Abstract
AGO/RISC-mediated antiviral RNA silencing, an important component of the plant’s immune response against RNA virus infections, was recapitulated in vitro. Cytoplasmic extracts of tobacco protoplasts were applied that supported Tombusvirus RNA replication, as well as the formation of RNA-induced silencing complexes (RISC) that could be functionally reconstituted with various plant ARGONAUTE (AGO) proteins. For example, when RISC containing AGO1, 2, 3 or 5 were programmed with exogenous siRNAs that specifically targeted the viral RNA, endonucleolytic cleavages occurred and viral replication was inhibited. Antiviral RNA silencing was disabled by the viral silencing suppressor p19 when this was present early during RISC formation. Notably, with replicating viral RNA, only (+)RNA molecules were accessible to RISC, whereas (−)RNA replication intermediates were not. The vulnerability of viral RNAs to RISC activity also depended on the RNA structure of the target sequence. This was most evident when we characterized viral siRNAs (vsiRNAs) that were particularly effective in silencing with AGO1- or AGO2/RISC. These vsiRNAs targeted similar sites, suggesting that accessible parts of the viral (+)RNA may be collectively attacked by different AGO/RISC. The in vitro system was, hence, established as a valuable tool to define and characterize individual molecular determinants of antiviral RNA silencing.
INTRODUCTION
RNA silencing is a small RNA-mediated repression mechanism of gene regulation in eukaryotes that plays important roles in various biological processes, including the defense against viruses (1,2). The majority of plant viruses have a (+) stranded RNA genome (3) that acts as an mRNA but also as a template for RNA replication in the host cell’s cytoplasm (4). (+)RNA viruses are not only strong inducers but also targets of RNA silencing (5–7). The key inductors are assumed to be either highly structured parts of the viral genomes or double-stranded (ds) RNAs that consist of (+)RNAs and (−)RNA intermediates that are generated during RNA replication (8). The dsRNAs are processed by Dicer-like proteins (DCL) (9,10) into different types of viral small interfering RNA duplexes (vsiRNAs), among them are 21–24-nt long siRNAs that are thought to be involved in the antiviral defense (11–13). Thus, in the course of a viral infection, siRNAs accumulate in the plant, and this may correlate with a reduction in virus titer and local and systemic immunity (14–17). A major feature of antiviral RNA silencing involves that vsiRNAs are incorporated into effector complexes, such as RNA-induced silencing complexes (RISC) (5,9,18,19) that contain ARGONAUTE (AGO) nucleases and other yet incompletely characterized components. Ten AGO proteins were identified in the model plant Arabidopsis thaliana, from which AGO1, AGO2, AGO5 and AGO7 were indicated to contribute to the protection against virulent viruses (13,20–26). In antivirally acting RISC, the so-called passenger strand of the siRNA is removed, whereas the guide strand directs the effector complex to the cognate viral RNA that is inactivated by endonucleolytic cleavage in the middle of the siRNA–RNA duplex (27,28). In turn, many plant viruses evolved proteins that counteract the antiviral silencing process (29,30). For example, the Tombusvirus protein p19 sequesters siRNAs, and thus prevents their incorporation into RISC (31,32).
Members of the Tombusvirus genus of plant (+)RNA viruses, such as tomato bushy stunt virus (TBSV) and cymbidium ringspot virus (CymRSV) are intensively investigated to define the molecular determinants of viral replication (33) and of antiviral RNA silencing, respectively (19,34–37). The single-stranded ∼5–kb long TBSV genome (gRNA) consists of two non-translated regions (NTRs) at the 5′- and 3′-ends and several open reading frames (ORF; Figure 2A). Only the 5′-terminal ORF (5′-ORF) of the RNA is directly translated from the gRNA. Translation initiation is mediated by the so-called 3′-CITE (cap independent translational enhancer) element in the 3′-NTR and involves 5′–3′ interactions of the viral RNA (38,39). The 5′-ORF encodes the viral replicase proteins p33 and p92. p33 acts as an anchor of the viral replication complex and as an RNA chaperon; p92, which is generated by translational read-through of an ORF-internal stop codon, represents the RNA-dependent RNA polymerase. The residual ORFs are translated from two subgenomic mRNAs that are produced from the (−)RNA. They encode the coat protein p41, the viral movement protein p22 and the RNA silencing suppressor p19 (33).
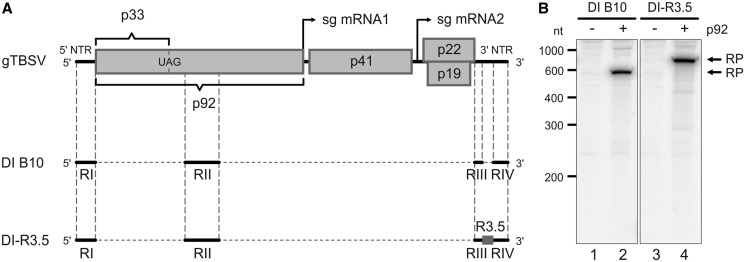
Figure 2.
Composition and in vitro replication of TBSV DI RNAs. (A) Schematic representations of TBSV gRNA, DI B10 RNA and DI-R3.5 RNA. The NTRs of the viral genome are depicted as lines, coding regions as boxes. Arrows indicate the transcriptional start of the two subgenomic (sg) mRNAs that are generated in the course of the TBSV life cycle. DI B10 (48) that was used in this study is essentially composed of the regions RI-RIV. DI-R3.5 also encloses the genomic R3.5 region inserted between RIII and RIV. (B) DI-R3.5 RNA replicates with similar efficiency as the DI B10 RNA in vitro. Using the protocol of Gursinsky et al. (46), the viral proteins p33 and p92 were produced by in vitro translation of the corresponding mRNAs in the BYL. Viral RNA replication was then started by the addition of a replication mix that included [α-32P]CTP and DI B10 RNA (lanes 1 and 2) or DI-R3.5 RNA (lanes 3 and 4). Total RNA was isolated from the reaction and analyzed by denaturing PAGE and autoradiography. The replication products (RP) are indicated. Lanes 1 and 3; replication assays performed in the absence of p92 (negative controls). Lanes 2 and 4; replication assays performed in the presence of p33 and p92.
Besides of storing the genetic information, the TBSV RNA acts as an assembly platform for the membrane-associated viral replication complex that contains p33 and p92, as well as co-opted cellular factors (40–43). The replication-related functions of the viral RNA are guided by defined cis-acting elements that are located in four regions of the genome, RI, RII, RIII and RIV. Importantly, RI–IV are all retained in defective interfering (DI) RNAs (Figure 2A). DI RNAs are small virus-derived RNAs that spontaneously emerge in infected plants and that are replicated by the TBSV replicase in trans (44).
By applying cytoplasmic extract of evacuolated Nicotiana tabacum BY2 protoplasts (termed BYL), a system which was earlier shown to support the replication of other plant (+)RNA viruses (45), we recently established an in vitro replication assay for TBSV. In one experimental variant, p33 and p92 are first synthesized by in vitro translation of separate mRNA transcripts in the BYL, and viral RNA replication is then initiated by the addition of a DI RNA or a gRNA template (46). BYL containing additionally expressed AGO1 protein was recently observed to also reproduce RISC activity on a target mRNA in vitro (47) (Figure 1).
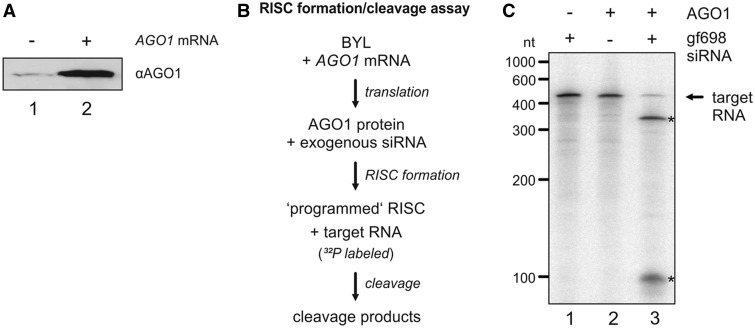
Figure 1.
RISC activity in BYL. (A) Quantity of AGO1 protein in BYL. Identical volumes of naive BYL (lane 1) and BYL that contained additional AGO1 (generated by in vitro translation; lane 2) were probed by immunoblot against AGO1. (B) Schematic representation of the ‘RISC formation/cleavage assay’ (47). AGO1 mRNA was translated in BYL in the presence of exogenous siRNA(s). Subsequently, a 32P-labeled target RNA was added and analyzed for cleavage. (C) ‘RISC formation/cleavage assay’ performed with ‘gf698’ siRNA and GFP mRNA as a target. The RISC cleavage products (indicated with asterisks) were analyzed by denaturing PAGE and autoradiography (lane 3). As negative controls, the reaction was carried out in the absence of additionally expressed (in vitro translated) AGO1 (lane 1) and in the absence of siRNA (lane 2), respectively.
Combining both findings, we here established a novel experimental set-up to reproduce AGO/RISC-mediated antiviral RNA silencing, i.e. RISC-mediated inhibition of viral RNA replication, in vitro. The in vitro system was applied in a comprehensive study to define those AGO proteins that support effective RISC-mediated RNA cleavage, and thus have the capacity to inhibit the replication of (+)RNA viruses, such as TBSV. Notably, functional AGO/RISC exclusively attacked the viral (+)RNA, whereas the (−)RNA replication intermediate was found to be protected. AGO/RISC-mediated inhibition of viral replication was further indicated to mainly involve a small subset of effective vsiRNAs that target accessible RNA sites.
MATERIALS AND METHODS
Cell culture and preparation of cytoplasmic BY-2 cell extract
Nicotiana tabacum BY-2 cells were cultured as described previously (46) at 23°C in Murashige–Skoog liquid medium. Evacuolated BY-2 protoplasts to prepare cytoplasmic extract (BYL) were obtained by percoll gradient centrifugation (45,46).
siRNAs
RNA oligonucleotides were purchased from Biomers (Ulm, Germany). The sequences of the 22 nt variant of ‘gf698’ siRNA were 5′-uaguucauccaugccaugugua-3′ (guide strand, gs) and 5′-cacauggcauggaugaacuaua-3′ (passenger strand, ps). The sequences of the 21 nt variant of ‘gf698’ siRNA were 5′-uaguucauccaugccaugugu-3′ (gs) and 5′-acauggcauggaugaacuaua-3′ (ps). For variants of ‘gf698’ siRNA that possessed a different 5′-terminal nucleotide in the guide strand, the passenger strand was adapted in each case to provide the complementary nucleotide at the respective position.
