Abstract
Transcription factors that belong to the same family typically have similar, but not identical, binding specificities. As such, they can be expected to compete differentially for binding to different variants of their binding sites. Pho4 is a yeast factor whose nuclear concentration is up-regulated in low phosphate, while the related factor, Cbf1, is constitutively expressed. We constructed 16 GFP-reporter genes containing all palindromic variants of the motif NNCACGTGNN, and determined their activities at a range of phosphate concentrations. Pho4 affinity did not explain expression data well except under fully induced conditions. However, reporter activity was quantitatively well explained under all conditions by a model in which Cbf1 itself has modest activating activity, and Pho4 and Cbf1 compete with one another. Chromatin immunoprecipitation and computational analyses of natural Pho4 target genes, along with the activities of the reporter constructs, indicates that genes differ in their sensitivity to intermediate induction signals in part because of differences in their affinity for Cbf1. The induction sensitivity of both natural Pho4 target genes and reporter genes was well explained only by a model that assumes a role for Cbf1 in remodeling chromatin. Our analyses highlight the importance of taking into account the activities of related transcription factors in explaining system-wide gene expression data.
INTRODUCTION
Most transcription factors belong to a relatively small number of families that are distinguished by the structures and sequences of their DNA-binding domains. In most of these families, members bind to similar DNA sequences (1,2). Because their DNA-binding specificities are similar, these proteins can be expected to compete with one another for binding to their cognate binding sites. Competition is avoided in many cases by the expression of family members in different cell types or under different conditions, or by member-specific interaction with other DNA-binding proteins that result in complexes of different binding specificity. Nevertheless, competition can occur, and it can be an important part of regulatory switches, a classic example being the lysis/lysogeny decision of phage lambda, mediated by the Repressor/Cro pair of helix–turn–helix proteins (3). The control of meiosis in yeast by Ndt80 and Sum1 is another well studied example, albeit involving factors from different families (4). Computational modeling of the Ndt80/Sum1 system has highlighted how competition enhances the specificity of target gene selection beyond the specificity intrinsic to either one of the factors by itself (5,6). One of the most widespread families of transcription factors is the basic helix–loop–helix (bHLH) family, a family typically found with multiple variants in animals, plants and fungi (7). The binding specificities of yeast bHLH proteins have been determined in vitro (1,2,8) and through motif enrichment analysis at in vivo binding sites defined by chromatin immunoprecipitation (ChIP) (9). These studies all show that bHLH proteins bind to short palindromic, or nearly palindromic, motifs, typically of the form CANNTG. Members of the family differ from one another, often quite subtly, through additional specificity determinants within the core hexamer and at flanking positions (8).
One member of the yeast bHLH family, Pho4, has a well-defined role in phosphate metabolism. In low phosphate, Pho4 becomes nuclear localized, binds to its cognate binding sites and activates transcription of a set of genes relevant to phosphate physiology (10–12). In contrast, another yeast bHLH protein, Cbf1, lacks a single well-defined function. It is a much more abundant protein than Pho4 (13), and there is no evidence for regulation of its expression or activity. The name CBF1 stands for ‘Centromere binding factor 1’ and mutants defective in Cbf1 grow poorly, apparently because of defects in chromosome segregation (14). However, Cbf1 also appears to have a direct role in regulating sulfur metabolism, and cbf1 mutants are methionine auxotrophs (14,15). Despite their differences in function, the methionine auxotrophy of a cbf1 mutant can be complemented by overexpression of Pho4, implying that Pho4 can bind and activate genes at sites normally bound by Cbf1 (16). Furthermore, cells that grow poorly on low phosphate due to a defect in phosphate transport can be suppressed by a cbf1 null mutation (16). This suggests that Cbf1 might ordinarily compete for Pho4 binding, so that in the absence of Cbf1, Pho4 better activates its target genes, mitigating the effects of low phosphate.
Pho4 and Cbf1 bind to many promoters in common (9) (Supplementary Figure S1), and recent ChIP-seq experiments demonstrate that the two proteins bind to some of the very same binding sites (17). Many CACGTG motifs in the genome are bound more by Pho4 in a cbf1Δ strain than in a wild-type strain, providing direct evidence for competition. Even more striking, in the cbf1Δ strain, some genes that are not normally regulated by Pho4 become inducible by low phosphate, and the newly regulatable promoters have Pho4 bound at sites normally occupied by Cbf1 (17).
Pho4 and Cbf1 differ subtly in their binding specificities. These differences have been studied most thoroughly in vitro using an assay that rapidly traps protein–DNA complexes in a microfluidic device (8). This technique was used to measure the affinities of Pho4 and Cbf1 to each of the 16 possible dinucleotides flanking one side of the CACGTG core. Assuming that each half of the binding site contributes independently to affinity, these values allow the calculation of relative affinities for all 256 variants of the NNCACGTGNN motif (Figure 1). The richness and presumptive accuracy of these data offer a unique opportunity to ask how well in vitro binding affinity data can explain expression in vivo. We show that they do, in fact, do a remarkably good job explaining gene expression data for 16 reporter constructs over a range of phosphate concentrations. However, to explain the data at phosphate concentrations higher than ∼80 μM, we find that a modest, but significant, contribution to transcriptional activation is required from Cbf1; also required to explain the data is competition for binding between Pho4 and Cbf1. Finally, we show that differences in the affinity for Cbf1 can explain much of the differences among natural Pho4 target genes in terms of their sensitivity to induction at moderate phosphate concentrations.
Figure 1.
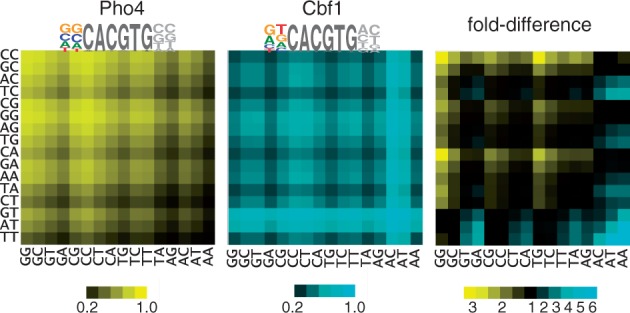
Heatmaps showing relative occupancy by Pho4 and Cbf1 at all 256 variants of the binding site NNCACGTGNN; rows indicate the 5′ dinucleotide, columns the 3′ dinucleotide. Occupancies for Pho4 and Cbf1 (left and center, respectively) are based on the relative free energies of binding for dinucleotides flanking the CACGTG core (8), with the assumed protein concentration adjusted so the mean occupancy among all sites is 0.5. The fold-difference between Pho4 and Cbf1 occupancies is shown in the panel on the right, with motifs having higher occupancy for Pho4 shown in blue and those with higher occupancy for Cbf1 shown in yellow. The sequence logos for Pho4 and Cbf1 are based on a consensus CACGTG core plus flanking base preferences that are derived from a linear regression of the experimentally measured free energy differences for the 16 dinucleotide free energy values (8). The result is a best-fit estimation for the relative contributions to binding of each of the four bases at each of the two positions.
