Key Points
Foxp3 expression is increased by DNMT inhibitors and may have potential utility in efforts to develop Foxp3+ Tregs for cellular therapy.
Dnmt1 deletion impairs Treg function and results in lethal autoimmunity, such that use of Dnmt inhibitors may warrant careful consideration.
Abstract
Protocols to use Foxp3+ T-regulatory (Treg) cells for cellular therapy, especially postallogeneic stem cell transplantation, are currently being developed and tested by various groups. Inhibitors of DNA methyltransferase (Dnmt) enzymes have been advocated as a means to promote and stabilize Foxp3 expression in Tregs undergoing expansion in vitro before their injection in vivo. We investigated the effects of conditionally deleting two Dnmt enzymes that co-immunoprecipitated with Foxp3 in Treg isolates. Deletion of Dnmt1, but not Dnmt3a, decreased the numbers and function of peripheral Tregs and impaired conversion of conventional T cells into Foxp3+ Tregs under polarizing conditions. Importantly, mice with conditional deletion of Dnmt1 in their Tregs died of autoimmunity by 3 to 4 weeks of age unless they were rescued by perinatal transfer of wild-type Tregs. Conditional Dnmt1 deletion did not affect methylation of CpG sites within Foxp3 but decreased global DNA methylation and altered Treg expression of several hundred pro-inflammatory and other genes. Hence, Dnmt1 is necessary for maintenance of the core gene program underlying Treg development and function, and its deletion within the Treg lineage leads to lethal autoimmunity. These data suggest that caution may be warranted when considering the use of DNMT inhibitors in development of Treg-based cellular therapies.
Introduction
Epigenetic regulation of gene expression encompasses DNA methylation, chromatin remodeling, histone variants, posttranslational modifications of histone tails, and the actions of small, noncoding RNAs.1 These pathways allow cells to develop and differentiate from the zygote stage of life, without changes in DNA sequence occurring. DNA methylation is the best-established epigenetic mechanism and is important in processes ranging from parental imprinting to X-chromosome inactivation.2 Methylation of cytosines located in CpG-rich regions upstream of transcriptional start sites provides a stable and tissue-specific mechanism for regulation of gene expression. DNA methylation is catalyzed by one or more DNA methyltransferase (Dnmt) enzymes: Dnmt1, Dnmt3a, and Dnmt3b. The dual Dnmt3 enzymes establish DNA methylation in germ cells and in early development, whereas Dnmt1 binds preferentially to hemimethylated DNA and reestablishes DNA methylation after DNA replication.3,4 The targeting of Dnmt1 to hemimethylated DNA is promoted by PCNA, UHRF1, and USP7.5-7 Dnmt1 also interacts with Dnmt3a and Dnmt3b and multiple additional silencing proteins, including the HDAC1 and HDAC2, heterochromatin protein-1, and histone lysine methyltransferases and methyl-binding domain proteins.5-7 Dnmt1 stability and function are regulated by several posttranslational modifications, including phosphorylation, acetylation, ubiquitylation, methylation, and sumoylation.8,9
Foxp3+ T-regulatory (Treg) cells are key to immune regulation. Foxp3 is located on the X-chromosome, and Foxp3-deficient mice or male Scurfy mice, with a frameshift mutation that results in disruption in Foxp3 DNA-binding, lack functional Tregs and succumb to fatal autoimmunity within a month after birth,10,11 unless adoptively transferred with normal CD4+CD25+ Tregs.12 Similarly, mice deficient in IL-2 or CD25 have few Foxp3+ Tregs and succumb to lethal autoimmunity.13 Like Dnmt1, Foxp3 expression is subject to epigenetic regulation.14-18 The CpG island in the first intron of Foxp3 DNA is demethylated in naturally occurring human and murine Treg, and demethylation of this site correlates with Foxp3 expression and Treg function.19,20 Deletion of Dnmt1 in conventional T (Tcon) cells increases their Foxp3 expression upon TCR stimulation.21 Likewise, use of the Dnmt inhibitor, 5-aza-2′-deoxycytidine, increases Foxp3 expression in WT Tcon cells and promotes their conversion into induced Treg (iTreg) cells.18
Thus, it is clear that Dnmt1 limits the ability of CD4 T cells to express Foxp3 and develop into functional Treg cells. However, the role of Dnmt1 in the development and function of natural Tregs has not been explored. To investigate the contribution of Dnmt1 to Treg biology, we conditionally deleted Dnmt1 within Foxp3+ Treg cells. Unexpectedly, we found that Treg development was impaired and that extra-thymic Foxp3+ Tregs lost their suppressive function, leading to lethal autoimmunity. Loss of Dnmt1 was accompanied by marked upregulation of inflammatory genes within Tregs, widespread mononuclear cell infiltration of host tissues, and exuberant T- and B-cell responses. Thus, although modulation of Dnmt1 therapeutically may contribute to the conversion of T cells into Tregs in some systems, our data suggest that this may potentially be counterproductive in the long term because Dnmt1 appears to be indispensable for the inhibitory function of Foxp3+ Treg cells.
