Abstract
PEA3, ERM and ER81 belong to the PEA3 subfamily of Ets transcription factors and play important roles in a number of tissue-specific processes. Transcriptional activation by PEA3 subfamily factors requires their characteristic amino-terminal acidic transactivation domain (TAD). However, the cellular targets of this domain remain largely unknown. Using ERM as a prototype, we show that the minimal N-terminal TAD activates transcription by contacting the activator interacting domain (ACID)/Prostate tumor overexpressed protein 1 (PTOV) domain of the Mediator complex subunit MED25. We further show that depletion of MED25 disrupts the association of ERM with the Mediator in vitro. Small interfering RNA-mediated knockdown of MED25 as well as the overexpression of MED25-ACID and MED25-VWA domains efficiently inhibit the transcriptional activity of ERM. Moreover, mutations of amino acid residues that prevent binding of MED25 to ERM strongly reduce transactivation by ERM. Finally we show that siRNA depletion of MED25 diminishes PEA3-driven expression of MMP-1 and Mediator recruitment. In conclusion, this study identifies the PEA3 group members as the first human transcriptional factors that interact with the MED25 ACID/PTOV domain and establishes MED25 as a crucial transducer of their transactivation potential.
INTRODUCTION
The Ets transcription factors are regulatory proteins involved in cancer, cell growth and differentiation. All Ets proteins contain a DNA-binding domain (ETS domain) of ∼85 amino acids that enables them to bind to GGAA/T sites (1). Functional domains and post-translational modifications lying outside the highly conserved ETS domain provide the potential for individual Ets proteins to exhibit unique properties (1).
The human ERM protein belongs to the PEA3 subfamily of Ets proteins (2) and contains at least four functional domains (Figure 1A): an amino-terminal transactivation domain (TAD; residues 1–72) (3,4), a central negative regulatory domain (NRD; residues 73–298) (5,6), the carboxy-terminal ETS domain (residues 363–451) and a carboxy-terminal TAD (residues 452–510) (4). Initial experiments demonstrated that the first 72 residues and the carboxy-terminal tail constituted transferrable activation domains (4). Subsequent experiments demonstrated that the amino-terminal TAD is regulated by a flanking NRD, which functions in a sumoylation-dependent manner (5,6). The amino-terminal activation domain is highly conserved (∼85% sequence identity) among PEA3 subfamily members and represents the main activation domain of all three proteins (3,4,7,8). The TAD from the protein ERM displays minimal stable tertiary structure (9), and the combination of acidic and hydrophobic amino acids within this domain appears similar to that found in the TADs of other activators such as the herpes simplex viral protein 16 (VP16) (10,11).
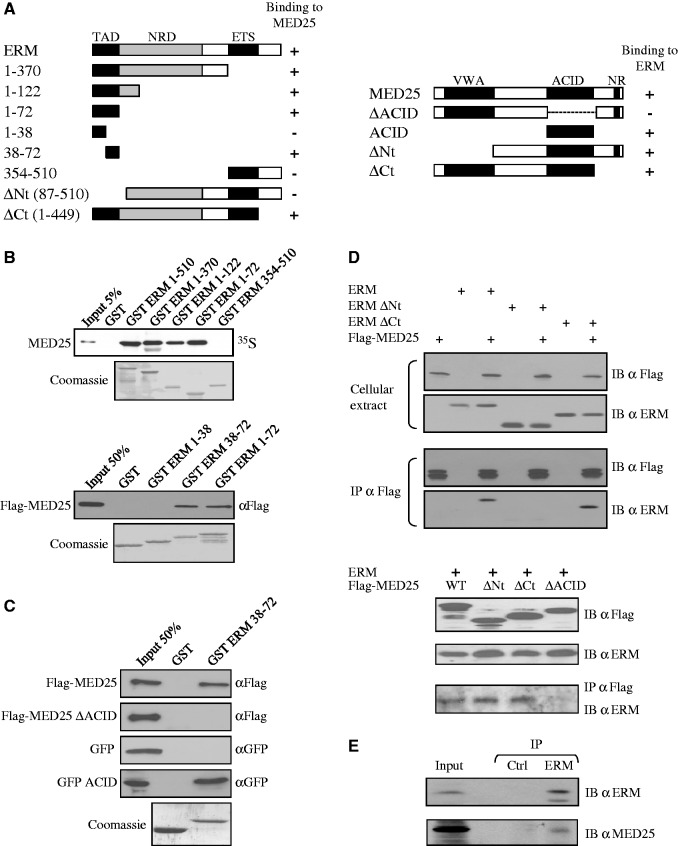
Figure 1.
ERM binds to MED25. (A) Schematic summary of the interaction between ERM and MED25 proteins. The N-terminal TAD of ERM interacts with the ACID of MED25. NR: Nuclear receptor box. Numbers refer to amino acid. (B) Deletion analysis of ERM shows that the N-terminal 38–72 domain is sufficient for binding MED25 in vitro. GST fusion proteins of the indicated ERM fragments were used to assess the binding to full-length rabbit reticulocyte lysate in vitro generated Flag-MED25. Binding was detected by autoradiography (upper panel) or immunoblotting with anti-Flag (bottom panel). An SDS gel stained with Coomassie showing the expression of the GST fusion proteins is shown. (C) Deletion analysis of MED25 shows that the ACID domain is sufficient for binding to ERM 38–72 in vitro. GST and GST-ERM 38–72 were incubated with the indicated MED25 fragments produced in reticulocyte lysate. Binding was detected by immunoblotting with anti-Flag or anti-GFP. (D) Co-immunoprecipitation of MED25 with ERM. Flag-MED25 and wild-type or mutants ERM (upper panel) or mutants Flag-MED25 together with full-length ERM (bottom panel) were expressed in RK13 cells. Cellular extracts were immunoprecipitated with anti-Flag antibody (IP α Flag) and immunoblotted with anti-Flag (IB α Flag) and anti-ERM (IB α ERM) antibodies. Aliquots of the same extracts were analysed with the same antibodies to detect exogenous proteins (cellular extract). (E) Interaction between endogenous proteins. MDA-MB 231 nuclear extracts were subjected to immunoprecipitation with anti-Gal4 DBD (IP Ctrl) or anti-ERM (IP ERM) antibodies. Interactions were detected by western blot using polyclonal anti-MED25 (IB α MED25) or polyclonal anti-ERM (IB α ERM).
Despite the importance of the ERM TAD, mechanistic details of how it interacts with other proteins and operates at a molecular level are limited. It is thought that the ERM TAD participates in interactions with transcriptional cofactors (3). However, a direct ERM TAD interacting protein within the RNA polymerase II general transcription machinery has not yet been reported. In this article, we identify the Mediator complex subunit MED25 as a direct physical target and functional partner of the ERM TAD. The Mediator is an evolutionary conserved multi-subunit RNA polymerase II transcriptional regulator complex critical for the tight control of gene expression (12). The MED25 subunit is found exclusively in higher eukaryotes (13) and has been shown to link the Mediator to many viral proteins such as VP16, IE62 and Lana-1 (14–17). In mammals, MED25 supports the activation of transcription by several transcription factors, including the retinoic acid receptor RARα (18), the orphan receptor HNF4α (19) and the chondrogenic factor Sox9 (20). Our findings identify ERM as the first nuclear transcription factor partner of the MED25 activator interacting domain (ACID) and suggest that the Mediator complex is an important cofactor for the PEA3 group member activity.