MATERIALS AND METHODS
Strains
Diploid strains containing a single vtc4::Green Fluorescent Protein (GFP) allele, and differing in the promoter sequence driving GFP expression, were constructed as follows. BY4741 (Mata his3Δ leu2Δ met15Δ ura3Δ) was first modified to contain a Myc tag fused to the 3′ end of the PHO4 coding sequence and a triple HA tag (3HA) at the 3′ end of the CBF1 coding sequence. The epitope tags were introduced by homologous integration of PCR products, using URA3 as a selectable marker for the Myc tag and LEU2 for the 3HA tag. This strain was used as the parent for construction of vtc4::GFP reporter variants, using HIS4-tagged PCR products integrated by homologous recombination. For each of the vtc4::GFP variants, bridging PCR was used to produce a single PCR product from two input PCR products, one that was common to all the constructs and one that was unique to each variant. The one in common contains yeGFP and HIS4, flanked at the 5′ end by VTC4 5′ UTR sequences adjacent to the initiation codon and at the 3′ end by VTC4 3′ UTR sequences. The unique input consisted of the desired Pho4/Cbf1-binding site motifs, with sequences at the 3′ end overlapping the yeGFP-HIS4 product. The two inputs were then amplified with primers that extended the flanking regions of identity to the genome. The vtc4::GFP haploid strains were mated to BY4742 PHO4-myc producing the diploids that were used in the FACS expression analysis. For ChIP analyses, two epitope-tagged variants of strain W303 were constructed in which the epitope tags for Pho4 and Cbf1 were switched: W303 PHO4-myc::TRP1 CBF1-3HA::LEU2 and W303 PHO4-3HA::LEU2 CBF1-myc::TRP1. W303 PHO4-myc::TRP1 was used in the gene expression microarray analyses, demonstrating that a C-terminal epitope tag does not destroy PHO4’s regulatory activity.
Flow cytometry
vtc4::GFP bearing diploid strains were grown in minimal media supplemented with high phosphate (10 mM). Overnight cultures were spun down, washed twice in water, resuspended again in water and then diluted into 3 ml of minimal media supplemented with 0 μM, 20 μM, 50 μM, 65 μM, 80 μM, 100 μM, 1 mM or 10 mM phosphate. Because yeast grows faster in high phosphate, the cells were diluted to ∼0.3 K cells/ml for the two highest concentrations of phosphate, ∼1–2 K cells/ml for the 50–100 μM phosphate samples, ∼5 K/ml for the 20 μM sample and ∼25 K/ml for the 0 μM sample. Following overnight rocking at 30°, cell fluorescence was analyzed on a FacsCalibur flow cytometer (Becton-Dickinson). At least three biological replicates, each consisting of at least three technical replicates, were performed for each combination of reporter and phosphate concentration. For each technical replicate, the median fluorescence signal from 5000–10 000 cells was used for further analysis. Values reported here are the average of these median values, taken over the set of biological replicates. The reasonable precision of these data is evident from the way the averaged median values vary smoothly and monotonically across phosphate concentrations. The values and their standard deviations, as well as the means and standard deviations for the first and third quartiles of the fluorescence signals, are provided in Supplemental Dataset S1.
Expression microarrays
W303 PHO4-myc::TRP1 was grown to mid-log phase in Yeast extract, bacto Peptone, Dextrose (YPD), washed in water, resuspended in minimal media lacking phosphate and incubated at 30° with shaking for 3 h. The culture was split and phosphate added to final concentrations of 10 mM, 1 mM, 100 μM, 10 μM, or 0 μM. The cultures were incubated a further 80 min. Biotinylated cRNA were prepared according to the Affymetrix protocol. After fragmentation, 10 μg of cRNA were hybridized for 16 h at 45°C on the S98 Yeast Genome Array. GeneChips were washed and stained in the Affymetrix Fluidics Station 450. Data were analyzed with Microarray Suite version 5.0 using Affymetrix default settings and global scaling as normalization method. Data are available at GEO: GSE26770.
Chromatin immunoprecipitation–qPCR
For ChIP–qPCR analysis, a pair of haploid strains was used that contained either myc-tagged Pho4 and 3HA-tagged Cbf1, or myc-tagged Cbf1 and 3HA-tagged Pho4 (see section on Strains for details). Each strain was grown in minimal media containing 0 or 10 mM phosphate as described in the section on flow cytometry, and ChIP was performed essentially as previously described (18). Two biological replicates were performed for each of the eight combinations of transcription factor (Pho4/Cbf1), epitope tag (myc/3HA) and phosphate concentration (0/10 mM), and each biological replicate was analyzed by qPCR twice, each time with three technical replicates on the plate. Along with the eight Pho4-target genes, two genomic regions lacking Pho4/Cbf1 motifs were used as controls, along with the input DNA. Ct values for the triplicate on-plate replicates were first averaged, and then used to calculate the ΔCt values for each locus relative to the input DNA. These data are provided as Supplemental Dataset S2. Differences in the efficiency of crosslinking or immunoprecipitation ΔCt can lead to systematic variation in ΔCt values across biological replicates, so we first median-normalized all of the sets of ΔCt values, then averaged these values across the four replicates (two biological × two technical) and, finally, subtracted from each normalized averaged ΔCt value the lowest of the ΔCt values for the two control sequences. These ΔΔCt values were converted into fold-enrichment values. There are eight sets of fold-enrichment values for each promoter (two phosphate concentrations × two transcription factors × two epitope tags for each factor). The average enrichment for the myc-tagged proteins was ∼3× lower than for 3HA, so to merge the myc- and 3HA-tagged data, we scaled the fold-enrichment values for each gene in the myc-tagged dataset, Egene,myc, according to 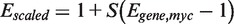 where the scale factor S was defined as
where the scale factor S was defined as 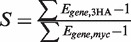 .
.
Computational modeling of binding and expression
For each of the 16 motif variants analyzed in this work, we calculated relative equilibrium constants based on the ΔΔG values for flanking dinucleotides affinities determined in vitro (8) under the assumption that the two flanking regions contribute additively to the free energy of binding (Supplementary Table S1). The affinity values are expressed relative to the mean Ka of all 256 variants. Binding site occupancies, θ, were determined by standard binding isotherms: the fractional occupancy for factor A is defined as 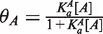 where [A] is the concentration of factor and
where [A] is the concentration of factor and 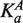 is the equilibrium association constant of factor A for the site. In this context, the association constant is the predicted affinity for the NNCACGTGNN variant found at the site of interest, normalized to the average for all NNCACGTGNN variants. This equation was used to infer the effective concentration of the factor in the cell by fitting gene expression values to binding site affinity, under the assumption that gene expression is directly proportional to transcription factor occupancy. The protein concentration in turn can be used to calculate occupancy, yielding a linear relationship between expression and occupancy. To accommodate competition from a second factor, B, the binding isotherm formula is modified to:
is the equilibrium association constant of factor A for the site. In this context, the association constant is the predicted affinity for the NNCACGTGNN variant found at the site of interest, normalized to the average for all NNCACGTGNN variants. This equation was used to infer the effective concentration of the factor in the cell by fitting gene expression values to binding site affinity, under the assumption that gene expression is directly proportional to transcription factor occupancy. The protein concentration in turn can be used to calculate occupancy, yielding a linear relationship between expression and occupancy. To accommodate competition from a second factor, B, the binding isotherm formula is modified to: 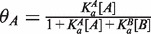 where [B] is the concentration of the competing factor and
where [B] is the concentration of the competing factor and 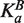 is its binding affinity. Reporter gene expression levels, E, in arbitrary units, were predicted by summing the weighted contributions of Cbf1 and Pho4 occupancies:
is its binding affinity. Reporter gene expression levels, E, in arbitrary units, were predicted by summing the weighted contributions of Cbf1 and Pho4 occupancies: 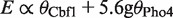 . The weighting factor of 5.6 for Pho4-bound sites was determined from the ratio of slopes for the expression versus occupancy fits for Pho4 and Cbf1 under conditions when each dominates the expression differences (i.e., 0 mM phosphate for Pho4 and 10 mM phosphate for Cbf1). A fixed concentration of [Cbf1] = 1 was used to fit expression data at all phosphate concentrations; [Pho4] was varied systematically to maximize the fit between experimental and predicted expression levels. For calculations involving reporter constructs, which differ at a single site, these calculations were performed in Excel. Excel was also used for calculating expression sensitivity values for the natural promoters and for modeling Cbf1 as a chromatin remodeling activity. For this purpose, if the promoter had more than one CACGTG motifs, calculations were performed independently for each motif, and the total expression activity was based on the probability that one (or more) Pho4 was bound and the probability that one (or more) Cbf1 was bound. For other purposes, occupancies of the natural promoter were predicted with the program GOMER, which uses a Position Weight Matrix (PWM) and an assumed protein concentration to score a genomic region for the probability of being bound at least one location (5). To generate PWMs for Pho4 and Cbf1, we started with a shared CACGTG core. The weights for this core were obtained by averaging the relative free energy terms from several PWMs for Pho4 and Cbf1, and then symmetricizing the PWM around the palindromic center (9,19,20). We then added weights for the flanking dinucleotides based on experimental binding affinities (21). To derive the PWM weights from the 16 experimental values for dinucleotide-binding affinity, a linear regression was performed to estimate the six independent parameters required (three at each position; the weight for the fourth base is determined by the other three). The free energy differences predicted by the PWMs are well correlated with the experimental free energy differences (R = 0.95 for Pho4 and R = 0.85 for Cbf1). The PWMs are available as Supplementary Tables S2 and S3.
