Abstract
The olfactory system integrates signals from receptors expressed in olfactory sensory neurons. Each sensory neuron expresses only one of many similar olfactory receptors (ORs). The choice of receptor is made stochastically early in the differentiation process and is maintained throughout the life of the neuron. The underlying mechanism of this stochastic commitment to one of multiple similar OR genes remains elusive. We present a theoretical analysis of a mechanism that invokes important epigenetic properties of the system. The proposed model combines nucleosomes and associated read–write enzymes as mediators of a cis-acting positive feedback with a trans-acting negative feedback, thereby coupling the local epigenetic landscape of the individual OR genes in a way that allow one and only one gene to be active at any time. The model pinpoint that singular gene selection does not require transient mechanisms, enhancer elements or transcription factors to separate choice from maintenance. In addition, our hypothesis allow us to combine all reported characteristics of singular OR gene selection, in particular that OR genes are silenced from OR transgenes. Intriguingly, it predicts that OR transgenes placed in close proximity should always be expressed simultaneously, though rarely.
INTRODUCTION
Mutually exclusive gene expression is described in several organisms, from allelic exclusion in X-chromosome inactivation (1) and imprinting in mammals (2), to antigenic variation by selective Variant Selective Glycoprotein (VSG) expression in African trypanosomes (3). The present topic of olfactory neuron differentiation is yet an example.
The first step in odour reception in mammals is effectuated in the almost canonical one neuron-one receptor rule (4,5). Each olfactory neuron expresses only one allele out of a large and highly homologous gene family, comprising almost 1400 olfactory receptor (OR) genes in mice (6–9). Millions of olfactory neurons comprise the olfactory epithelium (OE). Olfactory neurons expressing a particular OR gene are confined to zones along the dorsal-ventral axis of the OE in mice, with possible overlaps between differently segregated zones (10–12). The coding region of a general OR gene is just 1 kb. However, transgenes must include a much larger part of the OR gene region to reproduce expression patterns parallel to endogenous genes (13–15). During development, the neuron extends its axon to a receptor defined glomerulus of the olfactory bulb, ensuring the essential conversion of olfactory signals to a typographical map in the bulb (16–18). Main characteristics of the olfactory neuron differentiation are summarized in Table 1 [reviewed recently by (7,19,20)].
Table 1.
Characteristics of olfactory neuron differentiation
|
In choosing to express just one among thousands of OR genes, each olfactory neuron represents a system with multiple stable states. Numerous mechanisms have been put forth to describe the underlying nature of this seemingly stochastic multi-stability. So far, all proposals fail to fully encompass both the choice and the maintenance of the expression of a single OR gene. Most descriptions like limiting transcriptional complexes or singular enhancer elements picture plausible ways of stochastically choosing a single OR gene for expression but resort to transient mechanisms to fix the memory of the chosen OR gene (14,27,28,31,35–38). Recent reviews that hypothesize the involvement of chromatin re-modelling in upholding OR gene selection implement similar transient mechanisms or some shielding of the chosen OR gene in establishing the choice (7,39). Moreover, though based on experimental observations, all proposals remain descriptive and lack a mean for reliably testing if the dynamics of the system would in fact be as envisioned.
In this article, we present a quantitative analysis of a model of OR gene choice inspired by the proposed involvement of epigenetic modifications. We revisit a theoretical approach on epigenetic cell memory by nucleosome modification, initially taken by Dodd et al. (40,41) and explored in the context of vernalization in plants by Angel et al. (42). In the two state version of this theoretical analysis, a DNA region containing L nucleosomes is considered (43,44). Each nucleosome may be in one of the two states—silent or active. Transitions between states are made randomly or by active recruitments. In a random event, a nucleosome spontaneously converts to the other state. During recruitment a histone-modifying enzyme is recruited by nearby nucleosomes and is thought to modify the nucleosome at hand to match the modification of the recruiting nucleosomes. When these recruitment processes include either implicit or explicit cooperativity, the state of the DNA region can be bistable (40,44), with the majority of the nucleosomes being in either the silent or active state, see Figure 1B.
Figure 1.
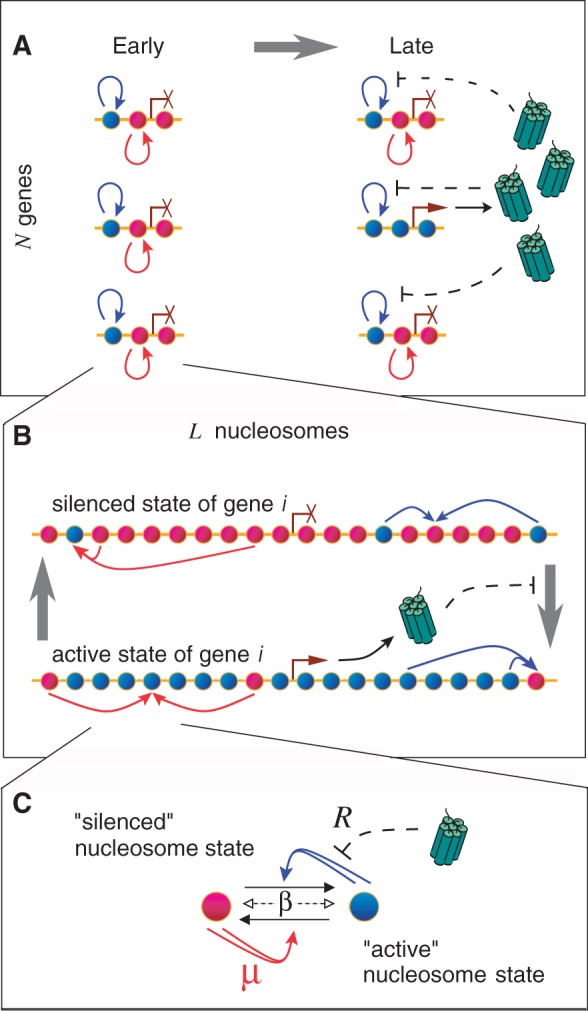
Model for maintenance of selected gene expression in an olfactory neuronal cell. (A) Local positive feedback and global negative feedback. N genes each covered by nucleosomes (red for silently marked nucleosomes, blue for actively marked). The nucleosomes on each gene form positive feedback systems that maintain the selected expression state (red and blue arrows). The active gene expresses receptor proteins, which direct enzymes that favour nucleosomes to stay in a silenced state by repressing the positive feedback towards the active nucleosome state (black arrow and blunted lines, respectively). (B) Switching property of a single gene. Each gene is covered by L nucleosomes that each can be in an active state that is open for transcription (blue) or in a silenced state (red). The nucleosomes together with associated ‘read-write’ enzymes form positive feedback systems that allow a gene to maintain a previously selected state (blue and red arrows). Enzymatic activity effectuated by the activity of all active genes captures genes in the silent state (black arrow and dashed blunted line). (C) Detailed nucleosome conversions. Nucleosome modifications recruit read-write enzymes within each gene, as in (40), but for simplicity, we consider only two nucleosome states, which each can modify the states of other nucleosomes cooperatively. In the model implementation, β is the noise on nucleosome conversions and  parameterizes the bias in recruited conversions. The feedback between genes, described in (A) and (B), acts by reducing conversions of nucleosomes to the active state, parameterized by R in our model.
parameterizes the bias in recruited conversions. The feedback between genes, described in (A) and (B), acts by reducing conversions of nucleosomes to the active state, parameterized by R in our model.
One may associate the two nucleosome states of the model with permissive and repressive methylation marks as recently reported for the active and inactive OR gene regions by Magklara et al. (34). Alternatively, the states could be DNA methylation marks (45) or increasing levels of histone acetylation. Histone de-acetylases are reported in immature and mature olfactory sensory neurons (46,47), and in vitro-cultured vomeronasal progenitor cells of adult rats only develop the adult neuronal phenotype when subjected to histone de-acetylase inhibitors (48). We use a compressed description of these multi-step scenarios where different levels of histone modifications dictate activity of the genes.