MATERIALS AND METHODS
Plasmid construction
The pSG5 expression vectors encoding full-length ERM, ETV1, ER81, PEA3 as well as ERM ΔNt (ERM 87–510) and ERM ΔCt (ERM 1–449) have been described previously (21–23), as have the GST fusion protein expression vectors PGEX ERM 1–510, ERM 1–370, ERM 1–122, ERM 1–72, ERM 354–510 (21,24,25). ERM 1–38, ERM 38–72 and ERM 38–68 were amplified by PCR and cloned into the PGEX-6P1 vector. The pCI-neo Flag human MED25 expression plasmid was a gift from M. Meisterernst (14). The MED25 mouse cDNA ORF was obtained from OriGene. The human and mouse MED25 cDNAs were subcloned into the pCDNA3 vector with an N-terminal Flag epitope tag. All other derivatives of MED25 correspond to the human factor unless otherwise indicated and were generated by PCR using MED25 cDNA as template. The siRNA-resistant MED25 expression vectors were generated by replacing the ACID/PTOV domain of human origin by its murine homologue. The pcDNA3-NLS-EFGP vector was constructed by inserting the open reading frame of NLS-GFP from the pNLS-EGFP vector into the pcDNA3 vector. The ACID/PTOV domain of human MED25 was subcloned into pcDNA3-NLS-EGFP. The ERM activation domain derivatives (1–72, 1–38, 38–72), E1A 13S and E2F were subcloned into pBIND (Gal4 DNA-binding domain) vector (Promega). The reporter plasmid (Gal4)5-E1B luc has been described (26). The bacterial expression plasmid encoding Halo-Tag ERM 1–72 (pET24d-Halo-Tag ERM 1–72) was generated by co-ligating the PCR amplified Halo-Tag [template pFN22K (Promega)] and ERM 1–72. To insert point mutations in our respective vectors (MED25 Q451E, MED25 M470A, ERM F47L and ERM 1–72 F47L), we used the Quickchange site-directed mutagenesis kit from Stratagene following the manufacturer’s directions. All clones were verified by sequencing. Primers sequences and detailed procedures are available on request.
Protein expression and purification
The MED25 ACID/PTOV domain was purified as previously described (27). GST, GST ERM derivatives (1–510, 1–370, 1–122, 1–72, 354–510, 1–38, 38–72) or GST ACID were expressed in Escherichia coli strain BL21 (DE3), and soluble lysates were prepared as described previously (4,21,28). Recombinant Halo-Tag, Halo-Tag ERM 1–72 and Halo-Tag ERM 1–72 F47L were purified in E. coli according to the manufacturer’s instructions (Promega). The purification of the ERM 38–68 peptide will be described elsewhere. Recombinant MED25 was expressed and radiolabelled with 35S-methionine by coupled in vitro transcription–translation reactions (TNT T7 quick coupled transcription–translation system Promega). Flag-MED25, Flag-MED25 ΔACID, GFP, GFP ACID, Flag-MED25 Q451E, Flag-MED25 M470A, Flag-ERM and Flag-ERM F47L were expressed by coupled in vitro transcription–translation reactions and detected by western blot with anti-Flag, anti-GFP or anti-ERM antibodies.
Pull-down assay
The interaction between MED25 and ERM was measured in GST or Halo-Tag pull-down assays. In vitro translated proteins were incubated with GST derivatives immobilized on glutathione-sepharose beads or Halo-Tag derivatives immobilized on HaloLink Magnetics beads (Promega), washed, eluted and bound proteins resolved by 10% SDS-PAGE followed by autoradiography or immunoblotting as described previously (28).
Antibodies
Anti-Flag M2 antibody was purchased from Sigma and anti-GFP monoclonal from Roche. Anti-MED14 (anti-CRSP2/DRIP150, A301-044A), anti-MED24 (anti-TRAP100/MED24, A301-472A) and anti-MED1 (anti-CRSP1/TRAP220, A300-793A) were purchased from Bethyl Laboratories, anti-MED6 (sc-9434) and anti-Gal4 DBD (sc-577) from Santa Cruz. Rabbit antisera were generated against the human MED25 ACID domain by Covalab. Anti-ERM has been previously described (21).
Cell culture and transfection
RK13, U2OS, MDA-MB 231 and MCF-7 cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% FCS (Gibco BRL). DAMI (HEL) cells were cultured in IDMEM (Iscove’s modification of DMEM) supplemented with 10% horse serum. In six-well plates, 1.5 × 105 cells/well were plated, and the next day, transfections were performed using the PEI Exgen 500 procedure (Euromedex, France). For co-immunoprecipitation experiments, 150–250 ng of PEA3 group expression plasmid and/or 200–300 ng of MED25 plasmid were used. For MED25 overexpression assays, cells were cotransfected with 10 ng of ERM expression vector with increasing concentrations (0.5, 1, 2.5 and 5 ng) of MED25 expression vectors [MED25 full-length, ACID domain or Von Willebrand factor A domain (VWA domain)]. Nuclear extracts of DAMI were prepared as previously described (28).
Luciferase assay
The activities of GAL-ERM derivatives were assayed using the dual-luciferase reporter assay system (Promega) as described previously (29).
Co-immunoprecipitation
Transfected cells were lysed in co-immunoprecipitation buffer (50 mM Tris/HCl, pH 7.5, 150 mM NaCl, 0.2 mM EDTA, 1 mM DTT, 1 mM PMSF, 0.5% Triton X100). Proteins were immunoprecipitated overnight with the anti-FLAG M2 affinity gel (Sigma) at 4°C. Detection of immunoblotted target bands was performed with anti-ERM (21) and anti-Flag (Sigma) antibody. For interaction between endogenous proteins, MDA-MB 231 nuclear extracts were subjected to immunoprecipitation with anti-Gal4 DBD (Santa Cruz sc-577) or anti-ERM (Santa Cruz sc-100941) antibodies. Interactions were detected by western blot using polyclonal anti-MED25 or polyclonal anti-ERM (21).
Chromatin immunoprecipitations
Chromatin immunoprecipitation (ChIP) assays with MDA-MB 231 cells (∼2 × 106) were performed as described (26) using the EZ ChIp Kit (Millipore). Sonicated chromatin was incubated at 4°C overnight with 5 µg of rabbit control IgG or specific antibodies. Antibodies used were as follows: MED1 (IHC-00149 Bethyl) and MED18 (IHC-00181 Bethyl). Immunoprecipitated and input material was analysed by quantitative PCR. ChIP signal was normalized to total input. The primers used for amplification of the −1607 Ets element in the human MMP-1 promoter were previously described (30).
Isothermal titration calorimetry
The equilibrium dissociation constant of the MED25 ACID-ERM 38–68 interaction was determined using an iTC200 calorimeter (MicroCal). The binding enthalpies were measured at 25°C in 20 mM sodium phosphate pH 6.5, 150 mM NaCl, 5 mM β-mercaptoethanol, 0.25 mM EDTA. The concentration of the MED25 ACID domain in the cell (10 µM) was roughly 10 times the estimated Kd, and the concentration of the ERM 38–68 peptide (100 µM) in the syringe was ten times that in the cell. Data were processed using Origin 7 software (OriginLab) and fitted the single-binding site mechanism with 1:1 stoichiometry.
siRNA
Cells were transfected with siRNA using INTERFERin (Polyplus) and/or retransfected with expression vector 18 h later with PEI or Fugene HD according to the manufacturer’s instructions. SMART pools (Dharmacon) were used to knockdown MED25, PEA3, ER81 and ERM.
Quantitative real-time PCR analysis
Total cellular RNA was isolated by Illustra TriplePrep extraction kit (GE Healthcare) or by Trizol (Molecular research Center)/RNeasy kit (Qiagen) following the manufacturer’s instructions. Reverse-transcription was performed with High Archive cDNA kit (Applied Biosystems) on extracted RNA after DNase treatment as previously described (28). This cDNA template was PCR amplified and copy number was determined with the SYBRgreen qPCR master mix (Applied) on Stratagene Mx3005P instrument. Primers sequences are available on request.
RESULTS
ERM interacts with the Mediator complex subunit MED25
Recently, we and others determined the solution structure of the Mediator subunit MED25 ACID/PTOV domain and defined the region contacted by the VP16 TAD (27,31,32). Interestingly, the VP16 TAD contains a region with significant homology to the TAD of ERM (Supplementary Figure S1A). Based on this finding, we hypothesized that ERM may interact with MED25. Figure 1A shows the structure of ERM deletion mutants and summarizes their behaviour in pull-down and transfection assays. Binding studies were first performed using resin bound GST and GST-ERM deletion mutants with a panel of MED25 truncation fragments (Figure 1B and 1C). Full-length MED25 was retained with full-length, 1–370, 1–122, 1–72 and 38–72 ERM GST fusion proteins but not with 1–38, 354–510 or the GST control (Figure 1B). Conversely, deletion of the MED25 ACID domain completely abolished the interaction with GST-ERM 38–72, while the ACID domain alone was sufficient to mediate the interaction (Figure 1C). Taken together, these experiments clearly demonstrated that the ERM TAD, like the VP16 TAD, can interact with MED25 and mapped their reciprocal binding domains to regions that include the ACID/PTOV domain of MED25 and the ERM 38–72 region.