The sequences of vsiRNAs that were deduced from cloned RNA fragments were the following: vsiRNA1, 5′-uauccgaccauaggcccaugu-3′ (gs) and 5′-augggccuauggucggauaag-3′ (ps) in case of the 21-nt siRNA variant and 5′-uauccgaccauaggcccauguu-3′ (gs) and caugggccuauggucggauaag (ps) in case of the 22 nt siRNA variant; vsiRNA2, 5′-auccgaccauaggcccauguu-3′ (gs) and 5′-caugggccuauggucggauaa-3′ (ps); vsiRNA3, 5′-cuuauccgaccauaggcccau-3′ (gs) and 5′-gggccuauggucggauaaguc-3′ (ps); vsiRNA4, 5′-uuaggaugacgagucgacccg-3′ (gs) and 5′-ggucgacucgucauccuaaca-3′ (ps).
To produce siRNA duplexes, the single-stranded RNAs were incubated in annealing buffer (30 mM HEPES–KOH, pH 7.4, 100 mM potassium acetate and 2 mM magnesium acetate) for 1 min at 90°C and annealed for 60 min at 37°C. The synthetic RNAs were used mostly non-phosphorylated: compared with 5′-phosphorylated siRNAs, the non-phosphorylated siRNAs were found to be similarly effective in the BYL, which indicates the presence of a suitable kinase in the extract (data not shown). For the generation of vsiRNAs from the TBSV R3.5 region, equimolar amounts of the sense and antisense transcripts were heat denatured for 2 min at 94°C in STE buffer (10 mM Tris, pH 8.0, 100 mM NaCl and 1 mM EDTA) and then annealed at 25°C. The double-stranded RNA was subsequently treated with ShortCut® RNase III (New England Biolabs) according to the manufacturer’s instructions. The resulting RNAs were separated on a 12% Tris–borate polyacrylamide gel and extracted in TNES buffer (10 mM Tris–HCl, pH 7.5, 300 mM NaCl, 1 mM EDTA and 0.1% SDS).
AGO expression
In vitro translation of AGO mRNAs was performed in 50% (v/v) BYL at previously described conditions (46). Unless differently stated, 1.5 µg of the mRNA was translated in a 20-µl reaction for 60 min at 25°C. To visualize the translation products, 10 µCi of l-[35S]-methionine (1000 Ci/mmol, Hartmann Analytic) was added; the proteins were separated on 10% SDS–PAGE and detected by phosphor-imaging (Storm 860, Molecular Dynamics). For the immunodetection of N. tabacum AGO1, western blotting was performed at standard conditions. The monoclonal antibody directed against AGO1 from Nicotiana benthamiana was used at a 1:1000 dilution and detected by chemiluminescence staining using horseradish peroxidase-linked anti-mouse IgG (Sigma) and the SuperSignal West Pico Substrate (Pierce).
‘RISC formation/cleavage assay’
In vitro translation of AGO mRNAs was performed in the presence of 50 nM synthetic siRNA or 500 nM RNase III-generated siRNA pool for 60 min. Then the same amount of siRNA was added again and the reaction continued for other 90 min. Two micrograms of firefly luciferase (competitor) mRNA and the 32P-labeled target RNA (50 fmol) was added, and the cleavage reaction was performed for other 15 min. Total RNA was isolated from the reaction by treatment with 20 µg proteinase K in the presence of 0.5% SDS for 30 min at 37°C, followed by extraction with one volume chloroform and ethanol precipitation. 32P-labeled products were separated on 5% Tris–borate polyacrylamide gels containing 8 M urea and visualized by phosphor-imaging.
In vitro replication and ‘replication inhibition assays’
The in vitro replication assay was performed essentially as described previously (46). That is, the TBSV p33 and p92 proteins were generated by in vitro translation in the BYL using the described conditions and 5 pmol of p33 mRNA and 0.25 pmol of p92 mRNA in a 50-µl reaction. For replication, 40 µl of the translation reaction was mixed with 10 µl of 5× RdRp buffer (50 mM DTT, 500 µg/ml of actinomycin D, 17 mM magnesium acetate, 5 mM of each ATP, GTP and UTP and 0.250 mM of CTP containing 15 µCi of [α-32P]CTP). In all, 0.5 pmol of TBSV DI RNA or of the respective DI RNA variants was added as a template, and the reaction was performed for 3 h at 25°C. Total RNA was purified, and the 32P-labeled RNA products were analyzed as described previously. To test for RISC-mediated antiviral RNA silencing, the reaction volume of the in vitro translation reaction that generated p33 and p92 was reduced to 25 µl. In a parallel 25-µl reaction, AGO mRNA (2 µg) was translated in the presence of the siRNA that was used to ‘program’ the assembling RISC. If not indicated differently (see text and Figure 3B), both translation reactions were combined and replication initiated via the addition of RdRp buffer and DI RNA. The replication products were analyzed as described previously. Suppression of antiviral RNA silencing was achieved via the addition of 25 U of purified p19 (New England Biolabs) to the translation reaction that generated AGO1 and RISC.
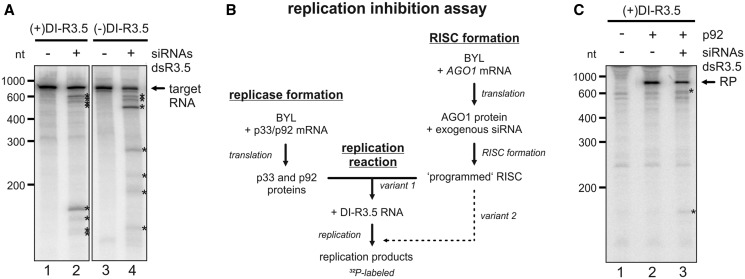
Figure 3.
Antiviral RNA silencing with a virus-derived siRNA pool. (A) ‘RISC formation/cleavage assay’ with DI-R3.5 RNA. A pool of siRNAs was generated by RNase III (ShortCut®) cleavage of dsR3.5 RNA. Using BYL where AGO1 was overexpressed by in vitro translation, RISC was formed with this siRNA pool. 32P-labeled (+) or (−)DI-R3.5 RNA transcripts were added to the extract that contained the programmed RISC, and the reaction products were subsequently analyzed by denaturing PAGE and autoradiography (lanes 2 and 4). The analogous experiments performed with a non-related (‘gf698’) siRNA served as negative controls (lanes 1 and 3). The positions of the labeled target RNAs are indicated; most prominent cleavage products are indicated by asterisks. (B) Schematic representation of the in vitro ‘replication inhibition assay’. A BYL reaction mixture that contained in vitro translated AGO1 and RISC that was ‘programmed’ with the siRNA(s) of choice was added to a second BYL reaction mixture that contained the in vitro translated TBSV replicase proteins p33 and p92. In experimental ‘variant 1’, RNA replication was initiated by combining both reactions and by the subsequent addition of replication mix and DI-R3.5 RNA template. In experimental ‘variant 2’, the mixture that contained the programmed RISC was added at a later time point to the replication reaction (Figure 4E). (C) ‘replication inhibition assay’. The reaction described in (B) was performed with (+)DI-R3.5 RNA. Lane 1; in the absence of p92 (no replication). Lane 2; with an unrelated (‘gf698’) siRNA (negative control). Lane 3; with the dsR3.5-generated vsiRNA pool. The RNA replication products (RP) are indicated, as well as the most prominent RNA cleavage products (asterisks).
Analysis of dsRNA processing in BYL
Double-stranded 32P-labeled R3.5 RNA (1.5 pmol) was incubated in a 20-µl reaction containing 50% (v/v) BYL under translation conditions. Total RNA was isolated from the reaction as described previously and separated on 15% denaturing Tris–borate polyacrylamide gels. 32P-labeled products were visualized by phosphor-imaging.
Information on plasmids constructs, on the in vitro transcription procedure and on the procedure to generate and clone cDNAs from RNA cleavage products is provided as Supplementary Data.
RESULTS
RISC-mediated antiviral RNA silencing reconstituted in vitro
The initial aim of this study was to understand whether the BYL system that supports TBSV RNA replication in vitro would also reproduce the siRNA-directed antiviral immune response of a plant cell. To this end, it was first important to test our BYL preparations for slicer/RISC activity. Following the findings of Iki et al. (47), we increased the concentration of AGO1 protein (AGO1 gene cloned from N. tabacum, Nt) in the BYL by in vitro translation of the corresponding mRNA (Figure 1A). Then the extract was ‘programmed’ via the addition of a synthetic siRNA (siRNA ‘gf698’), the guide strand of which was complementary to a certain site in the mRNA encoding GFP (green fluorescent protein). When we exposed the 32P-labeled target mRNA to the extract, a site-specific endonucleolytic cleavage of the RNA was detectable, which indicated the formation of active AGO1/RISC (47) (Figure 1C, lane 3). This reaction, which is schematically depicted in Figure 1B, was subsequently termed as ‘RISC formation/cleavage assay’. A ‘gf698’-directed RISC activity was absent in BYL that contained no additionally expressed AGO1 (Figure 1C, lane 1).
Because of their stability and high replication rate, TBSV DI RNAs were earlier shown to be most suitable to perform in vitro replication studies with BYL (46). Tombusvirus DI RNAs are suggested to attenuate infections by competing for viral and host replication factors and to modulate the antiviral immune response through the production of massive amounts of vsiRNAs. Thus, most wild-type (wt) DI RNAs turned out to be poor targets of antiviral RNA silencing (16). To obtain an RNA substrate that could be equally well used in replication and in silencing experiments, we constructed a modified version of the TBSV DI B10 (48) that included the so-called R3.5 region (DI-R3.5; Figures 2A and 7B). R3.5 is part of the TBSV gRNA’s 3′NTR; it is located between the RIII and RIV elements and contains the 3′CITE. R3.5 was chosen because in plants infected with the TBSV-related CymRSV, this region was found to be a hot spot for vsiRNA-mediated cleavage (19,37). The DI-R3.5 RNA showed the same replication competence as the DI B10 (Figure 2B, lanes 2 and 4).