We study whether olfactory differentiation can be described by combining several subsystems with internal architectures similar to those of the nucleosome modification model of cell memory. Each OR gene should then be in one of the two general states, either activated or silenced. To mimic the mutual exclusion of expression amongst OR genes, we couple the sub-systems through a hypothesized factor, effectively governed by active OR genes, see Figure 1A. The factor is thought to prevent the conversion of silent nucleosome marks to active ones, perhaps through binding and shielding of the silent marks or by inhibiting the activating enzymes. Thus effectively, the factor represses the positive feedback towards active nucleosomes, see Figure 1C.
Formulating our model of OR gene selection as a stochastic simulation, we examine how the model captures the properties of the system and identify the effect of the involved mechanisms. We demonstrate that combining a cis-acting positive feedback mediated by nucleosomes and associated read–write enzymes, with a transacting negative feedback, encompass or reproduce all the characteristics of OR gene selection summarized in Table 1. In particular, we reproduce functional differentiation without the need for transient mechanisms to fix the memory of the chosen OR.
MATERIALS AND METHODS
In general, our model describes the OR neuron differentiation as a combination of cis-acting positive feedback that stabilize the state of the individual OR gene and a trans-acting negative feedback governed by active OR genes.
The model of epigenetic cell memory through nucleosome modification allows for sensitive gene regulation of individual genes by positive feedback loops (40,41). This provides us with a predefined bistable sub-system to describe each OR gene. The individual OR genes are coupled through modification reactions that increase with the activity of all OR genes. This global negative feedback favours the silent state. The model, outlined in Figure 1, is formulated as a stochastic simulation as in (40,41), and analysed in terms of deterministic equations. The real OR gene family is one of the largest mammalian gene families. In modelling, we consider system sizes of N = 10, 100 or 1000 similar genes consisting of L = 50 nucleosomes.
Governing equations
In the limit of an infinite number of nucleosomes per gene, the governing equation for the fraction of active nucleosomes, ai, in gene i, reads:
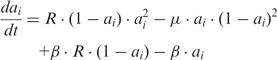 |
(1) |
where 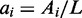 , with Ai being the number of active nucleosomes of gene i,
, with Ai being the number of active nucleosomes of gene i, 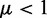 is the bias that favour the active state of all genes, and R is the reduction in conversion activity of nucleosomes in the silent state resulting from the negative feedback that couples the OR genes. β resembles nucleosome conversions owing to noise, where we reduce the noise on silent nucleosomes by R.
is the bias that favour the active state of all genes, and R is the reduction in conversion activity of nucleosomes in the silent state resulting from the negative feedback that couples the OR genes. β resembles nucleosome conversions owing to noise, where we reduce the noise on silent nucleosomes by R.
The negative feedback reflects the gene activity of all OR genes. One may imagine the feedback as some factors that effectively are governed by the gene activity of all genes:
| (2) |
The ‘Hill coefficient’, h, in Equation (2) quantifies the threshold function for activity of the individual gene. If h = 1, the activity is simply proportional to having one particular nucleosome in the active state, e.g. the nucleosome at the promoter. Larger h values represent an increased threshold for production from the individual genes. Throughout the article, we use h = 2 but emphasize that our results are robust to sharper threshold functions.
The feedback from the activity of all genes to the inhibition of each individual gene is solely captured by P, which however may act on the nucleosome conversions in different ways. For example, P may act through a protein binding to silenced nucleosomes and preventing their conversion to active ones. Alternatively, the factor could act by inhibiting/sequestering enzymes that favour the active nucleosome state. Finally, P may act by activating enzymes that favour the silenced state. The only requirement is that the coupling feedback must favour the silent nucleosome state. For simplicity, we here concentrate on P acting as an inhibitor.
With P describing a binding and sequestering of an enzyme, e, required for recruited conversion to the active state, the bound and free fraction of this enzyme eb and efree is determined by
 |
(3) |
where we assume fast rates for this sequestering process and, for completeness, have included a parameter r that parameterizes the ratio between feedback factor P and the enzyme in the cell e. We set r = 1 in our standard modelling but demonstrate in the supplementary that other r values works. The recruited conversion rate to active nucleosomes is accordingly reduced by a factor
 |
(4) |
This form of the feedback is the same in case the feedback instead works through a shielding of silenced nucleosomes. In that case, only a silenced nucleosome fraction given also by Equation (4), is accessible for conversions.
Stochastic implementation of model
The stochastic version of the model mimics a system of N genes each covered by L nucleosomes. Let at each time 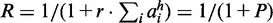 where
where  with Ai being the number of active nucleosomes in gene i.
with Ai being the number of active nucleosomes in gene i.
At each step of the simulation one
Select 2 random nucleosomes in one random gene. If both are active, one with probability R, then select another nucleosome in this gene and set its state to active. If the two nucleosomes are in the silenced state, one with probability μ, the select another nucleosome and set its state to silenced.
Select with probability β a nucleosome among all genes in the system. If the chosen nucleosome is active, then change it to silenced. If the chosen nucleosome is silenced, then change it with probability R to active.
One time unit corresponds to one update per nucleosome in the entire system of 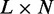 nucleosomes. Thus, in each time unit of a simulation, every nucleosome is on average attempted converted towards the active state, with actual conversions happening much less frequently.
nucleosomes. Thus, in each time unit of a simulation, every nucleosome is on average attempted converted towards the active state, with actual conversions happening much less frequently.
In our standard simulations, we assume that  , which in effect implies that a gene that is half covered by silenced nucleosomes, still retain 1/4 of its maximal activity. This rather soft repression limits the functional range of our model considerably. A much more robust differentiation is obtained by using a higher value of h or by simply assuming that there is only activity from genes i where the fraction of active nucleosomes, ai, has reached a finite threshold, say
, which in effect implies that a gene that is half covered by silenced nucleosomes, still retain 1/4 of its maximal activity. This rather soft repression limits the functional range of our model considerably. A much more robust differentiation is obtained by using a higher value of h or by simply assuming that there is only activity from genes i where the fraction of active nucleosomes, ai, has reached a finite threshold, say 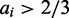 . The latter case is examined in Figure 4C.
. The latter case is examined in Figure 4C.
Figure 4.
