Abstract
MeCP2 is a methyl-CpG-binding protein that is a main component of brain chromatin in vertebrates. In vitro studies have determined that in addition to its specific methyl-CpG-binding domain (MBD) MeCP2 also has several chromatin association domains. However, the specific interactions of MeCP2 with methylated or non-methylated chromatin regions and the structural characteristics of the resulting DNA associations in vivo remain poorly understood. We analysed the role of the MBD in MeCP2–chromatin associations in vivo using an MeCP2 mutant Rett syndrome mouse model (Mecp2tm1.1Jae) in which exon 3 deletion results in an N-terminal truncation of the protein, including most of the MBD. Our results show that in mutant mice, the truncated form of MeCP2 (ΔMeCP2) is expressed in different regions of the brain and liver, albeit at 50% of its wild-type (wt) counterpart. In contrast to the punctate nuclear distribution characteristic of wt MeCP2, ΔMeCP2 exhibits both diffuse nuclear localization and a substantial retention in the cytoplasm, suggesting a dysfunction of nuclear transport. In mutant brain tissue, neuronal nuclei are smaller, and ΔMeCP2 chromatin is digested faster by nucleases, producing a characteristic nuclease-resistant dinucleosome. Although a fraction of ΔMeCP2 is found associated with nucleosomes, its interaction with chromatin is transient and weak. Thus, our results unequivocally demonstrate that in vivo the MBD of MeCP2 together with its adjacent region in the N-terminal domain are critical for the proper interaction of the protein with chromatin, which cannot be replaced by any other of its protein domains.
INTRODUCTION
Methyl-CpG-binding protein 2 (MeCP2) is a basic chromosomal protein that binds to symmetrical methylated 5′CpG dinucleotide sequences (1). Although the protein consists of several well-defined structural domains (2) (Figure 1A), it is an excellent example of an intrinsically disordered protein (5) because of a very low content of secondary structure organization (40%) (5,6). Intrinsically disordered proteins contain low levels of secondary structure that can increase on interaction with other binding partners (7). In addition to DNA, MeCP2 has numerous protein interaction partners (8) and has been shown to interact with RNA (9). It should thus not come as a surprise that mutations throughout the whole protein have potentially deleterious consequences.
Figure 1.
Expression of ΔMeCP2 in Mecp2tm1.1Jae mutant mice. (A) Schematic representation of mouse MeCP2 E1. The N-terminal domain (NTD), MBD, the ID, the C-terminal domains (2), NLS (32) and the two PEST sequences (33) are indicated. (B) SDS–PAGE and western blot of nuclear extracts from Mecp2tm1.1Jae mutant (Δ) and wt mouse brain. CM, chicken erythrocyte histone marker. Top: SDS–PAGE loadings were normalized based on histone H4 contents. Bottom: Western blots were performed using C-terminal MeCP2 (left) or N-terminal MeCP2 (right) antibodies. Both of them label a higher MW band in wt nuclear extracts but only the C-terminal antibody labels a lower MW band in the mutant (Δ), consistent with the presence of a truncated MeCP2 protein with an N-terminal deletion in the MBD. (C) RT–PCR analysis of wtMeCP2 and ΔMeCP2 expression in brain.
The protein is encoded by a single-copy gene located in the X-chromosome and contains 4 exons. Alternative splicing of exon 2 results in two isoforms, E1 and E2, that differ only in the first 24 N-terminal amino acids (10). For historical reasons, many of the structural features of MeCP2 are referred to the shorter E2 isoform, which was identified first in rat (11). Although discovered later, the longer E1 isoform is by far the most abundant (10-fold higher in brain tissue) (10,12,13) and will be used as a reference throughout this article (Figure 1A).
The structural organization resulting from interactions of MeCP2 with DNA and chromatin templates has been studied extensively in vitro (14–20) and to a lesser extent in situ (20–22). However, interpreting conclusions from in vitro studies is complicated in part by the difficulty in reproducing the conditions occurring within the native nuclear environment. In addition to the difficulty in replicating the exact ionic composition and concentrations in the nucleus, the methyl binding preference of MeCP2 by DNA and nucleosome templates can only be mimicked in the presence of competitor DNA (20,23). MeCP2 is a highly basic protein that will bind indiscriminately to DNA under conditions of sufficiently low ionic strength. As a result, the preference of MeCP2 for methylated DNA templates, as assessed from the in vitro work, has remained controversial [reviewed in (2)]. Although the interactions of MeCP2 with methylated and non-methylated regions of DNA in vivo remain an issue for discussion [reviewed in (8)], high-throughput DNA sequencing has demonstrated a strong correlation between DNA methylation and the MeCP2 distribution (24,25).
MeCP2 is an X-linked gene, and mutations are associated with >85% of the cases of Rett syndrome (RTT) (26). RTT is a neurodevelopmental disease that affects mainly females, as males hemizygous for the mutation are severely affected (27). As a result of this association, a large amount of the initial research on this protein has focused on this disease. An important part of this effort has been directed towards the development of several mouse models that recapitulate phenotypic features of the human disorder (8). Two well-studied lines are Mecp2tm1.1Bird (28) and Mecp2tm1.1Jae (29), both originally described as protein-null mutations. In the Mecp2tm1.1Bird line, exons 3 and 4 were deleted, resulting in a complete knockout of protein expression. Mecp2tm1.1Jae mice contain an in-frame deletion of exon 3. The status of Mecp2tm1.1Jae mice as protein-null has remained contentious, although subsequent characterizations have confirmed the presence of stable mRNA (30) and suggested that a truncated protein may be present (31). Direct phenotypic comparisons of the two lines has supported the presence of a partially functional protein in Mecp2tm1.1Jae mice, which have shown a different and often milder phenotype for many parameters, including brain weight, brain region volumes, dendritic spine morphology and cerebellar gene expression profiles (30,32,33).
As a result of the exon 3 deletion in the Mecp2tm1.1Jae model (29), the truncated version of MeCP2 lacks 116 N-terminal amino acids, including the 48 N-terminal amino acids of the methyl-CpG-binding domain (MBD), as well as the first ‘enriched in proline, glutamic, serine and threonine’ (PEST) sequence. However, within exon 4, it retains the complete transcriptional repressor domain, the nuclear localization signal (NLS) (3) and the PEST 2 sequence (4) (Figure 1A). Consequently, this mouse model provides a valuable tool in trying to address the question about the effect of a disrupted MBD on MeCP2–chromatin interactions in vivo. Furthermore, several MeCP2 mutations in patients suffering from RTT occur within the MBD, including R106W, R1333C, P152R, F155S and T158M. Of those, R106W takes place within the region of MBD truncated in the Mecp2tm1.1Jae mouse model.
Our results show that the N-terminal truncated ΔMeCP2 is still capable of binding to DNA and chromatin, but that the binding specificity is significantly impaired, suggesting that interactions with methylated DNA are necessary for binding specificity. The trafficking of the protein from the cytoplasm to the nucleus is also altered, despite the retained NLS. Our results demonstrate that the RTT-like phenotypic characteristics of the Mecp2tm1.1Jae mutant mouse model are a consequence of the truncated protein’s inability to properly recognize features of methylated chromatin, impaired nuclear import and accelerated protein turnover.
MATERIALS AND METHODS
Animal breeding
Mecp2 mutant mice (Mecp2tm1.1Jae/Mmcd) (MMRRC, UC Davis) (29) and yellow fluorescent protein (YFP)-H transgenic mice [B6.Cg-Tg(Thy1-YFPH)2Jrs/J, Jackson Laboratory Sacramento, CA, USA) (34) were bred and genotyped as described elsewhere (35).
Chromatin preparation and fractionation
Chromatin purification and fractionation was carried out as described elsewhere (22).
Time course micrococcal nuclease digestions of mouse liver and brain nuclei were carried out in 50 mM NaCl, 10 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES), pH 6.8, 5 mM MgCl2, 1 mM CaCl2 buffer with 2 U of micrococcal nuclease per milligram of DNA for 8, 16, 24 and 32 min.
Sucrose gradient fractionation
Chromatin fractions S1 and SE were prepared as described previously (22) and were run on 5–20% sucrose gradients in 25 mM NaCl, 10 mM Tris, pH 7.5, and 5 mM EDTA buffer for 21 h at 96 000g and 4°C and collected thereafter in 0.5-ml aliquots.
Cytoplasmic and nuclear fractionation
Briefly, wild-type (wt) and mutant brain and liver tissues were homogenized in 1× phosphate-buffered saline (PBS) (137 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4 and 1.47 mM KH2PO4) and centrifuged at 4500g for 10 min at 4°C. The pellet was added to 1× PBS, and the sample was split in two aliquots. One half was kept (starting MeCP2), while the other half was centrifuged at 4500g for 10 min. The cellular pellet was homogenized in 0.25 M sucrose, 0.5% NP 40 buffer 1, incubated for 10 min on ice and centrifuged at 4500g to produce a supernatant cytoplasmic MeCP2 fraction and a pellet nuclear MeCP2 fraction. The samples were mixed with 2× SDS sample loading buffer and analysed by western blot.
