Abstract
The leucine-specific domain (LSD) is a compact well-ordered module that participates in positioning of the conserved KMSKS catalytic loop in most leucyl-tRNA synthetases (LeuRSs). However, the LeuRS from Mycoplasma mobile (MmLeuRS) has a tetrapeptide GKDG instead of the LSD. Here, we show that the tetrapeptide GKDG can confer tRNA charging and post-transfer editing activity when transplanted into an inactive Escherichia coli LeuRS (EcLeuRS) that has had its LSD deleted. Reciprocally, the LSD, together with the CP1-editing domain of EcLeuRS, can cooperate when inserted into the scaffold of the minimal MmLeuRS, and this generates an enzyme nearly as active as EcLeuRS. Further, we show that LSD participates in tRNALeu recognition and favours the binding of tRNAs harbouring a large loop in the variable arm. Additional analysis established that the Lys598 in the LSD is the critical residue for tRNA binding. Conversion of Lys598 to Ala simultaneously reduces the tRNA-binding strength and aminoacylation and editing capacities, indicating that these factors are subtly connected and controlled at the level of the LSD. The present work provides a novel framework of co-evolution between LeuRS and its cognate tRNA through LSD.
INTRODUCTION
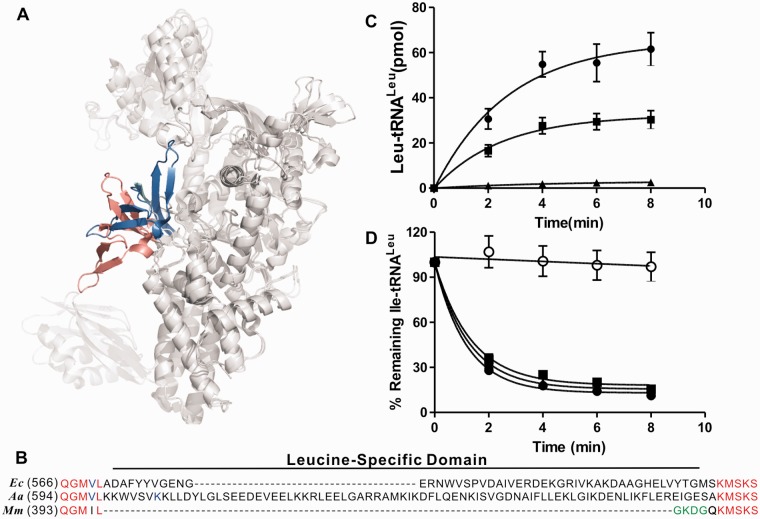
Aminoacyl-tRNA synthetases (aaRS) are a large and diverse family of enzymes that catalyze the attachment of amino acids to their cognate tRNAs. Each aaRS specifically attaches its cognate amino acid to its corresponding tRNA isoacceptor. A two-step process is performed by the aaRS during aminoacylation: (i) activation of the amino acid by ATP hydrolysis to form an aminoacyl-adenylate intermediate; (ii) transfer of the aminoacyl moiety from the intermediate to the cognate tRNA isoacceptor to make the aminoacyl-tRNA (1). Based on sequence homology and the structures of the catalytic active sites, aaRSs are divided into two classes (2). Leucyl-tRNA synthetase (LeuRS) is a class I aaRS that has an active site folded to form a typical Rossmann dinucleotide-binding fold. According to evolutionary models, the primitive catalytic core of LeuRS was extended by the insertion and appendage of additional domains (also called modules) (3). Most LeuRSs carry a large insert called the connective polypeptide 1 (CP1) domain that is responsible for the amino acid-editing function. LeuRSs also exhibit tRNA-binding domains that recognize and bind tRNALeu isoacceptors (4–7). A well-ordered module inserted into the catalytic domain, named the leucine-specific domain (LSD), is also found in most bacterial and some eukaryotic LeuRSs. LSD is connected to the KMSKS motif via a β-ribbon. The three-dimensional structure of the Thermus thermophilus LeuRS (TtLeuRS) shows that the LSD contains five β-strands and two short α-helices (3,5). In comparison, the LSD of Escherichia coli LeuRS (EcLeuRS) exhibits an additional extended β-hairpin (4). Crystal studies have also revealed that the LSD plays a critical role in positioning the conserved catalytic KMSKS loop during aminoacylation reactions (4).
Although the LSD is mainly found in prokaryotic LeuRSs, it is not highly conserved in sequence or length (3,4,8). The heterodimeric αβ-LeuRS from Aquifex aeolicus (AaLeuRS) has one of the largest LSDs, and this also serves to split the enzyme into two subunits (3,9). The LSD can also be missing completely in some species, such as Bacillus subtilis or Mycoplasma mobile, in which LeuRS is remarkable for the complete absence of a CP1-editing domain (8,10). In addition, sequence alignment has shown that the LSD in the LeuRS from M. mobile (MmLeuRS) is replaced by the tetrapeptide 398GKDG401 (4,10).
A recent study revealed that the CP1 domain and LSD of EcLeuRS both undergo large rotations when tRNA shifts from the synthetic site to the editing active site (Figure 1A) (4). The CP1 domain rotates by 12° to open up a passage for the translocation of the 3′ end of the tRNA, while the more dynamic LSD, together with the adjacent catalytically crucial KMSKS loop, is rotated by about 33° between the aminoacylation and editing conformations. Consistently, both the CP1 domain and LSD positions move by about 19° and 35° in the TtLeuRS when comparing the aminoacylation and editing conformations (5). Another study indicated that the tRNA-triggered conformational rearrangement leads to inter-domain communication between the editing and synthetic domains of EcLeuRS (11). All these data strongly suggest that both the CP1 domain and LSD are functionally connected and cooperate during the aminoacylation and editing reactions.
Figure 1.
Impact of LSD mutations on aminoacylation and editing of EcLeuRS. (A) Three-dimensional view of EcLeuRS showing the LSD motion in the aminoacylation (blue) and editing state (red) (PDB entry 4AQ7 and 4ARC). (B) Sequence alignment based on structural elements of the LeuRS LSD; the tetrapeptide linker is highlighted in green. Ec, Escherichia coli; Aa, Aquifex aeolicus; Mm, Mycoplasma mobile. (C) Aminoacylation of 10 µM EctRNALeu by 5 nM of EcLeuRS (black circle), EcLeuRS-GKDG (black square) and EcLeuRS-AAAA (black triangle). (D) Hydrolysis of 1 µM [3H]-Ile-EctRNALeu by 5 nM of EcLeuRS (black circle), EcLeuRS-GKDG (black square), EcLeuRS-AAAA (black triangle) and no enzyme (open circle) .
tRNALeu, together with tRNASer and tRNATyr, are class II tRNAs which are characterized by the presence of both a long variable stem and loop (12). Interactions between LeuRS and tRNALeu have been extensively investigated, and the conserved A73 nucleotide is considered to be the main element for identification. The amino acid-accepting end (CCA76) of EctRNALeu is critical for both the aminoacylation and the editing processes (13). The tertiary interactions between the D- and T-loops that determine the tRNA folding are additional critical elements of the leucine identity (14). In addition, tRNA elements that are critical during the editing process have been detected in the anticodon arms of tRNALeu from A. aeolicus and Saccharomyces cerevisiae (15,16).