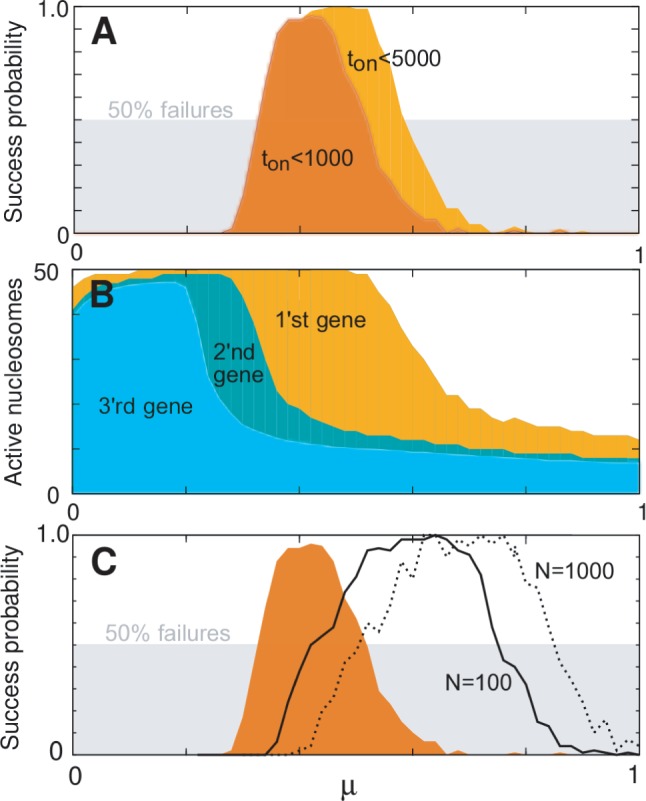
Model sensitivity to activation bias parameter μ, for 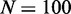 genes, L = 50 nucleosomes system with repression factor r = 1, hill h = 2 and noise
genes, L = 50 nucleosomes system with repression factor r = 1, hill h = 2 and noise 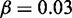 fixed. Data are averaged over 200 simulations. (A) Orange area shows the probability that one and only one gene becomes active within 5000 time units as function of asymmetry μ. Concurrent dark orange area marks success with the additional constraint that one gene becomes active within the first 1000 time units. Grey area marks the cut-off at 50% successful simulations. (B) Orange area marks the largest number of active nucleosomes within one gene during a 5000 time-unit simulation. When no genes reach full activity during the first 5000 time-units, no ORs have turned on. Cyan and dark cyan show maximal activity of the second and third most active genes, respectively. Where the second most active gene has many active nucleosomes, the two genes have shown simultaneous activity. (C) Orange area as in panel A. The two black curves refer to simulations with the sharper threshold function for gene activity described in ‘Materials and Methods’ section, and gene copy numbers as indicated. Only assigning feedback activity to genes where more than two-third of their nucleosomes are marked as active, even N = 1000 genes can differentiate successfully. Data for black curves are averaged over 50 simulations. See also Supplementary Figure S2 for sensitivity to parameters r, h, and β.
fixed. Data are averaged over 200 simulations. (A) Orange area shows the probability that one and only one gene becomes active within 5000 time units as function of asymmetry μ. Concurrent dark orange area marks success with the additional constraint that one gene becomes active within the first 1000 time units. Grey area marks the cut-off at 50% successful simulations. (B) Orange area marks the largest number of active nucleosomes within one gene during a 5000 time-unit simulation. When no genes reach full activity during the first 5000 time-units, no ORs have turned on. Cyan and dark cyan show maximal activity of the second and third most active genes, respectively. Where the second most active gene has many active nucleosomes, the two genes have shown simultaneous activity. (C) Orange area as in panel A. The two black curves refer to simulations with the sharper threshold function for gene activity described in ‘Materials and Methods’ section, and gene copy numbers as indicated. Only assigning feedback activity to genes where more than two-third of their nucleosomes are marked as active, even N = 1000 genes can differentiate successfully. Data for black curves are averaged over 50 simulations. See also Supplementary Figure S2 for sensitivity to parameters r, h, and β.
The OE is constantly renewed through neurogenesis (49–51). In adult rats, regeneration of OR neurons from immature progenitors is achieved within 2 weeks (29). Similar timescales of neurogenesis are observed in embryonic development (30,52). In contrast, reported lifetimes for neurons in rodents are very variable, ranging from 1 month to almost a year (29,53,54). With a turnover rate of histone modifications in the order of 10 min reported for the de-acetylation processes (55–58), we thus want our model to choose one subsystem within 1 week  time units in the simulations and contain exclusive expression of that subsystem for at least 1 month
time units in the simulations and contain exclusive expression of that subsystem for at least 1 month 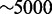 time units. Other modification processes may well be slower (59–61), but such change of timescale does not change our overall results. It would only move the parameter range for which we have acceptable differentiation properties.
time units. Other modification processes may well be slower (59–61), but such change of timescale does not change our overall results. It would only move the parameter range for which we have acceptable differentiation properties.
For each simulation, we hence run 5000 time units, starting from an all silenced state and requiring that only one gene turns on within this time window and that only one gene is active in the time window. In tread with previous nucleosome models, we score a subsystem as activated when the fraction of actively marked nucleosomes exceeds 2/3. Activation is considered lost when the fraction falls below 1/3 (40), but in fact an insignificant fraction of the genes that switch to become the single dominating gene will switch back to the silent state within the plotted time frame. The model was implemented in both C++ and fortran and are available on request.
RESULTS
Stable activity of one and only one subsystem
Our model of OR gene expression captures the basic property of the system, namely, the exclusive expression of just one gene out a large highly homologous gene population. We initiate the system with all nucleosomes in the silent state consistent with the observations by Magklara et al. (34). Within the simulations, we identify the state of the individual gene, i, as active, when the active fraction of nucleosomes, ai, exceeds 2/3. Figure 2 shows a simulation of N = 10 genes covered by L = 50 nucleosomes each. Gene 5 achieves activation as the number of active nucleosomes exceeds 33 at time 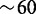 . Stochastic fluctuation and internal local bias towards the active state move the subsystem into a dominant active state. Activation increases the globally acting negative feedback, thus reinforcing the dominant gene by decreasing the probability for local activation of other genes.
. Stochastic fluctuation and internal local bias towards the active state move the subsystem into a dominant active state. Activation increases the globally acting negative feedback, thus reinforcing the dominant gene by decreasing the probability for local activation of other genes.
Figure 2.
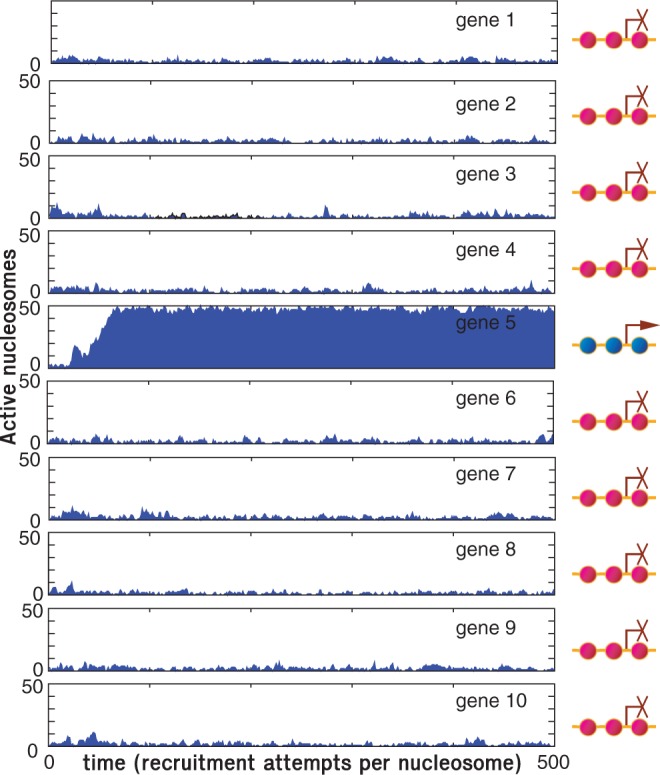
Simulation of  genes, each covered by
genes, each covered by 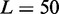 nucleosomes. The simulation shows that one gene is turned on early, whereas all other genes remain silenced throughout the simulation. In fact, with these parameters, all these genes stay silenced up to at least t = 5000 time units. Sketches in the right panel illustrate the OR gene state at the final time of the simulation. Crossed promoters indicate silent genes. In color version nucleosomes with silent and active modifications are shown as red and blue, respectively. Other parameters of the simulation are
nucleosomes. The simulation shows that one gene is turned on early, whereas all other genes remain silenced throughout the simulation. In fact, with these parameters, all these genes stay silenced up to at least t = 5000 time units. Sketches in the right panel illustrate the OR gene state at the final time of the simulation. Crossed promoters indicate silent genes. In color version nucleosomes with silent and active modifications are shown as red and blue, respectively. Other parameters of the simulation are 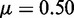 , overall repression factor r = 1, hill coefficient of repression h = 2 and noise conversion
, overall repression factor r = 1, hill coefficient of repression h = 2 and noise conversion 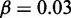 .