Gel electrophoresis
SDS–(15%) PAGE (36), acetic acid–urea (AU)–PAGE (37), acetic acid–urea–Triton X-100 (AUT)–(10%) PAGE (38), native-(4%) PAGE (39) and 1% agarose gels were performed as described earlier (40,41).
Histological preparation
Animals were anaesthetized using urethane (2 g/kg i.p.) (Sigma-Aldrich, St. Louis, MO, USA) and transcardially perfused with 10 ml of 0.1 M PBS (pH 7.4), 10 ml of 4% formaldehyde at room temperature in 0.1 M phosphate buffer (PFA), pH 6.5, followed by 10 ml of PFA, pH 11, with a 2-h post-fix in PFA, pH 11. Brain tissue was dissected out and fixed in 4% PFA at 4°C overnight, washed three times in PBS and sunk in 30% sucrose–PBS. Agar-embedded 200-μm coronal sections were cut on a Pelco Vibratome 1000 and coverslipped with Shandon Immunomount (Thermo Scientific Rockford, IL, USA). Animals used for MeCP2 immunofluorescence were processed in the same manner except for fixation conditions: 10 ml of PFA, pH 6.5, followed by 10 ml of PFA, pH 11, with a 2-h post-fix in PFA, pH 11. Cryopreserved brains were frozen on dry ice, and 50-μm coronal sections were cut on a Leica CM1850 UV cryostat.
Western blotting
Whole brain homogenates from 8-week-old wt and MeCP2 mutant littermates were prepared in 4 volumes of solution A [0.32 M sucrose, 1 mM MgCl2, 0.5% (w/v) NP-40], incubated for 15 min on ice and centrifuged for 10 min at 5000g at 4°C. Pellets were resuspended in solution A and recentrifuged as described above. The pellets were then taken through two rounds of resuspension and centrifugation in 4 volumes of solution B [50 mM NaCl, 5 mM MgCl2, 1 mm CaCl2, 10 mM PIPES (pH 6.8)] at 8000g for 10 min at 4°C. The final pellets were resuspended in equal volumes of solution B and 2× SDS–PAGE buffer to a DNA concentration of 2 mg/ml, boiled for 3 min at 100°C, loaded on a 15% SDS–PAGE gel and blotted onto a polyvinylidene difluoride (PVDF) membrane. All samples contained Complete Protease Cocktail Inhibitor (Roche Molecular Biochemicals, Laval, Quebec, Canada) at a 1:100 dilution. The primary antibodies used were a polyclonal rabbit α-MeCP2 (M9317) targeting C-terminal amino acids 465–478 (Sigma-Aldrich St. Louis, MO, USA) at 1:2000 dilution, and monoclonal mouse α-MeCP2 against N-terminal amino acids 81–170 (Clone 4B6) (Sigma-Aldrich St. Louis, MO) at 1:5000 dilution. Secondary antibodies used were horseradish peroxidase-conjugated anti-rabbit (Jackson ImmunoResearch Inc., West Grove, PA, USA) at 1:10 000 dilution and anti-mouse (Rockland Immunochemicals, Gilbertsville, PA, USA) at 1:3000 dilution. Visualization was performed using enhanced chemiluminescence (ECL) (PerkinElmer, Santa Clara, CA, USA).
For quantitative western analysis, the proteins of the SDS–PAGE were blotted onto a 0.2-µm nitrocellulose membrane, using 25 mM Tris base with 192 mM glycine as transfer buffer. The antibodies used were SIGMA M 9317 α-MeCP2 at a 1:2000 dilution, mouse polyclonal anti-histone H1 [0.T.60] Cat no. ab4269 (Abcam, Cambridge, MA, USA) at a 1:500 dilution and an anti-histone H4 (prepared in-house) at a 1:10 000 dilution. The secondary antibodies used were as follows: anti-rabbit IRDye® 800-conjugated antibody, cat no. 611-132-122 (Rockland, Gilbertsville, PA, USA) at a 1:5000 dilution and goat anti-mouse IRDye® 680, Cat no.926-32220 (LI-COR Biosciences, Lincoln, NE, USA) at a 1:10 000 dilution. Membrane was incubated with secondary antibodies in dark conditions for 1 h and visualized using Odyssey Licor Developer v.2.1. Band intensity was quantified using Licor software.
Immunohistochemistry
Tissue sections were permeabilized in 1% PBST (PBS, pH 7.4, with 1% Triton X-100) and blocked overnight in 5% goat serum. All antibody incubations were done for 24 h at room temperature in 0.1% PBST. A chicken α-MeCP2 antibody targeting C-terminal amino acids 466–484 (gift from Janine LaSalle, UC Davis) and mouse α-MeCP2 4B6 were used as primary antibodies at a 1:5000 dilution. Alexa 555-conjugated goat anti-chicken and Alexa 647-conjugated goat anti-mouse (Invitrogen, Carlsbad, CA, USA) were used as secondary antibodies (1:500). YO-PRO-1 (1:10 000) (Invitrogen, Carlsbad, CA, USA) was used as a nuclear counter-stain. Sections were mounted on Vectashield Hard Set medium (Vector Laboratories, Burlingame, CA, USA), coverslipped and allowed to set for 24 h before imaging.
Plasmid constructs
Wild-type and mutant MeCP2 cDNAs (20) were used in conjunction with the following primers: 5′-GCGCGAATTCAGTAGCTGGGATGTTAGGGCT-3′/5′-GCGCGGATCCTCAGCT AACTCTCTCGGTCAC-3′ and cloned into the pEGFP-C1 plasmid (Clontech, Mountain View, CA, USA) to allow expression of C-terminal EGFP-tagged wt and mutant MeCP2 under the control of the hCMV IE promoter.
Cell culture and plasmid transfection
The Neuro-2A (N2A) mouse neuroblastoma cell line (ATCC) was used for live-cell plasmid transfection experiments. N2A cells were cultured at 37°C and 5% CO2 in equal parts high-glucose Dulbecco’s modified Eagle’s medium and minimal essential medium (MEM) + glutamax (GIBCO/Invitrogen, Carlsbad, CA, USA), supplemented with 10% fetal bovine serum (Invitrogen, Carlsbad, CA, USA), 2 mM l-glutamine (Sigma-Aldrich, St. Louis, MO, USA), 100 U/ml of penicillin (Sigma-Aldrich, St. Louis, MO, USA) and 100 µg/ml of streptomycin (Sigma-Aldrich, St. Louis, MO, USA). The day before plasmid transfections, 1.0 × 106 N2A cells were seeded into 35-mm glass bottom culture dishes (No. 1.5 coverglass; MatTek). Coverslips were coated with 1% gelatin for 2.5 h before seeding. Cells were transfected with 2 µg of plasmid DNA/plate using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) following manufacturer’s instructions, incubated for 18 h in serum- and antibiotic-free culture medium and then cultured for 48 h in complete medium. Hoechst 33342 TriHCl (Molecular Probes/Invitrogen, Carlsbad, CA, USA) was added to culture media 30 min before imaging. Immediately prior to imaging, cells were washed twice with pre-warmed HEPES-buffered extracellular solution, pH 7.4 (150 mM NaCl, 4 mM KCl, 2 mM MgCl2, 2 mM CaCl2, 10 mM HEPES, 10 mM glucose and 2 mM ascorbic acid). Cells were maintained in extracellular solution at 37°C during imaging using a TC-344B Dual Automatic Temperature Controller (Warner Instrument Corp., Hamden, CT, USA).
Imaging and image analysis
For MeCP2 immunofluorescence, 1024 × 1024 pixel 3D confocal fluorescence image stacks were obtained using an Olympus Fluoview FV1000 with Fluoview software (v. 1.7 c) with 488-, 543- and 635-nm lasers, an Olympus Plan SApo NA 0.95 40× oil objective with 10 × 2-µm z-steps and pinhole size set automatically (85–120 µm). Multi-channel imaging was done sequentially. All images were averaged twice with scan speeds of 1.68–3.94 µs/pixel (Olympus, Center Valley, PA, USA).
For Neuro-2 A live-cell imaging, monochromatic 2560 × 1920 pixel wide-field fluorescence images were obtained using a Nikon 100×/1.49 NA Apo TIRF objective (Nikon, Melville, NY, USA), 1.5× zoom, with a Nikon DS-U1 camera and ACT-2U software. Images were converted to 8-bit .tif files, processed and analysed using ImageJ software (NIH). Green fluorescent protein (GFP)/Hoechst image pairs were converted to stacks, and noise pixels were smoothed using a 2-pixel median filter. The means and standard deviations of nuclear greyscale intensity values were obtained over the same outlined area for both channels. Nuclei from 27 wt and 22 mutant cells were outlined based on the Hoechst-staining pattern. Coefficients of variation were generated for MeCP2-GFP and Hoechst staining by dividing the greyscale standard deviation by the mean intensity for each cell. A ratio of the coefficients of variations (MeCP2-GFP/Hoechst) was generated for each cell. Genotypes were compared using the Mann–Whitney U-test. Images used for the pixel intensity line plots through the soma and nucleus were converted to 32-bit .tif files, and greyscale values were squared to enhance contrast. Images were adjusted for brightness and contrast and tiled using Adobe Photoshop CS4.