In this present study, the LSD of EcLeuRS was substituted with the minimal tetrapeptide linker GKDG from the MmLeuRS, and this created a chimeric mutant named EcLeuRS-GKDG. In addition, the LSD and CP1 domain of EcLeuRS were inserted into the minimal MmLeuRS, and this produced another chimera termed MmLeuRS-CP1/LSD. By comparing the catalytic performances of these chimeric enzymes, we found that the LSD is essential for neither aminoacylation nor editing functions of LeuRSs. However, LSD participates in tRNA binding, and it is able to discriminate between different tRNALeu isoacceptors. Indeed, the LSD acts as a sensor that can measure the size of the V-arm loop and identify the nucleotide at position 20 of the tRNALeu. These results highlight the role of the LSD during tRNA recognition and suggest that interactions between the LSD and tRNALeu might favour binding in both aminoacylation and editing catalytic steps. Altogether, these results emphasize the modular nature of the LSD as well as the important contribution played by the other synthetase modules in enhancing catalytic efficiency and tRNA specificity.
MATERIALS AND METHODS
Materials
l-leucine, l-norvaline (Nva), ATP, Tris-HCl buffer, MgCl2 solution and dithiothreitol (DTT) were purchased from Sigma (USA). [3H] l-leucine, [3H] l-isoleucine and adenosine 5′-[α-32P] triphosphate were obtained from PerkinElmer Life Sciences (USA). PEI Cellulose F plates for thin layer chromatography (TLC) were purchased from Merck (Germany). T4 DNA ligase and other restriction endonucleases were from MBI Fermentas (Lithuania). DEAE-Sepharose CL-6B and SuperdexTM 75 were purchased from GE Healthcare (USA). Ni2+-NTA Superflow was purchased from Qiagen, Inc. (Germany). Plasmid pET30a was obtained from Novagen (USA), and E. coli strain BL21 (DE3) was from Invitrogen (USA). The expression vector pTrc99B and E. coli strain MT102 were gifts from Dr. J. Gangloff of the Institut de Biologie Moléculaire et Cellulaire du CNRS, Strasbourg, France.
Expression and purification of MmLeuRS, EcLeuRS and their mutants
The definition of the LSD in EcLeuRS was based on the crystal structure of EcLeuRS (PDB entry 4ARC) and sequence alignment. The LSD of EcLeuRS spans from A571 to M617. Each of the enzymes was expressed in E. coli BL21 (DE3) with a His6-tag fused at the N-terminus. The enzymes were purified by affinity chromatography using Ni-NTA (Ni2+nitrilotriacetate) Superflow resin, followed by gel-filtration chromatography with SuperdexTM 75. The final concentration was determined using a Bradford protein assay as described in the manufacturer’s protocol (Bio-Rad, Hercules, CA, USA).
The genes encoding the various mutants were constructed using the KOD Plus Mutagenesis Kit (Toyobo Life Science) and confirmed by DNA sequencing (BioSun Bioscience). Insertion of the CP1 domain of EcLeuRS into MmLeuRS was performed as described previously (17). Insertion of the LSD of EcLeuRS into MmLeuRS-CP1 was performed in several steps. First, the 398GKDG401 peptide was deleted from the MmLeuRS-CP1, and then the 47 amino acid residues from the LSD of the EcLeuRS (from A571 to M617) were added progressively by five rounds of mutagenesis.
Preparation of RNA substrates
E. coli tRNALeuGAG (EctRNALeu) with an accepting activity of 1400 pM/A260 was prepared from overproducing strains constructed in our laboratory (18). In vitro transcription of MmtRNALeu and mutated derivatives was performed as described previously (17). The accepting activities of the MmtRNALeuUAA and MmtRNALeuUAG transcripts and the mutated derivatives (A6G, C20U, C67U, V-arm-4 nt, V-arm-5 nt, C20U + V-arm-5 nt, A6G + V-arm-5 nt) were all between 1200–1500 pM/A260. [3H]Ile-EctRNALeu, [3H]Ile-MmtRNALeu and its mutants were obtained using the editing-deficient EcLeuRS-Y330D mutant as described previously (19).
tRNA charging, misacylation and deacylation
Aminoacylation activities of MmLeuRS, EcLeuRS and their mutants were measured in a reaction mixture containing 100 mM Tris-HCl (pH 7.8), 30 mM KCl, 12 mM MgCl2, 0.5 mM DTT, 4 mM ATP, 10 μM tRNALeu, 40 μM [3H]Leu (11 Ci/mM) and the enzyme (5 nM EcLeuRS or 20 nM MmLeuRS and their mutants). Reactions were carried out at 30°C for MmLeuRS and the mutants, while EcLeuRS and derivatives were assayed at 37°C. For Km determinations, tRNA concentrations ranged 0.5–30 µM. Misacylation assays were performed under similar conditions, except that 40 µM [3H]Ile (30 Ci/mM; PerkinElmer) and 1 µM of enzyme were used. The deacylation reaction was measured by determining hydrolytic rates, and this was performed at 30°C in 100 mM Tris-HCl (pH 7.5), 30 mM KCl, 12 mM MgCl2, 0.5 mM MgCl2, 0.5 mM DTT and 1 μM [3H]Ile-tRNALeu. Reactions were initiated with enzyme diluted to 20 nM. Because radioactive Nva is commercially unavailable, [3H]Ile was used as a source to prepare mischarged tRNALeu.
AMP formation
The net effect of the editing reaction is the consumption of ATP. Therefore, editing can be measured through AMP formation in the presence of a non-cognate amino acid. AMP formation rates of MmLeuRS, EcLeuRS and their mutants were measured as described previously (19). The reaction mixture contained 100 mM Tris-HCl (pH 7.8), 30 mM KCl, 12 mM MgCl2, 5 mM DTT, 5 U/ml pyrophosphatase (Roche), 3 mM ATP, 20 nM [α-32P] ATP (3000 Ci/mM; PerkinElmer), 15 mM Nva and the presence or absence of 5 μM tRNALeu. The reaction was initiated by the addition of 1 μM MmLeuRS-CP1/LSD (at 30°C), or 0.2 μM for EcLeuRS and the mutant enzymes (at 37°C). At regular time intervals, aliquots of 1.5 µl were quenched in 6 µl of 200 mM sodium acetate (pH 5.0). Quenched aliquots (1.5 µl each) were spotted in duplicate on polyethyleneimine cellulose plates (PEI, Merck) that had been pre-washed with water. Separation of [32P] aminoacyl-adenylate, [32P]AMP and [32P]ATP was performed by developing TLC plates in the presence of 0.1 M ammonium acetate and 5% acetic acid. Plates were visualized by phosphorimaging, and data were analyzed using Multi Gauge V3.0 software (Fujifilm). The grey densities of [32P]AMP spots were compared with those of known [32P]ATP concentrations. Rate constants (kobs) were obtained from graphs of [32P]AMP formation plotted against time.