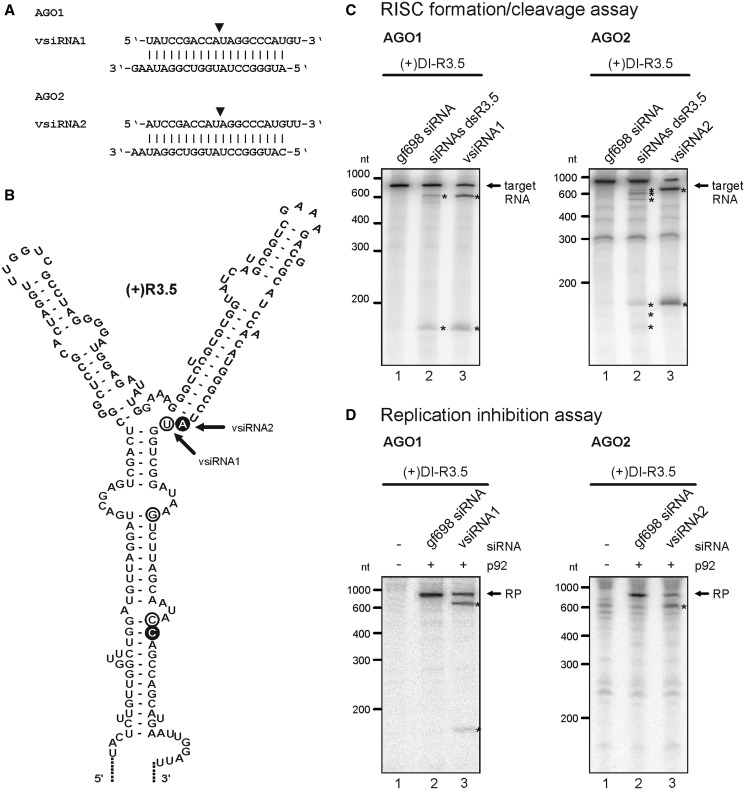
Figure 7.
Identification and characterization of effective vsiRNAs. (A) Sequences of vsiRNAs1 and 2 that were identified to effectively target the TBSV R3.5 element. The siRNAs were deduced from cleavage products of ‘RISC formation/cleavage assays’ that applied the indicated AGO proteins, a pool of dsR3.5-derived siRNAs, and (+)DI-R3.5 as a target RNA. The position between the guide strand’s nucleotides 10 and 11 that is opposite the cleavage site in the target RNA (52, 53) is indicated by a triangle. (B) RNA secondary structure of the TBSV R3.5 region [modified from a previous study (38)]. Circles indicate the 5′-ends of RNA cleavage products, the cDNAs of which were cloned from ‘RISC formation/cleavage assays’ that applied AGO1 (white circles, Figure 3A, left panel) or AGO2 (black circles). Corresponding vsiRNAs were deduced via nucleotide 10 of the guide strand that should be complementary to these cleavage sites (indicated by the triangle in A). Arrows indicate the 5′-ends of the most frequently cloned cleavage products that led to the identification of vsiRNAs1 and 2. (C) ‘RISC formation/cleavage assays’ that were performed with AGO1, AGO2 and labeled (+)DI-R3.5 target RNA. The assays were performed as described in Figure 3A and tested the dsR3.5-derived siRNA pool side-by-side with the synthetic vsiRNA1 (left panel) and vsiRNA2 (right panel), respectively. Target RNA and cleavage products are indicated in the same way as in the previous figures. Lanes 1; assays performed with a non-specific (‘gf698’) siRNA (negative control). Lanes 2; assays performed with the dsR3.5-derived siRNA pool (siRNAs dsR3.5). Lanes 3; assays performed with vsiRNA1 or vsiRNA2. (D) ‘Replication inhibition assays’ with AGO1 and vsiRNA1 or AGO2 and vsiRNA2. The assays were carried out as described in Figure 3B (variant 1) using (+)DI-R3.5 RNA. RP and cleavage products (asterisks) are indicated. Lanes 1; assays performed in the absence of p92 (no replication). Lanes 2; assays performed in the presence of a non-specific (‘gf698’) siRNA (negative control). Lane 3; assays performed in the presence of vsiRNA1 or vsiRNA2.
To gain an initial idea on whether DI-R3.5 RNA was targeted by RISC that was formed in the BYL (containing additional AGO1, see earlier in the text), we first tested a whole pool of vsiRNAs. For this purpose, sense and antisense in vitro transcripts of the R3.5 region were hybridized to generate double-stranded (ds) RNA molecules (dsR3.5 RNA). These RNAs were digested with an RNase III variant (ShortCut®), which has an activity comparable with that of DCL as it produces 18–25-nt long siRNAs from dsRNA (Supplementary Figure S1 and also later in the text). The siRNAs were purified and added to the same translation reaction in BYL that also generated AGO1. Thus, we expected RISC formed with the overexpressed AGO1 to incorporate the exogenous siRNAs and to be specifically ‘programmed’ towards the R3.5 element in the ‘RISC formation/cleavage assay’ (Figure 1B). Accordingly, we next added in vitro transcribed 32P-labeled DI-R3.5 target RNAs in (+) or (−) orientation to the reaction mixture and tested for endonucleoytic cleavage. SiRNA-mediated cleavage was observed with both, the target (+) and (−)RNAs, whereas this was not the case in control experiments with the unrelated (‘gf698’) siRNA (Figure 3A, lanes 2 and 4 versus lanes 1 and 3). Notably, in the experiments that applied the vsiRNAs, we obtained a distinct number rather than a broad variety of cleavage products as one would expect this to be the case with a whole pool of vsiRNAs. Moreover, considerable amounts of the target RNAs remained intact. These observations indicated that the majority of the vsiRNAs that were generated by RNase III from the dsR3.5 region were inefficient in guiding the formed RISC. However, 3–4 vsiRNAs, the number which could be deduced from the detectable cleavage products, seemed to be highly effective (see also later in the text).
Next, we tested whether the dsR3.5-derived siRNAs were capable to inhibit viral replication. For this purpose, the following experimental strategy was applied. In one reaction, p33 and p92 were in vitro translated to permit the formation of TBSV replication complexes (‘replicase formation’). In a second, separate reaction, RISC were formed with in vitro synthesized AGO1 and programmed with the exogenous siRNA pool (‘RISC formation’). Both reaction mixtures were then combined, the conditions switched to replication and (+)DI-R3.5 RNA added as a replication template. Replication of the RNA was measured via the incorporation of a 32P-labeled nucleotide into newly synthesized progeny RNA (Figure 3B; variant 1). Interestingly, when we performed the reaction in the presence of the pool of dsR3.5-derived vsiRNAs (Figure 3C, lane 3), the newly generated viral replication product was cleaved into defined cleavage products, and we observed a slight inhibitory effect on viral RNA replication. Hence, with this experiment (hereafter termed as ‘replication inhibition assay’), we obtained initial indications that the BYL system reproduced RISC-mediated antiviral RNA silencing in vitro.
In vitro formed RISC targets (+)RNA
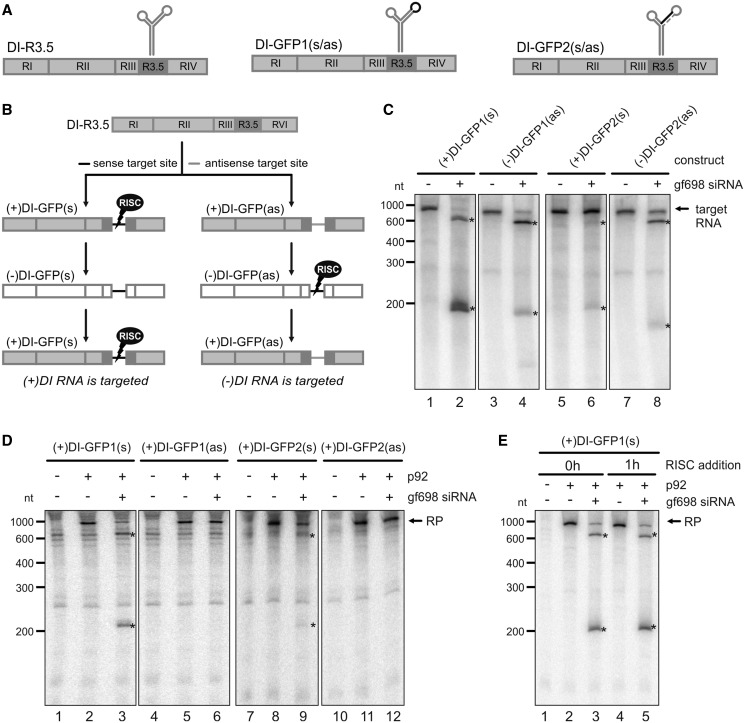
The subsequent set of experiments served two major purposes, to considerably improve the system and to understand whether the in vitro formed AGO1/RISC targeted (+) and (−)RNA molecules similarly or differently during the viral replication process. For this, we used two DI-R3.5 RNA variants, DI-GFP1 and DI-GFP2, which were confirmed to be as replication competent as the original DI-R3.5 (Figure 4, and data not shown). These RNAs contained the ‘gf698’ siRNA-specific target site of the GFP mRNA, which was shown to be particularly accessible to RISC (47) (Figure 1), in a single- or double-stranded region of the R3.5 element (Figure 4A). As in vitro TBSV RNA replication solely initiates with (+)RNA (T. Gursinsky, unpublished data), we generated two forms of the DI-GFP1 and 2 (+)RNAs that contained the ‘gf698’ target site in sense (s) and antisense (as) orientation, respectively. During viral replication, RISC-mediated cleavage via the ‘gf698’ siRNA was accordingly supposed to occur in either the (+)- or the (−)-oriented RNA replication products (Figure 4B).