Fluorescence recovery after photobleaching
WtMeCP2-GFP and ΔMeCP2-GFP plasmids with C-terminal GFP were constructed (42) and analysed by fluorescence recovery after photobleaching (FRAP) as described elsewhere (43).
Preparation of tissue samples for real-time quantitative polymerase chain reaction
Brain tissue samples from 8-week-old animals (wt n = 5, ΔMeCP2 n = 4) were rapidly dissected, flash-frozen in isopentane chilled to −20°C and stored at −80°C. Tissues were transferred to chilled RNAlater® ice (Life Technologies) at −20°C for at least 16 h before RNA extraction. Approximately 100–150 mg of brain tissue was homogenized in 1 ml of TRIzol® (Invitrogen, Carlsbad, CA, USA) using 3-mm stainless steel beads and 1.5 ml Safe-Lock Eppendorf microcentrifuge tubes in a Retsch MM301 Mixer Mill (Fisher Scientific Ltd., Ottawa, Ontario, Canada) at 25 Hz for 9 min. RNA was isolated according to the manufacturer’s instructions. Extracted RNA was resuspended in 30 µl of RNase-free dH2O and stored at −80°C. QuantiTect Reverse Transcription cDNA Synthesis Kit (Qiagen, Germantown, MD, USA) was used to synthesize cDNA from 1 μg of total RNA as described by the manufacturer (Qiagen, Germantown, MD, USA). The cDNA reactions were diluted 20-fold for use in PCR amplification. Total RNA and cDNA quality was confirmed by 1.5% agarose gel electrophoresis.
The RT–PCR primer sequences used were: MECP2 primers: forward 5′-CCCAGGTTTTGTGCTTTGTT and reverse 5′-GGGAGTGCTCTTGTCTCCTG (104-bp amplicon); rp L8 primers: forward 5′-AAAGGCATCGTAAAGGACAT and reverse 5′-ACCAGCCACAACACCAAC (376-bp amplicon). rp L8 was used to normalize the target gene (MECP2) and to account for any reference gene expression variations that may be present between different samples (44). The suitability of the target and reference gene primers for RT–PCR was assessed by serial dilution validation assay. The slope of the ΔCt (MeCP2 gene primer minus rpL8 normalizer gene primer), for the MeCP2 primer, versus log cDNA concentration dilution was <0.1, and efficiency of the primer set was >99%.
RT–PCR was conducted using the First Strand Universal SYBR Green Master Mix with Rox (Roche Molecular Biochemicals, Laval, Quebec, Canada) with an Agilent Technologies Stratagene Mx3005P thermocycler. Transcript levels were analysed using MxPro-Mx3005P v4.10 Build 389, Schema 85 software (Stratagene, La Jolla, CA, USA). Cycling parameters were Segment 1 (1 cycle): 9 min. denaturation at 94°C; Segment 2 (40 cycles): 15 s denaturation at 95°C, 30 s annealing at 62°C, 45 s extension at 72°C; Segment 3 (1 cycle—for dissociation profile): 1 min at 95°C, 30 s at 55°C, 30 s at 95°C. Each cDNA 1/20 dilution was run in quadruplicates as were cDNA template-free controls to test for any DNA contamination. Quadruplicate Ct values were averaged for mutant and wt samples, and average Ct values were normalized to the rp L8 control using the comparative Ct method (ΔΔCt ). Data analysis was done using comparative Ct (2-ΔΔCt) method (45) with one wt sample as the comparator for the mutant samples to determine the relative fold changes in Mecp2 expression levels (45–47). The specificity of the amplification reactions was determined by sharpness of dissociation curves and matching of melting temperature (Tm) between samples.
Statistical analyses were conducted using Prism 5 v.5.0.b (Graphpad Software). All data were analysed using the two-tailed unpaired student’s t-test unless otherwise indicated. Results were considered significant at P < 0.05 and reported as the mean ± SEM. Data set distributions were tested for normality using the D’Agostino and Pearson omnibus normality test.
RESULTS
MeCP2 protein missing the sequence corresponding to exon 3 of Mecp2tm1.1Jae mutant mice is expressed at reduced levels in different regions of the brain and liver
The expression of ΔMeCP2 in the Mecp2tm1.1Jae mouse model of RTT has been controversial. Although the first report of the mutant phenotype showed expression of a faster electrophoretic band by western blot analysis (29), a later report found stable mRNA but was unable to detect the protein by western blot (30).
We therefore decided to revisit this issue. Nuclear fractions from 8- to 11-week-old Mecp2tm1.1Jae mutant and wt mice were probed by western blot using two antibodies, a mouse α-MeCP2 targeting the N-terminus and a rabbit α-MeCP2 targeting the C-terminus (Figure 1B). The first 45 of 89 amino acids of the N-terminal antibody epitope are deleted in the mutant protein. Consistent with the predicted labelling pattern, the C-terminal α-MeCP2 generated both a high MW band in the wt mouse and a fainter, faster band (corresponding to a lower molecular mass) in the mutant, whereas the N-terminal α-MeCP2 only labelled an identical high-molecular mass band in the wt mice. These results are in agreement with the results from the group that created the mouse model (29).
Western blot quantification of the amount of protein expressed in whole brain homogenates carried out as described previously (22) and using IRDye green fluorescence indicated that 35 ± 3% of the truncated protein is present in the Mecp2tm1.1Jae mice, a result that closely resembles the western shown in Figure 1E of Chen et al. (29). As seen in Figure 1C, the RT–PCR graph shows a trend towards a decrease in the relative abundance of ΔMeCP2 mRNA in agreement with Chen et al. (29), who reported a reduced ΔMeCP2 transcript in mutant brain using northern-blot analysis. However, statistical analysis of the data in Figure 1C (see ‘Materials and Methods’ section) revealed that, despite the trend, there was no significant difference (t7 = 0.9716, P = 0.3636) between wt MeCP2 and ΔMeCP2.
Wild-type MeCP2 has been shown to be differentially expressed in various brain regions, with the highest expression in cerebellum and cortex (48–50). Therefore, and because of the reduced expression of the protein, we decided to check its expression in different regions of the brain. As seen in Figure 2, ΔMeCP2 is expressed throughout different sections of the brain. However, although a noticeable increase in the expression of wtMeCP2 was detected in wt cortex as previously described, no significant difference could be observed for the expression of ΔMeCP2 among different regions of the Mecp2tm1.1Jae mutant brain.
Figure 2.

Distribution of wtMeCP2 and ΔMeCP2 in different sections of the brain. The upper and lower part show a western blot analysis using an MeCP2 and histone H3 antibody, respectively, of the SDS–PAGE analysis of the nuclear protein composition shown in the middle. CB: cerebellum; CT: cortex and MB: midbrain.
To assess whether the mutant protein is expressed in peripheral tissues, we also measured MeCP2 levels in the liver. ΔMeCP2 is expressed in Mecp2tm1.1Jae mutant mouse liver, but the amount of protein expressed is only 12 ± 1% of that of wtMeCP2 expressed in wt mice (results not shown). The reason for the further decrease of ΔMeCP2 in this tissue when compared with brain is not clear, but it could be related to the faster turnover rate of proteins in liver when compared with brain (51,52).
ΔMeCP2 exhibits an unusual cellular distribution
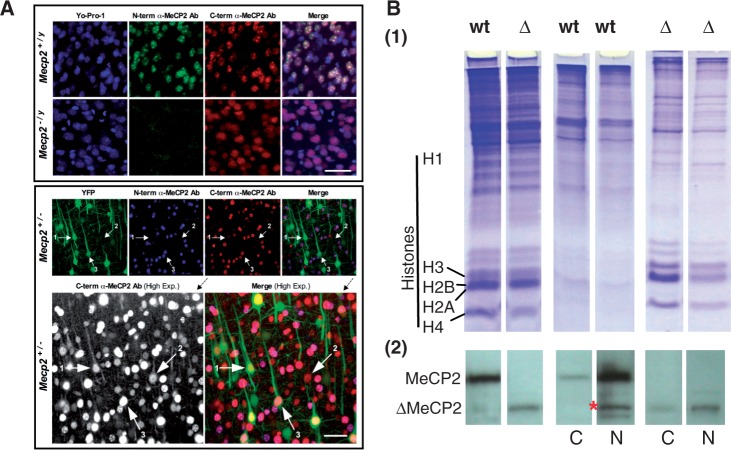
Previous immunohistochemical data from the Mecp2tm1.1Jae line have been contradictory, with the original study showing a loss of immunoreactivity in histological sections of hippocampus and cerebellum [see (29), Figure 1F], while a subsequent study found diffuse ΔMeCP2 staining in the hippocampus [see (31), Figure 2G]. We re-investigated this issue by double-immunolabelling cortical brain slices using the mouse N-terminal α-MeCP2 and a chicken C-terminal α-MeCP2 to compare wt males, mutant males and heterozygous Mecp2+/− females (Figure 3A). We found that successful double-immunolabelling was highly sensitive to tissue fixation conditions, which may account for the discrepant results in previous reports. The immunoreactivity of the C-terminal antibody was robust under most fixation conditions, but the N-terminal antibody immunoreactivity was extremely sensitive to overfixation, and was only reliably obtained after a brief formaldehyde fixation using a pH-shift protocol (53). In wt males, both antibodies labelled all neurons with a typical staining pattern showing bright puncta against a diffuse nuclear background (Figure 3A, top). Immunolabelled puncta co-localized with heterochromatin regions, as previously described (3,54). In hemizygous Mecp2−/y mutant males, only the C-terminal antibody was immunoreactive, and nuclear labelling was faint and diffuse, with no puncta observed (Figure 3A, bottom). To facilitate comparison of the subcellular distribution patterns of wt with mutant MeCP2 in vivo, heterozygous females were obtained from a cross of the Mecp2tm1.1Jae line with the B6.Cg-Tg(Thy1-YFPH)2Jrs/J line, which express YFP in a subset of cortical neurons. Heterozygous Mecp2+/− females showed a mixture of the two patterns of immunoreactivity observed in males. When imaged under high-exposure conditions to amplify the fluorescence signal, wt MeCP2 labelled with both antibodies was strictly localized to the nucleus. In neurons labelled by the C-terminal antibody alone, immunoreactivity was also observed in the cytoplasm, extending up into the primary apical dendrite, albeit at much lower levels. The marked difference in nuclear staining at the border of the nucleus relative to adjacent cytoplasm indicated that ΔMeCP2 still retained a predominantly nuclear localization.