Methods
Mice
We purchased wild-type (WT) and Rag1−/− C57BL/6 mice from The Jackson Laboratory, and floxed Dnmt122 and Dnmt3a23 mice from the Mutant Mouse Regional Resource Centers. CD4-Cre24 and Foxp3-YFP-Cre25 mice were kind gifts of their originators. Animal studies were approved by the Institutional Animal Care and Use Committee of The Children’s Hospital of Philadelphia (##2010-6-561).
Treg purification and Treg suppression assays
Cells from lymph nodes and spleens were pooled and Tregs, Tcon, and APC isolated using magnetic beads (Miltenyi Biotech) or cell sorting (Aria, BD Biosciences); Treg purity was greater than 80% using beads and > 98% using cell sorting. To test Treg suppression in vitro, equal numbers of crystal field stabilization energy–labeled CD4+CD25– Tcon cells and γ-irradiated APC (5 × 104/well) were incubated for 3 days (5% CO2, 37°C) along with CD3 mAb (1 μg/mL) and serial dilutions of Treg (Treg:Tcon ratios of 1:1 to 0.125:1), followed by flow cytometric assessment of Tcon proliferation.16
Hematology and autoantibody detection
Citrated blood samples were assessed using an automated hematology analyzer modified and calibrated for murine samples. Sera from male and female 3- to 4-week-old Dnmt1−/− or WT mice, with or without WT Treg adoptive transfer, were serially diluted, incubated with cryosections of tissues from normal C57BL/6 mice for 30 minutes, washed in phosphate-buffered saline, and bound autoantibodies detected using fluorescein isothyocyanate–conjugated goat anti-mouse IgG (Jackson ImmunoResearch Laboratories, 1/200). Negative controls included use of pooled healthy sera and use of conjugated-antibody alone.
Flow cytometry
Cells from thymus, lymph nodes (LN), and spleen were labeled using CD4-pacific blue, CD8-alexa 647, CD44-PE, CD25-PE-cy7, CD90.1-PE, Foxp3-PE-cy5, and CD62L-APC-cy7 and were analyzed by flow cytometry (Cyan, DakoCytomation).