.
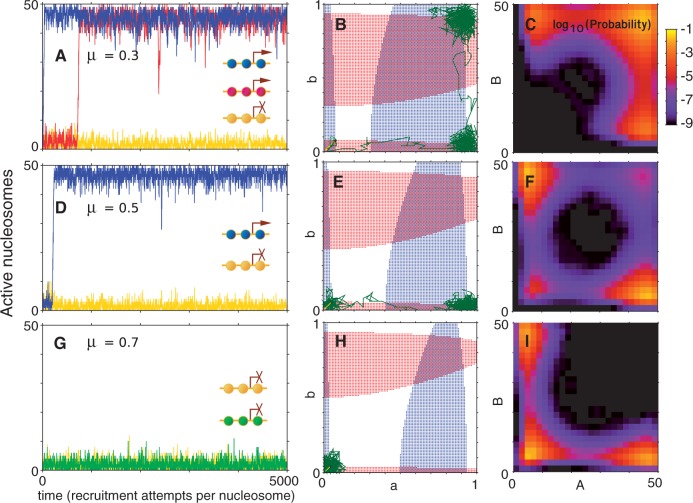
Success of a simulation may be accessed on three criteria. First, the system needs to selectively activate a single OR gene within a given time window. Second, the chosen gene should remain active for a considerable time. Third, no other OR genes may be activated while the initial OR gene is active. Figure 3A, D and G compactly show time courses like those of Figure 2 for a system of 100 genes at three different values of the local activation bias parameter μ. It is clear that increasing μ takes the system from defying the first criteria by switching on more than one gene, to fulfilling all criteria with a single active gene, and to failure owing to lack of turn on of any genes.
Figure 3.
Simulation of N = 100 genes, each covered by L = 50 nucleosomes. Left panels (A, D, G) show time course of the first activated gene (blue), second activated (red) and a few examples of other genes (yellow and green). Inserts show the promoter status of the correspondingly colored genes at the final simulation time. Crossed arrows indicate silent promoters. Middle panels (B, E, H) follow the trajectory of the two most expressed genes, identified by their active nucleosome fraction a and b, in a 2-d plane that illustrates deterministic drift of two individual genes, provided all other genes are assumed to act synchronously (see ‘Materials and Methods’ section and Supplementary Material). In the blue region, 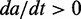 , whereas the red region shows where
, whereas the red region shows where 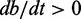 . (C, F, I) show the probability (lighter colour for higher) for the two most active genes in the system, obtained by stochastic simulation over 108 time-units. The negative logarithm of this probability may be interpreted as an epigenetic landscape (62,63), with states that to varying degree prefer to be in the corners, see also Supplementary Figure S1. Parameters are (A–C)
. (C, F, I) show the probability (lighter colour for higher) for the two most active genes in the system, obtained by stochastic simulation over 108 time-units. The negative logarithm of this probability may be interpreted as an epigenetic landscape (62,63), with states that to varying degree prefer to be in the corners, see also Supplementary Figure S1. Parameters are (A–C) 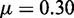 , r = 1, h = 2 and
, r = 1, h = 2 and 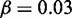 . (D–F)
. (D–F) 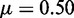 , r = 1, h = 2 and
, r = 1, h = 2 and 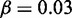 . (G–I)
. (G–I) 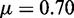 , r = 1, h = 2 and
, r = 1, h = 2 and 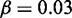 .
.
Activation barriers capture genes in silent state
Although simulations present a proof of concept for our model in reproducing the exclusive gene expression, the theoretical formulation of the model also let us examine in detail how the dominant gene expression is achieved. Arguing that all but the two most active genes are found at the same low level of activation, we graphically identify three stable fixed points for the states of the two genes (see ‘Materials and Methods’ section). The coloured regions in Figure 3B, E and H show where the net variation in gene activity fraction is positive for the two most active genes. Intersections between the regional limits are fixed points of the governing equations where the net variation in the gene activity fraction is zero. Fix points closest to the corners of the 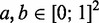 are stable as local variation in activity in gene ‘a’ and ‘b’ deterministically will return the system to the states of the fixed points (see ‘Materials and Methods’ section and Supplementary Section S1).
are stable as local variation in activity in gene ‘a’ and ‘b’ deterministically will return the system to the states of the fixed points (see ‘Materials and Methods’ section and Supplementary Section S1).
The 2-d planes in Figure 3B, E and H, include trajectories of the first versus the second most active gene from the simulations to the left and show how the success relies on two barriers. At low μ, examined in Figure 3B, the ‘first’ barrier stalls the system at the state where all genes are silenced until a random fluctuation causes activation of one of the genes. Subsequently, the system remains unstable against the passage of a second barrier to the state in the rightmost upper corner of Figure 3B. At larger μ, only one gene switches, as the second and larger barrier along the vertical axis in Figure 3E prevents a second gene from turning on once an initial switch has been made. At even larger μ, values the first barrier is so high that no gene may pass it, essentially stalling the system with all OR genes in the silent state (see Figure 3H). Thus, for a range of μ values, the system is successfully stalled in the lower right corner of the 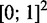 phase space with just one activated OR gene.
phase space with just one activated OR gene.
Interestingly, when the system is stalled in the lower right corner of Figure 3B, E or H, the repressed genes have a small fraction of their nucleosomes in the active state. This ‘nucleosomal noise’ should cause a residual production of receptors from each repressed gene of about 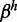 (see Supplementary Section S1). Sensitivity of the cell sensing system to the chosen gene implies that activity of this gene should be in excess of all the remaining N-1 genes together,
(see Supplementary Section S1). Sensitivity of the cell sensing system to the chosen gene implies that activity of this gene should be in excess of all the remaining N-1 genes together, 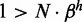 . A demand fulfilled for all examined parameters. Experimentally, the fraction of active nucleosomes in repressed genes could be measured and provide information about the size of β. The contrast between their gene product and the product of the chosen gene, on the other hand, provides information on the threshold for protein production, here parameterized by the hill coefficient h.
. A demand fulfilled for all examined parameters. Experimentally, the fraction of active nucleosomes in repressed genes could be measured and provide information about the size of β. The contrast between their gene product and the product of the chosen gene, on the other hand, provides information on the threshold for protein production, here parameterized by the hill coefficient h.
The differentiation of OR genes is also examined in terms of epigenetic landscapes generated from stochastic simulations. Using L = 50, the rightmost panels in Figure 3C, F and I examine the probability for the two most active genes in the system to be covered by ‘active’ nucleosomes. Light colours are associated with preferred states, whereas blue and black represent disfavoured combinations of activity. In Figure 3C, one can see that both the state where one gene is active, and the state where two genes are active represents ‘probability peaks’, corresponding to valleys of favoured states in an epigenetic landscape (62). In contrast, for the parameters of Figure 3F and I, the simultaneous activity of two genes is unlikely. Supplementary Figure S1 shows that the landscapes of Figure 3C, F and I are robust, with ‘passages’ between the valleys in the landscape being modulated by both L and an eventual time delay in the negative feedback.
Parameter sensitivity
Even for the best of parameter combinations, a fraction of the simulations will fail to fulfill the success criteria, see Figure 4. In the olfactory neuron, the loss of OR memory after axon extension to the pre-defined glomerulus of the olfactory bulb would be catastrophic. However, it is seen that ∼50% of neurons born in the epithelium of adult rats are lost within 5 days and 2 weeks (29,30). There is no explanation to this loss, but it has been argued that it could be a control mechanism to get rid of OR neuron expressing more than one receptor (39). Thus, for the model to successfully represent differentiation of olfactory neurons, only 50% of the simulations need to be successful, indicated by the grey-shaded area in Figure 4A. We initially set the criteria that the first OR gene should activate within 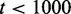 time units (see ‘Materials and Methods’ section). However, this restriction is somewhat conservative and relaxing it widens the success rate of the model, shown as concurrent orange areas in Figure 4A.
time units (see ‘Materials and Methods’ section). However, this restriction is somewhat conservative and relaxing it widens the success rate of the model, shown as concurrent orange areas in Figure 4A.