To shed more light on the biochemical interaction between ERM and MED25, we next determined by isothermal titration calorimetry the dissociation constant (Kd) for the complex formed between an ERM 38–68 peptide and the MED25 ACID/PTOV domain. In this assay, ERM bound MED25 with an apparent Kd value of 543 nM ± 40 (Supplementary Figure S2), confirming that this is a direct interaction. Interestingly, this binding constant lies between what has been previously reported for VP16 H1 binding to MED25 (1.6 µM) and that reported for VP16 H1 + H2 (50 nM) (31).
We then sought to test whether the interaction between ERM and MED25 occurred in the context of mammalian cells using co-immunoprecipitation experiments. When MED25 was recovered with Flag antibody, ERM was efficiently retained (Figure 1D). Interestingly, an ERM derivative (ERM ΔCt) containing the TAD retained the ability to interact with MED25, whereas a truncated ERM that lacked the TAD (ERM ΔNt) did not (Figure 1D, upper panel). Conversely, deletion of the N-terminal or the C-terminal domains of MED25 (MED25 ΔNt and MED25 ΔCt, respectively) had no effect on the interaction, whereas deletion of the ACID domain (MED25 ΔACID) completely abolished the interaction with ERM (Figure 1D, bottom panel). These results are consistent with the in vitro pull-down assays showing that full-length ERM associates with full-length MED25 and that the ERM TAD is primarily responsible for the association with the ACID domain of MED25. Importantly, this interaction was further verified by showing that endogenous MED25 co-immunoprecipitates with an ERM antibody from MDA-MB 231 nuclear extracts (Figure 1E). Moreover, as the amino-terminal activation domain of all three PEA3 family members are highly conserved, we tested and confirmed that MED25 could also bind to PEA3, ETV1 and its spliced variant ER81 (Supplementary Figure S3).
Based on deletion analysis, the N-terminal TAD of ERM lies within the first 72 amino acids (Figure 1A) (3,4). To test if this domain can be divided into subdomains, we determined if ERM 1–38 and ERM 38–72 are capable of activating transcription when tethered to the Gal4 DNA-binding domain. We found that only ERM 38–72 retained the ability to stimulate transcription (Supplementary Figure S1B), suggesting that the minimal TAD at the N-terminal portion of ERM maps between amino acids 38 and 72. Taken together, these results indicate that the MED25 binding interface colocalizes well with the minimal TAD of ERM.
Mutational studies of the interaction between ERM and MED25
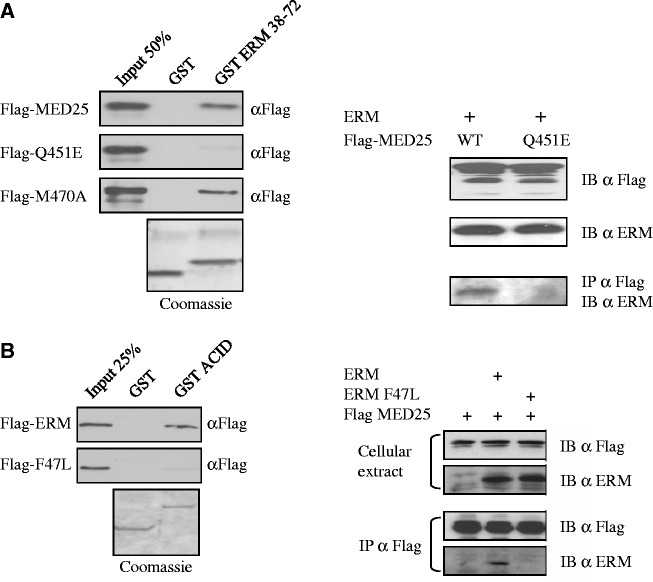
Because the residues of ERM 38–72 that form the MED25 interface display sequence homology with the VP16 H1 subdomain (Supplementary Figure S1A), we anticipated that mutations affecting the MED25/ERM and MED25/VP16 complexes would exhibit similar behaviours and indeed, found this to be the case. First, a MED25 point mutant (Q451E) that has been shown previously to abolish MED25/VP16 interaction (31) also greatly compromised the ability of MED25 to interact with ERM TAD in GST pull-down assay and in co-immunoprecipitation experiments (Figure 2A). Conversely, an M470A mutation that had no effect on ERM binding (Figure 2A), similarly failed to impair VP16 binding (data not shown). In addition, the MED25 ACID domain by itself or full-length MED25 failed to bind a mutated ERM activation domain where the F47 residue was changed to leucine (Figure 2B). It is notable that the mutation of this residue strongly impaired ERM transactivation (3) (Supplementary Figure S1B). Remarkably, the corresponding residue in VP16 H1 (F442, see Supplementary Figure S1A) has previously been shown to be critical for the transactivation function of VP16 (10) and for the recruitment of MED25 (14,15,33). Our results thus demonstrate that MED25 binds only to a functional ERM TAD.
Figure 2.
Effect of mutations in the ERM/MED25 interface. (A) Mutation of Q451 of MED25 severely reduces its ability to bind to ERM. (Left) GST ERM 38–72 was used to analyse the binding to full-length Flag-MED25 WT and point mutants Q451E and M470A. Q451E shows weak binding to ERM 38–72. Binding was detected by immunoblotting with anti-Flag. An SDS gel stained with Coomassie showing the expression of the GST fusion proteins is shown. (Right) Wild-type Flag-MED25 or MED25 with a Q451E mutation were examined for their ability to interact with ERM in co-immunoprecipitation experiments. RK13 cells were transfected with the indicated expression vectors and cellular extracts were IP α Flag and IB α Flag. Expression of Flag-MED25, Flag-MED25 Q451E and ERM are shown in the top two panels. (B) Mutation of F47 of ERM abolishes the recruitment of MED25. (Left) GST MED25 ACID domain was examined for its ability to interact in vitro with ERM wild type or ERM F47L mutant. (Right) The ability of Flag-MED25 to interact with ERM wild type and with ERM F47L mutant was analysed in co-immunoprecipitation experiments. Cells were transfected with the indicated expression vectors and cellular extracts were IP α Flag and IB α Flag or IB α ERM (IB). Expression of Flag-MED25, ERM and ERM F47L are shown in the top panel (cellular extract).
ERM binds Mediator through MED25
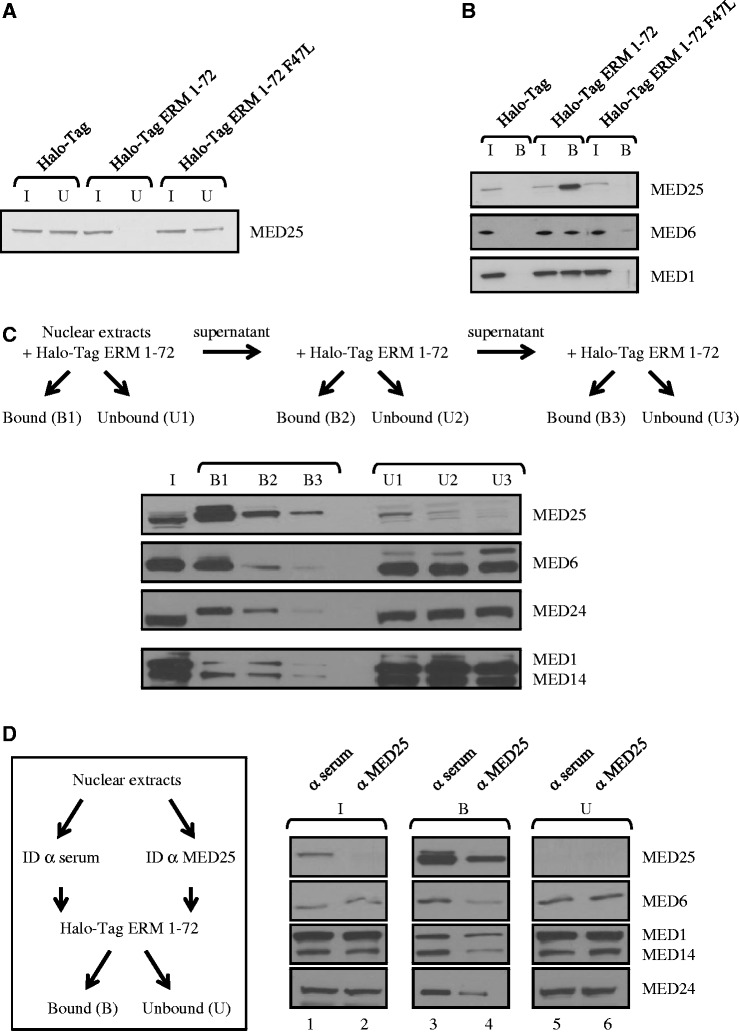
To determine whether the ERM TAD interacts with intact Mediator, we incubated nuclear extracts of DAMI cells with purified Halo-Tag ERM 1–72 coupled to magnetic beads. Unbound Mediator subunits present in the supernatant after incubation (U for unbound) and beads-bound Mediator subunits (B for bound) were then monitored by immunoblotting with antibodies to the subunits indicated in the figure (Figure 3). Firstly, endogenous MED25 could be almost completely depleted from the nuclear extracts after incubation with Halo-Tag ERM 1–72 but not with Halo-Tag alone, confirming that ERM interacts very efficiently with MED25 (Figure 3A). Notably this depletion was not seen when we introduced a mutation in ERM (ERM 1–72 F47L) that prevents MED25 binding (Figure 2B). Accordingly, endogenous MED25 was only detected in the fraction bound to wild-type ERM 1–72 (Figure 3B). Secondly, we observed that MED1 and MED6 bound Halo-Tag ERM 1–72 but not Halo-Tag alone, thus revealing a specific association of ERM with Mediator (Figure 3B). Importantly, as expected, incubation with the mutant ERM 1–72 F47L impaired the interaction between Mediator and ERM (Figure 3B). Finally, when the nuclear extracts were sequentially incubated with ERM 1–72, we observed a strong correlation between depletion of MED25 and decrease in MED1, MED6, MED14 and MED24 binding (Figure 3C, compare B1, B2 and B3).