DNA methylation
Global DNA methylation was assessed using a Methylated DNA Quantification kit (Sigma-Aldrich, St. Louis, MO). Briefly, Treg DNA was extracted (Puregene DNA isolation kit, Gentra Systems) and quantified with a spectrophotometer, and 200 ng of purified DNA in 30 μL of binding buffer was added per well. Methylated DNA was captured with anti–5-methylcytosine Ab and detected at 450 nm, and results were calculated with reference to a standard curve generated with methylated control DNA. For analysis of Treg-specific demethylated region (TSDR) methylation, 300 ng of DNA from Tcon or Treg was bisulfite-converted (Methyl detector, Active Motif) and used for two rounds of polymerase chain reaction (PCR) with primers designed by Dr. Andrew Wells and co-workers (Path & Laboratory Med, Children’s Hospital of Philadelphia [CHOP]). Primers sets were outer: forward 5′-GGGTTTTGG GATATTAATATATATAGTAAG-3′, reverse 5′-CCACTATATTAACTAACCCATATAACTAA-3′; and inner: forward 5′-TTGAGTTTTTGTTATTATAGTATTTGAAGAT-3′, reverse 5′-ACTAAAAACCTAAAA AACTAAACTAACCAA-3′. PCR products (502 bp) were separated on agarose gels, purified, cloned into pGEM-T easy vector (Promega) and transformed into bacterial clones. Plasmid DNA samples from bacterial colonies were sequenced at CHOP’s core facility. Pyrosequencing of TSDR methylation in isolated Tcon or Treg cells was performed by EpigenDx (assay-ID: ADS443FS2). Methylation of the IFN-γ enhancer-specific region was analyzed as reported.26
Methylated DNA immunoprecipitation (MeDIP)
Genomic DNA was extracted (Qiagen Dneasy kit), 6 μg of DNA was digested with 24 U of Mse I overnight, and methylated DNA was immunoprecipitated with anti–5-methyl-cytidine Ab (Abcam). Immunoprecipitated DNA was amplified using a genomic DNA amplification kit (WGA2, Sigma), purified, and sent for DNA methylation microarray (Roche). Resultant data were analyzed using Partek GS 6.6, NimbleScan 2.6, and Integrative Genomics Viewer 2.1.17 (www.broadinstitute.org/igv).27 Raw signal-intensity data underwent Loess normalization. Scaled log2 ratio data were tested for normal distribution (supplemental Figure 12) and for significance using Student’s t-test; significance was defined at a false discovery rate threshold of 0.5 (P < .00083). Probes with a significantly different scaled log2 ratio were fitted with a fixed 750-bp-long window, and a one-sided Kolmogorov-Smirnov test was applied to determine positive distribution of intensity log ratios compared with the rest of the array. The –log10 > 2 of the resulting P values were used for determining peak scores, which were mapped to overlapping features (5 kbp upstream and 1 kbp downstream).
Luminex
Serum immunoglobulin isotypes were assessed with a multiplex mouse immunoglobulin assay kit (Millipore) using LiquiChip Luminex 100 (Qiagen) according to the manufacturer’s instructions.
Homeostatic proliferation
One million Thy1.1+ CD4+ CD25– Tcon cells isolated using magnetic beads were injected intravenously into Rag1−/− C57BL/6 mice along with equal numbers of bead-isolated fl-Dnmt1/CD4cre or WT CD90.2+ Tregs, or with fluorescence-activated sorting of fl-Dnmt1/Foxp3cre vs Foxp3cre Tregs, as indicated. Seven days later, LN and spleens were harvested and CD90.1+CD4+ T cell numbers determined by flow cytometry.16
Cardiac transplantation
Hearts were harvested from BALB/c mice and engrafted using microvascular techniques into Rag1−/− C57BL/6 recipients.16 Immediately post transplant, 1 million CD4+CD25- WT Tcon cells and a half-million Dnmt1−/− or WT C57BL/6 Treg mice were injected via the tail vein, and allograft survival was monitored daily.
Real-time PCR
RNA from freshly sorted CD4+CD25– T cells and Foxp3/YFP+ Tregs, as well as RNA from corresponding cells activated for 24 hours with phorbol 12-myristate 13-acetate (PMA)/ionomycin, was prepared (Qiagen RNAse kit), first-strand cDNA was made by reverse transcription, and real-time PCR performed with commercial probes and primers (Applied Biosystems).
Microarrays
RNA was extracted using RNeasy kits (Qiagen), and RNA integrity and quantity were analyzed by photometry (DU640, Beckman-Coulter). Microarray experiments were performed using whole-mouse-genome oligoarrays (Mouse430a, Affymetrix), and array data were analyzed using MAYDAY 2.12 software.28 Array data were subjected to robust multiarray average normalization. Normalized data were used for calculating fold changes of up- and downregulated genes using Student’s t-test. Only data with a false discovery rate–adjusted P value of < .05 and at least 1.5× differential expression were included in the analysis. Data underwent z-score transformation for display. We deposited our data in the NCBI Gene Expression Omnibus (GEO) database (www.ncbi.nlm.nih.gov/geo) under accession #GSE27434.
