0.85, whereas the TR of H5C7152.4 reached 1.48 and 1.93 in A549 and HBE cells, respectively, evidencing a clear translational preference of this AST (Figure 6C). Collectively, the TR variance of the ASTs, either within a single cell type or across the two cell lines, displayed biased translation preference, suggesting a fine tuning in the genetic information transmission from mRNA to translation level.
Figure 6.
Biased TRs of BDP1 splice variants in A549 and HBE cells. (A and B) BDP1 splice variants detected in mRNA (A) and RNC-mRNA (B) of A549 and HBE cells, respectively. The bars represent the normalized number of reads that were mapped to specific splicing junctions of different variants. (C) TR of these splice variants in A549 and HBE cells, respectively.
DISCUSSION
We report here, for the first time, that a multivariate linear model integrating the full mRNA length can fit the relative abundances of RNC-mRNAs and proteins with significantly tight correlation. Given this correlation, we demonstrated that the translating mRNA represents an independent source for the prediction of protein abundances. The hypothesis of this study was reasoned from our previous findings, indicating the interplay of length-dependent decay of translating mRNAs and translational efficiency, suggesting that certain synergy exists between the amount of translating mRNAs and the mRNA lengths (41). As a favourable validation, we discovered that the mRNA length is one of the determinant factors in the translational control, according to its significant correlation with the TRs of a single cell type or relative TR fold changes. Although similar studies with full mRNA length has not been noted, Arava et al.(45) discovered in yeast cells that ribosome density decreases in mRNAs with longer open reading frames and underlined the rate-limiting role of translation initiation in translational control, which was confirmed in a human cell study (12). This is reinforced by the general trend observed in the regression assays of this study, suggesting that genes with shorter mRNA lengths tend to have higher TRs and vice versa. These reports and our findings suggest that the mRNA length plays an important role in connecting translatome to proteome in terms of its correlation with translation initiation and TR. As a negative control, if not taking translating mRNA into consideration, we reproduced low correlation between mRNAs and proteins, and the mRNA length does not contribute to this correlation. This is consistent with studies from other groups, showing low bivariate or partial correlations between mRNA-length and protein abundances (45–47) [reviewed in (21)]. Therefore, we could propose that the translation initiation preferentially occurs on the shorter mRNAs, which resulted in the increased fraction of these mRNA molecules that can be translated to proteins. This mechanism may serve as a possible explanation of why the mRNA length has significant contribution to the multivariate linear model reported in this study. Furthermore, with this model, we can propose a quantitative answer, with the relative abundance analyses, to what extent that the translational regulation involves in the flow of genetic information from translating mRNAs to proteins in human cells at steady-state and its relevance to cellular phenotypes.
To be noted, translatome sequencing used in this study did not consider the number of ribosomes that are attached to a single mRNA strand. Hence, it differs from a very similar method, namely, ribosome profiling that analyses ribosome protected fragments (48). The difference of these two techniques is illustrated in Supplementary Figure S6: ribosome profiling yields the ribosome protected mRNA regions, whereas translatome sequencing obtains the information of all RNC-mRNA regions, including ribosome protected and unprotected regions. Ribosome profiling detects the ribosome locations at nucleotide resolution, providing important information on ribosome density of genes. However, it provides insufficient TR information, owing to its systematic limitation in resolving the amount of translating mRNA (illustrated in Supplementary Figure S7). In contrast, translatome sequencing detects the entire translating mRNAs, suitable for TR determination; however, it does not output the ribosome density. These two techniques depict the translation scenario from two aspects and cannot replace each other.
We demonstrated that the TR changes can accurately discern phenotype-specific canonical pathways, providing an independent index other than widely used DEPs and/or differentially transcribed genes. Relevant to our study, it has been well known that hyper-activation of multiple signalling pathways in cancer cells results in global upregulation of translation [reviewed in (49)]. Our IPA on TR-changed genes indicated remarkable TR increase of p90RSK that is known to promote translation via Ras and cap-dependent protein synthesis (50). This partially explains the global TR upregulation in A549 cells, compared with HBE cells, as observed in this study. As such, TR-based information provides a unique strategic view and opens up a new avenue for functional investigations.