Figure 4.
RISC-mediated RNA silencing targets viral (+)RNA. (A) Schematic representations of the DI RNA variants DI-GFP1 and DI-GFP2 that contained the target sequence of the ‘gf698’ siRNA at different positions. The target sequence, which is indicated by a black line, corresponds to a short sequence of GFP mRNA and is complementary to the ‘gf698’ siRNA guide strand. The dashed line in DI-GFP2 represents a sequence that is complementary to the target sequence. (B) With both constructs shown in (A), the ‘gf698’ target sequence was introduced in either sense (s) or antisense (as) orientation. Accordingly, as it is shown in the schematic representation of DI RNA replication, RISC programmed with the ‘gf698’ siRNA was supposed to target either (+) or (−)RNA molecules. (C) ‘RISC formation/cleavage assay’ with DI-GFP RNAs. The assay was performed as described in Figure 1B using ‘gf698’ siRNA and 32P-labeled RNA transcripts of the respective (+) and (−)DI-GFP RNAs. The target RNA (indicated) and the cleavage products (asterisks) were analyzed by denaturing PAGE and autoradiography. Lanes 1, 3, 5 and 7; assays performed in the absence of siRNA (negative controls). Lanes 2, 4, 6 and 8; assays performed in the presence of ‘gf698’ siRNA. (D) ‘Replication inhibition assay’ with DI-GFP RNAs. The assays were performed as described in Figure 3B (variant 1), i.e. RISC programmed with ‘gf698’ siRNA was added to a translation/replication reaction performed with the (+)RNA of the different DI-GFP variants. RP and cleavage products (asterisks) are indicated. Lanes 1, 4, 7 and 10; assays in the absence of p92 (no replication). Lanes 2, 5, 8 and 11; assays in the absence of siRNA (negative controls). Lanes 3, 6, 9 and 12; assays in the presence of ‘gf698’ siRNA. (E) SiRNA-programmed RISC also interferes with ongoing viral replication. ‘Replication inhibition assays’ were performed with (+)DI-GFP1(s) and ‘gf698’ siRNA as depicted in Figure 3B following experimental variants 1 or 2. That is, the RISC- and replicase-forming reactions were combined either before the initiation of RNA replication (0 h) or 1 h (1 h) after starting the replication reaction. The RP and cleavage products (asterisks) are indicated. Lane 1; assay performed in the absence of p92 (no replication). Lanes 2 and 3; assays where the reaction mixtures were combined before the initiation of replication in the absence (lane 2) and presence (lane 3) of ‘gf698’ siRNA. Lanes 4 and 5; assays where the reaction mixtures were combined after 1 h of viral replication in the absence (lane 4) and presence (lane 5) of ‘gf698’ siRNA.
Evaluating first 32P-labeled (+) and (−) RNA transcripts of DI-GFP1 and DI-GFP2 in ‘gf698’-directed ‘RISC formation/cleavage assays’, all RNAs were found to be cleaved (Figure 4C, lanes 2, 4, 6 and 8). This demonstrated that the non-replicating (+) and (−)RNAs each were accessible to the in vitro formed AGO1/RISC. This was most apparent with the (−)-oriented DI-GFP1(as) and DI-GFP2(as) RNAs, both of which were efficiently cleaved (Figure 4C, lanes 4 and 8). The (+)DI-GFP2(s) RNA that contained the target sequence in a stem motif (Figure 4A) was less efficiently cleaved than variant (+)DI-GFP1(s) where the target site was single-stranded (Figure 4C, lane 2 versus lane 6).
Next, we tested the DI-GFP variants in the ‘replication inhibition assay’ by combining a viral replication reaction and a RISC programming reaction with ‘gf698’ siRNA. For the explained reason, the replication assay was performed with the (+)RNAs. With the DI variants that contained the siRNA target site in sense orientation [(+)DI-GFP1(s) and (+)DI-GFP2(s); Figure 4B], we observed in each case a cleavage of the newly synthesized and radioactively labeled replication product and an inhibition of RNA replication (Figure 4D, lanes 3 and 9). Yet, with variant (+)DI-GFP2(s) that had the target sequence in a stem region, RNA cleavage, as well as the inhibitory effect on replication, was less apparent (Figure 4D, lane 9). In contrast, when we tested the viral DI RNAs that contained the ‘gf698’ target sequence in antisense orientation and where the (−)RNA intermediate should be targeted by RISC [(+)DI-GFP1(as) and (+)DI-GFP2(as); Figure 4B], the newly synthesized RNA was not cleaved, and replication was not inhibited (Figure 4D, lanes 6 and 12).
So far, the ‘replication inhibition assay’ was carried out by combining two reaction mixtures that contained the programmed RISC and the preformed replicase, respectively. RNA replication was then started by the addition of the DI RNA template (Figure 3B; variant 1). However, this constellation included the possibility that the AGO1/RISC simply cleaved the RNA template and hereby interfered with viral replication. We, therefore, changed the protocol of the assay such that the preformed RISC was now added to a replication reaction that was ongoing for 1 h (Figure 3B; variant 2). As shown in Figure 4E, RISC-mediated antiviral RNA silencing was observed with both experimental variants (lanes 3 and 5).
From earlier replication assays with TBSV DI RNA, it was known that most template RNA are degraded within 1 h of incubation time in the BYL. Moreover, nearly all detectable (32P-labeled) RNA replication products were shown to correspond to progeny TBSV (+)RNA, i.e. newly synthesized (−)RNA is highly underrepresented in the assay and only measurable by RT–PCR (46). Considering these points and the fact that the observed cleavage products had the sizes that were expected with a ‘gf698’-mediated cleavage of the DI-GFP (+)RNA, these data indicated that the AGO1/RISC targeted directly the (+)RNA replication product. In summary, these results supported and extended our earlier findings with the vsiRNA pool and confirmed that RISC-mediated antiviral RNA silencing was capably reproduced under the in vitro conditions of the BYL system. Interestingly, silencing was not detectable on the level of the viral (−)RNA intermediate.
RISC-mediated antiviral RNA silencing in vitro is suppressed by p19
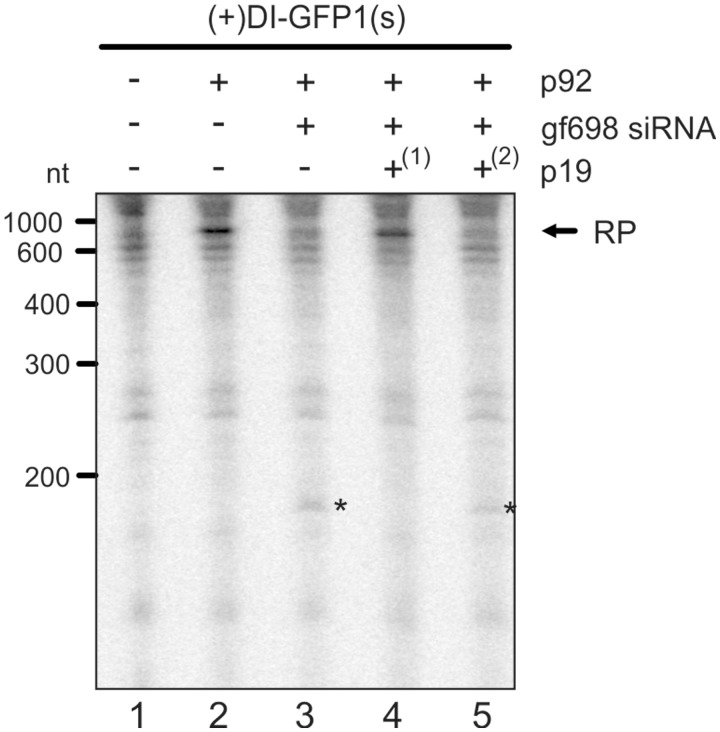
As outlined, the Tombusvirus p19 is an effective antagonist of antiviral RNA silencing. To test for the suppression of antiviral RNA silencing by p19 in our system, we added a defined amount of the protein at different time points to the assay, i.e. at the beginning and at the end of the translation reaction that generated AGO1 and RISC (Figure 3B). Early addition of p19 effectively counteracted antiviral RNA silencing, i.e. RISC-mediated cleavage of the viral RNA was less prominent, and viral replication was only marginally inhibited (Figure 5, lane 4). In contrast, late addition of the suppressor to the RISC-forming reaction had nearly no effect (Figure 5, lane 5). We, therefore, deduced that p19 suppressed RNA silencing in a similar way in the plant BYL system as this was early found in a Drosophila in vitro system, namely, by sequestration of unbound siRNA molecules (49). In the experiment shown here, we applied a significant excess of the p19 protein as compared with the amount of ‘gf698’ siRNA. Reducing the amount of p19 (10 instead of 25 U) resulted in a partial inhibition of silencing (data not shown).
Figure 5.
p19 counteracts antiviral RNA silencing in vitro. ‘Replication inhibition assays’ were performed with (+)DI-GFP1(s) and ‘gf698’ siRNA following experimental variant 1 (Figure 3B), and p19 was added at different time points of the RISC-forming reaction. Lane 1; assay performed in the absence of p92 (no replication). Lane 2; assay performed in the absence of siRNA and p19 (negative control). Lane 3; assay performed in the presence of ‘gf698’ siRNA and in the absence of p19 (positive control of silencing). Lane 4; assay performed in the presence of ‘gf698’ siRNA where p19 was added at the beginning(1) of the RISC formation reaction. Lane 5; assay performed in the presence of ‘gf698’ siRNA where p19 was added at the end(2) of the RISC formation reaction.