Statistics
Data were analyzed using GraphPad Prism software. All normally distributed data were displayed as means ± SD. Measurements between 2 groups were done with a Student’s t-test or Mann-Whitney U test. Groups of three or more were analyzed by one-way analysis of variance or Kruskal-Wallis test. Allograft survival was assessed using a log-rank (Mantel-Cox) test.
Results
Mice with Dnmt1−/− Tregs develop Scurfy-like lethal autoimmunity
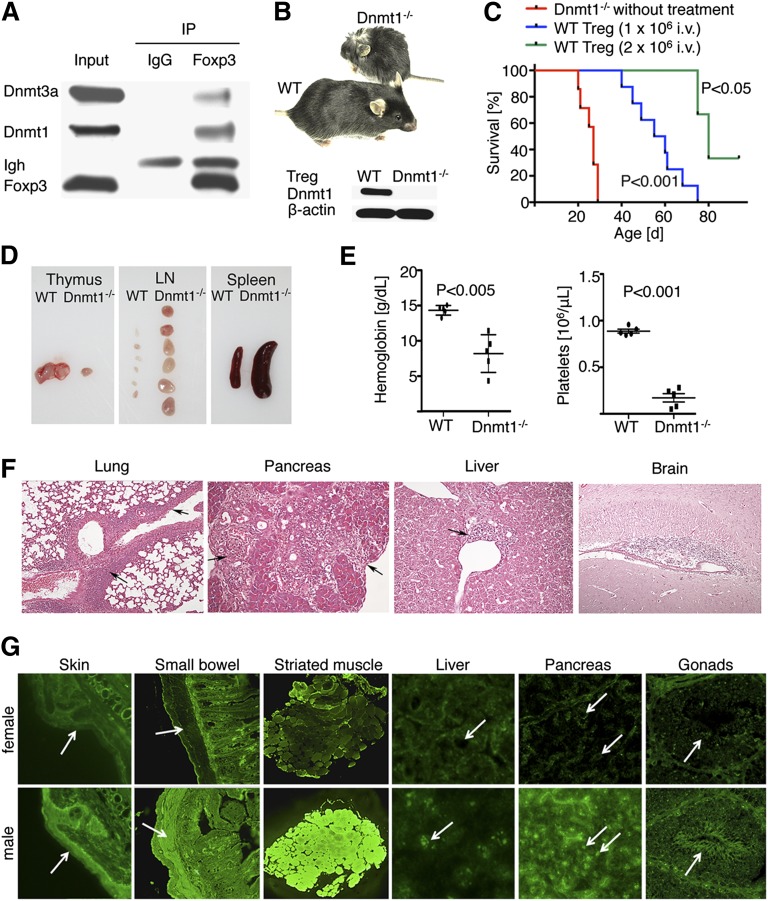
In many cells, Dnmt1 forms a complex with Dnmt3 and other proteins and represses gene transcription.29 We therefore investigated potential biochemical interactions of Dnmt1 and Foxp3. We purified Treg cells and found that immunoprecipitation of Foxp3 led to coprecipitation of Dnmt1 and Dnmt3a, but not of Dnmt3b (Figure 1A). Hence, Foxp3 is physically associated with enzymes that are thought to control de novo (Dnmt3a) and maintenance (Dnmt1) methylation of multiple genes in Treg cells. Foxp3-mediated deletion of floxed Dnmt3a had no significant impact on the absolute numbers of Treg cells in lymphoid tissues (supplemental Figure 1), and these mice displayed a normal phenotype and lifespan, without evidence of autoimmunity. By contrast, male mice in which the floxed dnmt1 gene was deleted just within Tregs (fl-Dnmt1/Foxp3cre, hereafter Dnmt1−/− mice unless specified otherwise) developed systemic autoimmunity and died by 3 to 4 weeks of age (Figure 1B), similar to male Scurfy mice,22,30 unless adoptively transferred with WT Treg cells at 2 to 3 days of life (Figure 1C). Dnmt1−/− male mice showed abnormally small thymii but markedly enlarged LN and spleens (Figure 1D). Male Dnmt1−/− mice also had abnormal bone marrow (supplemental Figure 2) and developed anemia, thrombocytopenia, and a leucopenia (Figure 1E and supplemental Figure 3) that arose primarily from reduced numbers of granulocytes (supplemental Figure 4). Male mice showed T-cell infiltration of skin and peripheral organs, including the lungs, pancreas, liver, brain, and other tissues (Figure 1F, supplemental Figure 5, and supplemental Table 1), and developed multiple autoantibodies (Figure 1G and supplemental Table 2). Hence, conditional deletion of Dnmt1 in Foxp3+ Tregs induces a lethal autoimmune disease comparable in severity and range of target organs affected with that seen in male Scurfy mice.