The biased TRs of different ASTs in a certain gene, such as BDP1 shown in this study, may represent a critical mechanism in translational modulation that commonly exists in various eukaryotes. We previously reported that BDP1 splice variants recognize various motifs in the transfer RNA (tRNA) gene upstream sequences, responsible for anticodon-specific regulation of tRNA expression in mammalian cells (35). TR regulation of BDP1 splice variants may influence the cellular tRNA composition, thus reshaping the cellular translation rate profiles and further globally altering co-translational protein folding [(51) and reviewed in (52)]. These factors can loop back to regulate the translational scenario that amplifies the effects of stimulation and finally drive the system to an altered steady state.
Our current work demonstrated that translatome sequencing can potentially add novel proteins to the proteome atlas. For example, we detected and validated potentially novel translating genes that have no protein and transcript evidence to date, such as HMGB3P1. This capacity of translatomics may confer greater impact on diverse biologies, allowing for investigations of proteins even in non-model species that have no available proteome knowledgebase. In addition, RNC-mRNA sequencing can exclude most of (if not all) non-coding transcripts from the transcriptome data and accurately quantify the translating mRNAs. This allows translatomics to generate expressing protein sequence databases on a genome-wide scale, serving as a solid base for next-step proteomic investigations and gene-centric annotations.
In conclusion, we provided the first direct translatome evidence, substantiating that global translation modulation is a key factor of phenotype formation in human cells at steady state. This is underscored by our discovery of a novel multivariate linear model to highly correlate the relative abundances of RNC-mRNAs and proteins by integrating the mRNA length as a critical factor. We demonstrated that TR regulation on genes and their ASTs is highly phenotype specific. Therefore, the translating mRNA and the TR can serve as independent research objectives to characterize cellular functionalities.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–7 and Supplementary Figures 1–7.
FUNDING
National Basic Research Program ‘973’ of China [2011CB910700 to Q.Y.H.]; National Natural and Science Foundation of China [81272185, 81000516 to T.W.]; Institutional Grant of Excellence of Jinan University, China [50625072 to G.Z.]; Fundamental Research Funds for the Central Universities of China [21612202, 21612406, 21610101 to G.Z., T.W. and Q.Y.H., respectively]. Funding for open access charge: National Basic Research Program ‘973’ of China [2011CB910700].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Li Sun, Liping Li and Zhipeng Chen, Jinan University, for their technical assistance. T.W. and G.Z. conceived the idea, designed the study, analysed the data and prepared the manuscript. Y.C. and J.J. performed the RNC isolation, RNA extraction, characterization and RNA-seq data analysis and also assisted on the mass spectrometry. J.G., G.W. and X.Y. performed proteomic experiments and analysed the data, supervised by Q.Y.H., who also contributed to manuscript composition.
REFERENCES
- 1.Gygi SP, Rochon Y, Franza BR, Aebersold R. Correlation between protein and mRNA abundance in yeast. Mol. Cell Biol. 1999;19:1720–1730. doi: 10.1128/mcb.19.3.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen G, Gharib TG, Huang CC, Taylor JM, Misek DE, Kardia SL, Giordano TJ, Iannettoni MD, Orringer MB, Hanash SM, et al. Discordant protein and mRNA expression in lung adenocarcinomas. Mol. Cell Proteomics. 2002;1:304–313. doi: 10.1074/mcp.m200008-mcp200. [DOI] [PubMed] [Google Scholar]
- 3.Lu P, Vogel C, Wang R, Yao X, Marcotte EM. Absolute protein expression profiling estimates the relative contributions of transcriptional and translational regulation. Nat. Biotechnol. 2007;25:117–124. doi: 10.1038/nbt1270. [DOI] [PubMed] [Google Scholar]