Additionally, the one-neuron one-receptor rule may not be as manifested as widely accepted within the field (7,64,65). To this end, we include in Figure 4B, the maximal activity of the second and third most active genes. These show that allowing a second or even third gene to activate further widens the μ range of successes. The model does not address how such tandem expression would affect axon guidance.
Robustness to gene copy number
The diversity and wide chromosomal position of OR gene clusters are generally considered a result of subsequent tandem gene duplications, gene conversions and recombination events and even conversions of entire coding regions. Accordingly, the mechanism of gene selection should be robust to drastic changes in gene numbers (7,22). The model successfully addresses this issue by selecting one OR gene, keeping the remaining genes silent. Provided the gene activity of silenced genes is strongly repressed, our model successfully addresses this issue. When this repression is parameterized by our standard value of h = 2, differentiation of a N = 500 system requires lower than our standard noise 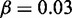 , see Supplementary Figure S2A. However, assuming a larger h value or a sharper threshold function, one easily obtain a larger functional range of differentiation for a fixed set of parameters, also when system size increases to N = 500 or N = 1000, see Figure 4C.
, see Supplementary Figure S2A. However, assuming a larger h value or a sharper threshold function, one easily obtain a larger functional range of differentiation for a fixed set of parameters, also when system size increases to N = 500 or N = 1000, see Figure 4C.
Pseudogenes defined as genes not able to effectuate feedback
Within the model settings that restrict the stable activation to a single OR gene, we considered the concept of pseudogenes. In this context, these are subsystems affected by feedback as previously described that do not produce the feedback them self. It will occasionally happen that first a pseudogene is activated, however, as the active pseudogene does not contribute to the negative feedback, R, another subsystem will eventually be activated and retain the dominant stable position. Thus, in accordance with experimental observations, our model predicts that one should often see co-expression of pseudogenes and functional OR genes (27), see Supplementary Figures S3 and S4.
Enhancers and modulation by transcription factors
Enhancer elements with features similar to the locus control region of the visual pigment genes have been identified for some subfamilies of OR genes (25,27,31). However, such elements are yet to be identified for all OR genes of the super gene family (31). In the context of our standard olfactory nucleosome modification model, we envision enhancer elements as a mean of disposing the OR gene for epigenetic activation. Decreasing the probability, μ, of a recruited event towards the silent state for one subsystem by as little as 15% greatly increase the frequency of that subsystem being activated, see Figure 5A. Such an increase in frequency for H-element associated genes has been reported (31,52,66).
Figure 5.
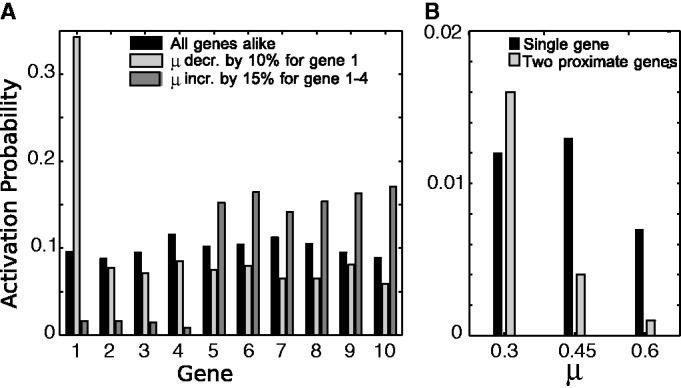
Predictions of sensitivity to enhancers and to double gene dosage: (A) Effect of small alterations in OR specific μ on the probability of switching on for the associated OR gene. Individual decrease (light grey bars) or zonal increase (dark grey bars) in the activation asymmetry μ might represent the effect of an enhancer element or the removal of zonal specific TFs. Each data set from 1000 simulations, with  genes, each covered by
genes, each covered by  nucleosomes,
nucleosomes,  ,
,  , r = 1, and h = 2. (B) Comparison between wild-type case, and an engineered situation where one gene contributed with double gene dosage to the feedback R. The likelihood of turning on the ‘doubled’ gene shown with light grey bars is smaller than for a normal gene to an extent that depend on μ. In case the ‘doubled’ gene is constructed as a tandem repeat, possibly with two reporter proteins inserted, the two identical promoters should become active together, but with smaller probability than the wild-type system. A reduction in probability that will pinpoint the effective value of μ. Each data set from 1000 simulations, with
, r = 1, and h = 2. (B) Comparison between wild-type case, and an engineered situation where one gene contributed with double gene dosage to the feedback R. The likelihood of turning on the ‘doubled’ gene shown with light grey bars is smaller than for a normal gene to an extent that depend on μ. In case the ‘doubled’ gene is constructed as a tandem repeat, possibly with two reporter proteins inserted, the two identical promoters should become active together, but with smaller probability than the wild-type system. A reduction in probability that will pinpoint the effective value of μ. Each data set from 1000 simulations, with 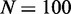 genes, each covered by
genes, each covered by 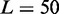 nucleosomes, noise
nucleosomes, noise 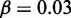 , r = 1, and h = 2.
, r = 1, and h = 2.
Conserved regions identified in alignments of OR genes include binding sites of known transcription factors (TFs) (13,32,33). To our knowledge, so far no single TF or TF-binding sites have been associated with the zonal exclusivity, even though TFs as Emx2 do alter the expression frequency of a large part of the OR genes (33). In our model, TFs may be incorporated, like enhancer elements, as chromatin modification control mechanisms (41), enhancing or reducing the probability of activation of their associated genes, see Figure 5A. The model shows that simple TF knockout experiments will not necessarily allow for a clear identification of such TFs as full exclusion of associated OR genes. Instead, the frequency by which the genes are chosen will decrease, whereas unrelated OR genes increase as reported for the Emx2 knockout mice (33).
Identification of regulatory elements
Reports on the size of the regulatory elements needed for a recapitulation of the punctuate OR gene expression in the OE, varies from 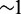 to >10 kb (13–15), with varying intrinsic levels of spatial restriction. The short and highly variable OR coding sequences may be sufficient to mark a transgene for the same feedback regulation as endogenous genes (15). In our model, we initially assume a regulatory size of ∼ kb or at least large enough to encompass 20–50 nucleosomes. However, in more general terms, the model presents a way of activating a single patch of the genome, including the feedback producing element, which is responsible for keeping the remaining patches silent.
to >10 kb (13–15), with varying intrinsic levels of spatial restriction. The short and highly variable OR coding sequences may be sufficient to mark a transgene for the same feedback regulation as endogenous genes (15). In our model, we initially assume a regulatory size of ∼ kb or at least large enough to encompass 20–50 nucleosomes. However, in more general terms, the model presents a way of activating a single patch of the genome, including the feedback producing element, which is responsible for keeping the remaining patches silent.
In our standard model, any early dominating OR gene will prevail. Thus, our model encompasses the experimental observations that OR transgene expression from a promoter that is active early in the development of the olfactory sensory neuron dominate the future differentiation (15).
The average OR genes of a locus are separated by 29 kb (6), allowing for separation between activated and silent regions, and implying that the activity of their respective nucleosomes is separated by substantial barriers. In transgene experiments, one may modify distances between genes to examine the spreading mechanisms associated to the local positive feedback. In particular, one may insert two genes just after each other, with identical promoters, and investigate whether such architecture facilitate non-synchronous turn-on of the genes, or synchronous activation as our model suggests. Notice that with standard promoter strength, the globally acting negative feedback will counteract any activity from such a coupled pair of genes (see Figure 5B), and thus it is recommended to reduce the strength of the two promoters.