Figure 1.
Foxp3cre/Dnmt1−/− mice developed lethal autoimmunity by 3 to 4 weeks of age. (A) In WT Tregs, immunoprecipitation of Foxp3 led to co-precipitation of Dnmt1 and Dnmt3a (immunoglobulin-heavy chain, Igh, is also detected). (B) Male mice with Dnmt1−/− Tregs became sick and runted, whereas WT littermate or female mice developed normally; (inset) representative Western blot showing the absence of Dnmt1 in the Tregs of targeted mice (developed by mating floxed Dnmt1 and Foxp3-Cre mice). (C) Kaplan-Meier survival plots showing death by 3 to 4 weeks of age of all mice with Dnmt1−/− Tregs, whereas their adoptive transfer with WT Tregs at 2 to 4 days of life rescued mice in a dose-dependent manner; data are from 3 to 8 mice per group. (D) Dnmt1−/− mice had small thymii but enlarged lymph nodes and spleens. (E) Dnmt1−/− mice developed anemia and thrombocytopenia; data are from 5 mice per group. (F) Histologic examination mononuclear cell infiltrates of multiple organs, including extensive bronchovascular bundles of the lung, periductular areas of the pancreas, periportal areas of the liver, and submeningeal areas of the brain. (G) Sera of male Dnmt1−/− mice contained autoantibodies directed against stratified squamous keratinizing epithelium in the skin (ear section, 1/40 to 1/80 titer), smooth muscle (eg, small bowel, titers of 1/40 to 1/80), striated muscle (titers of 1/320 to 1/640), nuclei of all tissues (nucleolar pattern, titers of 1/80 to 1/160, eg, in liver and pancreas in males vs unaffected nuclei in females), and antisperm antibodies (titers of 1/20 to 1/40 titers against mature sperm cells located in the central part of seminiferous tubules; arrows point to the tails of spermatozoa); sera from Dnmt1−/− females (upper row) had no autoantibodies and were indistinguishable from the sera of healthy C57BL/6 mice (n = 3/group).
