Abstract
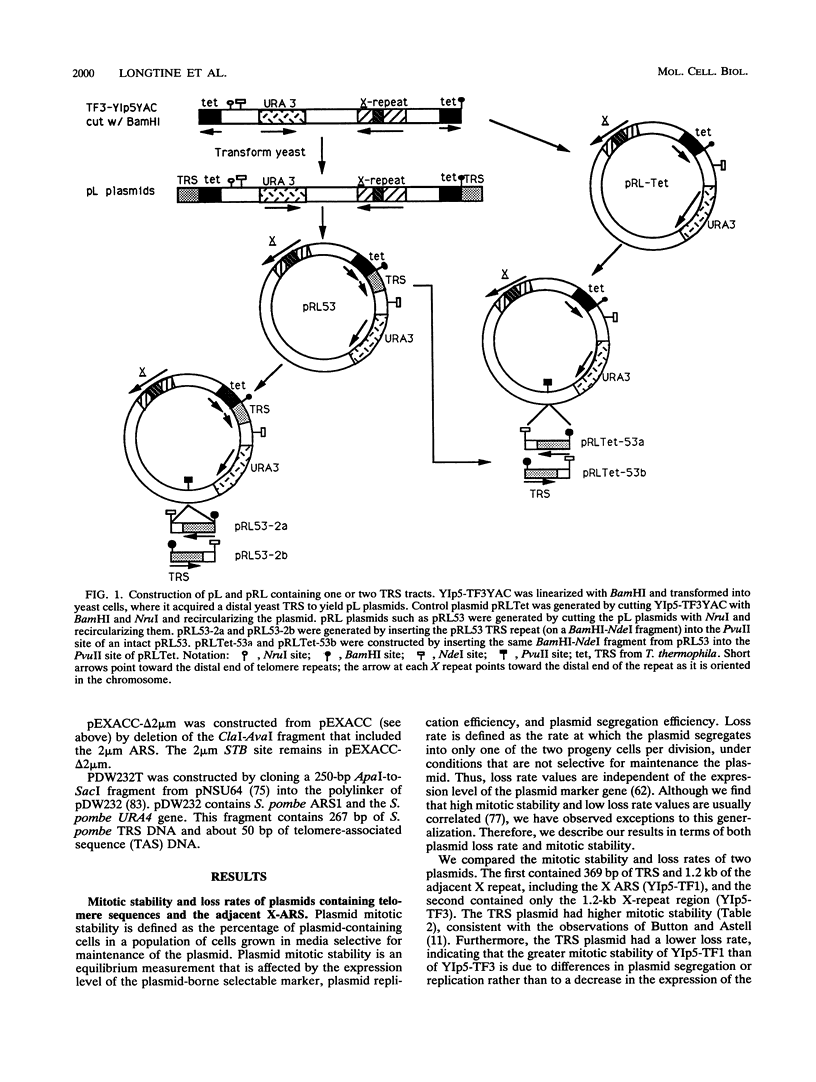
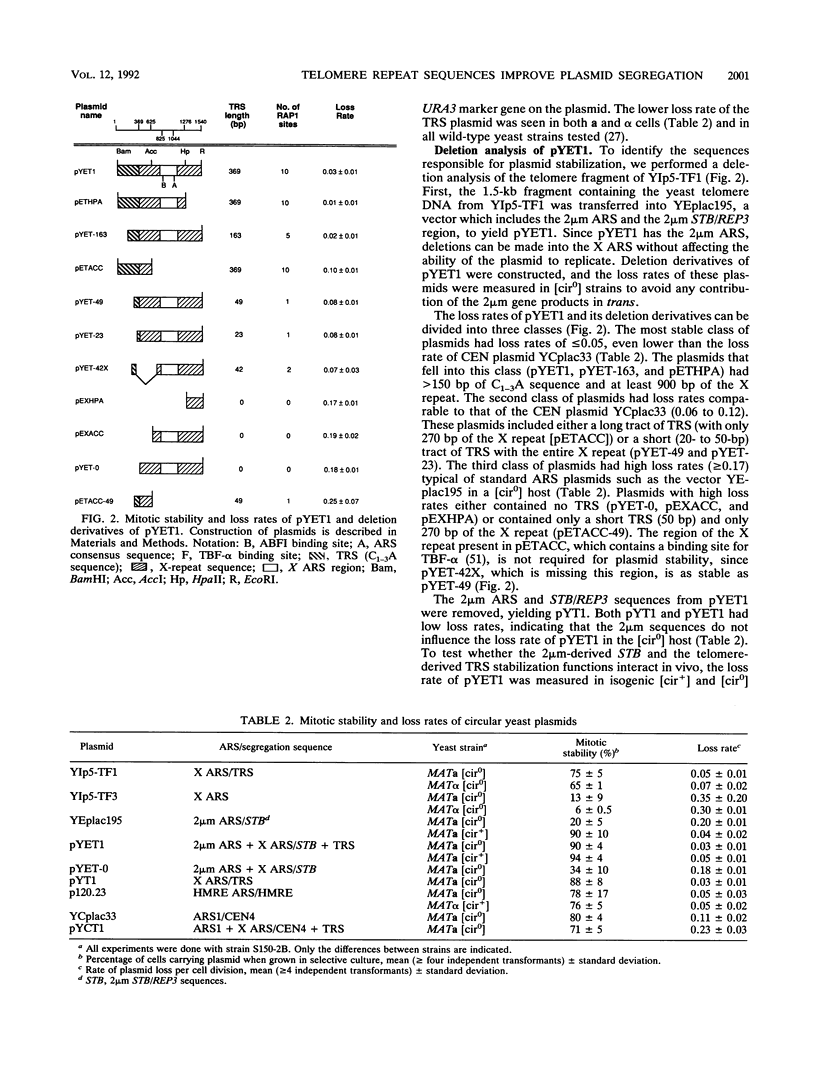
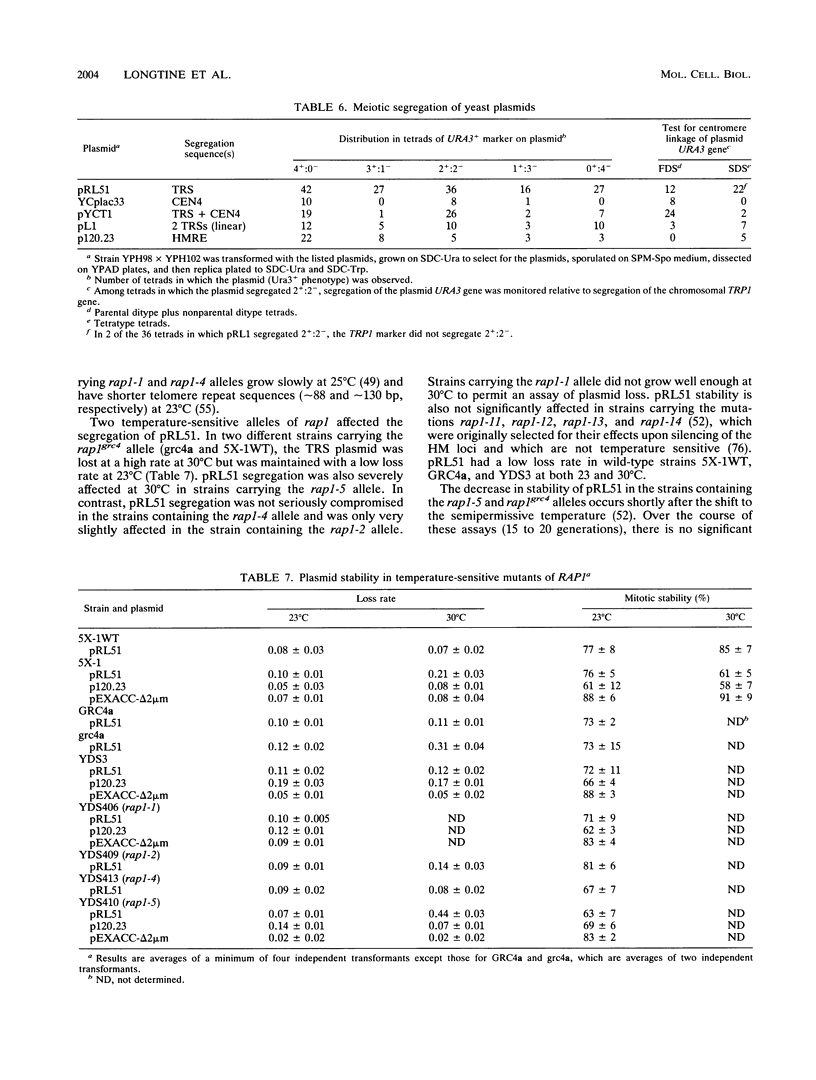
Telomere repeat sequences (TRSs) can dramatically improve the segregation of unstable circular autonomously replicating sequence (ARS) plasmids in Saccharomyces cerevisiae. Deletion analysis demonstrated that yeast TRSs, which conform to the general sequence (C(1-3)A)n, are able to stabilize circular ARS plasmids. A number of TRS clones of different primary sequence and C(1-3)A tract length confer the plasmid stabilization phenotype. TRS sequences do not appear to improve plasmid replication efficiency, as determined by plasmid copy number analysis and functional assays for ARS activity. Pedigree analysis confirms that TRS-containing plasmids are missegregated at low frequency and that missegregated TRS-containing plasmids, like ARS plasmids, are preferentially retained by the mother cell. Plasmids stabilized by TRSs have properties that distinguish them from centromere-containing plasmids and 2 microns-based recombinant plasmids. Linear ARS plasmids, which include two TRS tracts at their termini, segregate inefficiently, while circular plasmids with one or two TRS tracts segregate efficiently, suggesting that plasmid topology or TRS accessibility interferes with TRS segregation function on linear plasmids. In strains carrying the temperature-sensitive mutant alleles rap1grc4 and rap1-5, TRS plasmids are not stable at the semipermissive temperature, suggesting that RAP1 protein is involved in TRS plasmid stability. In Schizosaccharomyces pombe, an ARS plasmid was stabilized by the addition of S. pombe telomere sequence, suggesting that the ability to improve the segregation of ARS plasmids is a general property of telomere repeats.
Full text
PDF









Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Amati B. B., Gasser S. M. Chromosomal ARS and CEN elements bind specifically to the yeast nuclear scaffold. Cell. 1988 Sep 23;54(7):967–978. doi: 10.1016/0092-8674(88)90111-0. [DOI] [PubMed] [Google Scholar]
- Baker R. E., Masison D. C. Isolation of the gene encoding the Saccharomyces cerevisiae centromere-binding protein CP1. Mol Cell Biol. 1990 Jun;10(6):2458–2467. doi: 10.1128/mcb.10.6.2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr P. J. Mammalian subtilisins: the long-sought dibasic processing endoproteases. Cell. 1991 Jul 12;66(1):1–3. doi: 10.1016/0092-8674(91)90129-m. [DOI] [PubMed] [Google Scholar]
- Beach D., Rodgers L., Gould J. ran1+ controls the transition from mitotic division to meiosis in fission yeast. Curr Genet. 1985;10(4):297–311. doi: 10.1007/BF00365626. [DOI] [PubMed] [Google Scholar]