Figure 3.
MED25 mediates the interaction between ERM and Mediator. (A) Depletion of endogenous MED25 from DAMI cell nuclear extract using Halo-Tag fusion protein ERM 1–72. Nuclear extracts from DAMI cells were incubated with immobilized Halo-Tag, Halo-Tag ERM 1–72 and Halo-Tag ERM 1–72 F47L. After incubation and extensive washing, MED25 not associated with ERM 1–72 present in the supernatant was saved as the unbound fraction (U). Samples were separated by SDS-PAGE, and MED25 was detected by immunoblotting with anti-MED25 antibody. I: Input, 10% of the nuclear extracts used in binding reactions. U: Unbound, 10%. (B) MED25 and Mediator (here documented with MED1 and MED6) bind specifically to ERM 1–72. Nuclear extracts from DAMI cells were incubated with immobilized Halo-Tag, Halo-Tag ERM 1–72 and Halo-Tag ERM 1–72 F47L as in Figure 3A. ERM 1–72-bound Mediator complex (B) was eluted by boiling the beads in SDS buffer and was resolved by 12% SDS-PAGE before western blot analysis using the specified antibodies. I: input, 10% of the nuclear extracts used in binding reactions. B: bound material, 100%. (C) MED25 depletion of nuclear extract impairs recruitment of Mediator to ERM. Nuclear extracts from DAMI cells were sequentially incubated with Halo-Tag ERM 1–72 as indicated. After incubation and extensive washing, the supernatant (containing unbound material, U1, U2 and U3) and the affinity resin (containing bound material, B1, B2 and B3) were boiled in SDS buffer and resolved by SDS-PAGE. Specifically bound and unbound proteins were detected by western blot using the specified antibodies. I: input, 10%. U: unbound, 10%. B: bound, 100%. (D) MED25 immunodepletion reduces interaction between ERM and Mediator. Anti-MED25 antibody and non-specific anti-serum (control) were first coupled to Pureproteome protein A magnetic beads (Millipore). Nuclear extracts from DAMI cells were then incubated at 4°C with antibody-bound protein A beads. The resulting supernatant was incubated with immobilized Halo-Tag ERM 1–72 as indicated. ID: immunodepletion. Bound (B) and unbound (U) materials were resolved by 10% SDS-PAGE before western blot analysis using the specified antibodies. I: input, 10%. U: unbound, 10% and B: bound, 100%.
To exclude the possibility that ERM 1–72 may still be able to bind substoichiometric amounts of Mediator subunits, we also used immunodepletion assays (Figure 3D). Briefly, DAMI nuclear extracts were first depleted using polyclonal antibodies against MED25 followed by incubation with Halo-Tag ERM 1–72, and ERM-bound Mediator was detected as in Figure 3C. As expected, immunodepletion of MED25 specifically removed the majority of MED25 from the supernatant but left behind other Mediator subunits (Figure 3D, compare lane 1 with lane 2). Again, a good correlation between depletion of MED25 and decrease binding of Mediator subunits to ERM was observed (Figure 3D, compare lane 3 with lane 4). Importantly, as evidenced by the lack of change in the relative levels of Mediator subunits expression in the unbound fraction, the inhibition of Mediator recruitment appears to be specifically caused by MED25 depletion (Figure 3D, lanes 5 and 6). Taken together, these results clearly indicate that ERM physically associates with Mediator through the MED25 subunit.
ERM functionally interacts with MED25
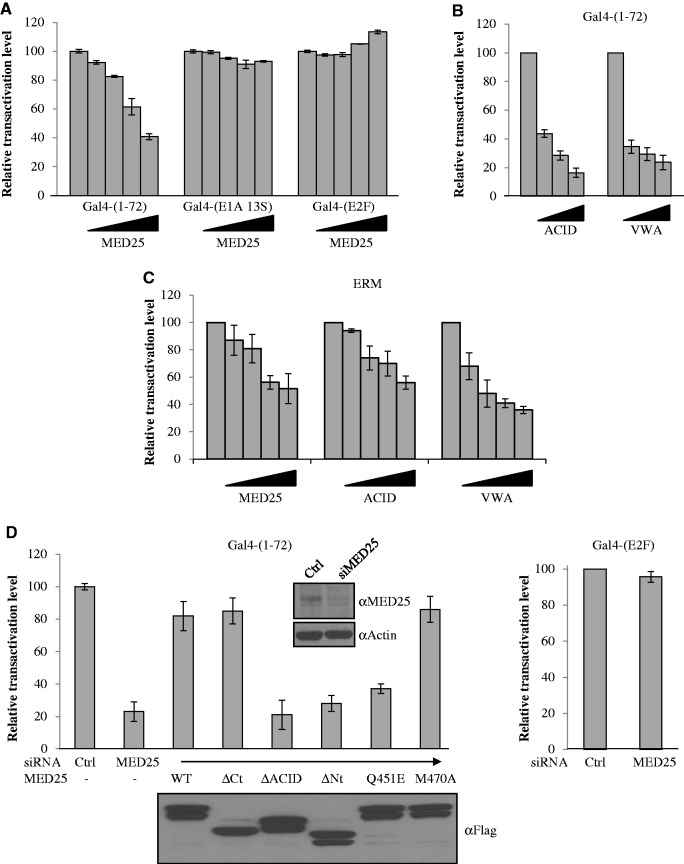
To investigate whether the interaction between ERM and MED25 is functionally important, we first examined the influence of MED25 expression on the transactivation activity of ERM TAD 1–72 tethered to the Gal4 DNA-binding domain. It has been reported that overexpression of Mediator subunits can result in the inhibition of transcription owing to competition between the overexpressed subunits and the Mediator complex for the transcriptional activator (15,17,34,35). In agreement with these results, ectopic expression of MED25 efficiently inhibited the transcriptional activity of ERM TAD (Figure 4A). By contrast, overexpression of MED25 had no influence on the transcriptional activity of the transcription factor E2F or the Adenovirus E1A protein (Figure 4A), the established target of which in Mediator is MED23 (34). Thus, the influence of MED25 in this context appears to be specific to ERM. Similar interference was observed with two MED25 truncated fragments VWA and ACID (Figure 4B), suggesting that the Mediator-binding domain (VWA) and the ERM-binding domain (ACID) of MED25 are functionally important for mediating the transcriptional activity of ERM. Interestingly, this behaviour of ERM is reminiscent of the reported effects of MED25 on VP16 transcriptional activity (14). We also sought to test if full-length ERM activity is affected by MED25 and chose the well-known Ets-responsive TORU-luciferase reporter construct (36). We found that full-length MED25 as well as MED25-VWA and MED25-ACID domains significantly reduced ERM transcriptional activity (Figure 4C). These results were in total agreement with those obtained with the Gal4 fusions used above.
Figure 4.