- 4.Guo Y, Xiao P, Lei S, Deng F, Xiao GG, Liu Y, Chen X, Li L, Wu S, Chen Y, et al. How is mRNA expression predictive for protein expression? A correlation study on human circulating monocytes. Acta. Biochim. Biophys. Sin. 2008;40:426–436. doi: 10.1111/j.1745-7270.2008.00418.x. [DOI] [PubMed] [Google Scholar]
- 5.Taniguchi Y, Choi PJ, Li GW, Chen H, Babu M, Hearn J, Emili A, Xie XS. Quantifying E. coli proteome and transcriptome with single-molecule sensitivity in single cells. Science. 2010;329:533–538. doi: 10.1126/science.1188308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lundberg E, Fagerberg L, Klevebring D, Matic I, Geiger T, Cox J, Algenas C, Lundeberg J, Mann M, Uhlen M. Defining the transcriptome and proteome in three functionally different human cell lines. Mol. Syst. Biol. 2010;6:450. doi: 10.1038/msb.2010.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nagaraj N, Wisniewski JR, Geiger T, Cox J, Kircher M, Kelso J, Paabo S, Mann M. Deep proteome and transcriptome mapping of a human cancer cell line. Mol. Syst. Biol. 2011;7:548. doi: 10.1038/msb.2011.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lackner DH, Schmidt MW, Wu S, Wolf DA, Bahler J. Regulation of transcriptome, translation, and proteome in response to environmental stress in fission yeast. Genome Biol. 2012;13:R25. doi: 10.1186/gb-2012-13-4-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maier T, Guell M, Serrano L. Correlation of mRNA and protein in complex biological samples. FEBS Lett. 2009;583:3966–3973. doi: 10.1016/j.febslet.2009.10.036. [DOI] [PubMed] [Google Scholar]
- 10.Pradet-Balade B, Boulme F, Beug H, Mullner EW, Garcia-Sanz JA. Translation control: bridging the gap between genomics and proteomics? Trends Biochem. Sci. 2001;26:225–229. doi: 10.1016/s0968-0004(00)01776-x. [DOI] [PubMed] [Google Scholar]
- 11.Greenbaum D, Colangelo C, Williams K, Gerstein M. Comparing protein abundance and mRNA expression levels on a genomic scale. Genome Biol. 2003;4:117. doi: 10.1186/gb-2003-4-9-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hendrickson DG, Hogan DJ, McCullough HL, Myers JW, Herschlag D, Ferrell JE, Brown PO. Concordant regulation of translation and mRNA abundance for hundreds of targets of a human microRNA. PLoS Biol. 2009;7:e1000238. doi: 10.1371/journal.pbio.1000238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Akan PD, Alexeyenko AD, Costea PI, Hedberg L, Solnestam BW, Lundin S, Hallman J, Lundberg ED, Uhlen MP, Lundeberg JP. Comprehensive analysis of the genome transcriptome and proteome landscapes of three tumor cell lines. Genome Med. 2012;4:86. doi: 10.1186/gm387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kong J, Lasko P. Translational control in cellular and developmental processes. Nat. Rev. Genet. 2012;13:383–394. doi: 10.1038/nrg3184. [DOI] [PubMed] [Google Scholar]
- 15.McCarthy JE. Posttranscriptional control of gene expression in yeast. Microbiol. Mol. Biol. Rev. 1998;62:1492–1553. doi: 10.1128/mmbr.62.4.1492-1553.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Picard F, Milhem H, Loubiere P, Laurent B, Cocaign-Bousquet M, Girbal L. Bacterial translational regulations: high diversity between all mRNAs and major role in gene expression. BMC Genomics. 2012;13:528. doi: 10.1186/1471-2164-13-528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beilharz TH, Preiss T. Translational profiling: the genome–wide measure of the nascent proteome. Brief Funct. Genomic. Proteomic. 2004;3:103111. doi: 10.1093/bfgp/3.2.103. [DOI] [PubMed] [Google Scholar]