- Berman J., Tachibana C. Y., Tye B. K. Identification of a telomere-binding activity from yeast. Proc Natl Acad Sci U S A. 1986 Jun;83(11):3713–3717. doi: 10.1073/pnas.83.11.3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bram R. J., Kornberg R. D. Isolation of a Saccharomyces cerevisiae centromere DNA-binding protein, its human homolog, and its possible role as a transcription factor. Mol Cell Biol. 1987 Jan;7(1):403–409. doi: 10.1128/mcb.7.1.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchman A. R., Lue N. F., Kornberg R. D. Connections between transcriptional activators, silencers, and telomeres as revealed by functional analysis of a yeast DNA-binding protein. Mol Cell Biol. 1988 Dec;8(12):5086–5099. doi: 10.1128/mcb.8.12.5086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke D. T., Carle G. F., Olson M. V. Cloning of large segments of exogenous DNA into yeast by means of artificial chromosome vectors. Science. 1987 May 15;236(4803):806–812. doi: 10.1126/science.3033825. [DOI] [PubMed] [Google Scholar]
- Button L. L., Astell C. R. The Saccharomyces cerevisiae chromosome III left telomere has a type X, but not a type Y', ARS region. Mol Cell Biol. 1986 Apr;6(4):1352–1356. doi: 10.1128/mcb.6.4.1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai M. J., Davis R. W. Purification of a yeast centromere-binding protein that is able to distinguish single base-pair mutations in its recognition site. Mol Cell Biol. 1989 Jun;9(6):2544–2550. doi: 10.1128/mcb.9.6.2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carson M. J., Hartwell L. CDC17: an essential gene that prevents telomere elongation in yeast. Cell. 1985 Aug;42(1):249–257. doi: 10.1016/s0092-8674(85)80120-3. [DOI] [PubMed] [Google Scholar]
- Casadaban M. J., Cohen S. N. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J Mol Biol. 1980 Apr;138(2):179–207. doi: 10.1016/0022-2836(80)90283-1. [DOI] [PubMed] [Google Scholar]
- Chan C. S., Tye B. K. A family of Saccharomyces cerevisiae repetitive autonomously replicating sequences that have very similar genomic environments. J Mol Biol. 1983 Aug 15;168(3):505–523. doi: 10.1016/s0022-2836(83)80299-x. [DOI] [PubMed] [Google Scholar]
- Chung H. M., Shea C., Fields S., Taub R. N., Van der Ploeg L. H., Tse D. B. Architectural organization in the interphase nucleus of the protozoan Trypanosoma brucei: location of telomeres and mini-chromosomes. EMBO J. 1990 Aug;9(8):2611–2619. doi: 10.1002/j.1460-2075.1990.tb07443.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke L., Carbon J. Isolation of a yeast centromere and construction of functional small circular chromosomes. Nature. 1980 Oct 9;287(5782):504–509. doi: 10.1038/287504a0. [DOI] [PubMed] [Google Scholar]