DISCUSSION
Early in our approach to the olfactory system, we recapitulated the need for multiple stable states in the system. Relying on mechanisms like TF or enhancer element binding to the chosen OR gene inevitably couples the stability of the choice to these associations. As a result, such models often incorporate a transient ‘feedback mechanism’ to lock the system to one OR (19). In contrast, within a considerable time window, our nucleosome modification model of the OR gene regulation does not require solidifying agents.
The model pinpoints the need for localizing the dependence of olfactory co-regulation on the size and status of chromosomal modifications of the individual olfactory gene locus. It also identifies the need for finding the common factor that facilitates the interplay between the different olfactory genes. In particular, we aim to direct the search for this later globally acting feedback factor within pathways associated with histone-modifying enzymes.
Important experiments addressing the former role of the local nucleosome mediated feedback in the model could be initiated without explicit knowledge of the players. As previously described, placing two OR genes close on the DNA should result in co-expression, provided a reduction in the strength of the associated promoters. A way to experimentally circumvent this need for reduced promoters would be replacing one of the proximate genes with a pseudogene. Lack of consistent co-expression would falsify our model. Alternatively, it should be possible to express two ORs from non-olfactory regions, provided that these regions of the genome in some way are protected from silenced nucleosomes. We again emphasize that our model does not address how the neuron will cope with such eventual co-expression.
The problem of possibly modelling mutually exclusive gene expression by TFs have been addressed in both the X-chromosome inactivation (67) and in the olfactory system (68). In both cases, the nucleation of many cooperatively binding activators was essential, which with the requirement of maintaining one of many hundred genes active leads to a modelling requirement of having essentially all activators in the cell bound to the same gene at all times. In contrast, our model does not require extreme binding properties but solely relies on on-going dynamics of read–write enzymes.
Perspectives for the model beyond the olfactory system
OR neuron differentiation is a well described case of mutually exclusive gene expression and thus presented a compact model system for our nucleosome modification model of epigenetic differentiation. Examples of exclusive gene expression are described in several other organisms. Similar to African trypanosomes, Plasmodium falciparum malaria avoids host antibody responses by switching between expressed surface antigens (69). At least, the mutually exclusive gene expression of the malaria var gene is associated with epigenetic alternations in chromatin structure and has been compared with the OR gene choice (70,71). Our model thus presents a formulation of epigenetic differentiation that stretches far beyond the considered model system of olfactory neurons.
Perspectives for olfactory differentiation as a model system for coupled epigenetic landscapes
A final speculation springs from the fact that olfactory neuronal cells and the sense of smell is associated to the oldest part of the cortex. Thus, their genetic regulatory design may be found more widespread. In fact, the olfactory bulb may well be the predecessor of the enlarged cortex of mammals (72), and differentiation of olfactory neurons could give insight into the way we store memories in the brain. In particular, Figure 4B discloses the possibility to activate two receptors in a cell, a possibility that opens for a large increase in the number of epigenetic states. As ORs are believed to direct the axon-to-glomerulus association at the olfactory bulb, similar receptor families could modulate physical connections between other neuronal cells. Thereby receptors could couple the intrinsic epigenetic state of cells to the neural network architecture. An architecture where neurons expressing a common receptor could be coupled, and build branching signalling pathways when cells are allowed to have more than one receptor. Accordingly, the ability for individual neuronal cells to differentiate their behaviour on the basis of expression of membrane bound receptors opens for a new perspective on memory. A perspective where the epigenetic state of individual neuronal cells may be coupled to the real memory storage in our cortex.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–5 and Supplementary Theory and Results 1–4.
FUNDING
Danish National Research Foundation through the Center for Models of Life. Funding for open access charge: Danish National Research Foundation.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Ian Dodd and Danesh Moazed for discussions and Szabolcs Semsey for valued comments on the manuscript.
REFERENCES
- 1.Okamoto I, Otte AP, Allis CD, Reinberg D, Heard E. Epigenetic dynamics of imprinted X inactivation during early mouse development. Science. 2004;303:644–649. doi: 10.1126/science.1092727. [DOI] [PubMed] [Google Scholar]
- 2.Arney KL, Erhardt S, Drewell RA, Surani MA. Epigenetic reprogramming of the genome - from the germ line to the embryo and back again. Int. J. Dev. Biol. 2001;45:533–540. [PubMed] [Google Scholar]
- 3.Borst P, Ulbert S. Control of VSG gene expression sites. Mol. Biochem. Parasitol. 2001;114:17–27. doi: 10.1016/s0166-6851(01)00243-2. [DOI] [PubMed] [Google Scholar]
- 4.Chess A, Simon I, Cedar H, Axel R. Allelic inactivation regulates olfactory receptor gene expression. Cell. 1994;78:823–834. doi: 10.1016/s0092-8674(94)90562-2. [DOI] [PubMed] [Google Scholar]
- 5.Serizawa S, Ishii T, Nakatani H, Tsuboi A, Nagawa F, Asano M, Sudo K, Sakagami J, Sakano H, Ijiri T, et al. Mutually exclusive expression of odorant receptor transgenes. Nat. Neurosci. 2000;3:687–693. doi: 10.1038/76641. [DOI] [PubMed] [Google Scholar]
- 6.Zhang X, Firestein S. The olfactory receptor gene superfamily of the mouse. Nat. Neurosci. 2002;5:124–133. doi: 10.1038/nn800. [DOI] [PubMed] [Google Scholar]
- 7.Kambere M, Lane R. Co-regulation of a large and rapidly evolving repertoire of odorant receptor genes. BMC Neurosci. 2007;8:S2. doi: 10.1186/1471-2202-8-S3-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gilad Y. Olfactory Receptor Genes: Human Loss During Evolution. Oxford: Academic Press; 2009. [Google Scholar]
- 9.Zhang X, Firestein S. Genomics of Olfactory Receptors. Heidelberg: Springer Berlin; 2007. [DOI] [PubMed] [Google Scholar]
- 10.Norlin EM, Alenius M, Gussing F, Hgglund M, Vedin V, Bohm S. Evidence for gradients of gene expression correlating with zonal topography of the olfactory sensory map. Mol. Cell. Neurosci. 2001;18:283–295. doi: 10.1006/mcne.2001.1019. [DOI] [PubMed] [Google Scholar]