RESULTS
The LSD is not essential for aminoacylation activity and post-transfer editing
MmLeuRS is an exceptionally small LeuRS that lacks both the CP1 domain and the LSD, and sequence alignment shows that these two domains are replaced by a nonapeptide linker 227KEEIDGKIT235 and a tetrapeptide linker 398GKDG401, respectively (Figure 1B). Previous studies have shown that the nonapeptide linker from MmLeuRS can replace the CP1 domain of EcLeuRS to permit aminoacylation (17). In this present study, we examined whether the tetrapeptide GKDG from MmLeuRS could replace the LSD of the EcLeuRS. The resulting mutant that lacked the LSD was called EcLeuRS-GKDG. The catalytic efficiency (kcat/Km) of EcLeuRS-GKDG for EctRNALeu aminoacylation was just more than half of that of the native EcLeuRS (Figure 1C, Table 1), indicating that the GKDG sequence of MmLeuRS could functionally replace the 47 amino acid residues of the LSD in EcLeuRS. In parallel, we constructed a similar mutant to contain a tetra Ala peptide instead of the GKDG insertion but the resulting mutant (EcLeuRS-AAAA) was inactive in the aminoacylation reaction (Figure 1C) despite intact folding as shown by CD-spectroscopy analysis (Supplementary Figure S1). Compared with results obtained in a previous study (8), the GKDG insertion led to much better recovery of activity in EcLeuRS (55% aminoacylation activity of the wild-type enzyme, Table 1). However, the EcLeuRS-AAAA mutant only displayed 0.55% of the aminoacylation activity of the wild-type enzyme (Table 1), and approximately 1/6 the activity of the previously reported ΔLSD-valRStt mutant (3.5%), which was obtained by using a seven-residue sequence (VLDEKGQ) from T. thermophilus ValRS instead of the LSD of EcLeuRS (8). These results indicate that the 398GKDG401 of MmLeuRS is a kind of minimal functional domain. In addition, both EcLeuRS-GKDG and EcLeuRS-AAAA exhibited intact deacylation activity for mischarged Ile-tRNALeu (Figure 1D), further proving that the native LSD does not play a critical role during the deacylation of tRNA (17).
Table 1.
Kinetic constants of various LeuRSs determined in the aminoacylation reaction
|
EctRNALeuGAG |
MmtRNALeuUAG |
D factor | |||||
|---|---|---|---|---|---|---|---|
| Enzyme | Km (µM) | kcat (s−1) | kcat/Km (s−1 µM−1) | Km (µM) | kcat (s−1) | kcat/Km (s−1 µM−1) | |
| EcLeuRSa | 2.2 ± 0.14 | 4.9 ± 0.30 | 2.2 | 10.6 ± 1.2 | 4.2 ± 0.40 | 0.40 | 5.5 |
| EcLeuRS-GKDG | 1.2 ± 0.10 | 1.4 ± 0.11 | 1.2 | 1.6 ± 0.12 | 2.1 ± 0.13 | 1.3 | 0.92 |
| EcLeuRS-AAAA | 1.6 ± 0.14 | (1.9 ± 0.21) × 10−2 | 0.012 | 0.60 ± 0.052 | (1.4 ± 0.12) × 10−2 | 0.023 | 0.52 |
| MmLeuRSa | 7.5 ± 0.90 | 1.8 ± 0.21 | 0.24 | 7.6 ± 0.80 | 2.0 ± 0.30 | 0.26 | 0.92 |
| MmLeuRS-CP1a | 4.3 ± 0.40 | 1.1 ± 0.15 | 0.25 | 4.6 ± 0.50 | 1.0 ± 0.17 | 0.22 | 1.1 |
| MmLeuRS-CP1/LSD | 1.3 ± 0.12 | 1.4 ± 0.11 | 1.1 | 1.7 ± 0.13 | 0.51 ± 0.049 | 0.30 | 3.7 |
D is the discrimination factor of the different LeuRSs for the two bacterial tRNAs, and this value was calculated as follows: D = kcat/Km(EctRNALeuGAG) / kcat/Km(MmtRNALeuUAG). Kinetic constants were determined using the tRNA charging assay described in the experimental section except the concentration was 100 nM for EcLeuRS-AAAA and from 0.2 to 20 μM for tRNAs. All parameters represent the average of three trials with the standard deviations indicated.
aData from Tan et al. (17).
A mutagenesis study was carried out to further explore the role in MmLeuRS of the residues of the GKDG peptide. Each of the residues in the tetrapeptide was mutated to Ala separately. All the mutants displayed altered tRNA-charging activity. Moreover, substitution of the flexible Gly398 and Gly401 to rigid Pro residues severely impaired aminoacylation activity to levels comparable with a full deletion of the tetrapeptide linker (Supplementary Table S1). These data suggest that the GKDG peptide of MmLeuRS plays a critical role in providing flexibility to the catalytic site.
As the two catalytic activities of EcLeuRS (aminoacylation and editing) do not require the presence of the 47-amino acid LSD, this raises questions concerning the conservation of this module in most prokaryotic LeuRS during evolution.
The LSD of EcLeuRS favours aminoacylation but inhibits tRNA-independent pre-transfer editing when inserted into MmLeuRS-CP1
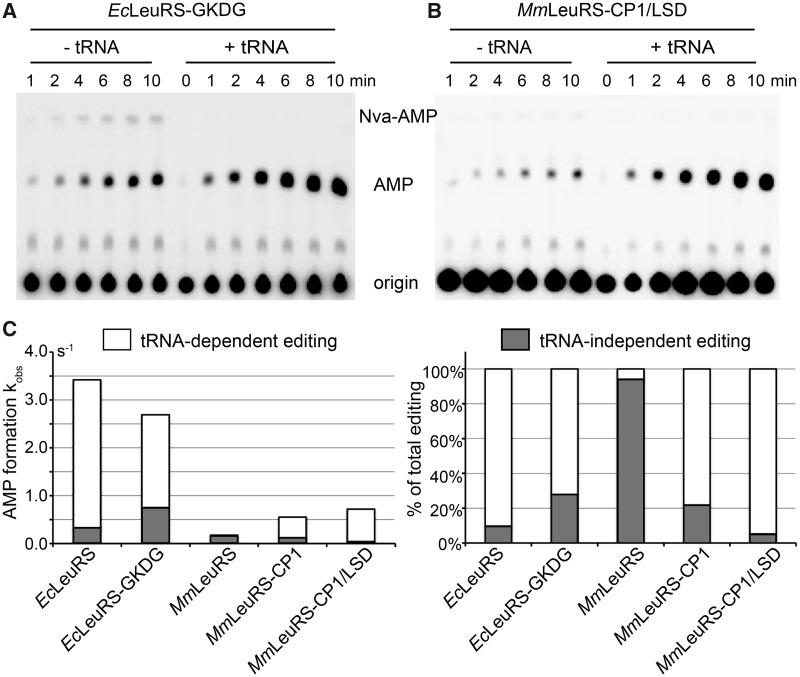
In the next experiments, a series of insertion mutants was constructed to mimic a possible evolutionary process. Chimeric proteins were constructed based on the MmLeuRS scaffold. First, the LSD of EcLeuRS was inserted in place of the tetrapeptide GKDG in the MmLeuRS (MmLeuRS-LSD). The resulting MmLeuRS-LSD mutant did not exhibit any detectable aminoacylation activity (data not shown). MmLeuRS-CP1 was constructed by inserting the CP1 domain of EcLeuRS into MmLeuRS, and this chimeric enzyme had both aminoacylation and editing activities (17). When MmLeuRS-CP1 was used as a scaffold to fuse the LSD of EcLeuRS into its catalytic core, the resulting chimera (MmLeuRS-CP1/LSD) had comparable aminoacylation activity to the native MmLeuRS but demonstrated better catalytic efficiency due to greater affinity with tRNA as indicated by a decrease in Km (Table 1). However, the LSD insertion severely decreased the tRNA-independent pre-transfer editing of MmLeuRS and MmLeuRS-CP1, and the observed rate constant for AMP formation in the presence of Nva (an analogue of Leu) dropped from 0.16 and 0.12 to 0.037 s−1 (Table 2). In the presence of EctRNALeu and Nva, the observed rate constant of MmLeuRS-CP1/LSD for AMP formation was comparable with that of MmLeuRS-CP1, and the rate was 3.6-fold (0.61 s−1) greater than that of MmLeuRS (0.17 s−1). This shows that the tRNA-dependent editing pathway became the main editing pathway of MmLeuRS-CP1/LSD, contributing to 94% of the total editing activity [(0.61 − 0.037)/0.61], whereas the corresponding value in MmLeuRS-CP1 was just 78% [(0.55 − 0.12)/0.55] (Figure 2C). On the other hand, when the LSD of EcLeuRS was replaced by the GKDG tetrapeptide of MmLeuRS to form EcLeuRS-GKDG, the observed rate constant for AMP formation in the presence of Nva of the mutant was 0.75 s−1 compared with 0.33 s−1 for the native EcLeuRS (Table 2), indicating that the tRNA-independent pre-transfer editing of EcLeuRS-GKDG contributed much more to total editing (28%; 0.75/2.69) than that of EcLeuRS (9.6%; 0.33/3.42).