. The weighting factor of 5.6 for Pho4-bound sites was determined from the ratio of slopes for the expression versus occupancy fits for Pho4 and Cbf1 under conditions when each dominates the expression differences (i.e., 0 mM phosphate for Pho4 and 10 mM phosphate for Cbf1). A fixed concentration of [Cbf1] = 1 was used to fit expression data at all phosphate concentrations; [Pho4] was varied systematically to maximize the fit between experimental and predicted expression levels. For calculations involving reporter constructs, which differ at a single site, these calculations were performed in Excel. Excel was also used for calculating expression sensitivity values for the natural promoters and for modeling Cbf1 as a chromatin remodeling activity. For this purpose, if the promoter had more than one CACGTG motifs, calculations were performed independently for each motif, and the total expression activity was based on the probability that one (or more) Pho4 was bound and the probability that one (or more) Cbf1 was bound. For other purposes, occupancies of the natural promoter were predicted with the program GOMER, which uses a Position Weight Matrix (PWM) and an assumed protein concentration to score a genomic region for the probability of being bound at least one location (5). To generate PWMs for Pho4 and Cbf1, we started with a shared CACGTG core. The weights for this core were obtained by averaging the relative free energy terms from several PWMs for Pho4 and Cbf1, and then symmetricizing the PWM around the palindromic center (9,19,20). We then added weights for the flanking dinucleotides based on experimental binding affinities (21). To derive the PWM weights from the 16 experimental values for dinucleotide-binding affinity, a linear regression was performed to estimate the six independent parameters required (three at each position; the weight for the fourth base is determined by the other three). The free energy differences predicted by the PWMs are well correlated with the experimental free energy differences (R = 0.95 for Pho4 and R = 0.85 for Cbf1). The PWMs are available as Supplementary Tables S2 and S3.
RESULTS
GFP reporter assays
The gene VTC4 is a direct regulatory target of Pho4 as defined by ChIP experiments and by a joint statistical analysis of sequence motifs and the effect of constitutive Pho4 expression on gene expression (Methods; Supplementary Figure S2) (5,22,23). We used the VTC4 promoter as a backbone for construction of 16 GFP fusions that differ at a single Pho4/Cbf1 binding site. The wild-type VTC4 promoter contains two perfect CACGTG motifs but, based on data from in vitro affinity measurements (8), one of these sites (caCACGTGaa) is predicted to bind Pho4 ∼30-fold less well than the other (cgCACGTGgc). Nevertheless, to avoid complications in interpretation, we knocked out the weaker site in all of our reporter constructs, using several substitutions in the core motif. The remaining site was replaced by all 16 possible palindromic variants of the sequence nnCACGTGnn. (The promoter sequences are provided as Supplementary Figure S3). The 16 reporter constructs were separately integrated by homologous recombination into a diploid yeast strain, replacing one of the VTC4 alleles with the gene fusion (Methods). Each of the 16 reporter strains was then assayed for GFP expression at eight different phosphate concentrations ranging from 0 to 10 mM (Figure 2).
Figure 2.
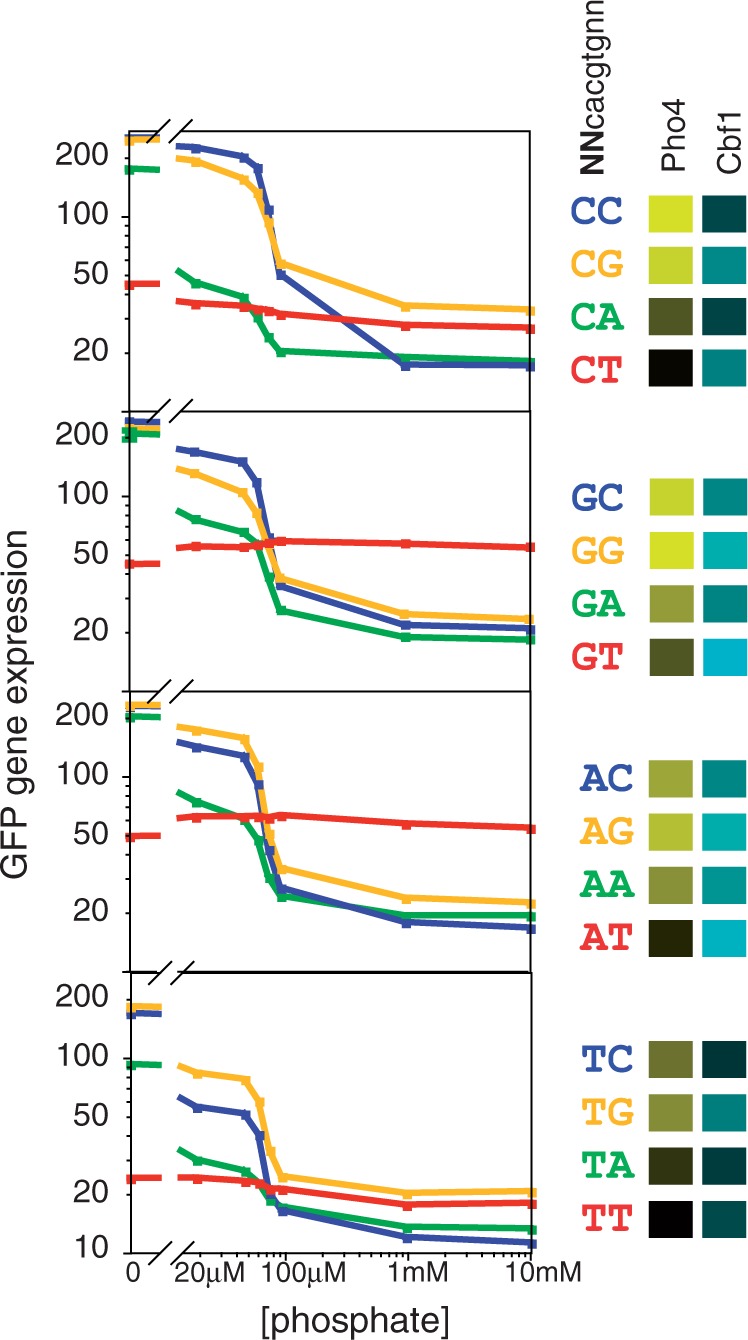
Phosphate-concentration dependence of gene expression from promoters containing each of the 16 possible palindromic Pho4/Cbf1 motifs (NNCACGTGNN). The four graphs group sites according to the identity of the distal flanking base; for example, the top graph shows expression for the four promoters that contain variants of CNCACGTGNG. Within each graph, the color denotes the base immediately 5′ to the core hexamer; for example, the blue line in each graph indicates a variant of NCCACGTGGN. Also shown in this panel are the relative affinities of the 16 sites for Pho4 and Cbf1, according to the color scheme in Figure 1.