- Cockerill P. N., Garrard W. T. Chromosomal loop anchorage of the kappa immunoglobulin gene occurs next to the enhancer in a region containing topoisomerase II sites. Cell. 1986 Jan 31;44(2):273–282. doi: 10.1016/0092-8674(86)90761-0. [DOI] [PubMed] [Google Scholar]
- Conrad M. N., Wright J. H., Wolf A. J., Zakian V. A. RAP1 protein interacts with yeast telomeres in vivo: overproduction alters telomere structure and decreases chromosome stability. Cell. 1990 Nov 16;63(4):739–750. doi: 10.1016/0092-8674(90)90140-a. [DOI] [PubMed] [Google Scholar]
- Conrad M. N., Zakian V. A. Plasmid associations with residual nuclear structures in Saccharomyces cerevisiae. Curr Genet. 1988 Apr;13(4):291–297. doi: 10.1007/BF00424422. [DOI] [PubMed] [Google Scholar]
- Cumberledge S., Carbon J. Mutational analysis of meiotic and mitotic centromere function in Saccharomyces cerevisiae. Genetics. 1987 Oct;117(2):203–212. doi: 10.1093/genetics/117.2.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dani G. M., Zakian V. A. Mitotic and meiotic stability of linear plasmids in yeast. Proc Natl Acad Sci U S A. 1983 Jun;80(11):3406–3410. doi: 10.1073/pnas.80.11.3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diffley J. F., Stillman B. Purification of a yeast protein that binds to origins of DNA replication and a transcriptional silencer. Proc Natl Acad Sci U S A. 1988 Apr;85(7):2120–2124. doi: 10.1073/pnas.85.7.2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dower W. J., Miller J. F., Ragsdale C. W. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 1988 Jul 11;16(13):6127–6145. doi: 10.1093/nar/16.13.6127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francesconi S. C., Eisenberg S. The multifunctional protein OBF1 is phosphorylated at serine and threonine residues in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1991 May 15;88(10):4089–4093. doi: 10.1073/pnas.88.10.4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futcher A. B., Cox B. S. Copy number and the stability of 2-micron circle-based artificial plasmids of Saccharomyces cerevisiae. J Bacteriol. 1984 Jan;157(1):283–290. doi: 10.1128/jb.157.1.283-290.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futcher A. B. The 2 micron circle plasmid of Saccharomyces cerevisiae. Yeast. 1988 Mar;4(1):27–40. doi: 10.1002/yea.320040104. [DOI] [PubMed] [Google Scholar]