Loss of Dnmt1 affects postthymic development of natural and induced Foxp3+ Tregs
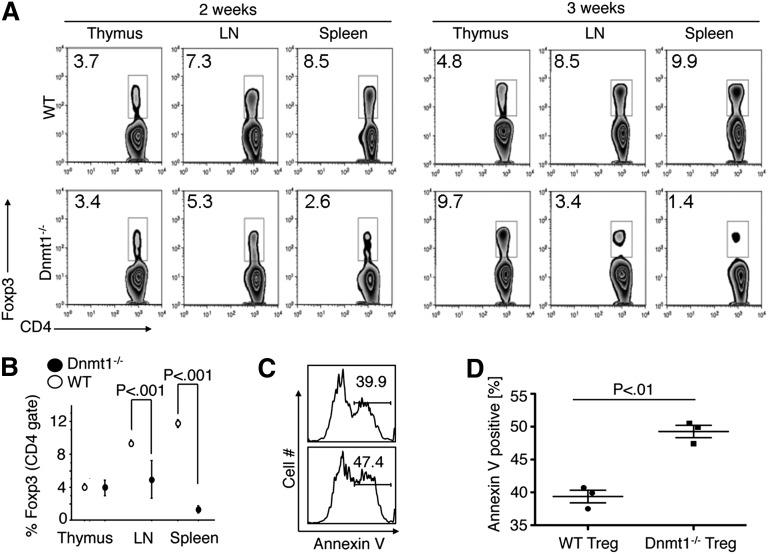
Flow cytometric analysis showed that male mice with Dnmt1−/− Tregs had markedly reduced proportions of peripheral Foxp3+ Tregs but increased proportions of intrathymic Foxp3+ Tregs; representative flow cytometric plots are shown in Figure 2A, and pooled data at 3 to 4 weeks of age are shown in Figure 2B and supplemental Figure 6. Dnmt1 deletion can cause DNA demethylation and p53-dependent cell apoptosis and death,22 but the findings that a lack of Dnmt1 did not affect thymic production of Tregs, but was accompanied by their marked decrease in the periphery, suggested that peripheral Tregs might have a shortened lifespan. Consistent with this, Dnmt1−/− Treg cells underwent increased apoptosis upon T-cell receptor (TCR) stimulation compared with WT Treg cells (Figure 2C-D). Hence, the postthymic lifespan of Dnmt1−/− Tregs and their peripheral induction from conventional T cells are impaired as a result of loss of Dnmt1.
Figure 2.
Development of Dnmt1−/− versus WT Foxp3+ Tregs. (A) Intranuclear flow cytometric staining for Foxp3 after gating on CD4 T cells, showing accumulation of Foxp3+ cells in thymii of Foxp3cre/Dnmt1−/− mice, but their diminution in LN and spleen samples compared with normal controls; proportion of labeled cells shown in each plot and data representative of 5 mice per group per time-point. (B) Pooled data (n = 5/ group) for proportions of Foxp3+ Tregs (mean ± SD) in each lymphoid tissue. (C) Compared with WT Tregs, Foxp3cre/Dnmt1−/− Tregs had increased susceptibility to apoptosis upon overnight TCR stimulation; annexin-V staining. (D) Pooled data to (C) from 3 independent experiments.
Dnmt1−/− Tregs lack suppressive function
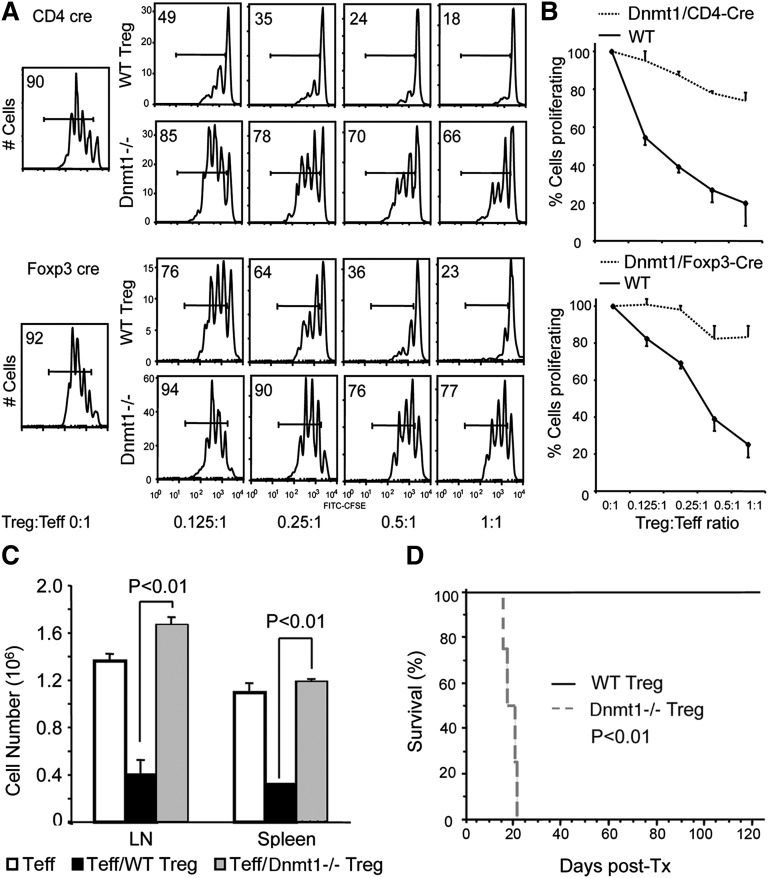
To analyze the development of lethal autoimmunity in mice whose Tregs lacked Dnmt1, we first tested the suppressive functions of Tregs isolated from floxed dnmt1/CD4-Cre mice using magnetic bead purification of CD4+CD25+ cells, and from floxed dnmt1/Foxp3-Cre mice using cell sorting based on YFP-Cre recombinase fusion protein present in the 3′UTR of their foxp3 locus.25 Compared with WT Tregs tested in standard Treg suppression assays in vitro,16 both Dnmt1−/− Treg populations showed a profound loss of suppressive function, as seen in flow cytometric plots of T cells proliferating in the presence of varying ratios of Tregs (Figure 3A), and in cumulative data (Figure 3B). Loss of suppressive activity by Dnmt1−/− Tregs was also evident in vivo. In a homeostatic proliferation model,16 Tcon cells expanded rapidly after adoptive transfer to immunodeficient (Rag1−/−) mice, whereas cotransfer of WT but not