For most of the promoter variants, there is a substantial increase in expression as the phosphate concentration is lowered, as is expected for a gene under the control of Pho4. Furthermore, there is a clear qualitative association between expression level and the affinity of the motif for Pho4 (as indicated by the brightness of the yellow color in the Pho4 column of the key) and expression under maximally induced conditions (0 mM phosphate) (Figure 2). However, for the variants that have the highest affinity for Cbf1 (brightness of blue color in the Cbf1 column of the key), the phosphate-dependent induction is more modest. In fact, in the extreme case, the two variants with the highest Cbf1 affinity (GT and AT in the 5′ flanking region), there is no increase in expression whatsoever in low phosphate. Interestingly, the expression level for each of these two promoters is actually higher than that of all others under non-inducing conditions (10 mM phosphate), when Pho4 activity is at its lowest level. As these two promoters are expected to bind Cbf1 most strongly, this result suggests that Cbf1 itself drives gene expression in the absence of strong competition from Pho4.
To quantify these effects, we first looked at the correlation between the Pho4 affinity of the binding site and the level of gene expression under maximally induced conditions. As affinity increases, expression asymptotically approaches a maximum, implying that occupancy of the site is becoming saturated (Figure 3A). The curve resembles the classic hyperbolic curve that describes occupancy of a binding site as a function of protein concentration. This is not a coincidence. The relationship between affinity, concentration and binding occupancy that allows us to determine the affinity of a site from the change in occupancy as a function of protein concentration can also be used to determine protein concentration from the change in occupancy as a function of affinity. In this case, we do not have values for the in vivo binding occupancy per se, but we can instead use gene expression as a proxy for binding occupancy. In addition, we do not know the absolute affinities of the binding sites under in vivo conditions, so the concentration that can be determined is not an absolute concentration but a concentration expressed relative to the Kd of the binding sites. Fitting the data in Figure 3A to two parameters, the maximum expression level and the concentration of Pho4, we find a good fit at a Pho4 concentration four times the mean Kd of the binding sites. Using this value of [Pho4] = 4, we can transform affinities into predicted fractional occupancies, yielding a linear relationship between expression and predicted occupancy (R = 0.94; Figure 3B). This is a remarkably strong correlation considering that the occupancy of binding sites in vivo was predicted on the basis of subtle differences in affinity in vitro; we conclude that the specificity in vivo must be very similar to the specificity in vitro (8). Furthermore, the experimental value that was correlated to predicted binding is the expression level of reporter genes. Expression levels need not have been related in some simple fashion to binding, but the strength and linearity of the correlation suggests that median single-cell GFP fluorescence values are, in fact, an accurate proxy for transcription factor binding in this system. The effective Pho4 concentration is also likely to be reasonably well estimated because both the linearity of the fit and the value of the correlation coefficient fall off as Pho4 concentration values vary from the optimum (Supplementary Figure S4). We have less confidence in the absolute Pho4 occupancy values because these values are based on the assumption that expression asymptotically approaches its limit entirely because Pho4 binding asymptotically approaches saturation. In reality, there may be additional factors, such as the saturation of RNA polymerase activity, that affect maximal expression level. However, these effects do not materially affect our interpretation, as the excellent fit that we find between in vitro affinity and expression requires none of the assumptions that are required for the estimation of occupancy.
Figure 3.
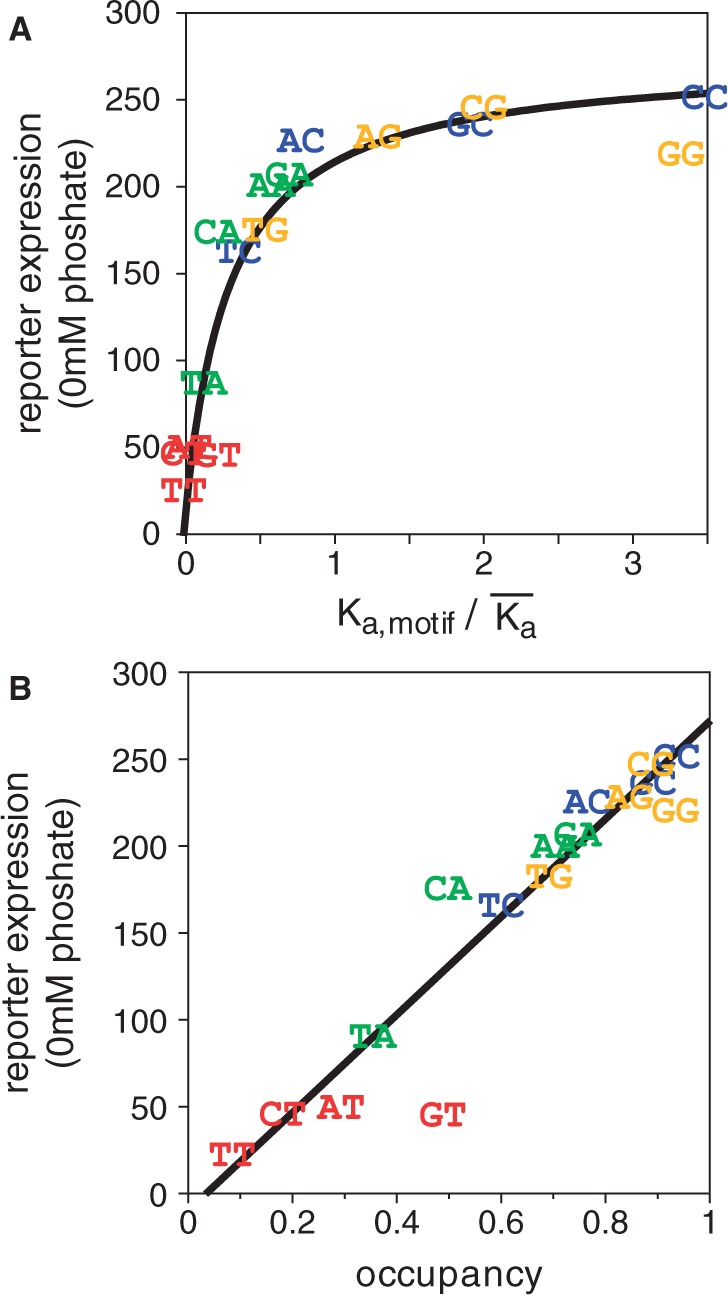
(A) Reporter expression levels under fully inducing conditions (0 mM phosphate) plotted against relative Pho4 binding affinity. Motifs are indicated by the same two bases used in Figure 2. The data have been fit to a standard binding isotherm, yielding an apparent protein concentration, relative to the mean affinity, of 4 (Methods). (B) Alternative representation of the data in panel A, based on calculating the predicted occupancy of each motif at a protein concentration 4 times the mean Kd. The line is a linear fit of expression versus predicted occupancy.
Cbf1-mediated activation and competition with Pho4 are required to explain expression under moderate-phosphate conditions
Under non-inducing conditions (10 mM phosphate), the differences in expression among the 16 promoter variants are much smaller than under inducing conditions, and the differences are uncorrelated with Pho4 affinity (Figure 4A). Surprisingly, though, they are well correlated with Cbf1 affinity (R = 0.90 for the linear fit to predicted occupancy [Cbf1] = 1). The strength of this correlation owes much to the GT- and AT- variants, which have unusually high Cbf1 affinities and are expressed at relatively high levels. However, there remains a substantial correlation even without these two promoters (R = 0.58). In contrast, there is no positive correlation whatsoever between Cbf1 affinity and expression under inducing conditions, just as there is no correlation between Pho4 affinity and expression under non-inducing conditions (Figure 4A). If anything, there is a slight negative correlation to each of these relationships. Thus Pho4 binding appears to dominate differences in gene expression when Pho4 concentrations levels are high (0 mM phosphate) while differences in Cbf1 binding dominate the (smaller) differences in expression when Pho4 levels are low (10 mM phosphate). The greater range of expression at 0 mM phosphate, where Pho4 dominates, than at 10 mM, where Cbf1 dominates, implies that Pho4 may be a more potent transcriptional activator. Based on the ratio of the correlation slopes at low and high phosphate, it appears that gene expression is 5.6 times more responsive to differences in Pho4 occupancy than it is to differences in Cbf1 occupancy. For purposes of modeling gene expression, then, we interpret this to mean that Pho4 is 5.6 times better as a transcriptional activator.
Figure 4.