Effects of MED25 on transcriptional activation by the ERM TAD. (A–D) U2OS cells were transfected with either Gal-ERM 1–72 (A, B and D) or full-length ERM (C) with (A and C) full length MED25, (B and C) MED25 ACID and MED25 VWA domains or (D) siRNA-resistant MED25 wild-type or derivatives. Cells collected 24 h after plasmid transfections were processed for luciferase activity. The relative luciferase activity of Gal-ERM 1–72 alone was assigned a transactivation level of 100%. Data represent the mean ± S.E.M. of at least three independent transfections performed in duplicate. (A) MED25 inhibits Gal4-ERM 1–72 transcriptional activation. Transient expression of MED25 in U2OS cells inhibits Gal4-ERM 1–72 transactivation but not activation by Gal4-E1A 13S or Gal4-E2F. (B) MED25 ACID and VWA domains inhibit Gal-ERM 1–72 transcriptional activation. (C) MED25 inhibits ERM transcriptional activity. (D) MED25 is required for full transcriptional activation by Gal4-ERM 1–72. siRNA-depleted cells were transfected with Gal-ERM 1–72 and reporters and analysed for luciferase activity. The expressed MED25 derivatives (WT, ΔACID, ΔCt, ΔNt, Q451E, M470A) were resistant to degradation induced by siRNA against wild-type MED25. The MED25 siRNA is directed against a sequence encoding the human ACID domain and therefore does not affect either the expression of MED25 ΔACID or the expression of MED25 WT, MED25 ΔCt, MED25 ΔNt, MED25 Q451E and MED25 M470A in which the human ACID domain has been replaced by its murine counterpart. (Inset) The levels of expression of MED25 after transient knockdown (as monitored by western blot with an antibody against MED25) and actin (loading control) are shown. The level of expression of the MED25 deletion mutants (western blot with anti-Flag antibody) after transfection into U2OS cells is also shown.
To further examine the functional role of MED25, we next used siRNA knockdown. Transient transfection of MED25-specific siRNA into U2OS cells resulted in an 80–90% reduction of MED25 mRNA (data not shown) and significant depletion of protein levels (Figure 4D, Inset). Remarkably, this depletion significantly reduced Gal4-ERM 1–72 mediated transactivation (Figure 4D, left panel), whereas activity of the Gal4-E2F construct was not affected (Figure 4D, right panel). To confirm that MED25 mediates transactivation through direct interaction with ERM, we assessed the respective abilities of siRNA-resistant version of MED25 derivatives (WT, ΔACID, ΔCt, ΔNt, Q451E and M470A) to rescue the transactivation potential of ERM TAD in MED25-depleted cells (Figure 4D). When expressed at equivalent levels, MED25 WT, MED25 M470A and MED25 ΔCt, but not ERM-binding defective MED25 (ΔACID and Q451E) and Mediator-binding defective MED25 (ΔNt), elicited efficient rescue of ERM-directed transactivation (Figure 4D). Taken together, these findings preclude a non-specific off-target effect and confirm that a MED25-containing Mediator complex is specifically required for full transactivation by ERM TAD.
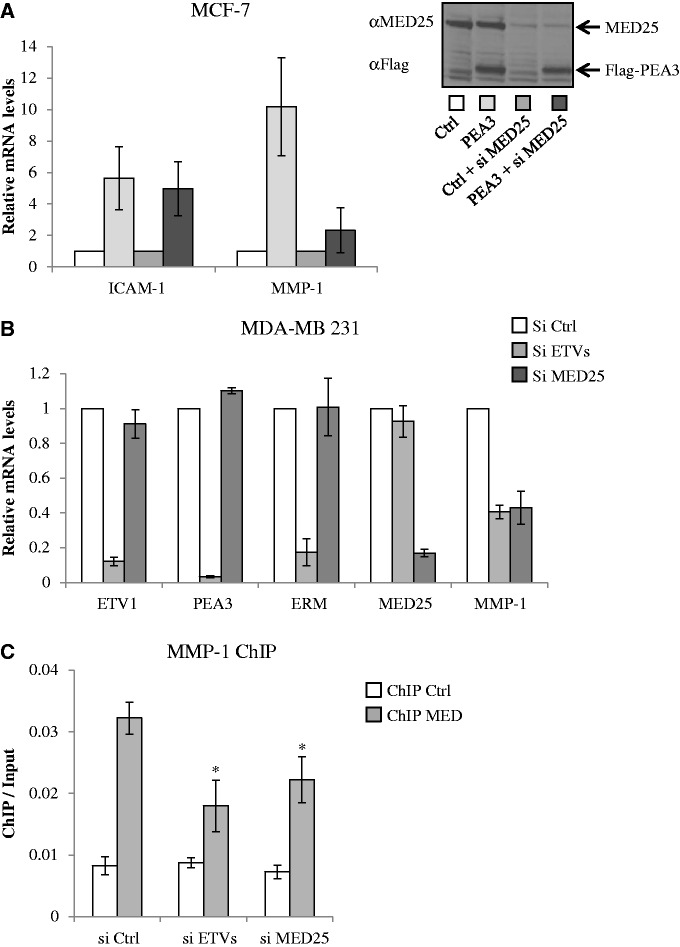
MED25 mediates PEA3-dependent gene expression
To evaluate the role of MED25 as a regulator of PEA3-dependent gene expression, we monitored the effect of MED25 depletion on PEA3-driven expression of two genes identified as targets of PEA3 members (24,37,38), namely MMP-1 and ICAM-1 (Figure 5). As a first functional approach, we used MCF-7 breast cancer cell line, which expresses low levels of PEA3 group members (21) as a model in which to overexpress PEA3. Expression of PEA3 was not affected by our MED25 siRNA, indicating the effect is specific to MED25 depletion (Figure 5A, Inset). Interestingly, silencing MED25 did not affect significantly ICAM-1 promoter activity or its activation by PEA3. However, it greatly diminished the transactivation of the MMP-1 promoter by PEA3. Importantly, similar results were obtained with ERM and ETV1 on MMP-1 and ICAM-1 activation (Supplementary Figure S4A), but in our hands, PEA3 was the most potent activator. This is in accordance with previous results (39).
Figure 5.
MED25 mediates PEA3-dependent target gene expression. (A) Silencing MED25 down-regulates the induction of MMP-1 mRNA by PEA3 in MCF-7 cells. RNA from cells transfected with control (Ctrl), control and MED25-specific siRNA (Ctrl + si MED25), PEA3 (PEA3) and PEA3 and MED25-specific siRNA (PEA3 + si MED25) was subjected to RT-qPCR. Relative expression levels were calculated using the ΔCt method using expression of GAPDH mRNA as a reference gene. Values are plotted relative to control set to 1. Data represent the mean ± S.E.M. of at least four independent transfections performed in duplicate. (Inset) The knockdown of endogenous MED25 and the expression of Flag-PEA3 were confirmed by western blot. (B) Effect of siRNA-mediated ETVs and MED25 depletion on MMP-1 expression in MDA-MB 231 cells. Data points are the average of three independent experiments. Error bars show standard deviation. (C) Effect of ETVs and MED25 depletion on Mediator occupancy at the MMP-1 promoter. ChIP experiments were performed using antibodies against MED1 and MED18 (ChIP MED) or IgG (ChIP Ctrl). ChIP/Input is average from two biological replicates. Error bars show standard deviation. Statistical significance was determined in paired Student’s t-tests (*P < 0.05).
In a converse experiment (Figure 5B), we used siRNA to knockdown endogenously expressed PEA3 members in MDA-MB 231 breast cancer cells (21). Consistent with the results in MCF-7 cells (Figure 5A), selective inhibition of individual PEA3, ERM and ETV1 in MDA-MB 231 cells resulted in significant but moderate reduction of MMP-1 expression (Supplementary Figure S4B). Interestingly, the most effective down-regulation of MMP-1 expression was achieved when all three PEA3 members were inhibited (Figure 5B and Supplementary Figure S4B). Thus our data suggest that there is functional redundancy among the three PEA3 factors with regard to MMP-1 regulation.
To finally explore the mechanistic relationship between the PEA3 group members and MED25, we used ChIP using antibodies against Mediator subunits MED1 and MED18 to monitor Mediator recruitment to the MMP-1 promoter in vivo (Figure 5C). In agreement with the above results, we observed Mediator complex at the MMP-1 promoter when MDA-MB 231 cells were transfected with control siRNA. Notably, depletion of MED25 levels or depletion of ERM, PEA3 and ETV1 levels reduced the Mediator ChIP signal, suggesting that the PEA3 members recruit Mediator to the MMP-1 promoter in a MED25-dependent manner (Figure 5C). Collectively, these data suggest that the PEA3 group members may play an important functional role in recruiting the Mediator complex to the MMP-1 gene.