- 18.Kuhn KM, DeRisi JL, Brown PO, Sarnow P. Global and specific translational regulation in the genomic response of Saccharomyces cerevisiae to a rapid transfer from a fermentable to a nonfermentable carbon source. Mol. Cell Biol. 2001;21:916–927. doi: 10.1128/MCB.21.3.916-927.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Causton HC, Ren B, Koh SS, Harbison CT, Kanin E, Jennings EG, Lee TI, True HL, Lander ES, Young RA. Remodeling of yeast genome expression in response to environmental changes. Mol. Biol. Cell. 2001;12:323–337. doi: 10.1091/mbc.12.2.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thomas A, Lee PJ, Dalton JE, Nomie KJ, Stoica L, Costa-Mattioli M, Chang P, Nuzhdin S, Arbeitman MN, Dierick HA. A versatile method for cell-specific profiling of translated mRNAs in drosophila. PLoS One. 2012;7:e40276. doi: 10.1371/journal.pone.0040276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vogel C, Marcotte EM. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat. Rev. Genet. 2012;13:227–232. doi: 10.1038/nrg3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jechlinger M, Grunert S, Tamir IH, Janda E, Ludemann S, Waerner T, Seither P, Weith A, Beug H, Kraut N. Expression profiling of epithelial plasticity in tumor progression. Oncogene. 2003;22:7155–7169. doi: 10.1038/sj.onc.1206887. [DOI] [PubMed] [Google Scholar]
- 23.Thireos G, Griffin-Shea R, Kafatos FC. Untranslated mRNA for a chorion protein of Drosophila melanogaster accumulates transiently at the onset of specific gene amplification. Proc. Natl Acad. Sci. USA. 1980;77:5789–5793. doi: 10.1073/pnas.77.10.5789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Standart N, Hunt T, Ruderman JV. Differential accumulation of ribonucleotide reductase subunits in clam oocytes: the large subunit is stored as a polypeptide, the small subunit as untranslated mRNA. J. Cell Biol. 1986;103:2129–2136. doi: 10.1083/jcb.103.6.2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nielsen FC, Ostergaard L, Nielsen J, Christiansen J. Growth-dependent translation of IGF-II mRNA by a rapamycin-sensitive pathway. Nature. 1995;377:358–362. doi: 10.1038/377358a0. [DOI] [PubMed] [Google Scholar]
- 26.Khan D, Sharathchandra A, Ponnuswamy A, Grover R, Das S. Effect of a natural mutation in the 5' untranslated region on the translational control of p53 mRNA. Oncogene. 2012 doi: 10.1038/onc.2012.422. October 1 (doi:10.1038/onc.2012.422; epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 27.Banfai B, Jia H, Khatun J, Wood E, Risk B, Gundling WE, Jr, Kundaje A, Gunawardena HP, Yu Y, Xie L, et al. Long noncoding RNAs are rarely translated in two human cell lines. Genome Res. 2012;22:1646–1657. doi: 10.1101/gr.134767.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paik YK, Jeong SK, Omenn GS, Uhlen M, Hanash S, Cho SY, Lee HJ, Na K, Choi EY, Yan F, et al. The Chromosome-Centric Human Proteome Project for cataloging proteins encoded in the genome. Nat. Biotechnol. 2012;30:221–223. doi: 10.1038/nbt.2152. [DOI] [PubMed] [Google Scholar]
- 29.Paik YK, Omenn GS, Uhlen M, Hanash S, Marko-Varga G, Aebersold R, Bairoch A, Yamamoto T, Legrain P, Lee HJ, et al. Standard guidelines for the chromosome-centric human proteome project. J. Proteome. Res. 2012;11:2005–2013. doi: 10.1021/pr200824a. [DOI] [PubMed] [Google Scholar]
- 30.Esposito AM, Mateyak M, He D, Lewis M, Sasikumar AN, Hutton J, Copeland PR, Kinzy TG. Eukaryotic polyribosome profile analysis. J. Vis. Exp. 2010;40:pii: 1948. doi: 10.3791/1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang G, Fedyunin I, Kirchner S, Xiao C, Valleriani A, Ignatova Z. FANSe: an accurate algorithm for quantitative mapping of large scale sequencing reads. Nucleic Acids Res. 2012;40:e83. doi: 10.1093/nar/gks196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 33.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bloom JS, Khan Z, Kruglyak L, Singh M, Caudy AA. Measuring differential gene expression by short read sequencing: quantitative comparison to 2-channel gene expression microarrays. BMC Genomics. 2009;10:221. doi: 10.1186/1471-2164-10-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang G, Lukoszek R, Mueller-Roeber B, Ignatova Z. Different sequence signatures in the upstream regions of plant and animal tRNA genes shape distinct modes of regulation. Nucleic Acids Res. 2011;39:3331–3339. doi: 10.1093/nar/gkq1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ge F, Xiao CL, Bi LJ, Tao SC, Xiong S, Yin XF, Li LP, Lu CH, Jia HT, He QY. Quantitative phosphoproteomics of proteasome inhibition in multiple myeloma cells. PLoS One. 2010;5 doi: 10.1371/journal.pone.0013095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 38.Cox J, Neuhauser N, Michalski A, Scheltema RA, Olsen JV, Mann M. Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 2011;10:1794–1805. doi: 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