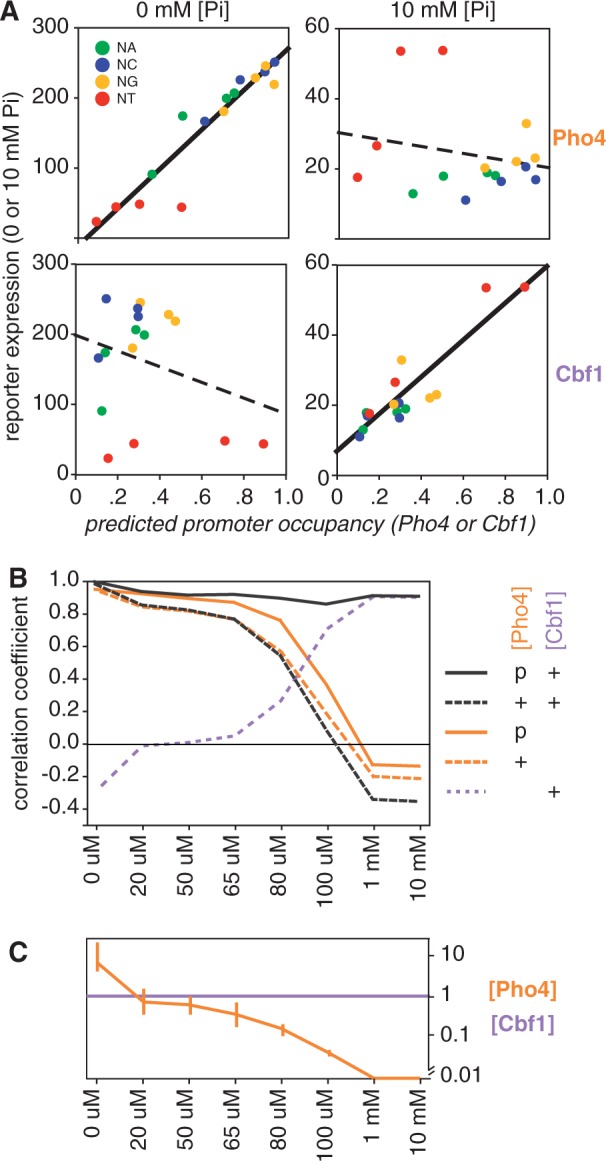
Expression and predicted binding. (A) Correlation between reporter gene expression at different phosphate concentrations and the predicted occupancy of Pho4 to 16 palindromic binding sites. (A: top left) Expression in 0 mM phosphate, reproducing data in Figure 2C. Solid line indicates a high linear correlation coefficient (R = 0.94). (A: top right) Expression at 10 mM phosphate. The dashed line indicates a weak inverse correlation between expression and Pho4 binding (R = −0.22). (A: bottom two panels) Analogous to the top panels, but using predicted Cbf1 occupancy rather than Pho4; a concentration of 1 was used (equivalent to the mean of the relative Kd’s). Note the strong correlation at 10 mM phosphate and the weak correlation at 0 mM phosphate, opposite to what is observed for Pho4. (B) Changes in correlation coefficient with predicted binding as expression levels change in response to different phosphate concentrations. Expression was predicted using five variations of the model as indicated in the key. ‘+’ Indicates a fixed concentration of [Pho4] = 4, as in panel A and in Figure 2, or a concentration of 1 in the case of Cbf1. ‘p’ indicates the parameterization of [Pho4], optimizing the fit at each phosphate concentration. (C) The transcription factor concentrations used at each phosphate concentration to obtain the best fit between predicted and observed expression (solid black line in panel B). The error bars on the Pho4 concentration line represent the range of concentrations that produce correlation coefficients within 0.015 of the optimal value shown in panel B (See Methods).
As Pho4 specificity is unable to explain expression at high phosphate concentrations, and Cbf1 specificity is unable to explain expression at low phosphate concentrations, it is not surprising that there are intermediate phosphate concentrations at which neither explains the data very well (Figure 4B). However, excellent correlations were achieved between observed and predicted expression across the full range of phosphate concentrations using a very simple model (Figure 4B; Methods). In brief, (i) Pho4 and Cbf1 compete for binding, (ii) Cbf1 concentration is constant while Pho4 increases with decreasing phosphate and (iii) Pho4 and Cbf1 each activate expression, albeit with different efficiencies. The efficiency of Pho4 as a transcriptional activator was fixed at a value 5.6× greater than Cbf1 for reasons described above. Cbf1 concentration was fixed at a value of 1 relative to the mean Cbf1 affinity of the 16 sites. The concentration of Pho4 was parameterized at each phosphate concentration to maximize the correlation between predicted occupancy and expression (Figure 4B and C). Interestingly, the inclusion of Cbf1 in the model improved the correlation to expression even under fully induced conditions, where Pho4 dominates, increasing from R = 0.94, for Pho4 only, to a value of 0.98 for Pho4 and Cbf1 together, and at the same concentration of Pho4 (Supplementary Figure S5). Both the hypothesized activation activity of Cbf1 and the increase in [Pho4] with decreasing phosphate are required to achieve a high correlation between predicted and observed expression at each phosphate concentration (Figure 4B).
Not surprisingly, the expression profiles for individual reporters show a similarly good improvement (Supplementary Figure S6). A particularly interesting example is the behavior of the CCCACGTGGG and GGCACGTGCC reporters. These motifs are nearly identical in Pho4 affinity, so a model that uses only Pho4 predicts the concentration dependence of their expression to be nearly identical (Supplementary Figure S7). Experimentally, however, the GG-flanked motif is more active than the CC-flanked motif in high phosphate, and less active in moderate phosphate. The model that incorporates Cbf1 correctly reproduces this behavior because the higher Cbf1 affinity of the GG site makes a modest positive contributor to expression in high phosphate (low [Pho4]), but it inhibits the binding of Pho4, the more potent activator, at moderate phosphate concentrations (Supplementary Figure S7).
Reporters with high Cbf1:Pho4 affinity ratios are unusually sensitive to low levels of induction signal
Thus far we have shown that the expression level for 16 reporters can be fit by a model in which Cbf1 has modest activation activity and competes with Pho4 for binding (Figure 4; Supplementary Figure S6). We also performed an additional second-order analysis of the expression profiles, examining the sensitivity of induction to the concentration of Pho4. Sensitivity is defined as the fraction of the full induction range that can be achieved at some intermediate concentration of the induction signal (Methods). Sensitivity to induction is of special interest because natural Pho4-regulated promoters differ in this regard, and these differences have been rationalized as being due to differences in chromatin structure (24–26). As our reporters are identical in sequence, except for the small differences flanking the CACGTG core motif, it is unlikely that will differ substantially in their intrinsic chromatin structure.
For a family of promoters with a single binding site, and which are otherwise identical, we can expect a monotonic relationship between the affinity for the binding site and the sensitivity to induction (Supplementary Figure S8). While the precise shape of this relationship depends on the concentrations of the transcription factor with respect to the affinities, the overall trend is robust to these differences. We therefore calculated sensitivity values from the experimental GFP expression values for the 16 reporter constructs and compared these values with the in vitro affinities of the binding sites (Methods). For 12 of the 16 promoters, those with sites flanked by [N][ACG], we find a strong linear correlation between Pho4 affinity and induction sensitivity (R = 0.85). The correlation is improved to R = 0.92 if, instead of using Pho4 affinity, we use predicted induction sensitivity (Figure 5). This can be done using the predicted expression levels obtained previously from the model, with no additional parameterization.
Figure 5.