Dnmt1−/− Tregs significantly inhibited Tcon cell proliferation (Figure 3C). Loss of Treg function in Dnmt1−/− was not associated with decreased Treg numbers at the end of the 3-day assay in vitro (supplemental Figure 7), whereas in vivo, Tregs lacking Dnmt1 were significantly reduced at the end of the 7-day homeostatic proliferation assay (supplemental Figure 8). We also assessed Treg function in a model in which fully major histocompatibility complex–disparate BALB/c cardiac allografts were engrafted into C57BL/6 (B6) Rag1−/− mice.16 The adoptive transfer of 1 × 106 CD4+ B6 Tcon cells induced acute rejection in this model, whereas cotransfer of 0.5 × 106 WT B6 Tregs inhibited effector cell function and allowed long-term allograft survival. However, adoptively transferred Dnmt1−/− B6 Treg cells were unable to suppress Tcon–cell activation and cardiac allografts were rejected by 3 weeks of transplantation (Figure 3D). Hence, loss of Dnmt1 in Foxp3+ cells led to almost complete loss of Treg suppressive function in vitro and in vivo. In contrast to mice with Dnmt1 deletion just within Tregs, T-cell deletion of Dnmt1 using CD4-Cre altered the normal distribution of Foxp3 (supplemental Figure 9A) but did not lead to the development of autoimmunity. TCR stimulation of these T cells in vitro resulted in reduced proliferation (supplemental Figure 9B) and increased rates of activation-induced apoptosis (supplemental Figure 9C) compared with WT cells, and they failed to undergo alloantigen-induced activation and proliferation in vivo (supplemental Figure 10). Collectively, these data point to important roles of Dnmt1 in both Treg and Tcon cells.
Figure 3.
Dnmt1−/− Tregs lack suppressive function in vitro and in vivo. (A) In vitro Treg assays assessing their ability to inhibit the TCR-induced proliferation of CFSE+ CD4+ CD25– Tcon cells. Upper panels compare Dnmt1−/− Tregs isolated from CD4-Cre/dnmt1−/− mice versus WT Tregs; lower panels compare Dnmt1−/− Tregs isolated from Foxp3-Cre/dnmt1−/− mice versus WT Tregs; the proportions of proliferating Tcon cells is shown at each ratio of Treg:Tcon cells. (B) Cumulative data (mean ± SD) from 4 such assays as shown in previous panel. (C) Lack of Dnmt1−/− Treg suppressive function in vivo in homeostatic proliferation assays involving transfer of WT Tcon cells plus WT or Dnmt1−/− Tregs to B6 RAG−/− mice; data (mean ± SD) from 4 mice per group, and comparable results were observed in 2 separate experiments. (D) Cardiac allograft survival in B6 Rag−/− mice engrafted with BALB/c hearts and reconstituted with B6 CD4+CD25– T con cells and either WT or Dnmt1−/− Treg cells; Kaplan-Meier plot with 4 animals per group, and comparable data were observed in 2 separate experiments.