DISCUSSION
During the past two decades, extensive efforts have been invested in identifying the targets of transcriptional activators within the transcriptional machinery. There is now considerable evidence that the Mediator is a bona fide target of many transcription factors (12,40). Interactions usually occur between TADs and one or more Mediator subunits. For example, the N-terminal TAD of p53 binds directly to MED17 (41,42), while the SREBP TAD recruits the MED15 subunit (43). In this manuscript, we have extended this list to the TAD of the PEA3 group members by demonstrating its ability to recruit the Mediator complex through the MED25 subunit. Analysis of point mutations in the ERM TAD resulted in significant loss of transactivating activity, which correlated with loss of Mediator interaction, suggesting that the transactivating activity of ERM and physical contact with the Mediator are linked.
Several transactivators have been shown to interact with MED25, such as the viral proteins VP16 (14,15), Lana-1 (16), IE62 (17) and the nuclear receptors RARα (18) and HNF4α (19). In light of these previous findings, it appears that the MED25 ACID/PTOV domain possesses a unique ability to preferentially target acidic TADs. This connection is further supported by our finding that the TAD of the PEA3 members directly interacts with this domain. However, to our knowledge, PEA3 represents the first example of a cellular gene-specific human transcription factor that interacts physically and functionally with the MED25 ACID/PTOV domain.
The findings presented here, as well as those of previous studies (14,15,17,31,32,44), allow us to make comparisons between three unrelated acidic TADs bound to a common target. In both the VP16/MED25 (31,32) and the IE62/MED25 complexes (44), a hydrophobic residue makes crucial contacts with the MED25 ACID/PTOV domain. Interestingly, a similar situation is observed with the TADs of p53, EKLF and Gcn4 in which hydrophobic residues nucleate binding to partner proteins (45–48). These findings illustrate how transactivators have evolved to target specific binding sites on cofactors using a small set of critical residues (49). Remarkably, the ERM mutant (F47L) is not only severely impaired in its ability to interact with MED25 but also showed compromised transactivation ability, confirming previous conclusions (3). Thus, the simplest explanation is that the interaction with MED25 is central to the ability of ERM to activate gene expression. Consistent with this result, we observed that the ERM-binding domain on MED25 (the ACID/PTOV domain) inhibits ERM transactivation via competition. We also found that the overexpression of the MED25 VWA domain significantly reduced the transcriptional activity of ERM. This observation is in accordance with a previous report showing the involvement of this domain for interaction of MED25 with Mediator (14).
Our study also points to another similarity between VP16, IE62 and PEA3 factors in binding to MED25 as they share the same binding site on the ACID/PTOV domain. The finding that mutation of Gln451 to glutamic acid (Q451E) abolished not only the binding of MED25 to ERM but also to VP16 (31) substantiates this conclusion. This is further supported by the observation that mutations across amino acids 447–450 of MED25 decreased IE62 binding to MED25 (44). Despite these similarities, there are also some significant differences between the results obtained with the three TADs. Unlike the VP16 TAD, which contains two subdomains H1 and H2 that can transactivate independently (33,50), the minimal IE62 (44) and ERM TADs appear to encompass a single domain. Moreover, although VP16 has been found to directly interact with not only MED25 but also MED17 (41) and MED15 (51), we have no evidence that ERM can directly target other Mediator subunits (data not shown). That is, we observed no decrease in relative MED1, MED6, MED14 and MED24 expression by western blot after incubation with ERM 1–72. This conclusion is further supported by the fact that the ERM mutation, which ablates interaction with MED25, also results in loss of capture of other Mediator subunits. We also noted that the stoichiometry of MED25, MED1, MED6, MED14 and MED24 recruitment by ERM is ∼10:1:1:1:1, respectively. This is in accordance with previous reports showing that MED25 is loosely (variably) associated with the Mediator (52–54). We thus propose that ERM recruits both Mediator-containing MED25 and MED25 in its free form. The existence of a free MED25 function (Mediator independent) remains to be assessed and will be the subject of future investigations.
Sequence comparisons indicate that the N-terminal TAD of the PEA3 group members is highly conserved (2,37). It is thus not surprising that ERM, PEA3, ETV1 and its splice variant ER81 are all capable of binding to MED25. Accordingly, we observed that RNAi-mediated MED25 depletion inhibited PEA3-driven expression of MMP-1 gene transcription but surprisingly not PEA3-induced ICAM-1 expression. The involvement of Mediator in the context of full-length PEA3-driven expression of MMP-1 was confirmed by ChIP experiments. A positive requirement for MED25 is further substantiated by our findings that MED25 depletion diminished Mediator recruitment at the MMP-1 promoter. We noticed that the MED25-PEA3 members interaction may not be solely responsible for Mediator recruitment because inhibition of their expression resulted in only a 50% reduction in MMP-1 expression and partial decrease in Mediator binding at the MMP-1 promoter. While these results do not rule out a direct role for MED25 in PEA3 target gene activation, they suggest the existence of a MED25-independent PEA3 regulation. This selectivity of MED25 action is reminiscent of the reported Mediator-dependent and -independent regulation by NF-κB p65 (MED17) and nuclear receptors (MED1) in mammals (55,56). Alternatively, the Mediator may also be recruited to PEA3 target genes through direct interactions with other DNA-binding transcription factors and/or coactivators that act synergistically with the PEA3 group members on specific genes. For example, the steroid receptor coactivator AIB1/SRC-3, which indirectly recruited the Mediator (57), has been shown to function cooperatively with PEA3 (38). Similarly, the PEA3 factors crosstalk with nuclear receptors such as the androgen receptor (58) and the oestrogen receptor beta (59), which are known to recruit the Mediator via MED1 (60). The functional importance of these potential alternative pathways for Mediator recruitment by the PEA3 factors remains to be established.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–4.
FUNDING
Nord-Pas de Calais Regional Council and FEDER through the ‘Contrat de Projets Etat Région (CPER) 2007–2013’ and the ‘Projets Emergents’, the BQR from Lille 1 University and the Ligue Nationale contre le cancer (comité Pas de Calais). K.V. was supported by fellowships from the Institut Pasteur de Lille/Région Nord-Pas de Calais and from the Association pour la Recherche sur le Cancer (ARC, France). Funding for open access charge: CNRS.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank M. Meisterernst for reagents and M. Crossley for his careful and critical reading.