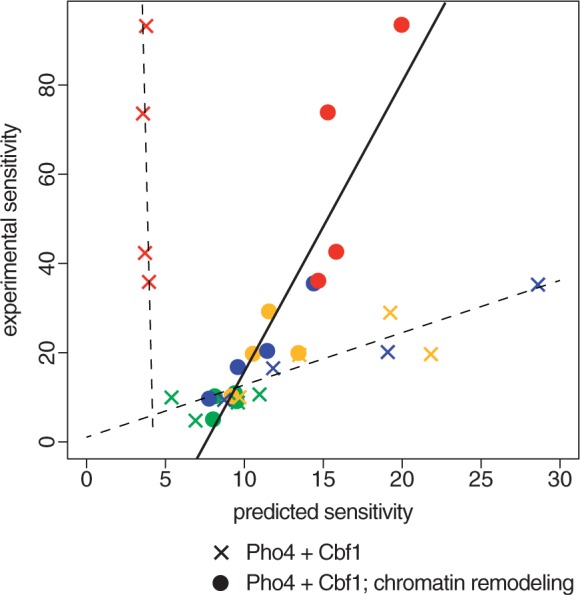
Reporters with high Cbf1: Pho4 affinity ratios show a sensitivity to induction that is anomalously high based on the simple Pho4 + Cbf1 model (X’s), but are explained well a model that assumes a role for bound Cbf1 in shifting promoter chromatin structures toward a more accessible state (circles). Sensitivities were calculated as described in the text. Colors indicate the NA (green), NC (blue), NG (yellow) and NT (red) reporters as in other figures.
Although most of the reporters adhere well to our expectations, the four sites with T adjacent to the core have experimental sensitivity values far above those predicted from the model (Figure 5). Two of these sites, those flanked by GT and AT, have exceptionally high Cbf1 affinities. That is not the case, however, for CT and AT, which rank 11th and 13th, respectively, in Cbf1 affinity. What does distinguish all four of these sites from the other twelve is that they have exceptionally high ratios of Cbf1:Pho4 affinity. This suggests that there is some aspect of Cbf1 binding, and in particular Cbf1/Pho4 competition, that is contributing to induction sensitivity but has not yet been captured by the model.
A quantitative model for high induction sensitivity: Cbf1 binding promotes conversion of ‘closed’ chromatin to ‘open’
Cbf1 has been reported to have a role in recruiting enzymes involved in chromatin remodeling (27,28). Thanks to positive feedback mechanisms involving histone modifications, chromatin states that are induced by a sequence-specific DNA-binding protein, like Cbf1, can persist even after the protein that induced that state dissociates. Thus, Cbf1 binding could shift the equilibrium from ‘closed’ chromatin to ‘open’ in a way that permits both Pho4 and Cbf1 itself to access binding sites more readily in the sub-population of cells in which the chromatin had been opened by a previous Cbf1-binding event. In this way, Cbf1, though competing with Pho4 for binding to the same sites, could, under certain conditions, function as if it were binding cooperatively. We modeled this process by assuming that Pho4 and Cbf1 each bind to sites in the open state with 10× higher affinity than in the closed state, that the equilibrium constant for chromatin favors the closed state by a factor of 10 and that sites occupied by Cbf1 are more likely to shift into the open state. The distribution between closed and open states was modeled by recursively calculating the fraction of each state at progressively lower concentrations of Pho4. In every other way, expression levels and the sensitivity to induction were calculated as already described. As shown in Figure 5, this modeling of Cbf1-induced chromatin opening dramatically improves the correlation between the predicted induction sensitivity and the observed. Although this does not, of course, prove that Cbf1-induced chromatin remodeling is responsible for differences in sensitivity, it does suggest that it is a plausible explanation.
Cbf1 binding appears to modulate the sensitivity of expression of natural promoters
It is well established that there are differences in the sensitivity to induction among Pho4-target genes. Lam et al. (24), using GFP fusions for seven Pho4 target genes, classified the genes into two groups on this basis. We validated this observation using a microarray gene expression data obtained at five phosphate concentrations (Figure 6A). Although the microarray and GFP reporter assays are measuring rather different things, the baseline expression in high phosphate for the seven genes that were assayed in common are well correlated (R = 0.95), as is the extent of their induction upon shifting to 0 mM phosphate (R = 0.95) (Supplementary Figure S9). We prefer to use the term expression sensitivity rather than threshold because induction is not an all or none phenomenon, but the concepts are related, and our data can readily be interpreted to produce the same two groups defined by GFP assays (Figure 6A). In addition to the seven genes studied previously, we included in our analysis VTC4, a Pho4 target gene that shows an induction sensitivity intermediate between the two classes defined earlier (Figure 6A).
Figure 6.
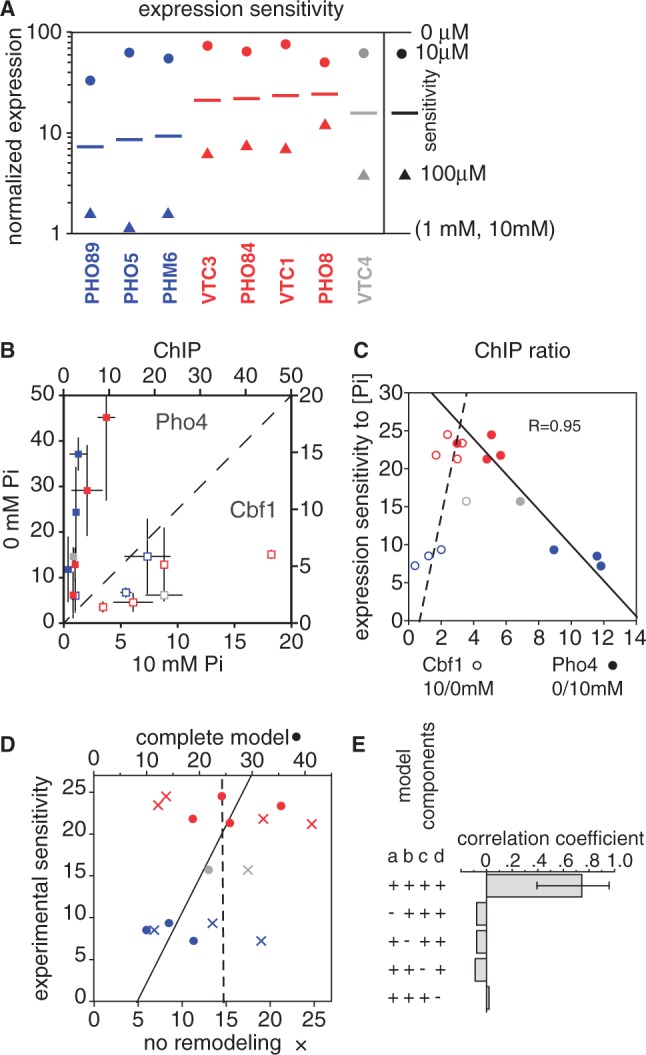
Sensitivity of natural promoters to intermediate phosphate concentrations is related to Cbf1 affinity. (A) Normalized gene expression levels at different phosphate concentrations. Expression was determined by microarray analysis at 0 μM, 10 μM, 100 μM, 1 mM and 10 mM [Pi], and was normalized between a value of 1 (average for 1 and 10 mM, as these were similarly low) and 100 (0 μM). As indicated in the legend, the colored circles represent the expression level in 10 μM phosphate, while triangles shows expression in 100 μM phosphate. The line midway between the two is the log-average of these values; we use this value as a measure of the sensitivity of a gene to intermediate phosphate. Blue and red genes are the same as those classified by Kim and O’Shea (25) as having low and high thresholds for induction, and are colored in the same way. [Some gene names are different, reflecting changes in the nomenclature used in the Saccharomyces Genome Database (SGD) (20)]. VTC4 (gray) has been included in this analysis, as its promoter was the basis for the reporter experiments described here. (B) ChIP of Pho4 (closed squares) and Cbf1 (open squares) in high and low phosphate. Values and error bars are the average and standard error of enrichment values for two sets of experiments using Myc-tagged and HA-tagged proteins (Methods). Colors are as in panel A. (C) Correlation of induction sensitivity with the change in transcription factor binding. The fold-difference in ChIP enrichment values are shown for both for Pho4 (closed squares; 0/10 mM phosphate) and Cbf1 (open squares; 10/0 mM). Note that the Pho4 and Cbf1 ChIP enrichment ratios can be compared for a given gene because the two values share the same y-axis values. (D) Correlation between the experimentally determined phosphate sensitivity of expression and two in silico predictions. One (X’s) is the model used for the reporter constructs in Figure 4, which takes into account competition between Cbf1 and Pho4 and ascribes a low level of transcriptional activity to bound Cbf1. The second model (circles) includes the same terms, but also models chromatin changes as reported for the reporter gene sensitivity values in Figure 5. The dashed and solid lines indicate fits to these predictions, respectively. The parameters used, and an analysis of the sensitivity to those parameters, are shown in Supplementary Figure S11. (E) Correlation coefficients for the chromatin remodeling model shown in panel D, along with the effect of leaving out certain terms. The error bars for the top histogram bar indicates the 95% confidence interval for the correlation as defined by bootstrap resampling. Leaving out any term that is required for modeling the cooperative effects of Cbf1 chromatin remodeling results in an insignificant correlation. The terms are labeled a–d: (a) are there two states for the promoter, ‘open’ and ‘closed’?; (b) is there tighter binding of Pho4 and Cbf1 to the ‘open’ state?; (c) does Cbf1 binding shift the equilibrium toward the open state?; (d) is Cbf1 included in the model at all?