Table 2.
Observed rate constants for AMP synthesis in the presence of Nva
| LeuRS | tRNALeu | Rate of AMP formation kobs(s−1) |
|---|---|---|
| EcLeuRSa | - | 0.33 ± 0.040 |
| +EctRNALeuGAG | 3.42 ± 0.51 | |
| +MmtRNALeuUAG | 2.22 ± 0.29 | |
| EcLeuRS-GKDG | - | 0.75 ± 0.080 |
| +EctRNALeuGAG | 2.69 ± 0.40 | |
| +MmtRNALeuUAG | 2.89 ± 0.47 | |
| MmLeuRSa | - | 0.16 ± 0.025 |
| +EctRNALeuGAG | 0.16 ± 0.022 | |
| +MmtRNALeuUAG | 0.17 ± 0.030 | |
| MmLeuRS-CP1a | - | 0.12 ± 0.020 |
| + EctRNALeuGAG | 0.55 ± 0.040 | |
| +MmtRNALeuUAG | 0.25 ± 0.032 | |
| MmLeuRS-CP1/LSD | - | (3.7 ± 0.75) × 10−2 |
| +EctRNALeuGAG | 0.61 ± 0.049 | |
| +MmtRNALeuUAG | 0.21 ± 0.030 | |
| MmLeuRS-CP1/LSD-K598A | - | (9.3 ± 1.5) × 10−2 |
| +EctRNALeuGAG | 0.14 ± 0.015 | |
| +MmtRNALeuUAG | 0.12 ± 0.013 |
All rates represent the average of three trials with the standard deviations indicated.
aData from Tan et al. (17).
Figure 2.
Effect of LSD mutations on tRNA-independent pre-transfer editing. (A) Total editing activity was measured using the AMP formation assay with 0.2 µM EcLeuRS-GKDG in the absence or presence of 5 µM EctRNALeu and 15 mM Nva. (B) A similar assay was performed with 1 µM MmLeuRS-CP1/LSD in the absence or presence of 5 µM EctRNALeu and 15 mM Nva. (C) Contributions of the different editing pathways for each protein: left, sum of the kobs of different editing pathways; right, relative contributions of each pathway. Percentages were calculated from kobs values of AMP formation reported in Table 1. tRNA-independent pre-transfer editing was measured in the absence of tRNA. tRNA-dependent editing was deduced by subtracting the tRNA-independent pre-transfer editing from total editing.
Taken together, these results show that LSD recruiting restricted internal tRNA-independent pre-transfer editing by the synthetic domain of LeuRS. As a consequence, the evolved LeuRS favoured tRNA-dependent pre-transfer editing, which was more effective in maintaining the catalytic fidelity. We propose that this is a possible reason why most prokaryotic LeuRSs have recruited and preserved LSD during their evolution.
The LSD is responsible for tRNA discrimination
In a previous study, it was found that MmLeuRS-CP1 cross-leucylates EctRNALeuGAG with efficiency comparable with that of the in vitro transcript of MmtRNALeuUAG (17). The present work showed that MmLeuRS-CP1/LSD aminoacylates more efficiently EctRNALeu than MmtRNALeuUAG with a discrimination factor (D factor) of 3.7 (according to kcat/Km) (Table 1). Therefore, MmLeuRS-CP1/LSD had similar discriminatory properties as the native EcLeuRS, which has a D factor of 5.5 (Table 1). These results suggest that the LSD may participate in tRNA binding and discrimination in some way. When the LSD of EcLeuRS was replaced by the tetrapeptide GKDG, the mutant EcLeuRS-GKDG leucylated EctRNALeuGAG and MmtRNALeuUAG with similar catalytic efficiency (Table 1). The editing activity of EcLeuRS-GKDG was also comparable in the presence of MmtRNALeuUAG or EctRNALeuGAG (Table 2, Supplementary Figure S2).
Furthermore, when the CP1 domain and LSD of EcLeuRS were inserted into MmLeuRS, the mutant MmLeuRS-CP1/LSD favoured EctRNALeuGAG not only in aminoacylation but also in editing. In the TLC-based AMP formation assay, MmLeuRS-CP1/LSD had a rate constant for AMP formation in the presence of MmtRNALeuUAG and Nva of 0.21 s−1, while it was 0.61 s−1 and 0.56 s−1 for EctRNALeuGAG and MmtRNALeuUAA, respectively (Supplementary Figure S2, Tables 2 and 3). Consistently, EcLeuRS also preferred EctRNALeu in the editing with AMP formation rate of 3.4 s−1, with a corresponding value of 2.2 s−1. However, LSD-deprived EcLeuRS-GKDG showed no preference towards these two tRNAs during editing (kobs 2.7 vs 2.9 s−1) (Table 2). These results show that the LSD could confer tRNA discrimination properties to LeuRS, and this raises questions about how the LSD can distinguish tRNAs during aminoacylation and editing.
Table 3.