- Giesman D., Best L., Tatchell K. The role of RAP1 in the regulation of the MAT alpha locus. Mol Cell Biol. 1991 Feb;11(2):1069–1079. doi: 10.1128/mcb.11.2.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz R. D., Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988 Dec 30;74(2):527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- Gottschling D. E., Aparicio O. M., Billington B. L., Zakian V. A. Position effect at S. cerevisiae telomeres: reversible repression of Pol II transcription. Cell. 1990 Nov 16;63(4):751–762. doi: 10.1016/0092-8674(90)90141-z. [DOI] [PubMed] [Google Scholar]
- Grimm C., Kohli J., Murray J., Maundrell K. Genetic engineering of Schizosaccharomyces pombe: a system for gene disruption and replacement using the ura4 gene as a selectable marker. Mol Gen Genet. 1988 Dec;215(1):81–86. doi: 10.1007/BF00331307. [DOI] [PubMed] [Google Scholar]
- Heyer W. D., Sipiczki M., Kohli J. Replicating plasmids in Schizosaccharomyces pombe: improvement of symmetric segregation by a new genetic element. Mol Cell Biol. 1986 Jan;6(1):80–89. doi: 10.1128/mcb.6.1.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill J. E., Myers A. M., Koerner T. J., Tzagoloff A. Yeast/E. coli shuttle vectors with multiple unique restriction sites. Yeast. 1986 Sep;2(3):163–167. doi: 10.1002/yea.320020304. [DOI] [PubMed] [Google Scholar]
- Hilliker A. J., Appels R. The arrangement of interphase chromosomes: structural and functional aspects. Exp Cell Res. 1989 Dec;185(2):267–318. doi: 10.1016/0014-4827(89)90301-7. [DOI] [PubMed] [Google Scholar]
- Hofmann J. F., Gasser S. M. Identification and purification of a protein that binds the yeast ARS consensus sequence. Cell. 1991 Mar 8;64(5):951–960. doi: 10.1016/0092-8674(91)90319-t. [DOI] [PubMed] [Google Scholar]
- Hofmann J. F., Laroche T., Brand A. H., Gasser S. M. RAP-1 factor is necessary for DNA loop formation in vitro at the silent mating type locus HML. Cell. 1989 Jun 2;57(5):725–737. doi: 10.1016/0092-8674(89)90788-5. [DOI] [PubMed] [Google Scholar]
- Hsiao C. L., Carbon J. Direct selection procedure for the isolation of functional centromeric DNA. Proc Natl Acad Sci U S A. 1981 Jun;78(6):3760–3764. doi: 10.1073/pnas.78.6.3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huet J., Sentenac A. TUF, the yeast DNA-binding factor specific for UASrpg upstream activating sequences: identification of the protein and its DNA-binding domain. Proc Natl Acad Sci U S A. 1987 Jun;84(11):3648–3652. doi: 10.1073/pnas.84.11.3648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jäger D., Philippsen P. Many yeast chromosomes lack the telomere-specific Y' sequence. Mol Cell Biol. 1989 Dec;9(12):5754–5757. doi: 10.1128/mcb.9.12.5754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmerly W. J., Rine J. Replication and segregation of plasmids