REFERENCES
- 1.Hollenhorst PC, McIntosh LP, Graves BJ. Genomic and biochemical insights into the specificity of ETS transcription factors. Annu. Rev. Biochem. 2011;80:437–471. doi: 10.1146/annurev.biochem.79.081507.103945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de Launoit Y, Baert JL, Chotteau-Lelievre A, Monte D, Coutte L, Mauen S, Firlej V, Degerny C, Verreman K. The Ets transcription factors of the PEA3 group: transcriptional regulators in metastasis. Biochim. Biophys. Acta. 2006;1766:79–87. doi: 10.1016/j.bbcan.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 3.Defossez PA, Baert JL, Monnot M, de Launoit Y. The ETS family member ERM contains an alpha-helical acidic activation domain that contacts TAFII60. Nucleic Acids Res. 1997;25:4455–4463. doi: 10.1093/nar/25.22.4455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Laget MP, Defossez PA, Albagli O, Baert JL, Dewitte F, Stéhelin D, de Launoit Y. Two functionally distinct domains responsible for transactivation by the Ets family member ERM. Oncogene. 1996;12:1325–1336. [PubMed] [Google Scholar]
- 5.Degerny C, de Launoit Y, Baert JL. ERM transcription factor contains an inhibitory domain which functions in sumoylation–dependent manner. Biochim. Biophys. Acta. 2008;1779:183–194. doi: 10.1016/j.bbagrm.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 6.Degerny C, Monte D, Beaudoin C, Jaffray E, Portois L, Hay RT, de Launoit Y, Baert JL. SUMO modification of the Ets-related transcription factor ERM inhibits its transcriptional activity. J. Biol. Chem. 2005;280:24330–24338. doi: 10.1074/jbc.M411250200. [DOI] [PubMed] [Google Scholar]
- 7.Bojovic BB, Hassell JA. The PEA3 Ets transcription factor comprises multiple domains that regulate transactivation and DNA binding. J. Biol. Chem. 2001;276:4509–4521. doi: 10.1074/jbc.M005509200. [DOI] [PubMed] [Google Scholar]
- 8.Janknecht R. Analysis of the ERK-stimulated ETS transcription factor ER81. Mol. Cell Biol. 1996;16:1550–1156. doi: 10.1128/mcb.16.4.1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lens Z, Dewitte F, Monté D, Baert JL, Bompard C, Sénéchal M, Van Lint C, de Launoit Y, Villeret V, Verger A. Solution structure of the N-terminal transactivation domain of ERM modified by SUMO-1. Biochem. Biophys. Res. Commun. 2010;399:104–110. doi: 10.1016/j.bbrc.2010.07.049. [DOI] [PubMed] [Google Scholar]
- 10.Cress WD, Triezenberg SJ. Critical structural elements of the VP16 transcriptional activation domain. Science. 1991;251:87–90. doi: 10.1126/science.1846049. [DOI] [PubMed] [Google Scholar]
- 11.Piskacek S, Gregor M, Nemethova M, Grabner M, Kovarik P, Piskacek M. Nine-amino-acid transactivation domain: establishment and prediction utilities. Genomics. 2007;89:756–768. doi: 10.1016/j.ygeno.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 12.Malik S, Roeder RG. The metazoan Mediator co-activator complex as an integrative hub for transcriptional regulation. Nat. Rev. Genet. 2010;11:761–772. doi: 10.1038/nrg2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bourbon HM. Comparative genomics supports a deep evolutionary origin for the large, four-module transcriptional mediator complex. Nucleic Acids Res. 2008;36:3993–4008. doi: 10.1093/nar/gkn349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mittler G, Stuhler T, Santolin L, Uhlmann T, Kremmer E, Lottspeich F, Berti L, Meisterernst M. A novel docking site on Mediator is critical for activation by VP16 in mammalian cells. EMBO J. 2003;22:6494–6504. doi: 10.1093/emboj/cdg619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang F, DeBeaumont R, Zhou S, Naar AM. The activator-recruited cofactor/Mediator coactivator subunit ARC92 is a functionally important target of the VP16 transcriptional activator. Proc. Natl Acad. Sci. USA. 2004;101:2339–2344. doi: 10.1073/pnas.0308676100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roupelieva M, Griffiths SJ, Kremmer E, Meisterernst M, Viejo-Borbolla A, Schulz T, Haas J. Kaposi's sarcoma-associated herpesvirus Lana-1 is a major activator of the serum response element and mitogen-activated protein kinase pathways via interactions with the Mediator complex. J. Gen. Virol. 2010;91:1138–1149. doi: 10.1099/vir.0.017715-0. [DOI] [PubMed] [Google Scholar]
- 17.Yang M, Hay J, Ruyechan WT. Varicella-zoster virus IE62 protein utilizes the human mediator complex in promoter activation. J. Virol. 2008;82:12154–12163. doi: 10.1128/JVI.01693-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee HK, Park UH, Kim EJ, Um SJ. MED25 is distinct from TRAP220/MED1 in cooperating with CBP for retinoid receptor activation. EMBO J. 2007;26:3545–3557. doi: 10.1038/sj.emboj.7601797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rana R, Surapureddi S, Kam W, Ferguson S, Goldstein JA. Med25 is required for RNA polymerase ii recruitment to specific promoters, thus regulating xenobiotic and lipid metabolism in human liver. Mol. Cell Biol. 2011;31:466–481. doi: 10.1128/MCB.00847-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakamura Y, Yamamoto K, He X, Otsuki B, Kim Y, Murao H, Soeda T, Tsumaki N, Deng JM, Zhang Z, et al. Wwp2 is essential for palatogenesis mediated by the interaction between Sox9 and mediator subunit 25. Nat. Commun. 2011;2:251. doi: 10.1038/ncomms1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baert JL, Monte D, Musgrove EA, Albagli O, Sutherland RL, de Launoit Y. Expression of the PEA3 group of ETS-related transcription factors in human breast-cancer cells. Int. J. Cancer. 1997;70:590–597. doi: 10.1002/(sici)1097-0215(19970304)70:5<590::aid-ijc17>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 22.Coutte L, Monte D, Baert JL, de Launoit Y. Genomic organization of the human e1af gene,a member of Ets transcription factors. Gene. 1999;240:201–207. doi: 10.1016/s0378-1119(99)00400-x. [DOI] [PubMed] [Google Scholar]
- 23.de Launoit Y, Audette M, Pelczar H, Plaza S, Baert JL. The transcription of the intercellular adhesion molecule-1 is regulated by Ets transcription factors. Oncogene. 1998;16:2065–2073. doi: 10.1038/sj.onc.1201726. [DOI] [PubMed] [Google Scholar]
- 24.Baert JL, Beaudoin C, Coutte L, de Launoit Y. ERM transactivation is up-regulated by the repression of DNA binding after the PKA phosphorylation of a consensus site at the edge of the ETS domain. J. Biol. Chem. 2002;277:1002–1012. doi: 10.1074/jbc.M107139200. [DOI] [PubMed] [Google Scholar]
- 25.Baert JL, Beaudoin C, Monte D, Degerny C, Mauen S, de Launoit Y. The 26S proteasome system degrades the ERM transcription factor and regulates its transcription-enhancing activity. Oncogene. 2007;26:415–424. doi: 10.1038/sj.onc.1209801. [DOI] [PubMed] [Google Scholar]
- 26.Blais A, Monté D, Pouliot F, Labrie C. Regulation of the human cyclin-dependent kinase inhibitor p18INK4c by the transcription factors E2F1 and Sp1. J. Biol. Chem. 2002;277:31679–31693. doi: 10.1074/jbc.M204554200. [DOI] [PubMed] [Google Scholar]
- 27.Bontems F, Verger A, Dewitte F, Lens Z, Baert JL, Ferreira E, Launoit YD, Sizun C, Guittet E, Villeret V, et al. NMR structure of the human Mediator MED25 ACID domain. J. Struct. Biol. 2011;174:245–251. doi: 10.1016/j.jsb.2010.10.011. [DOI] [PubMed] [Google Scholar]
- 28.Verreman K, Baert JL, Verger A, Drobecq H, Ferreira E, de Launoit Y, Monté D. The Coactivator Activator CoAA regulates PEA3 group member transcriptional activity. Biochem. J. 2011;439:469–477. doi: 10.1042/BJ20110728. [DOI] [PubMed] [Google Scholar]
- 29.Quinlan KG, Nardini M, Verger A, Francescato P, Yaswen P, Corda D, Bolognesi M, Crossley M. Specific recognition of ZNF217 and other zinc finger proteins at a surface groove of C-terminal binding proteins. Mol. Cell Biol. 2006;26:8159–8172. doi: 10.1128/MCB.00680-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Affara M, Dunmore BJ, Sanders DA, Johnson N, Print CG, Charnock-Jones DS. MMP1 bimodal expression and differential response to inflammatory mediators is linked to promoter polymorphisms. BMC Genomics. 2011;12:43. doi: 10.1186/1471-2164-12-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Milbradt AG, Kulkarni M, Yi T, Takeuchi K, Sun ZJ, Luna RE, Selenko P, Naar A, Wagner G. Structure of the VP16 transactivator target in ARC/Mediator. Nat. Struct. Biol. 2011;18:410–415. doi: 10.1038/nsmb.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vojnic E, Mourao A, Seizl M, Simon B, Wenzeck L, Lariviere L, Baumli S, Baumgart K, Meisterernest M, Sattler M, et al. The mediator MED25 activator interaction domain: structure and cooperative binding of VP16 subdomains. Nat. Struct. Biol. 2011;18:404–409. doi: 10.1038/nsmb.1997. [DOI] [PubMed] [Google Scholar]