Several AGO proteins support antiviral RNA silencing
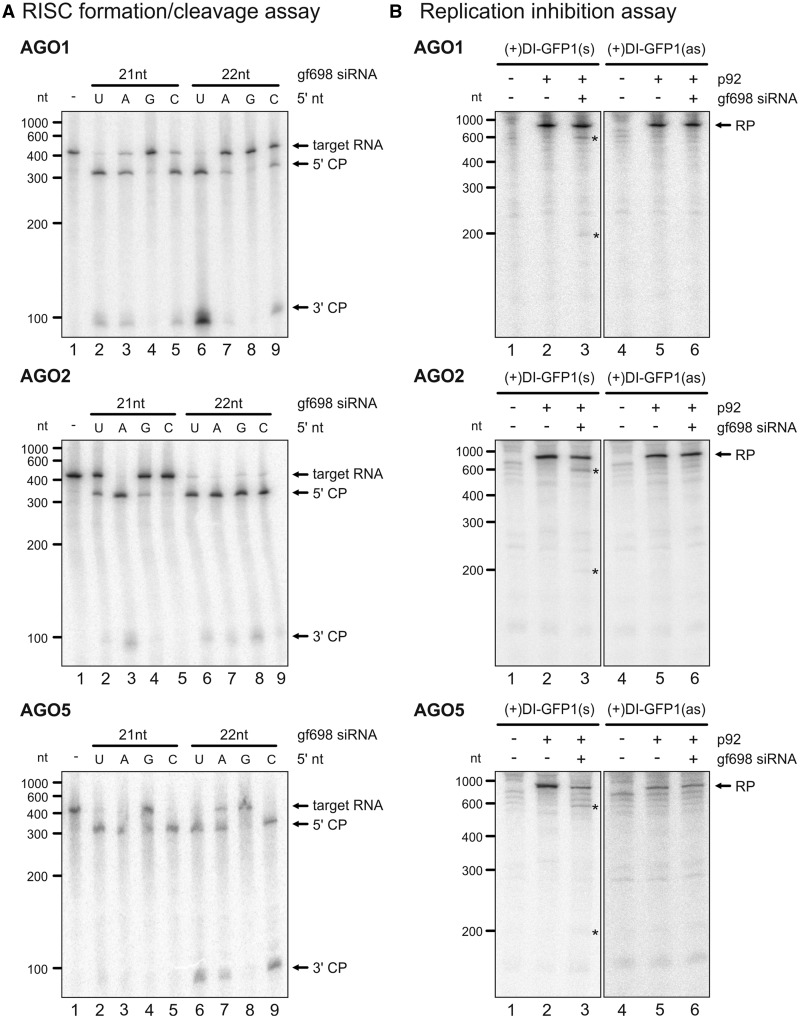
Having the ‘RISC formation/cleavage assay’ and the ‘replication inhibition assay’ in hands, we wanted to understand whether also other AGO proteins (besides AGO1) supported siRNA-directed cleavage of the TBSV RNA and potentially inhibited viral RNA replication in vitro. For this purpose, we cloned most AGO genes from A. thaliana (At). In fact, the subsequent experiments were performed with the At proteins, as comparative studies with the Nt AGO1 and At AGO1 yielded identical results (Table 1, and data not shown). In the BYL, all AGO proteins were expressed by in vitro translation of the corresponding mRNAs (Supplementary Figure S2). Using the 32P-labeled GFP mRNA as a target (Figure 1), we first performed ‘RISC formation/cleavage assays’ with the different AGO proteins and with 21 and 22 nt ‘gf698’ siRNAs, respectively. Moreover, considering that the sorting of siRNAs into AGO complexes was shown to be directed by the 5′-terminal nucleotide (21,50), we also tested ‘gf698’ siRNAs with different 5′-termini. Thus, we confirmed and extended earlier findings demonstrating that AGO1, 2, 3, 5, 7 and 10 had an evident slicer activity with 21 and 22 nt siRNAs. AGO4, 6 and 9 revealed no slicer activity with 21- and 22-nt siRNAs. RISC containing AGO1 or AGO10 were moreover confirmed having a clear preference for siRNAs with a 5′-U, AGO2 for siRNAs with a 5′-A and AGO5 for siRNAs with a 5′-C. AGO3 and AGO7 accepted the ‘gf698’ siRNAs only for cleavage if these had a 5′-terminal A. These data are summarized in Table 1, examples of cleavage assays are shown with AGO1, AGO2 and AGO5 in Figure 6A (cleavage data with AGO3, 4, 6, 7, 9 and 10 shown as Supplementary Figure S3). Next, we applied these findings to ‘replication inhibition assays’, which were performed with (+)DI-GFP1(s) and (+)DI-GFP1(as) RNAs (Figure 4), respectively. That is, RISC were reconstituted with the respective AGO proteins and programmed with the ‘gf698’ siRNA variant that turned out to be best-accepted by this AGO protein in the earlier cleavage assay (Table 1). We observed that those AGO proteins that had the most evident slicer activity, namely, AGO1, 2, 3 and 5, also had an inhibitory effect on replicating viral RNA (Figure 6B; replication data with AGO3, 4, 7 and 10 shown as Supplementary Figure S4). However, replication inhibition was observed only with the viral RNAs that contained the ‘gf698’ target site in sense orientation (Figure 6B and Supplementary Figure S4). This confirmed our initial findings and demonstrated that not only AGO1/RISC but also AGO2/RISC, AGO3/RISC and AGO5/RISC targeted viral replication exclusively on the level of the (+)RNA.
Table 1.
Slicer activity of ARGONAUTE proteins and inhibition of viral replication
| RISC formation/cleavage assay |
Inhibition of viral replication | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| siRNA | 21 nt |
22 nt |
|||||||
| 5′-nt | U | A | G | C | U | A | G | C | |
| AGO1_Nt | ++ | + | ± | + | ++ | + | ± | + | ✓ (21 nt, 5′-U) |
| AGO1_At | ++ | + | ± | + | ++ | + | ± | + | ✓ (21 nt, 5′-U) |
| AGO2_At | + | ++ | + | + | ++ | ++ | ++ | ++ | ✓ (21 nt, 5′-A) |
| AGO3_At | − | + | − | − | − | + | − | − | ✓ (21 nt, 5′-A) |
| AGO4_At | − | − | − | − | − | − | − | − | −(21 nt, 5′-A) |
| AGO5_At | + | + | + | ++ | ++ | + | ± | ++ | ✓ (21 nt, 5′-C) |
| AGO6_At | − | − | − | − | − | − | − | − | N/A |
| AGO7_At | − | + | − | − | − | ± | − | − | −(21 nt, 5′-A) |
| AGO9_At | − | − | − | − | − | − | − | − | N/A |
| AGO10_At | + | − | − | − | ± | − | − | − | −(21 nt, 5′-U) |
The results of multiple ‘RISC formation/cleavage assays’ and ‘replication inhibition assays’ are summarized (see examples in Figure 6 and Supplementary Figures S3 and S4). Left; results of ‘RISC formation/cleavage assays’ with the different A. thaliana (At) and N. tabacum (Nt) AGO proteins: (++) indicates strong slicer activity; (+) indicates detectable slicer activity; (±) indicates that slicer activity was detected only in some of several independent experiments; (−) indicates no detectable slicer activity. Right; results of ‘replication inhibition assays’ with the different AGO proteins: (✓) indicates inhibition of viral RNA replication; (−) indicates no detectable inhibition of viral RNA replication. The ‘gf698’ siRNA variants that were applied in the assays are indicated in brackets. AGO6 and AGO9 were not tested in ‘replication inhibition assays’ (N/A).
Figure 6.
Several AGO proteins support antiviral RNA silencing. (A) ‘RISC formation/cleavage assays’ performed with AGO1, AGO2 and AGO5 using different variants of ‘gf698’ siRNAs and 32P-labeled GFP mRNA as a target. The assay was performed essentially as described in Figure 1 (corresponding assays performed with AGO3, 4, 6, 7, 9 and 10 are provided as Supplementary Figure S3). That is, At AGO1, AGO2 or AGO5 were overexpressed in the BYL by in vitro translation of the corresponding mRNAs in the presence of 21 (lanes 2–5) or 22 nt (lanes 6–9) ‘gf698’ siRNA variants where the corresponding guide strands had different 5′-terminal nucleotides (as indicated). After addition of the target mRNA, the RISC cleavage products (marked by asterisks) were analyzed by denaturing PAGE and autoradiography. As a negative control, the reaction was carried out in the absence of siRNA (lane 1). (B) ‘Replication inhibition assays’ performed with (+)DI-GFP RNAs and RISC containing AGO1, AGO2 or AGO5 (corresponding assays performed with AGO3, 4, 7 and 10 are provided as Supplementary Figure S4). The assays were essentially performed as described in Figures 3B and 4D. Reactions where RISC were formed with AGO1 were performed with ‘gf698’ siRNA possessing a 5′-terminal U, reactions containing AGO2 or AGO5 were performed with ‘gf698’ siRNA variants that possessed a 5′-terminal A or C, respectively. The programmed RISC were added to a translation/replication reaction performed with (+)DI-GFP1(s) or (+)DI-GFP1(as). RP and cleavage products (asterisks) are indicated. Lanes 1 and 4; assays in the absence of p92 (no replication). Lanes 2 and 5; assays in the absence of siRNA (negative controls). Lanes 3 and 6; assays in the presence of ‘gf698’ siRNA.
Effective vsiRNAs specifically target accessible parts of the viral RNA
The following series of experiments were aimed at understanding whether antiviral RNA silencing in vitro was also efficiently induced by siRNAs that derived directly from the viral RNA. Along this line, we considered it first important to investigate whether the BYL also exhibited endogenous DCL activity. For this, we first exposed 32P-labeled double-stranded RNA of the R3.5 element (dsR3.5 RNA) directly to BYL and tested for the generation of siRNAs after a certain incubation time (see ‘Materials and Methods’ section). As shown in Supplementary Figure S1, the dsR3.5 RNA was indeed processed into a set of siRNAs. Primarily, these had an estimated size of 24 nt, but also smaller siRNA species of 21 or 22 nt were detectable, suggesting that the BYL contained not only active DCL3 but also DCL2 and/or DCL4 (51). Next, we added the dsR3.5 RNA to the BYL and simultaneously synthesized AGO2. The AGO2/RISC that was programmed with the endogenously formed vsiRNA pool, then was tested in a ‘RISC formation/cleavage assay’ with 32P-labeled (−)DI-R3.5 RNA, which in the earlier experiments was found to be particularly accessible to vsiRNA-mediated cleavages (Figure 3A). The same experiment was carried out side-by-side with the exogenous, RNase III-generated dsR3.5 vsiRNA pool. As also shown in Supplementary Figure S1, both cleavage assays yielded a similar pattern of products, demonstrating that the vsiRNAs that were generated by the BYL-contained endogenous DCL activity or by the commercial ShortCut® RNase III had comparable activities.