containing cis-acting regulatory sites of silent mating-type genes in Saccharomyces cerevisiae are controlled by the SIR genes. Mol Cell Biol. 1987 Dec;7(12):4225–4237. doi: 10.1128/mcb.7.12.4225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmerly W., Buchman A., Kornberg R., Rine J. Roles of two DNA-binding factors in replication, segregation and transcriptional repression mediated by a yeast silencer. EMBO J. 1988 Jul;7(7):2241–2253. doi: 10.1002/j.1460-2075.1988.tb03064.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshland D., Rutledge L., Fitzgerald-Hayes M., Hartwell L. H. A genetic analysis of dicentric minichromosomes in Saccharomyces cerevisiae. Cell. 1987 Mar 13;48(5):801–812. doi: 10.1016/0092-8674(87)90077-8. [DOI] [PubMed] [Google Scholar]
- Kurtz S., Shore D. RAP1 protein activates and silences transcription of mating-type genes in yeast. Genes Dev. 1991 Apr;5(4):616–628. doi: 10.1101/gad.5.4.616. [DOI] [PubMed] [Google Scholar]
- Lechner J., Carbon J. A 240 kd multisubunit protein complex, CBF3, is a major component of the budding yeast centromere. Cell. 1991 Feb 22;64(4):717–725. doi: 10.1016/0092-8674(91)90501-o. [DOI] [PubMed] [Google Scholar]
- Liu Z. P., Tye B. K. A yeast protein that binds to vertebrate telomeres and conserved yeast telomeric junctions. Genes Dev. 1991 Jan;5(1):49–59. doi: 10.1101/gad.5.1.49. [DOI] [PubMed] [Google Scholar]
- Longtine M. S., Wilson N. M., Petracek M. E., Berman J. A yeast telomere binding activity binds to two related telomere sequence motifs and is indistinguishable from RAP1. Curr Genet. 1989 Oct;16(4):225–239. doi: 10.1007/BF00422108. [DOI] [PubMed] [Google Scholar]
- Lustig A. J., Kurtz S., Shore D. Involvement of the silencer and UAS binding protein RAP1 in regulation of telomere length. Science. 1990 Oct 26;250(4980):549–553. doi: 10.1126/science.2237406. [DOI] [PubMed] [Google Scholar]
- Ma H., Kunes S., Schatz P. J., Botstein D. Plasmid construction by homologous recombination in yeast. Gene. 1987;58(2-3):201–216. doi: 10.1016/0378-1119(87)90376-3. [DOI] [PubMed] [Google Scholar]
- Mann C., Davis R. W. Instability of dicentric plasmids in yeast. Proc Natl Acad Sci U S A. 1983 Jan;80(1):228–232. doi: 10.1073/pnas.80.1.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally F. J., Rine J. A synthetic silencer mediates SIR-dependent functions in Saccharomyces cerevisiae. Mol Cell Biol. 1991 Nov;11(11):5648–5659. doi: 10.1128/mcb.11.11.5648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellor J., Jiang W., Funk M., Rathjen J., Barnes C. A., Hinz T., Hegemann J. H., Philippsen P. CPF1, a yeast protein which functions in centromeres and promoters. EMBO J. 1990 Dec;9(12):4017–4026. doi: 10.1002/j.1460-2075.1990.tb07623.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller C. A., Beaucage S. L., Cohen S. N. Role of DNA superhelicity in partitioning of the pSC101 plasmid. Cell. 1990 Jul 13;62(1):127–133. doi: 10.1016/0092-8674(90)90246-b. [DOI] [PubMed] [Google Scholar]