- 33.Ikeda K, Stuehler T, Meisterernst M. The H1 and H2 regions of the activation domain of herpes simplex virion protein 16 stimulate transcription through distinct molecular mechanisms. Genes Cells. 2002;7:49–58. doi: 10.1046/j.1356-9597.2001.00492.x. [DOI] [PubMed] [Google Scholar]
- 34.Boyer TG, Martin ME, Lees E, Ricciardi RP, Berk AJ. Mammalian Srb/Mediator complex is targeted by adenovirus E1A protein. Nature. 1999;399:276–279. doi: 10.1038/20466. [DOI] [PubMed] [Google Scholar]
- 35.Yuan CX, Ito M, Fondell JD, Fu ZY, Roeder RG. The TRAP220 component of a thyroid hormone receptor- associated protein (TRAP) coactivator complex interacts directly with nuclear receptors in a ligand-dependent fashion. Proc. Natl Acad. Sci. USA. 1998;95:7939–7944. doi: 10.1073/pnas.95.14.7939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Monté D, Coutte L, Baert JL, Angeli I, Stehelin D, de Launoit Y. Molecular characterization of the ets-related human transcription factor ER81. Oncogene. 1995;11:771–779. [PubMed] [Google Scholar]
- 37.Oh S, Shin S, Janknecht R. ETV1, 4 and 5: an oncogenic subfamily of ETS transcription factors. Biochim. Biophys. Acta. 2012;1826:1–12. doi: 10.1016/j.bbcan.2012.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Qin L, Liao L, Redmond A, Young L, Yuan Y, Chen H, O'Malley BW, Xu J. The AIB1 oncogene promotes breast cancer metastasis by activation of PEA3-mediated matrix metalloproteinase 2 (MMP2) and MMP9 expression. Mol. Cell Biol. 2008;28:5937–5950. doi: 10.1128/MCB.00579-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Howe LR, Crawford HC, Subbaramaiah K, Hassell JA, Dannenberg AJ, Brown AM. PEA3 is up-regulated in response to Wnt1 and activates the expression of cyclooxygenase-2. J. Biol. Chem. 2001;276:20108–20115. doi: 10.1074/jbc.M010692200. [DOI] [PubMed] [Google Scholar]
- 40.Borggrefe T, Yue X. Interactions between subunits of the Mediator complex with gene-specific transcription factors. Semin. Cell Dev. Biol. 2011;22:759–768. doi: 10.1016/j.semcdb.2011.07.022. [DOI] [PubMed] [Google Scholar]
- 41.Ito M, Yuan CX, Malik S, Gu W, Fondell JD, Yamamura S, Fu ZY, Zhang X, Qin J, Roeder RG. Identity between TRAP and SMCC complexes indicates novel pathways for the function of nuclear receptors and diverse mammalian activators. Mol. Cell. 1999;3:361–370. doi: 10.1016/s1097-2765(00)80463-3. [DOI] [PubMed] [Google Scholar]
- 42.Meyer KD, Lin SC, Bernecky C, Gao Y, Taatjes DJ. p53 activates transcription by directing structural shifts in Mediator. Nat. Struct. Mol. Biol. 2010;17:753–760. doi: 10.1038/nsmb.1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang F, Vought BW, Satterlee JS, Walker AK, Jim Sun ZY, Watts JL, DeBeaumont R, Saito RM, Hyberts SG, Yang S, et al. An ARC/Mediator subunit required for SREBP control of cholesterol and lipid homeostasis. Nature. 2006;442:700–704. doi: 10.1038/nature04942. [DOI] [PubMed] [Google Scholar]
- 44.Yamamoto S, Eletsky A, Szyperski T, Hay J, Ruyechan WT. Analysis of the varicella-zoster virus IE62 N-terminal acidic transactivating domain and its interaction with the human mediator complex. J. Virol. 2009;83:6300–6305. doi: 10.1128/JVI.00054-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brzovic PS, Heikaus CC, Kisselev L, Vernon R, Herbig E, Pacheco D, Warfield L, Littlefield P, Baker D, Klevit RE, et al. The acidic transcription activator gcn4 binds the mediator subunit gal11/med15 using a simple protein interface forming a fuzzy complex. Mol. Cell. 2011;44:942–953. doi: 10.1016/j.molcel.2011.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mas C, Lussier-Price M, Soni S, Morse T, Arseneault G, Di Lello P, Lafrance-Vanasse J, Bieker JJ, Omichinski JG. Structural and functional characterization of an atypical activation domain in erythroid Kruppel-like factor (EKLF) Proc. Natl Acad. Sci. USA. 2011;108:10484–10489. doi: 10.1073/pnas.1017029108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Di Lello P, Jenkins LM, Jones TN, Nguyen BD, Hara T, Yamaguchi H, Dikeakos JD, Appella E, Legault P, Omichinski JG. Structure of the Tfb1/p53 complex: insights into the interaction between the p62/Tfb1 subunit of TFIIH and the activation domain of p53. Mol. Cell. 2006;22:731–740. doi: 10.1016/j.molcel.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 48.Kussie PH, Gorina S, Marechal V, Elenbaas B, Moreau J, Levine AJ, Pavletich NP. Structure of the MDM2 oncoprotein bound to the p53 tumor suppressor transactivation domain. Science. 1996;274:948–953. doi: 10.1126/science.274.5289.948. [DOI] [PubMed] [Google Scholar]
- 49.Humphris EL, Kortemme T. Design of multi-specificity in protein interfaces. PLoS Comput. Biol. 2007;3:e164. doi: 10.1371/journal.pcbi.0030164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Walker S, Greaves R, O'Hare P. Transcriptional activation by the acidic domain of Vmw65 requires the integrity of the domain and involves additional determinants distinct from those necessary for TFIIB binding. Mol. Cell Biol. 1993;13:5233–5244. doi: 10.1128/mcb.13.9.5233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Park JM, Kim HS, Han SJ, Hwang MS, Lee YC, Kim YJ. In vivo requirement of activator-specific binding targets of mediator. Mol. Cell Biol. 2000;20:8709–8719. doi: 10.1128/mcb.20.23.8709-8719.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Paoletti AC, Parmely TJ, Tomomori-Sato C, Sato S, Zhu D, Conaway RC, Conaway JW, Florens L, Washburn MP. Quantitative proteomic analysis of distinct mammalian Mediator complexes using normalized spectral abundance factors. Proc. Natl Acad. Sci. USA. 2006;103:18928–18933. doi: 10.1073/pnas.0606379103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sato S, Tomomori-Sato C, Parmely TJ, Florens L, Zybailov B, Swanson SK, Banks CA, Jin J, Cai Y, Washburn MP, et al. A set of consensus mammalian mediator subunits identified by multidimensional protein identification technology. Mol. Cell. 2004;14:685–691. doi: 10.1016/j.molcel.2004.05.006. [DOI] [PubMed] [Google Scholar]
- 54.Uhlmann T, Boeing S, Lehmbacher M, Meisterernst M. The VP16 activation domain establishes an active mediator lacking CDK8 in vivo. J. Biol. Chem. 2007;282:2163–2173. doi: 10.1074/jbc.M608451200. [DOI] [PubMed] [Google Scholar]
- 55.Ge K, Cho YW, Guo H, Hong TB, Guermah M, Ito M, Yu H, Kalkum M, Roeder RG. Alternative mechanisms by which mediator subunit MED1/TRAP220 regulates peroxisome proliferator-activated receptor gamma-stimulated adipogenesis and target gene expression. Mol. Cell Biol. 2008;28:1081–1091. doi: 10.1128/MCB.00967-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.van Essen D, Engist B, Natoli G, Saccani S. Two modes of transcriptional activation at native promoters by NF-kappaB p65. PLoS Biol. 2009;7:e73. doi: 10.1371/journal.pbio.1000073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huang ZQ, Li J, Sachs LM, Cole PA, Wong J. A role for cofactor-cofactor and cofactor-histone interactions in targeting p300, SWI/SNF and Mediator for transcription. EMBO J. 2003;22:2146–2155. doi: 10.1093/emboj/cdg219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schneikert J, Peterziel H, Defossez PA, Klocker H, Launoit Y, Cato AC. Androgen receptor-Ets protein interaction is a novel mechanism for steroid hormone-mediated down-modulation of matrix metalloproteinase expression. J. Biol. Chem. 1996;271:23907–23913. doi: 10.1074/jbc.271.39.23907. [DOI] [PubMed] [Google Scholar]
- 59.Chen Y, Chen L, Li JY, Mukaida N, Wang Q, Yang C, Yin WJ, Zeng XH, Jin W, Shao ZM. ERbeta and PEA3 co-activate IL-8 expression and promote the invasion of breast cancer cells. Cancer Biol. Ther. 2011;11:497–511. doi: 10.4161/cbt.11.5.14667. [DOI] [PubMed] [Google Scholar]
- 60.Chen W, Roeder RG. Mediator-dependent nuclear receptor function. Semin. Cell Dev. Biol. 2011;22:749–758. doi: 10.1016/j.semcdb.2011.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.