Kinetic constants of MmLeuRS-CP1/LSD for mutants of MmtRNALeuUAG determined in the aminoacylation reaction
| MmtRNALeuUAG | Km (µM) | kcat (s−1) | kcat/Km (s−1 µM−1) | Relative catalytic efficiency |
|---|---|---|---|---|
| WT | 1.7 ± 0.13 | 0.51 ± 0.049 | 0.30 | 1.0 |
| A6G | 1.2 ± 0.10 | 0.53 ± 0.041 | 0.44 | 1.5 |
| C67U | 1.9 ± 0.21 | 0.48 ± 0.032 | 0.25 | 0.83 |
| C20U | 1.4 ± 0.13 | 1.1 ± 0.10 | 0.75 | 2.5 |
| V-arm-4nt | 3.4 ± 0.32 | 1.1 ± 0.11 | 0.34 | 1.1 |
| V-arm-5nt | 1.1 ± 0.09 | 0.79 ± 0.080 | 0.70 | 2.3 |
| A6G + V-arm-5nt | 2.1 ± 0.20 | 1.7 ± 0.13 | 0.78 | 2.6 |
| C20U + V-arm-5nt | 0.88 ± 0.091 | 1.0 ± 0.10 | 1.1 | 3.7 |
| MmtRNALeuUAA | 2.8 ± 0.29 | 2.1 ± 0.24 | 0.78 | 2.6 |
Kinetic constants were determined using the tRNA charging assay described in the experimental section. The concentration of the tRNALeus ranged from 0.5–30 μM. All parameters represent the average of three trials with the standard deviations indicated.
Identification of the critical nucleotides recognized by LSD
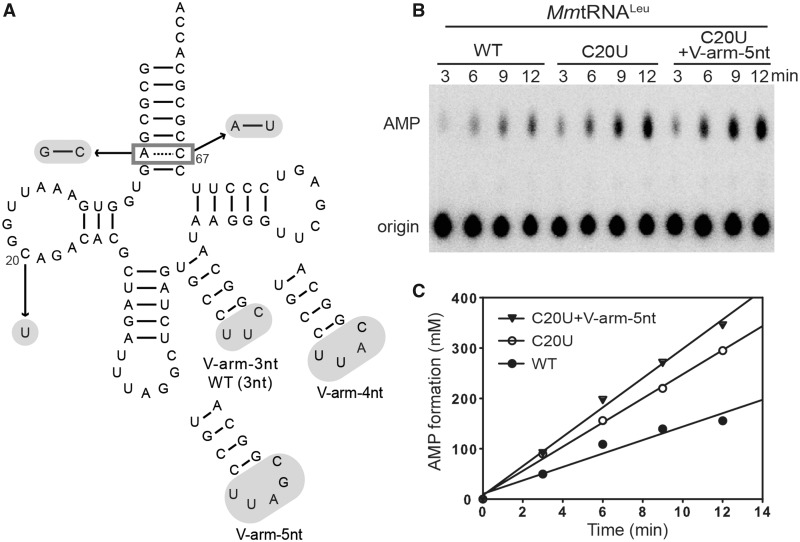
To identify the structural determinants of tRNALeu responsible for LeuRS ability to discriminate tRNALeus from various species, we compared the tRNALeu sequences from E. coli and M. mobile and focused our attention on three differences between them: (i) the sixth base-pair in the acceptor stem of MmtRNALeuUAG is a wobble base pair (A6•••C67), whereas it is a Watson Crick base pair G6–C67 in EctRNALeuGAG; (ii) the loop of the V-arm of MmtRNALeuUAG contains three nucleotides; however, EctRNALeuGAG has a 4-nucleotide loop; (iii) nucleotide 20, located in the ‘variable pocket’ (20) of the D-loop, is always a U in EctRNALeus but always a C in MmtRNALeuUAG (Figure 3A). Therefore, a series of mutants of MmtRNALeuUAG was constructed. Firstly, the A6•••C67 pair was mutated to a Watson Crick base pair by introducing A6G or C67U mutations. Secondly, in the ‘variable pocket’, nucleotide C20 was changed to a U. Thirdly, the loop of the V-arm was enlarged from three nucleotides to four (V-arm-4 nt) or five (V-arm-5 nt), which are usual sizes for these loops in tRNALeus. MmLeuRS-CP1/LSD leucylated the C20U and V-arm-5 nt mutants at more than twice the catalytic efficiency of the wild-type MmtRNALeuUAG (from 0.3 to 0.75 and 0.70 s−1 µM−1, respectively). MmLeuRS-CP1/LSD leucylated the double mutant (C20U + V-arm-5 nt), where the C20U mutation and V-arm-5 nt mutation were present, and catalytic efficiency (kcat/Km 1.1 s−1 µM−1) (Table 3) almost reached the level of EctRNALeuGAG (1.1 s−1 µM−1 in Table 1). Similarly, MmLeuRS-CP1/LSD charged another double mutant (A6G + V-arm-5 nt) and MmtRNALeuUAA (another MmtRNALeu isoacceptor) with the same catalytic efficiency (0.78 s−1 µM−1) (Table 3). Interestingly, we found that MmtRNALeuUAA naturally exhibits a large loop of 5 nucleotides in the V-arm according to the genomic tRNA database.
Figure 3.
Mutations in MmtRNALeuUAG that impact editing activity. (A) Cloverleaf structure of MmtRNALeuUAG showing the mutations tested during the study. (B) AMP formation assay in the presence of 15 mM Nva catalyzed by 1 µM MmLeuRS-CP1/LSD in the presence of 5 µM wild-type MmtRNALeuUAG, C20U and C20U + V-arm-5 nt. (C) Graphical representations of AMP formation as a function of time. kobs values of AMP formation were calculated from the slopes, and these are shown in Table 4.
In the editing reaction, the seven MmtRNALeu mutants showed various capacities to stimulate AMP formation. In the presence of Nva, MmLeuRS-CP1/LSD had a rate constant for AMP formation of 0.21 s−1 for wild-type MmtRNALeuUAG; however, for MmtRNALeu-C20U that was increased to 0.41 s−1. In addition, the most efficient mutant leucylated by MmLeuRS-CP1/LSD, MmtRNALeu-(C20U + V-arm-5 nt), showed very similar effects on editing activity as MmtRNALeuUAA in the presence of Nva (Figure 3B and C, Supplementary Figure S3, Table 4).
Table 4.
Observed rate constants for AMP synthesis of MmLeuRS-CP1/LSD in the presence of Nva
| MmtRNALeuUAG | Rate of AMP formation kobs (s−1) | Relative activity |
|---|---|---|
| WT | 0.21 ± 0.020 | 1.0 |
| A6G | 0.28 ± 0.026 | 1.3 |
| C67U | 0.31 ± 0.032 | 1.5 |
| C20U | 0.41 ± 0.051 | 2.0 |
| V-arm-4 nt | 0.12 ± 0.011 | 0.57 |
| V-arm-5 nt | 0.26 ± 0.029 | 1.2 |
| A6G + V-arm-5 nt | 0.34 ± 0.031 | 1.6 |
| C20U + V-arm-5 nt | 0.52 ± 0.063 | 2.5 |
| MmtRNALeuUAA | 0.56 ± 0.059 | 2.7 |
All rates represent the average of three trials with the standard deviations indicated.
A key Lys residue of the LSD is responsible for tRNA discriminatory activity
It has been reported that EcLeuRS contacts bases 10 and 27 of tRNALeu via the Arg595 and Arg600 residues located on the so-called β-hairpin of the LSD (4). To investigate whether these residues could be responsible of discrimination between EctRNALeuGAG and MmtRNALeuUAG, initially we mutated the Arg595 and Arg600 of EcLeuRS to Ala residues. Both mutants, EcLeuRS-R595A and EcLeuRS-R600A, showed high catalytic efficiency preference for EctRNALeuGAG but neither reached the value of the wild-type EcLeuRS (Table 5). However, another mutant on the β-hairpin, EcLeuRS-K598A, exhibited a stronger effect on aminoacylation activity. EcLeuRS-K598A displayed a considerably lower affinity for EctRNALeuGAG compared with wild-type EcLeuRS (Km increased about 4-fold), which resulted in a decrease of the catalytic efficiency by almost 3-fold from 2.2 to 0.84 s−1 µM−1. On the other hand, EcLeuRS-K598A bound more tightly with MmtRNALeuUAG, and this induced a significant increase in the catalytic efficiency for the leucylation of MmtRNALeuUAG (1.1 s−1 µM−1) compared with wild-type EcLeuRS (0.4 s−1 µM−1) (Table 5).