In contrast to the reporter constructs, for which the 12 [N][ACG] sites showed the expected correlation between Pho4 affinity and induction sensitivity, the natural promoters show no hint of a link between Pho4 affinity and sensitivity (Supplementary Figure S10). This lack of a correlation is consistent with the suggestion that differences in sensitivity may be dominated by differences in nucleosome occupancy at Pho4 sites (24–26). We also noted, though, that all four of the more sensitive genes have predicted occupancies for Cbf1 that are at least slightly higher than that of all the other genes (Supplementary Figure S10). Although the number of genes is too small for the predicted differences to be statistically significant, this is reminiscent of our observation that the reporter genes with high Cbf1:Pho4 affinity ratios have anomalously high induction sensitivities, and suggests that sensitivity of natural promoters might also be associated in some way with Cbf1 affinity.
To examine experimentally the relationship between expression sensitivity and transcription factor binding we performed ChIP-qPCR assays for both Pho4 and Cbf1 on all eight promoters in cells exposed to high and low phosphate (Figure 6B). As expected, Pho4 binds best in low phosphate while Cbf1 binds best in high phosphate because there is less Pho4 to compete with it. Although it is problematic to compare absolute enrichment values across different promoters because of possible differences in crosslinking efficiencies, fold-changes in enrichment can be more confidently compared (Figure 6C). We find that Pho4 generally increases in enrichment more than Cbf1 decreases, and that the changes are inversely correlated: genes that show a relatively large change in Pho4 enrichment tend to show a relatively small change in Cbf1 enrichment. This is an expected result because the total occupancy of the promoter increases under inducing conditions, when the sum of the concentrations of Pho4 and Cbf1 is highest. Perhaps less intuitive is the inverse relationship between Pho4 fold-changes and Cbf1 fold-changes. Nevertheless, this is also an expected result. This is because a high-affinity Pho4 site is further along the binding curve at low [Pho4] than a low-affinity site, and therefore will have a smaller fold-increase than the low-affinity site for the same change in [Pho4] (also, see Supplementary Figure S8); conversely, the higher occupancy of Pho4 at the high-affinity site means that there is less Cbf1 bound, so Cbf1 binding is effectively binding as if it were at a lower concentration, and thus has more room for increasing.
The more important result shown in Figure 6C is that there is a strong inverse correlation between the expression sensitivity of the promoters and the fold-change in Pho4 binding. Simulations of simple one-site promoters show that this is precisely the relationship that is expected between sensitivity and changes in transcription factor binding (Supplementary Figure S8). Thus, the microarray expression data and the Pho4 ChIP data are consistent with one another. Furthermore, simulations of simple promoters show that both the expression data and the ChIP data are consistent with high-sensitivity promoters (red) having higher effective affinities for Pho4 than do low sensitivity promoters (blue). However, as noted above, there is no correlation between induction sensitivity and the predicted affinity of promoters for Pho4. We reasoned, therefore, that there might be other features of the sequence, such as Cbf1 affinity, that modulate the effective affinities of the promoters for Pho4.
Based on the success of the chromatin remodeling model in reconciling the predicted and experimental sensitivities of the reporter constructs, we applied the same model to the eight natural promoters. Inclusion of the Cbf1 chromatin remodeling term yields a respectable correlation between predicted and experimental sensitivity values (R = 0.75) (Figure 6D). Bootstrap resampling implies that the correlation is significant (95% confidence interval: 0.39–0.95; Figure 6E), and sensitivity analyses show that the correlation is reasonably robust to the choice of parameters (Supplementary Figure S11). Furthermore, all of the terms relevant to the modeling of chromatin restructuring (i.e., open–closed equilibrium; preferential binding of Pho4 and Cbf1 to open DNA; Cbf1-mediated chromatin opening) results in correlation coefficients near zero (Figure 6E).
DISCUSSION
For reporter genes that contain single Pho4-binding sites, we found the maximum gene expression levels to be remarkably well correlated with the predicted occupancy of these sites by Pho4. We are not aware of another experiment of this type in which the affinity of so many sites, measured in vitro, have been shown to correlate so well with expression. The strength of the correlation implies that both the in vitro affinities and expression values were determined with a fair degree of accuracy. More importantly, it implies that expression in this system is an excellent proxy for transcription factor binding and that in vivo binding affinity differences are very similar to those in vitro.
The strong correlation between Pho4 affinity and expression under induced conditions is mirrored by a similarly good correlation between Cbf1 affinity and expression under non-inducing conditions. The simplest explanation for this is that Cbf1 itself has transcriptional activation activity. Excellent correlations were also achieved at intermediate conditions by modeling contributions from both Pho4 and Cbf1 in competition with each other. The one characteristic of reporter gene expression that was not well explained by this model was the anomalously high sensitivity to induction for the four reporters that have exceptionally high ratios of Cbf1:Pho4 affinity. These reporters are not well expressed under fully induced conditions, but to the degree that they are expressed, low levels of Pho4 are sufficient to achieve a substantial fraction of that expression (which is what we mean by sensitivity). We propose that high sensitivity derives from an activity of Cbf1 that leads to a remodeling of chromatin, allowing both Cbf1 itself and Pho4 to more readily bind their cognate sites. Incorporation of this feature into the model results in an excellent correlation between predicted and experimental sensitivities of the reporter constructs.
The discovery that natural Pho4 target genes can be classified into genes with low or high induction thresholds has spawned a number of articles that seek to explain the observation in terms of nucleosome occupancy and the competition of nucleosomes with Pho4 binding sites of varying affinities (24–26). More recently, Zhou and O’Shea (17) found that a cbf1Δ strain expresses Pho4 target genes at higher than wild-type levels in high phosphate. This suggests that Cbf1 binding itself could raise the threshold for induction of Pho4 target genes. Here, we have shown that we can achieve a significant correlation between predicted gene expression sensitivity values and experimental only by using the model that allowed us to reconcile the sensitivities of the 16 reporter constructs. That is, we propose that Cbf1 binds in competition with Pho4 at CACGTG motifs, that it has some transcriptional activation activity itself and that it shifts the equilibrium for nucleosome occupancy at CACGTG sites from high to low. The chromatin remodeling activity that is implied by the model’s ability to fit to the expression data is consistent with previous experimental characterizations of Cbf1 function (27,28).
It is not yet clear how much of the differences in the responsiveness to phosphate can be attributed to Cbf1 and how much is due to intrinsic nucleosome positioning, but the experiments reported here indicate that the contribution of Cbf1 binding is substantial. We suggest that at least some of the differences in Pho4 target gene sensitivity that have been attributed to differences in nucleosome occupancy are ultimately due to differences in Cbf1/Pho4 affinities. As it is very common for transcription factor family members to have related DNA-binding specificities, our analyses suggest that differential competition for binding sites, and differences in the contributions of the proteins to gene expression, could be a common mechanism for modulating the responsiveness of promoters to regulatory signals.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–3, Supplementary Figures 1–11 and Supplementary Datasets 1–2.