As the ShortCut®-based procedure generated the highest yields of siRNAs, it was also used in the following experiments. These aimed at identifying vsiRNAs from the dsR3.5-derived pool that were earlier indicated (Figure 3A) to be particularly effective in RNA cleavage and antiviral RNA silencing. To this end, we performed ‘RISC formation/cleavage assays’ with AGO1, AGO2 and the (+)DI-R3.5 RNA. Then, the predominant RNA cleavage products were isolated, and the corresponding cDNAs were cloned via 5′-RACE. Interestingly, with each of the two AGO/RISC, the vast majority of cDNAs derived from a distinct set of RNA cleavage products, and the vsiRNAs that originated these products were defined (Figure 7A and B). As expected, the guide strands of such deduced vsiRNAs that were supposed to be active with AGO1/RISC initiated with a 5′-U residue, whereas the corresponding vsiRNAs in AGO2/RISC initiated with an A (Figure 7A). Notably, the vsiRNAs that were deduced from the most prevailing cDNAs were indicated to target a similar site in the R3.5 target RNA. That is, the identified endonucleolytic cleavage sites and the sequences 10-nt downstream of the cleavage sites that were predicted to associate with the 5′-ends of the vsiRNA’s guide strands (27, 28) located in or near a defined bulged region of the RNA (Figure 7B).
With each of the different AGO/RISC, we could subsequently confirm that indeed some of the most efficient vsiRNAs were identified. Thus, with AGO1/RISC, one deduced vsiRNA, vsiRNA1, was predicted to direct the cleavage of the (+)DI-R3.5 target such that the resulting products had sizes of 607 and 172 nt, respectively. These sizes matched those RNA species that were most prominently detectable in the earlier performed ‘RISC formation/cleavage assays’ that applied the dsR3.5-derived vsiRNA pool and (+)DI-R3.5 (Figure 3A, lane 2). The assumption that highly effective vsiRNAs were identified was finally confirmed when we tested a synthetic 21-nt long version of vsiRNA1 side-by-side with the dsR3.5 vsiRNA pool in the ‘RISC formation cleavage assay’ and the ‘replication inhibition assay’, respectively. Thus, AGO1/RISC and vsiRNA1-directed cleavage of the (+)DI-R3.5 target RNA-generated products that had essentially the same sizes as the most prominent cleavage products in the corresponding experiment with the dsR3.5-derived vsiRNA pool (Figure 7C; left panel, lanes 2 and 3). Moreover, vsiRNA1-directed RNA silencing efficiently interfered with DI-R3.5 RNA replication (Figure 7D; left panel, lane 3). Analogous results were obtained with a 22-nt version of vsiRNA1 (data not shown). Efficient RNA cleavage and an evident inhibition of viral replication were also observed in experiments that applied AGO2/RISC and a synthetic 21-nt version of vsiRNA2, which was deduced to target the same region in the R3.5 element as vsiRNA1 (Figure 7C and D; right panel). As various other synthetic vsiRNAs that were designed to attack other parts of R3.5 remained inefficient (data not shown), the vsiRNAs1/vsiRNA2-targeted bulge-region was indicated as being particularly accessible to different AGO/RISC (see ‘Discussion’ section).
In a final experiment, we programmed AGO1/RISC with the dsR3.5-derived vsiRNA pool and applied this to (−)DI-R3.5 RNA in a ‘RISC formation/cleavage assay’ (Figure 3A, lane 4). As described earlier in the text, the most prominent RNA cleavage products were isolated, corresponding cDNAs cloned via 5′-RACE and a vsiRNA (vsiRNA3) deduced that originated these products (Supplementary Figure S5A and B). Although AGO1/RISC with synthetic vsiRNA3 efficiently cleaved non-replicating (−)DI-R3.5 RNA (Supplementary Figure S5C), it had no inhibitory effect on viral RNA replication (Supplementary Figure S5D). This supported our earlier data (Figures 4 and 6), demonstrating that also a virus-derived siRNA, which was highly efficient in the ‘RISC formation cleavage assay’, did not inhibit viral replication on the level of the (−)RNA intermediate.
DISCUSSION
Cytoplasmic extract of tobacco cells (BYL) was earlier shown to support mRNA silencing (47) and viral RNA replication (45, 46). However, it was uncertain whether in vitro formed RNA silencing complexes, such as RISC, also targeted replicative viral RNA. In this study, we integrated the two potentialities of the BYL system and reproduced AGO/RISC-directed antiviral RNA silencing with a TBSV RNA in vitro (Figure 3). The BYL also has endogenous DCL activity (Supplementary Figure S1); thus, it was confirmed as a valuable experimental system that recapitulates several elements of the plant’s immune response against viral infections under defined conditions.
In the course of this work, we applied whole pools of vsiRNAs, a specific siRNA (‘gf698’) directed against an engineered target site in the viral RNA, as well as individually characterized vsiRNAs to program RISC and to mediate antiviral RNA silencing. As expected, the antiviral silencing effect was effectively inhibited in the presence of the siRNA-sequestering viral protein p19. However, p19 suppressed silencing only at an early stage of RISC formation (Figure 5). This was consistent with findings in other experimental systems (49,52) and supported the earlier proposed concept that siRNAs that are incorporated into RISC remain unapproachable to a sequestration by p19.
As a key technical advantage, the established in vitro system enables the reconstitution of RNA silencing with a defined ARGONAUTE protein and (an) exogenous siRNA(s) of choice. Thus, in analogy to the situation with AGO1, the de novo synthesis of which is essentially needed to measure AGO1/RISC slicer activity in BYL (47) (Figure 1), we could reconstitute silencing complexes with various AGO proteins. This was made possible by overexpressing the proteins by in vitro translation of their mRNAs and in the presence of the RISC-‘programming’ siRNA. In the absence of AGO mRNA translation, we did not observe RISC activity in BYL, neither with synthetic siRNAs nor with RNase III or DCL-generated siRNA pools. The endogenous levels of many, if not of all, AGO proteins thus seem to be low in the BY-2 plant cell, and their expression may accordingly be subject of regulation processes, as it was observed to be the case with AGO1 (53–55).
The in vitro system enabled a comprehensive side-by-side testing of the known At AGO proteins [AGO8 is assumed to be a pseudogene (56) and was not investigated]. Thus, AGO1, 2, 3, 5, 7 and 10 belonging to two of the three phylogenetic clades of plant ARGONAUTE proteins (56), all were demonstrated to exhibit in vitro slicer activity. RISC containing AGO1, AGO2, the closely AGO2-related AGO3 or AGO5 was moreover shown to inhibit viral RNA replication in vitro (Table 1, Figures 6 and 7 and Supplementary Figures S3 and S4). Supporting earlier findings (12,57), it is important to note that with all these AGO/RISC (except for AGO7/RISC), 21- and 22-nt siRNAs were similarly effective in mediating slicer activity and to inhibit viral replication, respectively. AGO4, AGO6 and AGO9 belonging to the third phylogenetic clade, in contrast, revealed no slicer and replication inhibiting activity with 21- and 22-nt siRNAs (Supplementary Figures S3 and S4). This was not surprising considering that AGO4 and 6 preferentially bind 24-nt siRNAs and are supposed to be mainly involved in RNA-directed DNA methylation (58). In summary, these data suggest that a significant subset of the plant’s AGO proteins is involved in antiviral measures. Considering that the AGO-mediated slicer activity was found to be generally less effective with replicating than with non-replicating viral (+)RNA (Figures 3, 4, 6 and 7), this may explain why plants contain such a broad variety of AGO proteins. The fact that RISC containing AGO1- and AGO2-targeted neighboring sequences in the viral RNA (Figure 7) indicates that the proteins act redundantly and, perhaps, synergistically.
Our tests further confirmed and extended earlier data with other experimental systems that the siRNA’s 5′-terminal nucleotide has a strong impact on its sorting into the different AGO proteins (Table 1) (21,50). The particular sorting mechanism of AGO/RISC was again obvious when we investigated the TBSV R3.5 region as a supposed silencing hot spot and when we characterized individual viral RNA cleavage products and vsiRNAs. Thus, all vsiRNAs that were effective with AGO1/RISC contained a 5′-terminal U, whereas vsiRNAs that worked with AGO2 always had a 5′-terminal A (Figures 6 and 7).
What are the molecular determinants mediating an effective silencing of (+)-stranded RNA viruses such as TBSV? Our data revealed some initial valuable indications. Thus, it was obvious that those AGO proteins, namely, AGO1 and AGO2 that had the most evident slicer activity, also were most effective in inhibiting viral RNA replication (Figure 6). Interestingly, with each of these AGO/RISC and with different experimental approaches, we could demonstrate that replicating TBSV RNA is accessible to RISC, but only on the level of the (+)RNA (Figures 4 and 6, Supplementary Figure S5). The experiments further supported the notion that an important part of the antiviral silencing response involves the cleavage of newly generated (+)RNAs (Figure 4E). Taken together, these data suggest a straight model according to which the antiviral AGO/RISC does not target RNA that is entrapped in the active viral replication machinery but rather inhibits the formation of progeny replication complexes by cleaving accessible (non-replicating) viral (+)RNA. ‘Accessible’ RNA may be totally or partly uncoated viral genome in not only newly infected cells but also freshly synthesized progeny (+)RNA that may be attacked by RISC in statu nascendi. Along this line, it is again worth discussing that throughout the characterization of highly effective vsiRNAs that were directed against the R3.5 ‘silencing hot spot’, all antiviral AGO/RISC were found to target similar regions in this RNA element. Thus, in close homology to the situation that was described in the human system (59,60), the seed-sequences of the siRNA’s guide strands, as well as the endonucleolytic cleavage sites, in each case associated with single-stranded or weakly structured regions in the R3.5 element (Figure 7B). An evident inverse correlation of RNA structure and accessibility by RISC was also found with the viral RNA constructs that contained the ‘gf698’ siRNA target site in different RNA regions, i.e. RNA molecules that contained the target sequence in a stem-forming region (DI-GFP2) were evidently less vulnerable (Figure 4).