Table 5.
Kinetic constants of EcLeuRS and its mutants in the aminoacylation reaction
|
EctRNALeuGAG |
MmtRNALeuUAG |
D factor | |||||
|---|---|---|---|---|---|---|---|
| Enzyme | Km (µM) | kcat (s−1) | kcat/Km (s−1 µM−1) | Km (µM) | kcat (s−1) | kcat/Km (s−1 µM−1) | |
| EcLeuRSa | 2.2 ± 0.14 | 4.9 ± 0.30 | 2.2 | 10.6 ± 1.2 | 4.2 ± 0.40 | 0.40 | 5.5 |
| EcLeuRS-R595A | 3.4 ± 0.25 | 4.3 ± 0.31 | 1.3 | 4.2 ± 0.40 | 3.2 ± 0.33 | 0.75 | 1.7 |
| EcLeuRS-K598A | 8.2 ± 0.81 | 6.9 ± 0.57 | 0.84 | 5.7 ± 0.41 | 6.3 ± 0.55 | 1.1 | 0.76 |
| EcLeuRS-R600A | 3.9 ± 0.27 | 7.2 ± 0.65 | 1.9 | 8.6 ± 0.78 | 6.1 ± 0.59 | 0.70 | 2.7 |
| MmLeuRS-CP1/LSD | 1.3 ± 0.12 | 1.4 ± 0.11 | 1.1 | 1.7 ± 0.13 | 0.51 ± 0.049 | 0.30 | 3.7 |
| MmLeuRS-CP1/LSD-K598A | 6.6 ± 0.62 | 3.6 ± 0.37 | 0.54 | 5.7 ± 0.41 | 6.3 ± 0.55 | 1.4 | 0.39 |
D is the discrimination factor of the different LeuRSs for the two bacterial tRNAs, and this value was calculated as follows: D = kcat/Km(EctRNALeuGAG) / kcat/Km(MmtRNALeuUAG). All parameters represent the average of three trials with the standard deviations indicated.
aData from Tan et al.(17).
In the same way, when K598 in the LSD of chimeric MmLeuRS-CP1/LSD was replaced with an Ala residue, the catalytic efficiency of the mutant for MmtRNALeuUAG was greater than that for EctRNALeuGAG; however, the catalytic efficiency of MmLeuRS-CP1/LSD for MmtRNALeuUAG was lower than that for EctRNALeuGAG, indicating that mutant MmLeuRS-CP1/LSD-K598A prefers to charge MmtRNALeuUAG, while MmLeuRS-CP1/LSD prefers EctRNALeuGAG. Thus, mutation at K598 changed the species preference of these enzymes for their tRNALeu substrates (Table 5). The results show that residue K598 in the β-hairpin of the LSD of EcLeuRS contributes positively to the binding and aminoacylation of EctRNALeuGAG and acts as an antideterminant versus MmtRNALeuUAG. When K598 was mutated to an Ala residue, the antideterminant effect was suppressed and the specific recognition of EcLeuRS LSD for EctRNALeu was extended to MmtRNALeu. In parallel, there was a decrease of binding affinity of MmLeuRS-CP1/LSD-K598A for EctRNALeu, which reduced its catalytic efficiency to a lower level (kcat/Km 0.54 s−1 µM−1) than for MmtRNALeuUAG (kcat/Km 1.4 s−1 µM−1). These data show that K598 is a key residue that controls the cross-recognition of tRNALeus from different species.
Consistently, in the chimeric MmLeuRS-CP1/LSD, the K598A mutation controlled post-transfer editing, as there was a drop in the AMP synthesis rate (Figure 4A, Table 2) and an absence of deacylation activity towards Ile-EctRNALeuGAG (Figure 4B). The loss of deacylation properties was further confirmed by a loss of aminoacylation specificity as illustrated by the Ile mischarging of EctRNALeuGAG (Figure 4C). Both MmLeuRS-CP1/LSD-K598A and MmLeuRS were able to mischarge Ile in contrast to MmLeuRS-CP1/LSD and MmLeuRS-CP1 that could not catalyze this substrate. Taken together, these results suggested that the crucial K598 residue of LSD mediated the interaction with tRNA and involved in tRNA recognition.
Figure 4.
Editing and mischarging properties of MmLeuRS-CP1/LSD-K598A. (A) Total editing activity was measured by the AMP formation assay with 1 µM of MmLeuRS-CP1/LSD-K598A and 15 mM Nva in the absence or presence of 5 µM EctRNALeuGAG or MmtRNALeuUAG. (B) Deacylation of [3H]-Ile-EctRNALeu (1 µM) by 20 nM of MmLeuRS (black circle), MmLeuRS-CP1/LSD-K598A (inverted black triangle), MmLeuRS-CP1/LSD (black square) and MmLeuRS-CP1 (black triangle). (C) Mischarging of EctRNALeuGAG (20 µM) with Ile catalyzed by 1 µM of MmLeuRS (black circle), MmLeuRS-CP1/LSD-K598A (inverted black triangle), MmLeuRS-CP1/LSD (black square) and MmLeuRS-CP1(black triangle). (D) Crystal structure of tRNALeu (light blue in the cartoon model) in complex with EcLeuRS (grey) during the editing conformation (PDB ID code 4ARC, Ref.4). Residues R595, K598 and R600 of LSD (green) are numbered and shown in stick representation with labelling. Both G10 and G46 of tRNALeu were also highlighted with the stick model with their distances to K598 labelled.
In three of the four crystallographic structures that describe the aminoacylation and proofreading states of LeuRS (4), the ε-amino group of K598 is located in the vicinity of the phosphate group of G10. K598 approaches the tRNA bound in the editing conformation at distances from 4.6 to 4.9 Å according to the different tertiary structures (in 4ASI, 4ARC and 4ARI). In addition, in the editing complex bound with leucine (4ARC), the ε-amino group of K598 forms a potential interaction with the phosphate group of G46 at a distance of 3.9 Å (Figure 4D). However, these putative interactions with the phosphate backbone of tRNA can hardly explain the new discriminating properties of the K598A mutant for MmtRNALeu and EctRNALeu. Nevertheless, we cannot exclude the possibility that they play a role during the transition of the 3′ end of tRNA between the aminoacylation and the editing states, and thus favour the aminoacylation of one isoacceptor.