- Murray A. W., Szostak J. W. Construction and behavior of circularly permuted and telocentric chromosomes in Saccharomyces cerevisiae. Mol Cell Biol. 1986 Sep;6(9):3166–3172. doi: 10.1128/mcb.6.9.3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray A. W., Szostak J. W. Pedigree analysis of plasmid segregation in yeast. Cell. 1983 Oct;34(3):961–970. doi: 10.1016/0092-8674(83)90553-6. [DOI] [PubMed] [Google Scholar]
- Nordström K., Austin S. J. Mechanisms that contribute to the stable segregation of plasmids. Annu Rev Genet. 1989;23:37–69. doi: 10.1146/annurev.ge.23.120189.000345. [DOI] [PubMed] [Google Scholar]
- Panzeri L., Landonio L., Stotz A., Philippsen P. Role of conserved sequence elements in yeast centromere DNA. EMBO J. 1985 Jul;4(7):1867–1874. doi: 10.1002/j.1460-2075.1985.tb03862.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petracek M. E., Lefebvre P. A., Silflow C. D., Berman J. Chlamydomonas telomere sequences are A+T-rich but contain three consecutive G-C base pairs. Proc Natl Acad Sci U S A. 1990 Nov;87(21):8222–8226. doi: 10.1073/pnas.87.21.8222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhode P. R., Sweder K. S., Oegema K. F., Campbell J. L. The gene encoding ARS-binding factor I is essential for the viability of yeast. Genes Dev. 1989 Dec;3(12A):1926–1939. doi: 10.1101/gad.3.12a.1926. [DOI] [PubMed] [Google Scholar]
- Runge K. W., Wellinger R. J., Zakian V. A. Effects of excess centromeres and excess telomeres on chromosome loss rates. Mol Cell Biol. 1991 Jun;11(6):2919–2928. doi: 10.1128/mcb.11.6.2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runge K. W., Zakian V. A. Introduction of extra telomeric DNA sequences into Saccharomyces cerevisiae results in telomere elongation. Mol Cell Biol. 1989 Apr;9(4):1488–1497. doi: 10.1128/mcb.9.4.1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen D., Gilbert W. A sodium-potassium switch in the formation of four-stranded G4-DNA. Nature. 1990 Mar 29;344(6265):410–414. doi: 10.1038/344410a0. [DOI] [PubMed] [Google Scholar]
- Shore D., Nasmyth K. Purification and cloning of a DNA binding protein from yeast that binds to both silencer and activator elements. Cell. 1987 Dec 4;51(5):721–732. doi: 10.1016/0092-8674(87)90095-x. [DOI] [PubMed] [Google Scholar]