Interestingly, siRNAs that targeted the (−)RNA had no detectable effect on TBSV RNA replication (Figures 4 and 6 and Supplementary Figure S5). In view of the scenario that in the in vitro system, the ratio of TBSV progeny (+) versus (−)RNAs was ∼200:1 (46), the highly underrepresented (−)RNA molecules were supposed to be perfect targets for an inhibition of viral replication. The reasons why the (−)RNA is protected may be manifold. As the (−)RNA intermediates are believed to be predominantly present in active viral replication complexes and in specific membrane compartments (61), these may simply not be approachable by RISC. Alternatively, it is conceivable (and still a matter of debate) that during RNA replication the (−)RNA exists mainly as dsRNA.
The observation that (−)RNA was not accessible to antiviral RNA silencing was remarkable also in view of reports that showed that plants that were infected with different RNA viruses, e.g. the TBSV-related CymRSV, revealed a strong bias for the generation of vsiRNAs from the (+)RNA (35,37,62,63). The high level of (+)siRNAs was speculated to be caused by a preferential incorporation into RISC (13) or by a rapid degradation of effector complexes that contain (−)RNA targeting siRNAs (8). Moreover, there is significant evidence that dicing may also occur in structured RNA motifs and even in imperfect duplexes in the (+)RNA, and that (+)-derived siRNAs are generated from structural ‘hot spots’ of the viral RNA (35,37). Obviously, these observations fueled the idea that the excess of (+)siRNAs was directed against the underrepresented (see earlier in the text) and replication rate-limiting (−)RNA. Although this model of the (−)RNA as the Achilles heel of viral replication is questioned, an alternative explanation for the function of (+)RNA-derived siRNAs may be that some of these do not target (−)RNA but again structural motifs in the (+)RNA. The in vitro system will be a valuable tool to address this point.
Taken together, our data suggest that efficient antiviral RNA silencing involves the activity of different AGO/RISC. These complexes preferentially contain AGO proteins with a high slicer activity (e.g. AGO1 or AGO2) and siRNAs that target similar accessible regions of the viral (+)RNA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–5, Supplementary Methods and Supplementary References [46,64–66].
FUNDING
Sonderforschungsbereich (CRC) 648 of the Deutsche Forschungsgemeinschaft (DFG) at the Martin Luther University Halle-Wittenberg (project A7); the CRC 648 provided a Post-Doctoral position for T.G. and a short-term Fellowship for A.P. Graduiertenkolleg 1591 of the Deutsche Forschungsgemeinschaft (DFG) at the Martin Luther University Halle-Wittenberg (project B6) (to J.S.). Funding for open access charge: Sonderforschungsbereich (CRC) 648 of the Deutsche Forschungsgemeinschaft.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Mandy Hacke, Nancy Funk and Joanna Jagoda for valuable technical assistance, and they are grateful to Claudia Temme, Stefan Hüttelmaier and their laboratory members for helpful discussions.
REFERENCES
- 1.Baulcombe D. RNA silencing in plants. Nature. 2004;431:356–363. doi: 10.1038/nature02874. [DOI] [PubMed] [Google Scholar]
- 2.Malone CD, Hannon GJ. Small RNAs as guardians of the genome. Cell. 2009;136:656–668. doi: 10.1016/j.cell.2009.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hull R. Matthews' Plant Virology. 4th edn. San Diego, CA: Academic Press; 2002. p. 1001. [Google Scholar]
- 4.Ortín J, Parra F. Structure and function of RNA replication. Annu. Rev. Microbiol. 2006;60:305–326. doi: 10.1146/annurev.micro.60.080805.142248. [DOI] [PubMed] [Google Scholar]
- 5.Ding SW, Voinnet O. Antiviral immunity directed by small RNAs. Cell. 2007;130:413–426. doi: 10.1016/j.cell.2007.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Csorba T, Pantaleo V, Burgyán J. RNA silencing: an antiviral mechanism. Adv. Virus Res. 2009;75:35–71. doi: 10.1016/S0065-3527(09)07502-2. [DOI] [PubMed] [Google Scholar]
- 7.Pantaleo V. Plant RNA silencing in viral defence. Adv. Exp. Med. Biol. 2011;722:39–58. doi: 10.1007/978-1-4614-0332-6_3. [DOI] [PubMed] [Google Scholar]
- 8.Ding SW. RNA-based antiviral immunity. Nat. Rev. Immunol. 2010;10:632–644. doi: 10.1038/nri2824. [DOI] [PubMed] [Google Scholar]
- 9.Tomari Y, Zamore PD. Perspective: machines for RNAi. Genes Dev. 2005;19:517–529. doi: 10.1101/gad.1284105. [DOI] [PubMed] [Google Scholar]
- 10.Blevins T, Rajeswaran R, Shivaprasad PV, Beknazariants D, Si-Ammour A, Park HS, Vazquez F, Robertson D, Meins F, Jr, Hohn T, et al. Four plant Dicers mediate viral small RNA biogenesis and DNA virus induced silencing. Nucleic Acids Res. 2006;34:6233–6246. doi: 10.1093/nar/gkl886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hamilton A, Voinnet O, Chappell L, Baulcombe D. Two classes of short interfering RNA in RNA silencing. EMBO J. 2002;21:4671–4679. doi: 10.1093/emboj/cdf464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deleris A, Gallego-Bartolome J, Bao J, Kasschau KD, Carrington JC, Voinnet O. Hierarchical action and inhibition of plant Dicer-like proteins in antiviral defense. Science. 2006;313:68–71. doi: 10.1126/science.1128214. [DOI] [PubMed] [Google Scholar]
- 13.Wang XB, Jovel J, Udomporn P, Wang Y, Wu Q, Li WX, Gasciolli V, Vaucheret H, Ding SW. The 21-nucleotide, but not 22-nucleotide, viral secondary small interfering RNAs direct potent antiviral defense by two cooperative argonautes in Arabidopsis thaliana. Plant Cell. 2011;23:1625–1638. doi: 10.1105/tpc.110.082305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ratcliff F, Harrison BD, Baulcombe DC. A similarity between viral defense and gene silencing in plants. Science. 1997;276:1558–1560. doi: 10.1126/science.276.5318.1558. [DOI] [PubMed] [Google Scholar]
- 15.Hamilton AJ, Baulcombe DC. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science. 1999;286:950–952. doi: 10.1126/science.286.5441.950. [DOI] [PubMed] [Google Scholar]
- 16.Szittya G, Molnár A, Silhavy D, Hornyik C, Burgyán J. Short defective interfering RNAs of tombusviruses are not targeted but trigger post-transcriptional gene silencing against their helper virus. Plant Cell. 2002;14:359–372. doi: 10.1105/tpc.010366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moissiard G, Voinnet O. Viral suppression of RNA silencing in plants. Mol. Plant Pathol. 2004;5:71–82. doi: 10.1111/j.1364-3703.2004.00207.x. [DOI] [PubMed] [Google Scholar]
- 18.Omarov RT, Ciomperlik JJ, Scholthof HB. RNAi-associated ssRNA-specific ribonucleases in Tombusvirus P19 mutant-infected plants and evidence for a discrete siRNA-containing effector complex. Proc. Natl Acad. Sci. USA. 2007;104:1714–1719. doi: 10.1073/pnas.0608117104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pantaleo V, Szittya G, Burgyán J. Molecular bases of viral RNA targeting by viral small interfering RNA-programmed RISC. J. Virol. 2007;81:3797–3806. doi: 10.1128/JVI.02383-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morel JB, Godon C, Mourrain P, Béclin C, Boutet S, Feuerbach F, Proux F, Vaucheret H. Fertile hypomorphic ARGONAUTE (ago1) mutants impaired in post-transcriptional gene silencing and virus resistance. Plant Cell. 2002;14:629–639. doi: 10.1105/tpc.010358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Takeda A, Iwasaki S, Watanabe T, Utsumi M, Watanabe Y. The mechanism selecting the guide strand from small RNA duplexes is different among argonaute proteins. Plant Cell Physiol. 2008;49:493–500. doi: 10.1093/pcp/pcn043. [DOI] [PubMed] [Google Scholar]
- 22.Qu F, Ye X, Morris TJ. Arabidopsis DRB4, AGO1, AGO7, and RDR6 participate in a DCL4-initiated antiviral RNA silencing pathway negatively regulated by DCL1. Proc. Natl Acad. Sci. USA. 2008;105:14732–14737. doi: 10.1073/pnas.0805760105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Azevedo J, Garcia D, Pontier D, Ohnesorge S, Yu A, Garcia S, Braun L, Bergdoll M, Hakimi MA, Lagrange T, et al. Argonaute quenching and global changes in Dicer homeostasis caused by a pathogen-encoded GW repeat protein. Genes Dev. 2010;24:904–915. doi: 10.1101/gad.1908710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harvey JJ, Lewsey MG, Patel K, Westwood J, Heimstädt S, Carr JP, Baulcombe DC. An antiviral defense role of AGO2 in plants. PLoS One. 2011;6:e14639. doi: 10.1371/journal.pone.0014639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jaubert M, Bhattacharjee S, Mello AF, Perry KL, Moffett P. ARGONAUTE2 mediates RNA-silencing antiviral defenses against Potato virus X in Arabidopsis. Plant Physiol. 2011;156:1556–1564. doi: 10.1104/pp.111.178012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scholthof HB, Alvarado VY, Vega-Arreguin JC, Ciomperlik J, Odokonyero D, Brosseau C, Jaubert M, Zamora A, Moffett P. Identification of an ARGONAUTE for antiviral RNA silencing in Nicotiana benthamiana. Plant Physiol. 2011;156:1548–1555. doi: 10.1104/pp.111.178764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang X, Yuan YR, Pei Y, Lin SS, Tuschl T, Patel DJ, Chua NH. Cucumber mosaic virus-encoded 2b suppressor inhibits Arabidopsis Argonaute1 cleavage activity to counter plant defense. Genes Dev. 2006;20:3255–3268. doi: 10.1101/gad.1495506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carthew RW, Sontheimer EJ. Origins and Mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Voinnet O, Pinto YM, Baulcombe DC. Suppression of gene silencing: a general strategy used by diverse DNA and RNA viruses of plants. Proc. Natl Acad. Sci. USA. 1999;96:14147–14152. doi: 10.1073/pnas.96.24.14147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Silhavy D, Burgyán J. Effects and side-effects of viral RNA silencing suppressors on short RNAs. Trends Plant Sci. 2004;9:76–83. doi: 10.1016/j.tplants.2003.12.010. [DOI] [PubMed] [Google Scholar]