Figure 3.
ΔMeCP2 has altered nuclear distribution in fixed brain tissue and is present in the cytoplasm. (A) Confocal fluorescence images of MeCP2-immunolabelled coronal sections of motor cortex layer 5. Top eight panels (males): Nuclei are counter-stained with YO-PRO-1 (blue). Neuronal nuclei are larger, typically with bright punctate staining corresponding to dense heterochromatin, whereas glial nuclei are smaller and sometimes irregularly shaped. In wt (Mecp2+/y) males, both α-N-terminal (green) and α-C-terminal (red) MeCP2 immunolabelling show bright puncta against a diffuse nuclear background. In MeCP2J mutant (Mecp2-/y) males, α-N-terminal MeCP2 labelling is absent, whereas faint diffuse nuclear α-C-terminal MeCP2 labelling is still observed, consistent with the N-terminal deletion and absence of the MBD. Glial MeCP2 signal is not evident in either genotype. Scale bar = 40 µm. Bottom six (female): Heterozygous YFP+ Mecp2 +/− females have a mixture of both MeCP2 immunolabelling patterns (α-N-terminal, blue; α-C-terminal, red). α-C-terminal-MeCP2 labelling is comparatively faint in mutant cells, but can be seen in a high-exposure image (bottom left, greyscale). Under high exposure, immunolabelling in wt neurons for both antibodies is bright to oversaturated, but it remains exclusively nuclear (arrow #3). In mutant neurons, faint diffuse α-C-terminal MeCP2 immunolabelling is predominantly nuclear, but it also occurs in the cytoplasm extending into dendrites (arrows #1, #2). Scale bar = 40 µm. (B) SDS–PAGE (1) and western blot (2) analysis of overall MeCP2 present in wt and mutant brain in comparison with the cytoplasmic (C) and nuclear (N) fractions. The red asterisk indicates a degradation product of wt MeCP2.
To better characterize the cytoplasmic versus nuclear localization of MeCP2, the cytoplasmic and nuclear fractions of brain cells were obtained and analysed by western blot (Figure 3B). The quantification was carried out using anti-rabbit IRDye-conjugated fluorescent antibodies. In mutant brain tissue, 27 ± 3% of the ΔMeCP2 is detected in the cytoplasmic fraction as opposed to ∼3% of MeCP2, in agreement with the imunohistochemical data shown in Figure 3A. Interestingly, although the NLS is retained, the presence of mutant MeCP2 in the cytoplasm at low levels also suggests either a dysfunction or reversibility of nuclear transport.
ΔMeCP2 exhibits even distribution throughout nuclei and is associated with a significantly reduced nuclear size in the neuronal nuclei of Mecp2tm1.1Jae mutant mice
To look in more detail at the nuclear distribution of ΔMeCP2 with respect to wt MeCP2 in live cells, particularly in light of potential fixation artifacts, Neuro-2A cells were transfected with GFP-MeCP2 and GFP-ΔMeCP2 plasmid expression constructs. As seen in Figure 4A and B, the wt GFP-MeCP2 exhibits enrichment in the pericentric heterochomatin. By contrast, GFP-ΔMeCP2 exhibits pan-cellular distribution and seems to be homogeneous throughout the nucleoplasm unlike either wt protein or the distribution of DNA as revealed by Hoechst staining (Figure 4A). This is similar to what was observed immunohistochemically with the native ΔMeCP2 in brain tissue sections (Figure 3A). The absence of correlation between DNA distribution (Hoechst staining) and GFP-ΔMeCP2 is illustrated quantitatively in the line-scan (Figure 4B). Furthermore, the ratio of the variability of GFP-ΔMeCP2 staining relative to Hoechst staining is the inverse of that observed in the wt (Figure 4C). Moreover, in agreement with what was observed in cortical neurons in Figure 3, a substantial component of GFP-ΔMeCP2 is distributed in the cytoplasm.
Figure 4.
(A–C) ΔMeCP2 distribution is altered in Neuro-2A cells and does not concentrate in heterochromatin. (A) Live Neuro-2A cells expressing GFP-tagged wt or ΔMeCP2 from transfected plasmid constructs. Nuclei were counterstained with Hoechst 33 342. wt GFP-MeCP2 is restricted to the nucleus and co-localizes with heterochromatin puncta. Mutant GFP-ΔMeCP2 is diffuse throughout the cell but reaches its highest concentration in the nucleus. Dark patches in the nucleus co-localize with bright Hoechst puncta, indicating reduced levels in heterochromatin. Red arrows indicate end points of the line segments plotted in (B). Scale bar = 20 µm. (B) Line plots of greyscale intensities for GFP-MeCP2 (left axis) and Hoechst 33342 (right axis). Although GFP-MeCP2 peak intensities coincide with Hoechst staining and are brightest at heterochromatin puncta, this correlation is not observed with GFP-ΔMeCP2. Greyscale values were squared to highlight subtle differences in staining patterns. (C) The ratio of the variability of GFP-ΔMeCP2 staining relative to Hoechst staining is the inverse of that observed in the wt. The coefficient of variation was obtained for both GFP and Hoechst greyscale intensity over the entire nucleus. In the wt, a ratio value >1 is observed, indicating a greater variance in the distribution of GFP-MeCP2 relative to Hoechst staining. The inverse is observed in the mutant (Mann–Whitney U-test, P < 0.0001). (D and E) Nuclear size is reduced in cortical neurons of ΔMeCP2 mutant mice. (D) Wide field fluorescence images of Hoechst 33342-stained nuclei. Mutant brain tissue contains a greater proportion of small nuclei. Scale bar = 10 µm. (E) The average nuclear area is reduced by 20.2% in 6-week-old mutant males (85.00 ± 1.04 µm2 in wt versus 67.84 ± 1.71 µm2 in mutant, t4 = 9.284, P < 0.01). (F) Western blot analysis of histone H1 and histone H3 with an IRDye fluorescent antibody detection. CE: chicken erythrocyte histones used as a histone marker. Bwt: nuclear brain from wt mice; BΔ: nuclear brain from Mecp2tm1.1Jae mutant; Lwt: nuclear liver from wt mice; LΔ: nuclear liver from Mecp2tm1.1Jae mutant. The numbers underneath the image represent the percentile of MeCP2 with respect to brain wt taken as 100% (in red) and of H1 with respect to liver wt taken as 100% (in black).
The nuclear distribution alteration that we observed for GFP-ΔMeCP2 is similar to initial observations by Nan et al. (3) using mouse L cells. Similarly, Matsumura et al. (55) found that in mouse NIH 3T3 cells transfected with a truncated MeCP2 lacking the complete N-terminal region (including the MBD), the mutant protein also failed to efficiently retain in chromocenters. Moreover, this construct exhibited a diffuse nuclear distribution albeit with subtle enrichment in chromocenters (55). It is important to mention here that in both instances, transfections were carried out in the background of the native wt MeCP2, and the increased presence at chromocenters could arise from the binding of the wt form.
In the course of these experiments, we noticed that transfection of Neuro-2A cells with GFP-ΔMeCP2 systematically resulted in a decrease in the volume of the nuclei when compared with the cells transfected with GFP-MeCP2, similar to what has been observed in neurons of the Mecp2tm1.1Jae mouse (29). A quantification of this phenomenon carried out by comparison of the nuclear size in wt and Mecp2tm1.1Jae mouse cortical neurons showed a 21% decrease in the latter (Figure 4D and E). Qualitative observations of reduced neuronal nuclear size in the hippocampus were made in the original publication describing Mecp2tm1.1Jae line, but these were not quantified (29). Interestingly, the decreased nuclear size we observed is comparable with the 28% reduction in neuronal nuclear size in the brains of MeCP2-null Mecp2tm1.1Bird mice (56). An MeCP2-related size dependence was also observed in embryonic stem cells by the same authors, and it has attributed to an increase in histone H1 on MeCP2 depletion (24,56). Similarly, neurons from the Mecp2tm1.1Bird line were shown to have smaller chromocenters and an increase in the number of nucleoli that exhibit a smaller size when compared with the wt neurons (57). However, as we have described earlier, the brain cells still retain a significant amount (∼35%) of ΔMeCP2 in Mecp2tm1.1Jae (Figure 1B–3B), while retaining the exact same amount of histone H1 (Figure 4F). This suggests that the decrease in nuclear volume and nucleolar organization on MeCP2 depletion is not the direct result of a compensatory H1 increase but rather of an MBD-dependent change in MeCP2-mediated chromatin organization that is normally associated to an increase in size of the nucleus.