DISCUSSION
With genome sizes <1 Mb, bacteria from the genus Mycoplasma have been described as the ‘smallest free-living organisms’, and thus are considered to be the best representatives for the concept of a minimal cell. The M. mobile genome encodes only 635 proteins (21), and includes 27 tRNA genes, one of the lowest abundances reported for any organism. Strong evidence suggests that mycoplasmas evolved by a process of reductive evolution that was made possible by adopting a parasitic lifestyle. During this process, the mycoplasmas lost considerable portions of their ancestral chromosomes but retained the genes essential for life. Genome compaction in mycoplasmas is often reflected by the presence of reduced intergenic spacers and by the shortness of most putative proteins relative to their orthologues (22). aaRSs genes did not escape this size reduction, and several of these enzymes have lost key residues in their editing domains, and in the extreme case of LeuRS, the CP1-editing domain has been deleted completely (10,17). Therefore, mycoplasmas are following a kind of reverse evolution that consists of selecting minimalist proteins that mimic the primitive proteins. Primitive aaRSs have followed an opposite evolutionary pathway by progressively adding domains to improve efficiency and fidelity and to conserve the genetic code and proteome in its present form.
LeuRSs from various species are very complex enzymes that are amongst the largest aaRSs. These enzymes have an unusually high number of modules appended to the catalytic core that participate in a concerted way in tRNA binding, aminoacylation and proofreading. Recent X-ray analysis of tRNALeu–LeuRS complexes in the aminoacylation or editing conformation has provided the structural basis and dynamics of the aminoacylation and proofreading functional cycle (4). LeuRS produces error-free Leu-tRNALeu by coordinating the translocation of the CCA-end of mischarged tRNAs from its synthetic site to the separate proofreading site where the editing occurs. Such translocation involves correlated rotations of four LeuRS domains that are linked to the catalytic core. These motions drive the CCA sequence of the tRNA from the aminoacylation site to the editing site. During this process, the CP1-editing domain stabilizes the tRNA during aminoacylation, while a large rotation of the LSD positions the conserved KMSKS loop of the LeuRS to bind the CCA end of the tRNA, thereby promoting catalysis (4).
The absence of both CP1 and LSD in MmLeuRS offers the opportunity to investigate the mechanism of insertion of these additional modules and explore the plasticity of the catalytic core to acquire new functions. Previously, it was shown that insertion of the CP1 domain into the minimal MmLeuRS did not change synthetic efficiency (17). CP1 insertion does improve affinity for the tRNA but it decreases kcat, suggesting that the tighter binding of the substrate is deleterious for its subsequent reactivity or release. The fusion of the domains of EcLeuRS with MmLeuRS also provided the post-transfer editing function to the chimeric enzyme MmLeuRS-CP1, and this enzyme demonstrated greater activity for E. coli tRNALeu. Although the post-transfer editing activity of MmLeuRS-CP1 remained modest compared with that of EcLeuRS, this observation supports the theory that the aaRS evolved by fusion with additional modules (23).
Here, we showed that insertion of the LSD of EcLeuRS into the pre-existing chimeric protein MmLeuRS-CP1 further improved tRNA binding, leading to a protein with greater catalytic efficiency. In contrast, the editing activity of the double insertion mutant was increased only rather poorly, and a decrease of the pre-existing pre-transfer editing activity of MmLeuRS was observed. Therefore, fusion with the second insertion domain improved not only tRNA binding and the synthetic activity of the enzyme but it also conferred greater importance to post-transfer editing relative to pre-transfer editing. This change might be explained by adenylate molecules reacting faster with tRNA to synthesize aminoacyl-tRNAs, thereby reducing their opportunity to be edited by the pre-transfer editing process in the synthetic site.
These data provide evidence that the CP1 domain and LSD cooperate for greater synthetic and proofreading properties when inserted in the MmLeuRS framework, and these observations suggest how these enzymes could have evolved from primitive aaRSs. In this manner, the editing domain, or another domain, could have been distributed amongst different aaRSs before their fine adjustment to the new substrate through the accumulation of mutations. In this present work, we further simulate evolution and show that single mutation events could significantly improve enzyme activity. For instance, mutations could take place in trans in the genes of the corresponding tRNAs. We showed that a mutation at position 20 of MmtRNALeu (C20U) doubled the relative activity of MmLeuRS-CP1/LSD in the aminoacylation and proofreading compared with the wild-type MmtRNALeu (Tables 3 and 4). Residue 20 is located in the ‘variable pocket’, and it is known to be a recognition element in different aminoacylation systems (20,24–26). In LeuRS, the only putative interaction of nucleotide 20 occurs with Lys813 that is located in the C-terminal domain, and this can occur only during the editing state (PDB entry 4ARC). Therefore, modifying a specific interaction of the editing state with a distinct module of the enzyme may have improved both synthetic and editing activities. As these activities contribute to a unique functional cycle, any mutation impacting one step may have repercussions on other activities. A similar improvement of catalytic properties was also observed with a double mutant that contained mutations in the acceptor arm and variable arm (A6G + V-arm-5 nt). Here also restoration of activity may occur through the C-terminal domain of LeuRS, which interacts with several nucleotides of the V-arm. Enlarging the loop might have reorganized tRNA binding and pivoting during the catalytic cycle. In addition, MmtRNALeuUAA with the natural 5-nt loop exhibited much greater aminoacylation and editing activities and was endowed with the most codon usage in M. mobile. The second mutation (A6G; located at a 50-Å distance in the acceptor arm) might have amplified the first effect (4).
Additionally, we showed that the synthetic performance of the chimeric enzyme could be improved in cis by a single mutation in the inserted LSD. We have found that Lys598 is an antideterminant for MmtRNALeu, but negative effects could be cancelled by Ala mutation. Therefore, this mutant shows that there are at least two alternative ways to improve the aminoacylation–proofreading functional cycle: one way consists of adapting the enzyme by mutating critical amino acids residues, while the second way consists of adjusting the tRNA structure in keeping with the newly inserted modules and the resulting conformation changes that occur during the catalytic processes.
Altogether, our results support the theory that fusion of additional modules to the ancient catalytic core of aaRSs during evolution introduced new catalytic functions to improve fidelity and catalytic performance (27). Moreover, this present study shows that the minimalist MmLeuRS is an ideal platform for further studies to understand the evolution of the aaRSs family through the acquisition of complementary modules.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Table 1 and Supplementary Figures 1–3.
FUNDING
National Key Basic Research Foundation of China [2012CB911000]; The Natural Science Foundation of China [30930022 and 31130064]; Committee of Science and Technology in Shanghai [12JC1409700]; Visiting Professorship for Senior International Scientists from the Chinese Academy of Sciences [2011T2S10]. Funding for open access charge: The Natural Science Foundation of China [30930022 and 31130064].