- 31.Silhavy D, Molnár A, Lucioli A, Szittya G, Hornyik C, Tavazza M, Burgyán J. A viral protein suppresses RNA silencing and binds silencing-generated, 21- to 25-nucleotide double-stranded RNAs. EMBO J. 2002;21:3070–3080. doi: 10.1093/emboj/cdf312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Omarov R, Sparks K, Smith L, Zindovic J, Scholthof HB. Biological relevance of a stable biochemical interaction between the tombusvirus-encoded P19 and short interfering RNAs. J. Virol. 2006;80:3000–3008. doi: 10.1128/JVI.80.6.3000-3008.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.White KA, Nagy PD. Advances in the molecular biology of tombusviruses: gene expression, genome replication, and recombination. Prog. Nucleic Acid Res. Mol. Biol. 2004;78:187–226. doi: 10.1016/S0079-6603(04)78005-8. [DOI] [PubMed] [Google Scholar]
- 34.Omarov RT, Rezende JA, Scholthof HB. Host-specific generation and maintenance of Tomato bushy stunt virus defective interfering RNAs. Mol. Plant Microbe Interact. 2004;17:195–201. doi: 10.1094/MPMI.2004.17.2.195. [DOI] [PubMed] [Google Scholar]
- 35.Molnár A, Csorba T, Lakatos L, Várallyay E, Lacomme C, Burgyán J. Plant virus-derived small interfering RNAs originate predominantly from highly structured single-stranded viral RNAs. J. Virol. 2005;79:7812–7818. doi: 10.1128/JVI.79.12.7812-7818.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pantaleo V, Burgyán J. Cymbidium ringspot virus harnesses RNA silencing to control the accumulation of virus parasite satellite RNA. J. Virol. 2008;82:11851–11858. doi: 10.1128/JVI.01343-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Szittya G, Moxon S, Pantaleo V, Toth G, Rusholme Pilcher RL, Moulton V, Burgyan J, Dalmay T. Structural and functional analysis of viral siRNAs. PLoS Pathog. 2010;6:e1000838. doi: 10.1371/journal.ppat.1000838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fabian MR, White KA. 5′-3′ RNA-RNA interaction facilitates cap- and poly(A) tail-independent translation of tomato bushy stunt virus mRNA: a potential common mechanism for tombusviridae. J. Biol. Chem. 2004;279:28862–28872. doi: 10.1074/jbc.M401272200. [DOI] [PubMed] [Google Scholar]
- 39.Nicholson BL, Wu B, Chevtchenko I, White KA. Tombusvirus recruitment of host translational machinery via the 3′ UTR. RNA. 2010;16:1402–1419. doi: 10.1261/rna.2135210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Panaviene Z, Panavas T, Nagy PD. Role of an internal and two 3′-terminal RNA elements in assembly of tombusvirus replicase. J. Virol. 2005;79:10608–10618. doi: 10.1128/JVI.79.16.10608-10618.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pogany J, White KA, Nagy PD. Specific binding of tombusvirus replication protein p33 to an internal replication element in the viral RNA is essential for replication. J. Virol. 2005;79:4859–4869. doi: 10.1128/JVI.79.8.4859-4869.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pathak KB, Sasvari Z, Nagy PD. The host Pex19p plays a role in peroxisomal localization of tombusvirus replication proteins. Virology. 2008;379:294–305. doi: 10.1016/j.virol.2008.06.044. [DOI] [PubMed] [Google Scholar]
- 43.Wu B, Pogany J, Na H, Nicholson BL, Nagy PD, White KA. A discontinuous RNA platform mediates RNA virus replication: building an integrated model for RNA-based regulation of viral processes. PLoS Pathog. 2009;5:e1000323. doi: 10.1371/journal.ppat.1000323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pathak KB, Nagy PD. Defective Interfering RNAs: Foes of Viruses and Friends of Virologists. Viruses. 2009;1:895–919. doi: 10.3390/v1030895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Komoda K, Naito S, Ishikawa M. Replication of plant RNA virus genomes in a cell-free extract of evacuolated plant protoplasts. Proc. Natl Acad. Sci. USA. 2004;101:1863–1867. doi: 10.1073/pnas.0307131101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gursinsky T, Schulz B, Behrens SE. Replication of Tomato bushy stunt virus RNA in a plant in vitro system. Virology. 2009;390:250–260. doi: 10.1016/j.virol.2009.05.009. [DOI] [PubMed] [Google Scholar]
- 47.Iki T, Yoshikawa M, Nishikiori M, Jaudal MC, Matsumoto-Yokoyama E, Mitsuhara I, Meshi T, Ishikawa M. In vitro assembly of plant RNA-induced silencing complexes facilitated by molecular chaperone HSP90. Mol. Cell. 2010;39:282–291. doi: 10.1016/j.molcel.2010.05.014. [DOI] [PubMed] [Google Scholar]
- 48.Knorr DA, Mullin RH, Hearne PQ, Morris TJ. De novo generation of defective interfering RNAs of tomato bushy stunt virus by high multiplicity passage. Virology. 1991;181:193–202. doi: 10.1016/0042-6822(91)90484-S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lakatos L, Szittya G, Silhavy D, Burgyán J. Molecular mechanism of RNA silencing suppression mediated by p19 protein of tombusviruses. EMBO J. 2004;23:876–884. doi: 10.1038/sj.emboj.7600096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mi S, Cai T, Hu Y, Chen Y, Hodges E, Ni F, Wu L, Li S, Zhou H, Long C, et al. Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5′ terminal nucleotide. Cell. 2008;133:116–127. doi: 10.1016/j.cell.2008.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Aliyari R, Ding SW. RNA-based viral immunity initiated by the Dicer family of host immune receptors. Immunol. Rev. 2009;227:176–188. doi: 10.1111/j.1600-065X.2008.00722.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lakatos L, Csorba T, Pantaleo V, Chapman EJ, Carrington JC, Liu YP, Dolja VV, Calvino LF, López-Moya JJ, Burgyán J. Small RNA binding is a common strategy to suppress RNA silencing by several viral suppressors. EMBO J. 2006;25:2768–2780. doi: 10.1038/sj.emboj.7601164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rhoades MW, Reinhart BJ, Lim LP, Burge CB, Bartel B, Bartel DP. Prediction of plant microRNA targets. Cell. 2002;110:513–520. doi: 10.1016/s0092-8674(02)00863-2. [DOI] [PubMed] [Google Scholar]
- 54.Vaucheret H, Vazquez F, Crété P, Bartel DP. The action of ARGONAUTE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Genes Dev. 2004;18:1187–1197. doi: 10.1101/gad.1201404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vaucheret H, Mallory AC, Bartel DP. AGO1 homeostasis entails coexpression of MIR168 and AGO1 and preferential stabilization of miR168 by AGO1. Mol. Cell. 2006;22:129–136. doi: 10.1016/j.molcel.2006.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vaucheret H. Plant ARGONAUTES. Trends Plant Sci. 2008;13:350–358. doi: 10.1016/j.tplants.2008.04.007. [DOI] [PubMed] [Google Scholar]
- 57.Bouché N, Lauressergues D, Gasciolli V, Vaucheret H. An antagonistic function for Arabidopsis DCL2 in development and a new function for DCL4 in generating viral siRNAs. EMBO J. 2006;25:3347–3356. doi: 10.1038/sj.emboj.7601217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang H, Zhu JK. RNA-directed DNA methylation. Curr. Opin. Plant Biol. 2011;14:142–147. doi: 10.1016/j.pbi.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ameres SL, Martinez J, Schroeder R. Molecular basis for target RNA recognition and cleavage by human RISC. Cell. 2007;130:101–112. doi: 10.1016/j.cell.2007.04.037. [DOI] [PubMed] [Google Scholar]
- 60.Westerhout EM, Berkhout B. A systematic analysis of the effect of target RNA structure on RNA interference. Nucleic Acids Res. 2007;35:4322–4330. doi: 10.1093/nar/gkm437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.den Boon JA, Ahlquist P. Organelle-like membrane compartmentalization of positive-strand RNA virus replication factories. Annu. Rev. Microbiol. 2010;64:241–256. doi: 10.1146/annurev.micro.112408.134012. [DOI] [PubMed] [Google Scholar]
- 62.Ho T, Pallett D, Rusholme R, Dalmay T, Wang H. A simplified method for cloning of short interfering RNAs from Brassica juncea infected with Turnip mosaic potyvirus and Turnip crinkle carmovirus. J. Virol. Methods. 2006;136:217–223. doi: 10.1016/j.jviromet.2006.05.016. [DOI] [PubMed] [Google Scholar]
- 63.Donaire L, Wang Y, Gonzalez-Ibeas D, Mayer KF, Aranda MA, Llave C. Deep-sequencing of plant viral small RNAs reveals effective and widespread targeting of viral genomes. Virology. 2009;392:203–214. doi: 10.1016/j.virol.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 64.Elbashir SM, Lendeckel W, Tuschl T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 2001;15:188–200. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang Y, Juranek S, Li H, Sheng G, Wardle GS, Tuschl T, Patel DJ. Nucleation, propagation and cleavage of target RNAs in Ago silencing complexes. Nature. 2009;461:754–761. doi: 10.1038/nature08434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.