ΔMeCP2 exhibits decreased nucleosome-binding affinity
Although all the results described so far indicate that ΔMeCP2 is able to localize to the nucleus, its binding to chromatin remained to be established. To this end, nuclei from wt and Mecp2tm1.1Jae mutant mice were isolated, digested with micrococcal nuclease and fractionated as described earlier (22). As shown in Figure 5A despite the reduced amount of ΔMeCP2 present in the Mecp2tm1.1Jae mouse and the lack of a functional MBD, mutant MeCP2 exhibits a similar partitioning to the wt on nuclease digestion in both the wt and mutant mice. The fractions exhibited a similar biochemical composition (Figure 5B and C), although a larger quantity of di-nucleosomes was consistently observed in nuclei from Mecp2tm1.1Jae mouse brain as compared with the wt (see Figure 5C SE-Δ). Fraction SI, which is highly enriched in Pol2 (58), contained the largest amount of MeCP2 in both instances (Figure 5A). On sucrose gradient fractionation (Figure 5D), both MeCP2 and ΔMeCP2 remained bound to mono-nucleosomes of the SI fraction and in both instances a similar amount of free (non-nucleosome-bound) MeCP2 (sedimenting at the upper part of the gradient) is present in this fraction as described previously (22). MeCP2 is also bound to the mono-nucleosome and di-nucleosomes of the SE fraction, although no free sedimenting fraction was observed for ΔMeCP2 (Figure 5E).
Figure 5.
Brain chromatin distribution of wtMeCP2 and ΔMeCP2 in wt and Mecp2tm1.1Jae mutant mice (Δ). (A) Western blots of nuclear MeCP2 partitioning in the SI, SE and P chromatin fractions on digestion with micrococcal nuclease (20). (B) SDS–PAGE analysis of the SI and SE chromatin fractions. Note the absence of histone H1 (square) in SI. (C) Native PAGE analysis of the DNA composition of the SI and SE chromatin fractions. Cf: pBR322-CfoI–digested DNA marker (the numbers on the left hand side of the gel correspond to the size of the electrophoretic bands in base pairs); D: di-nucleosomes; M: mono-nucleosomes. The arrow head points to the increased presence of di-nucleosomes in Mecp2tm1.1Jae mutant mice. (D) Sucrose gradient fractionation of the SI chromatin fraction. The graph (upper panel) depicts the absorbance (260 nm) profile of the numbered fractions collected along the gradient, shown in the lower panel by western blot analysis using MeCP2 antibody. (E) Sucrose gradient fractionation of the SE chromatin fraction [as described in (D)].
A time course of micrococcal nuclease digestion of cell nuclei (Figure 6A) showed that chromatin from Mecp2tm1.1Jae mutant brain tissue digested much faster than chromatin from wt brains, but at a similar rate as wt liver nuclei. Interestingly, a di-nucleosome–‘resistant’ particle is observed at high-digestion times (Figure 6A, SE, 24 and 32 min), as it was also observed in Figure 5C, SE. Although the differences in the micrococcal nuclease digestion rates could be attributed to the differences in the amount of MeCP2 present in the different cell types (being the lowest in the wt liver), the amount of histone H1 was almost identical in all of them (Figure 4F). It is possible that the slower rate of digestion observed in wt brain cell nuclei is the result of a higher MeCP2 content in these cells, which is associated with a preference for binding the linker regions connecting adjacent nucleosomes (17,20). The preference of wt MeCP2 for linker DNA in proximity to the nucleosome, similar to histone H1, is likely the result of the highest levels of DNA methylation within these chromatin regions (59) and the affinity of MeCP2 for four-way junction DNA (60), a structure that resembles the DNA organization at its site of entry and exit from the nucleosome. Accordingly, histone H1, which also binds at this nucleosome site, exhibits a similar preference for four-way junctions (61).
Figure 6.
Nuclear size, histone H1 composition and chromatin accessibility of wt and Mecp2tm1.1Jae mutant mice (Δ). (A) Agarose gel electrophoresis of the DNA fragments obtained by micrococcal nuclease time course digestions of mouse brain and liver at the times indicated on top of the gel. M1 and M2 are the GeneRuler 50 bp and GeneRuler 1 kb DNA ladder markers (Fermentas), respectively. The arrow points to the higher intensity of the band corresponding to the di-nucleosome in the SE fraction of the Mecp2tm1.1Jae mutant mice nuclei at high times of nuclease digestion. Bwt, BΔ and Lwt same as in Figure 4F. D: di-nucleosome; M: mono-nucleosome; T: tri-nucleosome. The numbers under the lanes shown in SE indicate the ratios D: M as determined by densitometry. (B) FRAP of wtMeCP2-GFP and ΔMeCP2-GFP.
Interestingly, the presence of a structurally and functionally impaired MBD in ΔMeCP2 results in a highly transient weak chromatin binding as indicated by the FRAP experiments (Figure 6B). As shown, at almost any time, most of GFP-ΔMeCP2 is free in solution, and the half time residence (t50) of the protein was not measurable (below detection limits). GFP-ΔMeCP2 had a t50 of 1.3 s in comparison with 21 s for the wt GFP-MeCP2 (Figure 6B, blue diamonds), in agreement with similar results that used GFP-MeCP2 constructs lacking the whole MBD (62) [compare Figure 6A of Kumar et al. (62) with Figure 6B]. Thus, although we have shown that ΔMeCP2 is able to bind to chromatin in the cells of the Mecp2tm1.1Jae mouse model, the binding is weak and dynamic. This finding suggests that in vivo the contribution of the MBD of MeCP2 is critical for proper chromatin binding, and it largely overrides its electrostatic interaction potential. The in vivo relevance of the FRAP data is validated by the similar nuclear/cytoplasmic localization (Figure 4A and B) and decreased nuclear size (Figure 4A) of cells transfected with similar expression vectors of GFP-tagged MeCP2 when compared with neurons from the Mecp2tm1.1Jae mouse (Figure 3A and 4D). Moreover, a single-point mutation in the region of MBD truncated in the Mecp2tm1.1Jae mouse model exhibits a FRAP that is almost identical to MeCP2 lacking MBD (62) or to shown in Figure 6B.
DISCUSSION
Although the interactions of MeCP2 with different chromatin templates have been extensively characterized in vitro (2), its binding to chromatin in the native in vivo setting is less well understood. Wild-type MeCP2 E1 has a pI of 9.9, and removal of the protein sequence encoded by exon 3 [which removes part of the MBD and part of the N-terminal region (Figure 1A)] renders a protein (ΔMeCP2) with a 10.1 pI, which is close to that of histone H1 (i.e. human H1.2, pI = 10.9). Hence, the interaction of MeCP2 in vitro with the nucleosome or nucleosome array substrate in the absence of competitor DNA can be largely electrostatically driven (20). The extent to which this mimics physiological conditions and the prevalence of the ionic interactions over those of the MBD domain within the cell is not well known. The in vitro studies of MeCP2 and reconstituted nucleosome complexes have provided evidence for the existence of several specific sites of interaction of MeCP2 with the nucleosome (18,63) in addition to its MBD. Little is known about the role these sites play in the interaction of MeCP2 with the native chromatin template in vivo.
To address some of these issues in the in vivo setting, we have taken advantage of the Mecp2tm1.1Jae RTT mouse model, as shown in Figure 1 and 2, which expresses ΔMeCP2 in all tissues examined, albeit at a lower amount than MeCP2 in wt mice (∼50% between cytoplasm and nuclei). The data obtained with this model nicely complement previous experiments directed to understand the role of the MBD in MeCP2–chromatin binding in vitro, based on transfection of plasmids encoding wt and truncated MeCP2 constructs (3,55,62). The cause of reduced levels of ΔMeCP2 [Figure 1 B and (29)] in the Mecp2tm1.1Jae mouse model is not obvious, however. Although the data shown in Figure 1C could suggest a decrease in Mecp2 expression or mRNA instability, the statistical significance for the observed difference is not strong. A lower level of expression would be in agreement with the lower presence of ΔMeCP2 RNA detected by northern blot analysis (29). However, the high variability of the data (Figure 1C) could also be in agreement with a previous study that found no difference in mRNA abundance using the same technique in another characterization of the same mouse model (30). A potential alternative and/or complementary explanation could be a higher cellular turnover of ΔMeCP2 that binds transiently to the chromatin template (Figure 6B) and partitions itself between the cytoplasmic and nuclear compartments differently than native MeCP2 in wt mouse (Figure 3) and in transfected cells (Figure 4). In this regard, it is interesting to note that removal of exon 3 in ΔMeCP2 results in the loss of one of the PEST sequences (4) of the protein but does not affect the NLS (Figure 1A). PEST sequences are hydrophilic, at least 12 amino acids in length, and typically signal the protein containing them for rapid proteolytic degradation by the 26S ubiquitin proteasome system after ubiquitination at lysine residues within the PEST sequence (64,65). Notably, phosphorylation within PEST motifs precedes ubiquitination of proteins (66); one of the best characterized sites of MeCP2 phosphorylation (S80 in MeCP2 E2; S97 in MeCP2 E1) (67), as well as one of the recently identified ubiquitination sites (K82/K99) (58), both fall within one of these regions. Therefore, it is reasonable to assume that the decrease in expression and the unusual cytoplasmic/nuclear partitioning ratio are just a reflection of an enhanced or altered protein turnover.