FUNDING
Agency for Science, Technology and Research (A*STAR, Singapore); A*STAR National Science Scholarship (to J.S.Z.A.). Funding for open access charge: Operating funds from Genome Institute of Singapore.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Badis G, Chan ET, van Bakel H, Pena-Castillo L, Tillo D, Tsui K, Carlson CD, Gossett AJ, Hasinoff MJ, Warren CL, et al. A library of yeast transcription factor motifs reveals a widespread function for Rsc3 in targeting nucleosome exclusion at promoters. Mol. Cell. 2008;32:878–887. doi: 10.1016/j.molcel.2008.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Badis G, Berger MF, Philippakis AA, Talukder S, Gehrke AR, Jaeger SA, Chan ET, Metzler G, Vedenko A, Chen X, et al. Diversity and complexity in DNA recognition by transcription factors. Science. 2009;324:1720–1723. doi: 10.1126/science.1162327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Johnson AD, Poteete AR, Lauer G, Sauer RT, Ackers GK, Ptashne M. λ Repressor and cro—components of an efficient molecular switch. Nature. 1981;294:217–223. doi: 10.1038/294217a0. [DOI] [PubMed] [Google Scholar]
- 4.Pierce M, Benjamin KR, Montano SP, Georgiadis MM, Winter E, Vershon AK. Sum1 and Ndt80 proteins compete for binding to middle sporulation element sequences that control meiotic gene expression. Mol. Cell. Biol. 2003;23:4814–4825. doi: 10.1128/MCB.23.14.4814-4825.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Granek JA, Clarke ND. Explicit equilibrium modeling of transcription-factor binding and gene regulation. Genome Biol. 2005;6:R87. doi: 10.1186/gb-2005-6-10-r87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang W, Cherry JM, Nochomovitz Y, Jolly E, Botstein D, Li H. Inference of combinatorial regulation in yeast transcriptional networks: a case study of sporulation. Proc. Natl Acad. Sci. USA. 2005;102:1998–2003. doi: 10.1073/pnas.0405537102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Massari ME, Murre C. Helix-loop-helix proteins: regulators of transcription in eucaryotic organisms. Mol. Cell. Biol. 2000;20:429–440. doi: 10.1128/mcb.20.2.429-440.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maerkl SJ, Quake SR. A systems approach to measuring the binding energy landscapes of transcription factors. Science. 2007;315:233–237. doi: 10.1126/science.1131007. [DOI] [PubMed] [Google Scholar]
- 9.Harbison CT, Gordon DB, Lee TI, Rinaldi NJ, Macisaac KD, Danford TW, Hannett NM, Tagne J-B, Reynolds DB, Yoo J, et al. Transcriptional regulatory code of a eukaryotic genome. Nature. 2004;431:99–104. doi: 10.1038/nature02800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaffman A, Herskowitz I, Tjian R, O'Shea EK. Phosphorylation of the transcription factor PHO4 by a cyclin-CDK complex, PHO80-PHO85. Science. 1994;263:1153–1156. doi: 10.1126/science.8108735. [DOI] [PubMed] [Google Scholar]
- 11.O'Neill EM, Kaffman A, Jolly ER, O'Shea EK. Regulation of PHO4 nuclear localization by the PHO80-PHO85 cyclin-CDK complex. Science. 1996;271:209–212. doi: 10.1126/science.271.5246.209. [DOI] [PubMed] [Google Scholar]
- 12.Springer M, Wykoff DD, Miller N, O'Shea EK. Partially phosphorylated Pho4 activates transcription of a subset of phosphate-responsive genes. PLoS Biol. 2003;1:E28. doi: 10.1371/journal.pbio.0000028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ghaemmaghami S, Huh W-K, Bower K, Howson RW, Belle A, Dephoure N, O'Shea EK, Weissman JS. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 14.Cai M, Davis RW. Yeast centromere binding protein CBF1, of the helix-loop-helix protein family, is required for chromosome stability and methionine prototrophy. Cell. 1990;61:437–446. doi: 10.1016/0092-8674(90)90525-j. [DOI] [PubMed] [Google Scholar]
- 15.Thomas D, Jacquemin I, Surdin-Kerjan Y. MET4, a leucine zipper protein, and centromere-binding factor 1 are both required for transcriptional activation of sulfur metabolism in Saccharomyces cerevisiae. Mol. Cell. Biol. 1992;12:1719–1727. doi: 10.1128/mcb.12.4.1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O'Connell KF, Baker RE. Possible cross-regulation of phosphate and sulfate metabolism in Saccharomyces cerevisiae. Genetics. 1992;132:63–73. doi: 10.1093/genetics/132.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou X, O'Shea EK. Integrated approaches reveal determinants of genome-wide binding and function of the transcription factor Pho4. Mol. Cell. 2011;42:826–836. doi: 10.1016/j.molcel.2011.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xue X, Lehming N. Nhp6p and Med3p regulate gene expression by controlling the local subunit composition of RNA polymerase II. J. Mol. Biol. 2008;379:212–230. doi: 10.1016/j.jmb.2008.03.069. [DOI] [PubMed] [Google Scholar]
- 19.Macisaac KD, Wang T, Gordon DB, Gifford DK, Stormo GD, Fraenkel E. An improved map of conserved regulatory sites for Saccharomyces cerevisiae. BMC Bioinformatics. 2006;7:113. doi: 10.1186/1471-2105-7-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu C, Byers KJRP, Mccord RP, Shi Z, Berger MF, Newburger DE, Saulrieta K, Smith Z, Shah MV, Radhakrishnan M, et al. High-resolution DNA-binding specificity analysis of yeast transcription factors. Genome Res. 2009;19:556–566. doi: 10.1101/gr.090233.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang W, Cherry JM, Botstein D, Li H. A systematic approach to reconstructing transcription networks in Saccharomycescerevisiae. Proc. Natl Acad. Sci. USA. 2002;99:16893–16898. doi: 10.1073/pnas.252638199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yeo ZX, Yeo HC, Yeo JKS, Yeo AL, Li Y, Clarke ND. Inferring transcription factor targets from gene expression changes and predicted promoter occupancy. J. Comput. Biol. 2009;16:357–368. doi: 10.1089/cmb.2008.19TT. [DOI] [PubMed] [Google Scholar]
- 23.Ogawa N, DeRisi J, Brown PO. New components of a system for phosphate accumulation and polyphosphate metabolism in Saccharomyces cerevisiae revealed by genomic expression analysis. Mol. Biol. Cell. 2000;11:4309–4321. doi: 10.1091/mbc.11.12.4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lam FH, Steger DJ, O'Shea EK. Chromatin decouples promoter threshold from dynamic range. Nature. 2008;453:246–250. doi: 10.1038/nature06867.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim HD, O'Shea EK. A quantitative model of transcription factor-activated gene expression. Nat. Struct. Mol. Biol. 2008;15:1192–1198. doi: 10.1038/nsmb.1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Raveh-Sadka T, Levo M, Segal E. Incorporating nucleosomes into thermodynamic models of transcription regulation. Genome Res. 2009;19:1480–1496. doi: 10.1101/gr.088260.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kent NA, Eibert SM, Mellor J. Cbf1p is required for chromatin remodeling at promoter-proximal CACGTG motifs in yeast. J. Biol. Chem. 2004;279:27116–27123. doi: 10.1074/jbc.M403818200. [DOI] [PubMed] [Google Scholar]
- 28.Moreau J-L, Lee M, Mahachi N, Vary J, Mellor J, Tsukiyama T, Goding CR. Regulated displacement of TBP from the PHO8 promoter in vivo requires Cbf1 and the Isw1 chromatin remodeling complex. Mol. Cell. 2003;11:1609–1620. doi: 10.1016/s1097-2765(03)00184-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