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Ling J, Reynolds N, Ibba M. Aminoacyl-tRNA synthesis and translational quality control. Annu. Rev. Microbiol. 2009;63:61–78. doi: 10.1146/annurev.micro.091208.073210. [DOI] [PubMed] [Google Scholar]
- 2.Eriani G, Delarue M, Poch O, Gangloff J, Moras D. Partition of tRNA synthetases into two classes based on mutually exclusive sets of sequence motifs. Nature. 1990;347:203–206. doi: 10.1038/347203a0. [DOI] [PubMed] [Google Scholar]
- 3.Cusack S, Yaremchuk A, Tukalo M. The 2 A crystal structure of leucyl-tRNA synthetase and its complex with a leucyl-adenylate analogue. EMBO J. 2000;19:2351–2361. doi: 10.1093/emboj/19.10.2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Palencia A, Crepin T, Vu MT, Lincecum TL, Jr, Martinis SA, Cusack S. Structural dynamics of the aminoacylation and proofreading functional cycle of bacterial leucyl-tRNA synthetase. Nat. Struct. Mol. Biol. 2012;19:677–684. doi: 10.1038/nsmb.2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tukalo M, Yaremchuk A, Fukunaga R, Yokoyama S, Cusack S. The crystal structure of leucyl-tRNA synthetase complexed with tRNALeu in the post-transfer-editing conformation. Nat. Struct. Mol. Biol. 2005;12:923–930. doi: 10.1038/nsmb986. [DOI] [PubMed] [Google Scholar]
- 6.Fukunaga R, Yokoyama S. Crystal structure of leucyl-tRNA synthetase from the archaeon Pyrococcus horikoshii reveals a novel editing domain orientation. J. Mol. Biol. 2005;346:57–71. doi: 10.1016/j.jmb.2004.11.060. [DOI] [PubMed] [Google Scholar]
- 7.Fukunaga R, Yokoyama S. Aminoacylation complex structures of leucyl-tRNA synthetase and tRNALeu reveal two modes of discriminator-base recognition. Nat. Struct. Mol. Biol. 2005;12:915–922. doi: 10.1038/nsmb985. [DOI] [PubMed] [Google Scholar]
- 8.Vu MT, Martinis SA. A unique insert of leucyl-tRNA synthetase is required for aminoacylation and not amino acid editing. Biochemistry. 2007;46:5170–5176. doi: 10.1021/bi062078j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ma JJ, Zhao MW, Wang ED. Split leucine-specific domain of leucyl-tRNA synthetase from the hyperthermophilic bacterium Aquifex aeolicus. Biochemistry. 2006;45:14809–14816. doi: 10.1021/bi061026r. [DOI] [PubMed] [Google Scholar]
- 10.Li L, Boniecki MT, Jaffe JD, Imai BS, Yau PM, Luthey-Schulten ZA, Martinis SA. Naturally occurring aminoacyl-tRNA synthetases editing-domain mutations that cause mistranslation in Mycoplasma parasites. Proc. Natl Acad. Sci. USA. 2011;108:9378–9383. doi: 10.1073/pnas.1016460108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tan M, Zhu B, Liu RJ, Chen X, Zhou XL, Wang ED. Inter-domain communication modulates the tRNA-dependent pre-transfer editing of leucyl-tRNA synthetase. Biochem. J. 2013;449:123–131. doi: 10.1042/BJ20121258. [DOI] [PubMed] [Google Scholar]
- 12.Steinberg S, Misch A, Sprinzl M. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 1993;21:3011–3015. doi: 10.1093/nar/21.13.3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou XL, Du DH, Tan M, Lei HY, Ruan LL, Eriani G, Wang ED. Role of tRNA amino acid-accepting end in aminoacylation and its quality control. Nucleic Acids Res. 2011;39:8857–8868. doi: 10.1093/nar/gkr595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Du X, Wang ED. Tertiary structure base pairs between D- and TpsiC-loops of Escherichia coli tRNA(Leu) play important roles in both aminoacylation and editing. Nucleic Acids Res. 2003;31:2865–2872. doi: 10.1093/nar/gkg382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yao P, Zhou XL, He R, Xue MQ, Zheng YG, Wang YF, Wang ED. Unique residues crucial for optimal editing in yeast cytoplasmic Leucyl-tRNA synthetase are revealed by using a novel knockout yeast strain. J. Biol. Chem. 2008;283:22591–22600. doi: 10.1074/jbc.M801181200. [DOI] [PubMed] [Google Scholar]
- 16.Huang Q, Yao P, Eriani G, Wang ED. In vivo identification of essential nucleotides in tRNALeu to its functions by using a constructed yeast tRNALeu knockout strain. Nucleic Acids Res. 2012;40:10463–10477. doi: 10.1093/nar/gks783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tan M, Yan W, Liu RJ, Wang M, Chen X, Zhou XL, Wang ED. A naturally occurring nonapeptide functionally compensates for the CP1 domain of leucyl-tRNA synthetase to modulate aminoacylation activity. Biochem. J. 2012;443:477–484. doi: 10.1042/BJ20111925. [DOI] [PubMed] [Google Scholar]
- 18.Li Y, Wang ED, Wang YL. Overproduction and purification of Escherichia coli tRNA(Leu) Sci. China C Life Sci. 1998;41:225–231. doi: 10.1007/BF02895095. [DOI] [PubMed] [Google Scholar]
- 19.Tan M, Zhu B, Zhou XL, He R, Chen X, Eriani G, Wang ED. tRNA-dependent pre-transfer editing by prokaryotic leucyl-tRNA synthetase. J. Biol. Chem. 2010;285:3235–3244. doi: 10.1074/jbc.M109.060616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McClain WH, Foss K. Changing the acceptor identity of a transfer RNA by altering nucleotides in a “variable pocket”. Science. 1988;241:1804–1807. doi: 10.1126/science.2459773. [DOI] [PubMed] [Google Scholar]
- 21.Jaffe JD, Stange-Thomann N, Smith C, DeCaprio D, Fisher S, Butler J, Calvo S, Elkins T, FitzGerald MG, Hafez N, et al. The complete genome and proteome of Mycoplasma mobile. Genome Res. 2004;14:1447–1461. doi: 10.1101/gr.2674004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Katinka MD, Duprat S, Cornillot E, Metenier G, Thomarat F, Prensier G, Barbe V, Peyretaillade E, Brottier P, Wincker P, et al. Genome sequence and gene compaction of the eukaryote parasite Encephalitozoon cuniculi. Nature. 2001;414:450–453. doi: 10.1038/35106579. [DOI] [PubMed] [Google Scholar]
- 23.Schimmel P, Ribas de Pouplana L. Transfer RNA: from minihelix to genetic code. Cell. 1995;81:983–986. doi: 10.1016/s0092-8674(05)80002-9. [DOI] [PubMed] [Google Scholar]
- 24.McClain WH, Foss K, Jenkins RA, Schneider J. Four sites in the acceptor helix and one site in the variable pocket of tRNA(Ala) determine the molecule’s acceptor identity. Proc. Natl Acad. Sci. USA. 1991;88:9272–9276. doi: 10.1073/pnas.88.20.9272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sampson JR, DiRenzo AB, Behlen LS, Uhlenbeck OC. Nucleotides in yeast tRNAPhe required for the specific recognition by its cognate synthetase. Science. 1989;243:1363–1366. doi: 10.1126/science.2646717. [DOI] [PubMed] [Google Scholar]
- 26.Liu W, Huang Y, Eriani G, Gangloff J, Wang E, Wang Y. A single base substitution in the variable pocket of yeast tRNA(Arg) eliminates species-specific aminoacylation. Biochim. Biophys. Acta. 1999;1473:356–362. doi: 10.1016/s0304-4165(99)00143-9. [DOI] [PubMed] [Google Scholar]
- 27.SternJohn J, Hati S, Siliciano PG, Musier-Forsyth K. Restoring species-specific posttransfer editing activity to a synthetase with a defunct editing domain. Proc. Natl Acad. Sci. USA. 2007;104:2127–2132. doi: 10.1073/pnas.0611110104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.