We observed a more diffuse distribution of ΔMeCP2 throughout the nucleus of the Mecp2tm1.1Jae mouse model (Figure 3 and 4), which coincides with a reduction in the nuclear size, both in the comparison of transfected N2A cells or in the native brain, without a change in the histone H1 content (Figure 4F). Such heterogeneous distribution of ΔMeCP2, when compared with the wt, is in good agreement with the results of Nan et al. (3), who used an MTaseS cell line that contains only ∼5% of the levels of methyl cytosine to show that the location of MeCP2 depends on DNA methylation.
It is possible also that the lower levels of ΔMeCP2 exhibited by the Mecp2tm1.1Jae mouse model are indirectly contributing to this model’s phenotype as opposed to ΔMeCP2 itself. In other words, a decrease in the nuclear volume could result in a concentration increase of all the nuclear proteins relative to wt, which could lead to all kinds of biological effects currently unpredictable from our data.
Histone H1 and MeCP2 have both been shown to bind cooperatively to DNA and to methylated DNA, respectively (18,68). The binding constant of MeCP2 to methylated DNA is ∼40 × 10−9 M (18,69). This is much lower than that corresponding to the binding of the SPKK motif characteristic of the C-terminal motif of some H1 histones (4 × 10−9 M) (70), or that determined for the binding of H1 to four-way junction DNA constructs (16 × 10−9 M) (71). The binding affinity of MeCP2 for methylated DNA is also lower than that of histone H1 to the nucleosome (10 × 10−9 M) (72). Consequently, the mechanisms involved in the competition of MeCP2 with histone H1 for binding to the methylated region of the genome are ambiguous (21). Interestingly, the amount of histone H1 remains the same in the brains of Mecp2tm1.1Jae mice (Figure 4F), suggesting that the reduced levels of ΔMeCP2 relative to wt do not result in a compensatory increase by histone H1, as compared with previous data from embryonic stem cells lacking MeCP2 (56). Furthermore, an increase in H1 should have resulted in a more compact chromatin organization that was more resilient to nuclease accessibility, in contrast to what we observe (Figure 6A). Moreover, this also indicates that the unusually large nuclear volume size of wt neurons where MeCP2 is present in large amounts (Figure 4D) is not the result of a histone H1 deficiency but of the inherent presence of native MeCP2.
Ultimately, any interpretation of the interactions of MeCP2 with chromatin needs to take into consideration the difference in the binding affinity of MBD proteins, including MeCP2, for specific methylated DNA compared with generic non-methylated DNA binding. The difference exhibits a decrease from the nanomolar to the micromolar range (73) (∼10-fold for MeCP2), which is likely exacerbated by the presence of chromatin and is far beyond the binding affinity of linker histones. Therefore, in addition to the lacking the ability to bind to methylated DNA, any binding of ΔMeCP2 to chromatin will be secondary to H1 binding and will have to rely on other chromatin-binding sites of the molecule.
The in vitro characterization of the interaction of MeCP2 with nucleosomal templates has shown that, in addition to the MBD, MeCP2 contains a methylation-independent chromatin-binding site at its C-terminal end [residues 196–355 (312–501 in wt)]. The weakest interaction of MeCP2 in the absence of a functional MBD observed by us (Figure 6B) and others (62) indicates that within the cell, MeCP2 binds primarily to methylated DNA through its MBD, and it is in turn further stabilized by the interaction of its C-terminal end with the non-methylated DNA. This occurs in the opposite order to an earlier proposal based on their in vitro analysis (18). The presence of binary chromatin-binding sites within the same terminal region of MeCP2, such as the 195–271 (311–387 in wt) and 272–354 (388–470 in wt) (18) or the 63–102 (179–218 in wt), DNA-binding domain of the intervening domain (ID) (63) may be responsible for the ‘resistant’ nucleosome dimers observed on nuclease digestion of brain chromatin (Figure 5E and 6A). Such structures may correspond to the existence of MeCP2 molecules sandwiched between two adjacent nucleosomes through interactions with these regions (63).
The MBD of MeCP2 thus seems to confer high specificity and affinity to chromatin binding within the cell. It is important to point out here that the removal of exon 3 of MeCP2 in the Mecp2tm1.1Jae model in addition to an important part of MBD also removes 114 amino acids adjacent to this domain in the N-terminal region (Figure 1A). It has been shown that this region structurally synergises with the MBD to increase its binding to DNA and to chromatin (63). The coupling between these two regions most likely allows for both recognition of the four-way junction-like DNA organization at the entry and exit site of the DNA in the nucleosome, as well as for DNA methylation sites, which preferentially occur in linker regions of chromatin (59). It is possible that in cell types such as neurons where MeCP2 is highly abundant (20,22) other methylation-independent chromatin interactions such as those observed here for ΔMeCP2 also take place. These are likely to involve the C-terminal end of MeCP2, and their more labile interaction with chromatin could account for the apparently unbound MeCP2 that appears on the SI and SE fractions that are quickly released on micrococcal nuclease digestion of wt brain nuclei (4). Overall, the results presented here clearly demonstrate that in the in vivo setting, the DNA methylation-independent interactions of MeCP2 with chromatin are transient, weak and secondary in nature.
FUNDING
Canadian Institutes of Health Research (CIHR) [MOP-97878 to J.A.]; International Rett Syndrome Foundation (IRSF) [2186 to K.D.]. Funding for open access charge: CIHR [MOP-97878].
Conflict of interest statement. None declared.
REFERENCES
- 1.Hendrich B, Bird A. Identification and characterization of a family of mammalian methyl-CpG binding proteins. Mol. Cell. Biol. 1998;18:6538–6547. doi: 10.1128/mcb.18.11.6538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hansen JC, Ghosh RP, Woodcock CL. Binding of the Rett syndrome protein, MeCP2, to methylated and unmethylated DNA and chromatin. IUBMB Life. 2010;62:732–738. doi: 10.1002/iub.386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nan X, Tate P, Li E, Bird A. DNA methylation specifies chromosomal localization of MeCP2. Mol. Cell. Biol. 1996;16:414–421. doi: 10.1128/mcb.16.1.414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thambirajah AA, Eubanks JH, Ausio J. MeCP2 post-translational regulation through PEST domains: two novel hypotheses: potential relevance and implications for Rett syndrome. Bioessays. 2009;31:561–569. doi: 10.1002/bies.200800220. [DOI] [PubMed] [Google Scholar]
- 5.Adams VH, McBryant SJ, Wade PA, Woodcock CL, Hansen JC. Intrinsic disorder and autonomous domain function in the multifunctional nuclear protein, MeCP2. J. Biol. Chem. 2007;282:15057–15064. doi: 10.1074/jbc.M700855200. [DOI] [PubMed] [Google Scholar]
- 6.Hite KC, Kalashnikova AA, Hansen JC. Coil-to-helix transitions in intrinsically disordered methyl CpG binding protein 2 and its isolated domains. Protein Sci. 2009;21:531–538. doi: 10.1002/pro.2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dunker AK, Lawson JD, Brown CJ, Williams RM, Romero P, Oh JS, Oldfield CJ, Campen AM, Ratliff CM, Hipps KW, et al. Intrinsically disordered protein. J. Mol. Graph Model. 2001;19:26–59. doi: 10.1016/s1093-3263(00)00138-8. [DOI] [PubMed] [Google Scholar]
- 8.Guy J, Cheval H, Selfridge J, Bird A. The role of MeCP2 in the brain. Annu. Rev. Cell Dev. Biol. 2011;27:631–652. doi: 10.1146/annurev-cellbio-092910-154121. [DOI] [PubMed] [Google Scholar]
- 9.Jeffery L, Nakielny S. Components of the DNA methylation system of chromatin control are RNA-binding proteins. J. Biol. Chem. 2004;279:49479–49487. doi: 10.1074/jbc.M409070200. [DOI] [PubMed] [Google Scholar]