- 39.Wang T, Gong N, Liu J, Kadiu I, Kraft-Terry SD, Mosley RL, Volsky DJ, Ciborowski P, Gendelman HE. Proteomic modeling for HIV-1 infected microglia-astrocyte crosstalk. PLoS One. 2008;3:e2507. doi: 10.1371/journal.pone.0002507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li LP, Lu CH, Chen ZP, Ge F, Wang T, Wang W, Xiao CL, Yin XF, Liu L, He JX, et al. Subcellular proteomics revealed the epithelial-mesenchymal transition phenotype in lung cancer. Proteomics. 2011;11:429–439. doi: 10.1002/pmic.200900819. [DOI] [PubMed] [Google Scholar]
- 41.Valleriani A, Zhang G, Nagar A, Ignatova Z, Lipowsky R. Lengthvdependent translation of messenger RNA by ribosomes. Phys. Rev. E. Stat. Nonlin. Soft Matter Phys. 2011;83:042903. doi: 10.1103/PhysRevE.83.042903. [DOI] [PubMed] [Google Scholar]
- 42.Dillies MA, Rau A, Aubert J, Hennequet-Antier C, Jeanmougin M, Servant N, Keime C, Marot G, Castel D, Estelle J, et al. A comprehensive evaluation of normalization methods for Illumina high-throughput RNA sequencing data analysis. Brief Bioinform. 2012 doi: 10.1093/bib/bbs046. September 17 (doi:10.1093/bib/bbs046; epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 43.Blagoev B, Ong SE, Kratchmarova I, Mann M. Temporal analysis of phosphotyrosine-dependent signaling networks by quantitative proteomics. Nat. Biotechnol. 2004;22:1139–1145. doi: 10.1038/nbt1005. [DOI] [PubMed] [Google Scholar]
- 44.Nilsen TW, Graveley BR. Expansion of the eukaryotic proteome by alternative splicing. Nature. 2010;463:457–463. doi: 10.1038/nature08909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Arava Y, Wang Y, Storey JD, Liu CL, Brown PO, Herschlag D. Genome-wide analysis of mRNA translation profiles in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA. 2003;100:3889–3894. doi: 10.1073/pnas.0635171100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Beyer A, Hollunder J, Nasheuer HP, Wilhelm T. Post-transcriptional expression regulation in the yeast Saccharomyces cerevisiae on a genomic scale. Mol. Cell Proteomics. 2004;3:1083–1092. doi: 10.1074/mcp.M400099-MCP200. [DOI] [PubMed] [Google Scholar]
- 47.Vogel C, Abreu Rde S, Ko D, Le SY, Shapiro BA, Burns SC, Sandhu D, Boutz DR, Marcotte EM, Penalva LO. Sequence signatures and mRNA concentration can explain two-thirds of protein abundance variation in a human cell line. Mol. Syst. Biol. 2010;6:400. doi: 10.1038/msb.2010.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Grzmil M, Hemmings BA. Translation regulation as a therapeutic target in cancer. Cancer Res. 2012;72:3891–3900. doi: 10.1158/0008-5472.CAN-12-0026. [DOI] [PubMed] [Google Scholar]
- 50.Versteeg HH, Sorensen BB, Slofstra SH, Van den Brande JH, Stam JC, van Bergen en Henegouwen PM, Richel DJ, Petersen LC, Peppelenbosch MP. VIIa/tissue factor interaction results in a tissue factor cytoplasmic domain-independent activation of protein synthesis, p70, and p90 S6 kinase phosphorylation. J. Biol. Chem. 2002;277:27065–27072. doi: 10.1074/jbc.M110325200. [DOI] [PubMed] [Google Scholar]
- 51.Fedyunin I, Lehnhardt L, Bohmer N, Kaufmann P, Zhang G, Ignatova Z. tRNA concentration fine tunes protein solubility. FEBS Lett. 2012;586:3336–3340. doi: 10.1016/j.febslet.2012.07.012. [DOI] [PubMed] [Google Scholar]
- 52.Zhang G, Ignatova Z. Folding at the birth of the nascent chain: coordinating translation with co-translational folding. Curr. Opin. Struct. Biol. 2011;21:25–31. doi: 10.1016/j.sbi.2010.10.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.