- 11.Zhang X, Rogers M, Tian H, Zhang X, Zou DJ, Liu J, Ma M, Shepherd GM, Firestein SJ. High-throughput microarray detection of olfactory receptor gene expression in the mouse. Proc. Natl Acad. Sci. USA. 2004;101:14168–14173. doi: 10.1073/pnas.0405350101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tietjen I, Rihel J, Dulac C. Single-cell transcriptional profiles and spatial patterning of the mammalian olfactory epithelium. Int. J. Dev. Biol. 2005;49:201–207. doi: 10.1387/ijdb.041939it. [DOI] [PubMed] [Google Scholar]
- 13.Glusman G, Sosinsky A, Ben-Asher E, Avidan N, Sonkin D, Bahar A, Rosenthal A, Clifton S, Roe B, Ferraz C, et al. Sequence, structure, and evolution of a complete human olfactory receptor gene cluster. Genomics. 2000;63:227–245. doi: 10.1006/geno.1999.6030. [DOI] [PubMed] [Google Scholar]
- 14.Vassalli A, Rothman A, Feinstein P, Zapotocky M, Mombaerts P. Minigenes impart odorant receptor-specific axon guidance in the olfactory bulb. Neuron. 2002;35:681–696. doi: 10.1016/s0896-6273(02)00793-6. [DOI] [PubMed] [Google Scholar]
- 15.Nguyen MQ, Zhou Z, Marks CA, Ryba NJ, Belluscio L. Prominent roles for odorant receptor coding sequences in allelic exclusion. Cell. 2007;131:1009–1017. doi: 10.1016/j.cell.2007.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ressler KJ, Sullivan SL, Buck LB. Information coding in the olfactory system: evidence for a stereotyped and highly organized epitope map in the olfactory bulb. Cell. 1994;79:1245–1255. doi: 10.1016/0092-8674(94)90015-9. [DOI] [PubMed] [Google Scholar]
- 17.Malnic B, Hirono J, Sato T, Buck LB. Combinatorialreceptor codes for odors. Cell. 1999;96:713–723. doi: 10.1016/s0092-8674(00)80581-4. [DOI] [PubMed] [Google Scholar]
- 18.Treloar HB, Feinstein P, Mombaerts P, Greer CA. Specificity of glomerular targeting by olfactory sensory axons. J. Neurosci. 2002;22:2469–2477. doi: 10.1523/JNEUROSCI.22-07-02469.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fuss SH, Ray A. Mechanisms of odorant receptor gene choice in Drosophila and vertebrates. Mol. Cell. Neurosci. 2009;41:101–112. doi: 10.1016/j.mcn.2009.02.014. [DOI] [PubMed] [Google Scholar]
- 20.DeMaria S, Ngai J. The cell biology of smell. J. Cell. Biol. 2010;191:443–452. doi: 10.1083/jcb.201008163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Godfrey PA, Malnic B, Buck LB. The mouse olfactory receptor gene family. Proc. Natl Acad. Sci. USA. 2004;101:2156–2161. doi: 10.1073/pnas.0308051100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Glusman G, Yanai I, Rubin I, Lancet D. The complete human olfactory subgenome. Genome Res. 2001;11:685–702. doi: 10.1101/gr.171001. [DOI] [PubMed] [Google Scholar]
- 23.Menashe I, Aloni R, Lancet D. A probabilistic classifier for olfactory receptor pseudogenes. BMC Bioinformatics. 2006;7:393. doi: 10.1186/1471-2105-7-393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shykind BM, Rohani S, O’Donnell S, Nemes A, Mendelsohn M, Sun Y, Axel R, Barnea G. Gene switching and the stability of odorant receptor gene choice. Cell. 2004;117:801–815. doi: 10.1016/j.cell.2004.05.015. [DOI] [PubMed] [Google Scholar]
- 25.Serizawa S, Miyamichi K, Sakano H. One neuron-one receptor rule in the mouse olfactory system. Trends Genet. 2004;20:648–653. doi: 10.1016/j.tig.2004.09.006. [DOI] [PubMed] [Google Scholar]
- 26.Qasba P, Reed RR. Tissue and zonal-specific expression of an olfactory receptor transgene. J. Neurosci. 1998;18:227–236. doi: 10.1523/JNEUROSCI.18-01-00227.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Serizawa S, Miyamichi K, Nakatani H, Suzuki M, Saito M, Yoshihara Y, Sakano H. Negative feedback regulation ensures the one receptor-one olfactory neuron rule in mouse. Science. 2003;302:2088–2094. doi: 10.1126/science.1089122. [DOI] [PubMed] [Google Scholar]
- 28.Lewcock JW, Reed RR. A feedback mechanism regulates monoallelic odorant receptor expression. Proc. Natl Acad. Sci. USA. 2004;101:1069–1074. doi: 10.1073/pnas.0307986100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schwob J, Szumowski K, Stasky A. Olfactory sensory neurons are trophically dependent on the olfactory bulb for their prolonged survival. J. Neurosci. 1992;12:3896–3919. doi: 10.1523/JNEUROSCI.12-10-03896.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Taniguchi K, Taniguchi K. Embryonic and postnatal differentiation of olfactory epithelium and vomeronasal organ in the syrian hamster. J Vet Med Sci. 2008;70:57–64. doi: 10.1292/jvms.70.57. [DOI] [PubMed] [Google Scholar]
- 31.Fuss SH, Omura M, Mombaerts P. Local and cis effects of the H element on expression of odorant receptor genes in mouse. Cell. 2007;130:373–384. doi: 10.1016/j.cell.2007.06.023. [DOI] [PubMed] [Google Scholar]
- 32.Hoppe R, Frank H, Breer H, Strotmann J. The clustered olfactory receptor gene family 262: genomic organization, pro- motor elements, and interacting transcription factors. Genome Res. 2003;13:2674–2685. doi: 10.1101/gr.1372203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McIntyre JC, Bose SC, Stromberg AJ, McClintock TS. Emx2 stimulates odorant receptor gene expression. Chem. Senses. 2008;33:825–837. doi: 10.1093/chemse/bjn061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Magklara A, Yen A, Colquitt B, Clowney E, Allen W, Markenscoff-Papadimitriou E, Evans Z, Kheradpour P, Mountoufaris G, Carey C, et al. An epigenetic signature for monoallelic olfactory receptor expression. Cell. 2011;145:555–570. doi: 10.1016/j.cell.2011.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eggan K, Baldwin K, Tackett M, Osborne J, Gogos J, Chess A, Axel R, Jaenisch R. Mice cloned from olfactory sensory neurons. Nature. 2004;428:44–49. doi: 10.1038/nature02375. [DOI] [PubMed] [Google Scholar]
- 36.Shykind BM. Regulation of odorant receptors: one allele at a time. Hum. Mol. Genet. 2005;14(Suppl. 1):R33–R39. doi: 10.1093/hmg/ddi105. [DOI] [PubMed] [Google Scholar]
- 37.Lomvardas S, Barnea G, Pisapia DJ, Mendelsohn M, Kirkland J, Axel R. Interchromosomal interactions and olfactory receptor choice. Cell. 2006;126:403–413. doi: 10.1016/j.cell.2006.06.035. [DOI] [PubMed] [Google Scholar]
- 38.Savarese F, Grosschedl R. Blurring cis and trans in gene regulation. Cell. 2006;126:248–250. doi: 10.1016/j.cell.2006.07.008. [DOI] [PubMed] [Google Scholar]
- 39.McClintock TS. Achieving singularity in mammalian odorant receptor gene choice. Chem. Senses. 2010;35:447–457. doi: 10.1093/chemse/bjq041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dodd IB, Micheelsen MA, Sneppen K, Thon G. Theoretical analysis of epigenetic cell memory by nucleosome modification. Cell. 2007;129:813–822. doi: 10.1016/j.cell.2007.02.053. [DOI] [PubMed] [Google Scholar]
- 41.Sneppen K, Micheelsen MA, Dodd IB. Ultrasensitive gene regulation by positive feedback loops in nucleosome modification. Mol. Syst. Biol. 2008;4:182. doi: 10.1038/msb.2008.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Angel A, Song J, Dean C, Howard M. A polycomb-based switch underlying quantitative epigenetic memory. Nature. 2011;476:105–108. doi: 10.1038/nature10241. [DOI] [PubMed] [Google Scholar]