- 10.Zachariah RM, Rastegar M. Linking epigenetics to human disease and Rett syndrome: the emerging novel and challenging concepts in MeCP2 research. Neural Plast. 2012;2012 doi: 10.1155/2012/415825. 415825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lewis JD, Meehan RR, Henzel WJ, Maurer-Fogy I, Jeppesen P, Klein F, Bird A. Purification, sequence, and cellular localization of a novel chromosomal protein that binds to methylated DNA. Cell. 1992;69:905–914. doi: 10.1016/0092-8674(92)90610-o. [DOI] [PubMed] [Google Scholar]
- 12.Mnatzakanian GN, Lohi H, Munteanu I, Alfred SE, Yamada T, MacLeod PJ, Jones JR, Scherer SW, Schanen NC, Friez MJ, et al. A previously unidentified MECP2 open reading frame defines a new protein isoform relevant to Rett syndrome. Nat. Genet. 2004;36:339–341. doi: 10.1038/ng1327. [DOI] [PubMed] [Google Scholar]
- 13.Kriaucionis S, Bird A. The major form of MeCP2 has a novel N-terminus generated by alternative splicing. Nucleic Acids Res. 2004;32:1818–1823. doi: 10.1093/nar/gkh349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chandler SP, Guschin D, Landsberger N, Wolffe AP. The methyl-CpG binding transcriptional repressor MeCP2 stably associates with nucleosomal DNA. Biochemistry. 1999;38:7008–7018. doi: 10.1021/bi990224y. [DOI] [PubMed] [Google Scholar]
- 15.Nan X, Campoy FJ, Bird A. MeCP2 is a transcriptional repressor with abundant binding sites in genomic chromatin. Cell. 1997;88:471–481. doi: 10.1016/s0092-8674(00)81887-5. [DOI] [PubMed] [Google Scholar]
- 16.Georgel PT, Horowitz-Scherer RA, Adkins N, Woodcock CL, Wade PA, Hansen JC. Chromatin compaction by human MeCP2. Assembly of novel secondary chromatin structures in the absence of DNA methylation. J. Biol. Chem. 2003;278:32181–32188. doi: 10.1074/jbc.M305308200. [DOI] [PubMed] [Google Scholar]
- 17.Nikitina T, Ghosh RP, Horowitz-Scherer RA, Hansen JC, Grigoryev SA, Woodcock CL. MeCP2-chromatin interactions include the formation of chromatosome-like structures and are altered in mutations causing Rett syndrome. J. Biol. Chem. 2007;282:28237–28245. doi: 10.1074/jbc.M704304200. [DOI] [PubMed] [Google Scholar]
- 18.Nikitina T, Shi X, Ghosh RP, Horowitz-Scherer RA, Hansen JC, Woodcock CL. Multiple modes of interaction between the methylated DNA binding protein MeCP2 and chromatin. Mol. Cell. Biol. 2007;27:864–877. doi: 10.1128/MCB.01593-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang C, van der Woerd MJ, Muthurajan UM, Hansen JC, Luger K. Biophysical analysis and small-angle X-ray scattering-derived structures of MeCP2-nucleosome complexes. Nucleic Acids Res. 2011;39:4122–4135. doi: 10.1093/nar/gkr005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ishibashi T, Thambirajah AA, Ausio J. MeCP2 preferentially binds to methylated linker DNA in the absence of the terminal tail of histone H3 and independently of histone acetylation. FEBS Lett. 2008;582:1157–1162. doi: 10.1016/j.febslet.2008.03.005. [DOI] [PubMed] [Google Scholar]
- 21.Ghosh RP, Horowitz-Scherer RA, Nikitina T, Shlyakhtenko LS, Woodcock CL. MeCP2 binds cooperatively to its substrate and competes with histone H1 for chromatin binding sites. Mol. Cell. Biol. 2010;30:4656–4670. doi: 10.1128/MCB.00379-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thambirajah AA, Ng MK, Frehlick LJ, Li A, Serpa JJ, Petrotchenko EV, Silva-Moreno B, Missiaen KK, Borchers CH, Adam Hall J, et al. MeCP2 binds to nucleosome free (linker DNA) regions and to H3K9/H3K27 methylated nucleosomes in the brain. Nucleic Acids Res. 2011;40:2884–2897. doi: 10.1093/nar/gkr1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meehan RR, Lewis JD, Bird AP. Characterization of MeCP2, a vertebrate DNA binding protein with affinity for methylated DNA. Nucleic Acids Res. 1992;20:5085–5092. doi: 10.1093/nar/20.19.5085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Skene PJ, Illingworth RS, Webb S, Kerr AR, James KD, Turner DJ, Andrews R, Bird AP. Neuronal MeCP2 is expressed at near histone-octamer levels and globally alters the chromatin state. Mol. Cell. 2010;37:457–468. doi: 10.1016/j.molcel.2010.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cohen S, Gabel HW, Hemberg M, Hutchinson AN, Sadacca LA, Ebert DH, Harmin DA, Greenberg RS, Verdine VK, Zhou Z, et al. Genome-wide activity-dependent MeCP2 phosphorylation regulates nervous system development and function. Neuron. 2011;72:72–85. doi: 10.1016/j.neuron.2011.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat. Genet. 1999;23:185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- 27.Bienvenu T, Chelly J. Molecular genetics of Rett syndrome: when DNA methylation goes unrecognized. Nat. Rev. Genet. 2006;7:415–426. doi: 10.1038/nrg1878. [DOI] [PubMed] [Google Scholar]
- 28.Guy J, Hendrich B, Holmes M, Martin JE, Bird A. A mouse Mecp2-null mutation causes neurological symptoms that mimic Rett syndrome. Nat. Genet. 2001;27:322–326. doi: 10.1038/85899. [DOI] [PubMed] [Google Scholar]
- 29.Chen RZ, Akbarian S, Tudor M, Jaenisch R. Deficiency of methyl-CpG binding protein-2 in CNS neurons results in a Rett-like phenotype in mice. Nat. Genet. 2001;27:327–331. doi: 10.1038/85906. [DOI] [PubMed] [Google Scholar]
- 30.Jordan C, Li HH, Kwan HC, Francke U. Cerebellar gene expression profiles of mouse models for Rett syndrome reveal novel MeCP2 targets. BMC Med. Genet. 2007;8:36. doi: 10.1186/1471-2350-8-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Luikenhuis S, Giacometti E, Beard CF, Jaenisch R. Expression of MeCP2 in postmitotic neurons rescues Rett syndrome in mice. Proc. Natl Acad. Sci. USA. 2004;101:6033–6038. doi: 10.1073/pnas.0401626101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Belichenko NP, Belichenko PV, Li HH, Mobley WC, Francke U. Comparative study of brain morphology in Mecp2 mutant mouse models of Rett syndrome. J. Comp. Neurol. 2008;508:184–195. doi: 10.1002/cne.21673. [DOI] [PubMed] [Google Scholar]
- 33.Belichenko PV, Wright EE, Belichenko NP, Masliah E, Li HH, Mobley WC, Francke U. Widespread changes in dendritic and axonal morphology in Mecp2-mutant mouse models of Rett syndrome: evidence for disruption of neuronal networks. J. Comp. Neurol. 2009;514:240–258. doi: 10.1002/cne.22009. [DOI] [PubMed] [Google Scholar]
- 34.Feng G, Mellor RH, Bernstein M, Keller-Peck C, Nguyen QT, Wallace M, Nerbonne JM, Lichtman JW, Sanes JR. Imaging neuronal subsets in transgenic mice expressing multiple spectral variants of GFP. Neuron. 2000;28:41–51. doi: 10.1016/s0896-6273(00)00084-2. [DOI] [PubMed] [Google Scholar]
- 35.Stuss DP, Boyd JD, Levin DB, Delaney KR. MeCP2 mutation results in compartment-specific reductions in dendritic branching and spine density in layer 5 motor cortical neurons of YFP-H mice. PLoS One. 2012;7:e31896. doi: 10.1371/journal.pone.0031896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 37.Ausió J. Presence of a highly specific histone H1-like protein in the chromatin of the sperm of the bivalve mollusks. Mol. Cell. Biochem. 1992;115:163–172. doi: 10.1007/BF00230327. [DOI] [PubMed] [Google Scholar]
- 38.Abbott DW, Ivanova VS, Wang X, Bonner WM, Ausió J. Characterization of the stability and folding of H2A.Z chromatin particles: implications for transcriptional activation. J. Biol. Chem. 2001;276:41945–41949. doi: 10.1074/jbc.M108217200. [DOI] [PubMed] [Google Scholar]
- 39.Yager TD, van Holde KE. Dynamics and equilibria of nucleosomes at elevated ionic strength. J. Biol. Chem. 1984;259:4212–4222. [PubMed] [Google Scholar]
- 40.Abbott DW, Chadwick BP, Thambirajah AA, Ausio J. Beyond the Xi: macroH2A chromatin distribution and post-translational modification in an avian system. J. Biol. Chem. 2005;280:16437–16445. doi: 10.1074/jbc.M500170200. [DOI] [PubMed] [Google Scholar]
- 41.Thambirajah AA, Dryhurst D, Ishibashi T, Li A, Maffey AH, Ausio J. H2A.Z stabilizes chromatin in a way that is dependent on core histone acetylation. J. Biol. Chem. 2006;281:20036–20044. doi: 10.1074/jbc.M601975200. [DOI] [PubMed] [Google Scholar]