- Stone E. M., Swanson M. J., Romeo A. M., Hicks J. B., Sternglanz R. The SIR1 gene of Saccharomyces cerevisiae and its role as an extragenic suppressor of several mating-defective mutants. Mol Cell Biol. 1991 Apr;11(4):2253–2262. doi: 10.1128/mcb.11.4.2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussel L., Shore D. Separation of transcriptional activation and silencing functions of the RAP1-encoded repressor/activator protein 1: isolation of viable mutants affecting both silencing and telomere length. Proc Natl Acad Sci U S A. 1991 Sep 1;88(17):7749–7753. doi: 10.1073/pnas.88.17.7749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Houten J. V., Newlon C. S. Mutational analysis of the consensus sequence of a replication origin from yeast chromosome III. Mol Cell Biol. 1990 Aug;10(8):3917–3925. doi: 10.1128/mcb.10.8.3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker C. L., Cargile C. B., Floy K. M., Delannoy M., Migeon B. R. The Barr body is a looped X chromosome formed by telomere association. Proc Natl Acad Sci U S A. 1991 Jul 15;88(14):6191–6195. doi: 10.1073/pnas.88.14.6191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walmsley R. M., Petes T. D. Genetic control of chromosome length in yeast. Proc Natl Acad Sci U S A. 1985 Jan;82(2):506–510. doi: 10.1073/pnas.82.2.506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walmsley R. W., Chan C. S., Tye B. K., Petes T. D. Unusual DNA sequences associated with the ends of yeast chromosomes. Nature. 1984 Jul 12;310(5973):157–160. doi: 10.1038/310157a0. [DOI] [PubMed] [Google Scholar]
- Wang S. S., Zakian V. A. Sequencing of Saccharomyces telomeres cloned using T4 DNA polymerase reveals two domains. Mol Cell Biol. 1990 Aug;10(8):4415–4419. doi: 10.1128/mcb.10.8.4415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weilguny D., Praetorius M., Carr A., Egel R., Nielsen O. New vectors in fission yeast: application for cloning the his2 gene. Gene. 1991 Mar 1;99(1):47–54. doi: 10.1016/0378-1119(91)90032-7. [DOI] [PubMed] [Google Scholar]
- Wu L. C., Fisher P. A., Broach J. R. A yeast plasmid partitioning protein is a karyoskeletal component. J Biol Chem. 1987 Jan 15;262(2):883–891. [PubMed] [Google Scholar]
- Zakian V. A., Blanton H. M. Distribution of telomere-associated sequences on natural chromosomes in Saccharomyces cerevisiae. Mol Cell Biol. 1988 May;8(5):2257–2260. doi: 10.1128/mcb.8.5.2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakian V. A., Runge K., Wang S. S. How does the end begin? Formation and maintenance of telomeres in ciliates and yeast. Trends Genet. 1990 Jan;6(1):12–16. doi: 10.1016/0168-9525(90)90043-6. [DOI] [PubMed] [Google Scholar]