- 43.Micheelsen MA, Mitarai N, Sneppen K, Dodd IB. Theory for the stability and regulation of epigenetic landscapes. Phys. Biol. 2010;7:026010. doi: 10.1088/1478-3975/7/2/026010. [DOI] [PubMed] [Google Scholar]
- 44.Dodd IB, Sneppen K. Barriers and silencers: a theoretical toolkit for control and containment of nucleosome-based epigenetic states. J. Mol. Biol. 2011;414:624–637. doi: 10.1016/j.jmb.2011.10.027. [DOI] [PubMed] [Google Scholar]
- 45.MacDonald JL, Gin CS, Roskams AJ. Stage-specific induction of DNA methyltransferases in olfactory receptor neuron development. Dev. Biol. 2005;288:461–473. doi: 10.1016/j.ydbio.2005.09.048. [DOI] [PubMed] [Google Scholar]
- 46.Sammeta N, Yu TT, Bose SC, McClintock TS. Mouse olfactory sensory neurons express 10,000 genes. J. Comp. Neurol. 2007;502:1138–1156. doi: 10.1002/cne.21365. [DOI] [PubMed] [Google Scholar]
- 47.Nickell MD, Breheny P, Stromberg AJ, McClintock TS. Genomics of mature and immature olfactory sensory neurons. J. Comp. Neurol. 2012;520:2608–2629. doi: 10.1002/cne.23052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xia J, Broad K, Emson P, Keverne E. Epigenetic modification of vomeronasal (V2r) precursor neurons by histone deacetylation. Neuroscience. 2010;169:1462–1472. doi: 10.1016/j.neuroscience.2010.05.071. [DOI] [PubMed] [Google Scholar]
- 49.Graziadei P, Graziadei GA. Neurogenesis and neuron regeneration in the olfactory system of mammals. Part I: morphological aspects of differentiation and structural organization of the olfactory sensory neurons. J. Neurocytol. 1979;8:1–18. doi: 10.1007/BF01206454. [DOI] [PubMed] [Google Scholar]
- 50.Samanen DW, Forbes WB. Replication and differentiation of olfactory receptor neurons following axotomy in the adult hamster: a morphometric analysis of postnatal neurogenesis. J. Comp. Neurol. 1984;225:201–211. doi: 10.1002/cne.902250206. [DOI] [PubMed] [Google Scholar]
- 51.Weiler E, Farbman AI. Proliferation in the rat olfactory epithelium: age-dependent changes. J. Neurosci. 1997;17:3610–3622. doi: 10.1523/JNEUROSCI.17-10-03610.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rodriguez-Gil DJ, Treloar HB, Zhang X, Miller AM, Two A, Iwema C, Firestein SJ, Greer CA. Chromosomal location-dependent nonstochastic onset of odor receptor expression. J. Neurosci. 2010;30:10067–10075. doi: 10.1523/JNEUROSCI.1776-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hinds J, Hinds PL, McNelly NA. An autoradiographic study of the mouse olfactory epithelium: evidence for long-lived receptors. Anat. Rec. 1984;210:375–383. doi: 10.1002/ar.1092100213. [DOI] [PubMed] [Google Scholar]
- 54.Mackay-Sim A, Kittel P. On the life span of olfactory receptor neurons. Eur. J. Neurosci. 1991;3:209–215. doi: 10.1111/j.1460-9568.1991.tb00081.x. [DOI] [PubMed] [Google Scholar]
- 55.Dose A, Liokatis S, Theillet FX, Selenko P, Schwarzer D. NMR profiling of histone deacetylase and acetyl-transferase activities in real time. ACS Chem. Biol. 2011;6:419–424. doi: 10.1021/cb1003866. [DOI] [PubMed] [Google Scholar]
- 56.Sun JM, Chen HY, Davie JR. Effect of estradiol on histone acetylation dynamics in human breast cancer cells. J. Biol. Chem. 2001;276:49435–49442. doi: 10.1074/jbc.M108364200. [DOI] [PubMed] [Google Scholar]
- 57.Sun JM, Spencer VA, Chen HY, Li L, Davie JR. Measurement of histone acetyltransferase and histone deacetylase activities and kinetics of histone acetylation. Methods. 2003;31:12–23. doi: 10.1016/s1046-2023(03)00083-5. [DOI] [PubMed] [Google Scholar]
- 58.Covault J, Chalkley R. The identification of distinct populations of acetylated histone. J. Biol. Chem. 1980;255:9110–9116. [PubMed] [Google Scholar]
- 59.Thomas G, Lange HW, Hempel K. Kinetics of histone methylation in vivo and its relation to the cell cycle in Ehrlich ascites tumor cells. Eur. J. Biochem. 1975;51:609–615. doi: 10.1111/j.1432-1033.1975.tb03963.x. [DOI] [PubMed] [Google Scholar]
- 60.Sweet SM, Li M, Thomas PM, Durbin KR, Kelleher NL. Kinetics of re-establishing H3K79 methylation marks in global human chromatin. J. Biol. Chem. 2010;285:32778–32786. doi: 10.1074/jbc.M110.145094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zee BM, Levin RS, Xu B, LeRoy G, Wingreen NS, Garcia BA. Kinetics of re-establishing H3K79 methylation marks in global human chromatin. J. Biol. Chem. 2010;285:3341–3350. doi: 10.1074/jbc.M110.145094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Waddington CH. The Strategy of the Genes: A Discussion of Some Aspects of Theoretical Biology. London: Allen & Unwin; 1957. [Google Scholar]
- 63.Goldberg AD, Allis CD, Bernstein E. Epigenetics: a landscape takes shape. Cell. 2007;128:635–638. doi: 10.1016/j.cell.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 64.Mombaerts P. Odorant receptor gene choice in olfactory sensory neurons: the one receptor-one neuron hypothesis revisited. Curr. Opin. Neurobiol. 2004;14:31–36. doi: 10.1016/j.conb.2004.01.014. [DOI] [PubMed] [Google Scholar]
- 65.Tian H, Ma M. Activity plays a role in eliminating olfactory sensory neurons expressing multiple odorant receptors in the mouse septal organ. Mol. Cell. Neurosci. 2008;38:484–488. doi: 10.1016/j.mcn.2008.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Khan M, Vaes E, Mombaerts P. Regulation of the probability of mouse odorant receptor gene choice. Cell. 2011;147:907–921. doi: 10.1016/j.cell.2011.09.049. [DOI] [PubMed] [Google Scholar]
- 67.Scialdone A, Cataudella I, Barbieri M, Prisco A, Nicodemi M. Conformation regulation of the X chromosome inactivation center: a model. PLoS Comput. Biol. 2011;7:e1002229. doi: 10.1371/journal.pcbi.1002229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kolterman BE, Iossifov I, Koulakov AA. A race model for singular olfactory receptor expression. 2012 darXiv 1201.233 [q-bio.MN] [Google Scholar]
- 69.Dzikowski R, Frank M, Deitsch K. Mutually exclusive expression of virulence genes by malaria parasites is regulated independently of antigen production. PLoS Pathog. 2006;2:e22. doi: 10.1371/journal.ppat.0020022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Freitas-Junior LH, Hernandez-Rivas R, Ralph SA, Montiel-Condado D, Ruvalcaba-Salazar OK, Rojas-Meza AP, Mâncio-Silva L, Leal-Silvestre RJ, Marques Gontijo A, Shorte S, et al. Telomeric heterochromatin propagation and histone acetylation control mutually exclusive expression of antigenic variation genes in malaria parasites. Cell. 2005;121:25–36. doi: 10.1016/j.cell.2005.01.037. [DOI] [PubMed] [Google Scholar]
- 71.Duraisingh MT, Voss TS, Marty AJ, Duffy MF, Good RT, Thompson JK, Freitas-Junior LH, Scherf A, Crabb BS, Cowman AF. Heterochromatin silencing and locus repositioning linked to regulation of virulence genes in plasmodium falciparum. Cell. 2005;121:13–24. doi: 10.1016/j.cell.2005.01.036. [DOI] [PubMed] [Google Scholar]
- 72.Rowe TB, Macrini TE, Luo ZX. Fossil evidence on origin of the mammalian brain. Science. 2011;332:955–957. doi: 10.1126/science.1203117. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.