- 42.Dryhurst D, Ishibashi T, Rose KL, Eirin-Lopez JM, McDonald D, Silva-Moreno B, Veldhoen N, Helbing CC, Hendzel MJ, Shabanowitz J, et al. Characterization of the histone H2A.Z-1 and H2A.Z-2 isoforms in vertebrates. BMC Biol. 2009;7:86. doi: 10.1186/1741-7007-7-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Carrero G, McDonald D, Crawford E, de_Vries G, Hendzel MJ. Using FRAP and mathematical modeling to determine the in vivo kinetics of nuclear proteins. Methods. 2003;29:14–28. doi: 10.1016/s1046-2023(02)00288-8. [DOI] [PubMed] [Google Scholar]
- 44.Kwon MJ, Oh E, Lee S, Roh MR, Kim SE, Lee Y, Choi YL, In YH, Park T, Koh SS, et al. Identification of novel reference genes using multiplatform expression data and their validation for quantitative gene expression analysis. PloS One. 2009;4:e6162. doi: 10.1371/journal.pone.0006162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2[-Delta Delta C(T)] Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 46.Walzak AA, Veldhoen N, Feng X, Riabowol K, Helbing CC. Expression profiles of mRNA transcript variants encoding the human inhibitor of growth tumor suppressor gene family in normal and neoplastic tissues. Exp. Cell Res. 2008;314:273–285. doi: 10.1016/j.yexcr.2007.07.029. [DOI] [PubMed] [Google Scholar]
- 47.Wong ML, Medrano JF. Real-time PCR for mRNA quantitation. Biotechniques. 2005;39:75–85. doi: 10.2144/05391RV01. [DOI] [PubMed] [Google Scholar]
- 48.Shahbazian MD, Antalffy B, Armstrong DL, Zoghbi HY. Insight into Rett syndrome: MeCP2 levels display tissue- and cell-specific differences and correlate with neuronal maturation. Hum. Mol. Genet. 2002;11:115–124. doi: 10.1093/hmg/11.2.115. [DOI] [PubMed] [Google Scholar]
- 49.Jung BP, Jugloff DG, Zhang G, Logan R, Brown S, Eubanks JH. The expression of methyl CpG binding factor MeCP2 correlates with cellular differentiation in the developing rat brain and in cultured cells. J. Neurobiol. 2003;55:86–96. doi: 10.1002/neu.10201. [DOI] [PubMed] [Google Scholar]
- 50.Zachariah RM, Olson CO, Ezeonwuka C, Rastegar M. Novel MeCP2 isoform-specific antibody reveals the endogenous MeCP2E1 expression in murine brain, primary neurons and astrocytes. PLoS One. 2012;7:e49763. doi: 10.1371/journal.pone.0049763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lajtha A, Latzkovits L, Toth J. Comparison of turnover rates of proteins of the brain, liver and kidney in mouse in vivo following long term labeling. Biochim. Biophys. Acta. 1976;425:511–520. doi: 10.1016/0005-2787(76)90015-0. [DOI] [PubMed] [Google Scholar]
- 52.Johnson HA, Baldwin RL, Klasing KC, France J, Calvert CC. A rodent model of protein turnover used to design an experiment for measuring the rates of channeling, recycling and protein synthesis. J. Nutr. 2000;130:3097–3102. doi: 10.1093/jn/130.12.3097. [DOI] [PubMed] [Google Scholar]
- 53.Berod A, Hartman BK, Pujol JF. Importance of fixation in immunohistochemistry: use of formaldehyde solutions at variable pH for the localization of tyrosine hydroxylase. J. Histochem. Cytochem. 1981;29:844–850. doi: 10.1177/29.7.6167611. [DOI] [PubMed] [Google Scholar]
- 54.Kishi N, Macklis JD. MECP2 is progressively expressed in post-migratory neurons and is involved in neuronal maturation rather than cell fate decisions. Mol. Cell. Neurosci. 2004;27:306–321. doi: 10.1016/j.mcn.2004.07.006. [DOI] [PubMed] [Google Scholar]
- 55.Matsumura S, Persson LM, Wong L, Wilson AC. The latency-associated nuclear antigen interacts with MeCP2 and nucleosomes through separate domains. J. Virol. 2010;84:2318–2330. doi: 10.1128/JVI.01097-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yazdani M, Deogracias R, Guy J, Poot RA, Bird A, Barde YA. Disease modeling using embryonic stem cells: MeCP2 regulates nuclear size and rna synthesis in neurons. Stem Cells. 2012;30:2128–2139. doi: 10.1002/stem.1180. [DOI] [PubMed] [Google Scholar]
- 57.Singleton MK, Gonzales ML, Leung KN, Yasui DH, Schroeder DI, Dunaway K, LaSalle JM. MeCP2 is required for global heterochromatic and nucleolar changes during activity-dependent neuronal maturation. Neurobiol. Dis. 2011;43:190–200. doi: 10.1016/j.nbd.2011.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gonzales ML, Adams S, Dunaway KW, LaSalle JM. Phosphorylation of distinct sites in MeCP2 modifies cofactor associations and the dynamics of transcriptional regulation. Mol. Cell. Biol. 2012;32:2894–2903. doi: 10.1128/MCB.06728-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Felle M, Hoffmeister H, Rothammer J, Fuchs A, Exler JH, Langst G. Nucleosomes protect DNA from DNA methylation in vivo and in vitro. Nucleic Acids Res. 2011;30:2128–2139. doi: 10.1093/nar/gkr263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Galvao TC, Thomas JO. Structure-specific binding of MeCP2 to four-way junction DNA through its methyl CpG-binding domain. Nucleic Acids Res. 2005;33:6603–6609. doi: 10.1093/nar/gki971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Varga-Weisz P, van Holde K, Zlatanova J. Preferential binding of histone H1 to four-way helical junction DNA. J. Biol. Chem. 1993;268:20699–20700. [PubMed] [Google Scholar]
- 62.Kumar A, Kamboj S, Malone BM, Kudo S, Twiss JL, Czymmek KJ, LaSalle JM, Schanen NC. Analysis of protein domains and Rett syndrome mutations indicate that multiple regions influence chromatin-binding dynamics of the chromatin-associated protein MECP2 in vivo. J. Cell Sci. 2008;121:1128–1137. doi: 10.1242/jcs.016865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ghosh RP, Nikitina T, Horowitz-Scherer RA, Gierasch LM, Uversky VN, Hite K, Hansen JC, Woodcock CL. Unique physical properties and interactions of the domains of methylated DNA binding protein 2. Biochemistry. 2010;49:4395–4410. doi: 10.1021/bi9019753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rechsteiner M, Rogers SW. PEST sequences and regulation by proteolysis. Trends Biochem. Sci. 1996;21:267–271. [PubMed] [Google Scholar]
- 65.Rogers S, Wells R, Rechsteiner M. Amino acid sequences common to rapidly degraded proteins: the PEST hypothesis. Science. 1986;234:364–368. doi: 10.1126/science.2876518. [DOI] [PubMed] [Google Scholar]
- 66.Spencer ML, Theodosiou M, Noonan DJ. NPDC-1, a novel regulator of neuronal proliferation, is degraded by the ubiquitin/proteasome system through a PEST degradation motif. J. Biol. Chem. 2004;279:37069–37078. doi: 10.1074/jbc.M402507200. [DOI] [PubMed] [Google Scholar]
- 67.Tao J, Hu K, Chang Q, Wu H, Sherman NE, Martinowich K, Klose RJ, Schanen C, Jaenisch R, Wang W, et al. Phosphorylation of MeCP2 at Serine 80 regulates its chromatin association and neurological function. Proc. Natl Acad. Sci. USA. 2009;106:4882–4887. doi: 10.1073/pnas.0811648106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Watanabe F. Cooperative interaction of histone H1 with DNA. Nucleic Acids Res. 1986;14:3573–3585. doi: 10.1093/nar/14.8.3573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ballestar E, Yusufzai TM, Wolffe AP. Effects of Rett syndrome mutations of the methyl-CpG binding domain of the transcriptional repressor MeCP2 on selectivity for association with methylated DNA. Biochemistry. 2000;39:7100–7106. doi: 10.1021/bi0001271. [DOI] [PubMed] [Google Scholar]
- 70.Suzuki M. SPKK, a new nucleic acid-binding unit of protein found in histone. EMBO J. 1989;8:797–804. doi: 10.1002/j.1460-2075.1989.tb03440.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hill DA, Reeves R. Competition between HMG-I(Y), HMG-1 and histone H1 on four-way junction DNA. Nucleic Acids Res. 1997;25:3523–3531. doi: 10.1093/nar/25.17.3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Caterino TL, Fang H, Hayes JJ. Nucleosome linker DNA contacts and induces specific folding of the intrinsically disordered H1 carboxyl-terminal domain. Mol. Cell. Biol. 2011;31:2341–2348. doi: 10.1128/MCB.05145-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fraga MF, Ballestar E, Montoya G, Taysavang P, Wade PA, Esteller M. The affinity of different MBD proteins for a specific methylated locus depends on their intrinsic binding properties. Nucleic Acids Res. 2003;31:1765–1774. doi: 10.1093/nar/gkg249. [DOI] [PMC free article] [PubMed] [Google Scholar]