Loss of Treg function leads to T- and B-cell activation
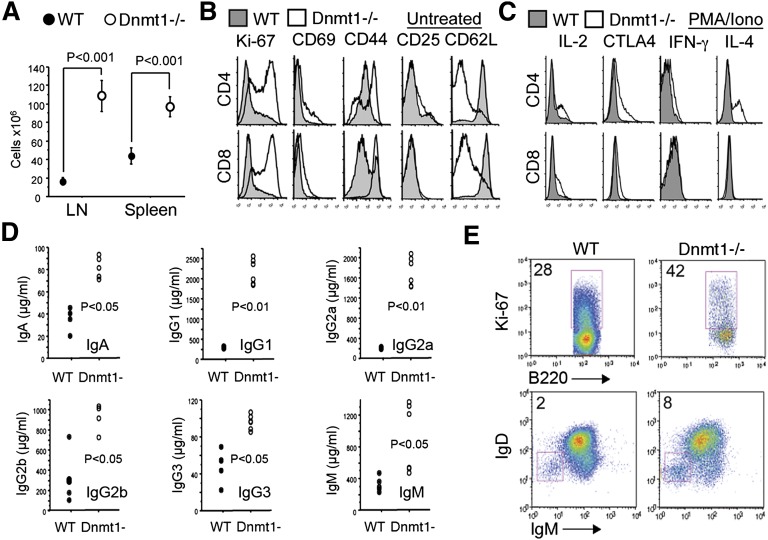
We assessed T- and B-cell activation in mice with Dnmt1−/− Treg cells, given a marked increase in cellularity of their peripheral lymph nodes and spleens compared with littermate controls (Figure 4A). Compared with WT controls, Tcon cells from mice with Dnmt1−/− Tregs showed immune activation in the absence of any exogenous stimulation, including increased expression of Ki-67, CD25, CD44, CD69, and reduced CD62L (Figure 4B). Likewise, after 4 hours of stimulation with PMA/ionomycin, T cells from Dnmt1−/− mice showed greater expression of CTLA4 and increased production of IL-2, IL-4, and IFN-γ cytokines than T cells from WT mice (Figure 4C). Treg deletion of Dnmt1 also affected B-cell activation, as reflected by the panoply of autoantibodies detected (Figure 1F and supplemental Table 2). Thus, loss of Dnmt1 in Tregs was accompanied by significant increases in the levels of all immunoglobulin classes assayed (Figure 4D), by increased proportions of Ki-67+ B cells (Figure 4E), and by increase of an activated IgD and IgM double-negative population of B cells (Figure 4E). Hence, loss of Dnmt1 in Treg cells resulted in increased activation of host T and B cells.
Figure 4.
Loss of Treg Dnmt1 expression leads to increased activation of T and B cells. (A) Total mononuclear cell counts of peripheral LN and spleens isolated from Dnmt1−/− versus WT mice (mean ± SD, n = 4 per group). (B) Flow cytometry showing increased expression of T-cell activation markers using CD4+CD25– YFP-Foxp3– Tcon cells isolated from mice with conditional targeting of Dnmt1 in their Tregs versus WT mice. (C) Effects of PMA/ionomycin activation (4 hours) of these cells showing increased cytokine expression by T cells from Dnmt1−/− versus WT mice. (D) Increased immunoglobulin levels in the sera of Dnmt1−/− versus WT mice (mean ± SD, n = 4 per group). (E) Increased B-cell activation and class switching in Dnmt1−/− versus WT mice; proportion of labeled cells is shown in each plot and data are representative of 4 mice analyzed.
Adoptive transfer of WT Tregs alleviates autoimmune disease in fl-Dnmt1/Foxp3cre mice
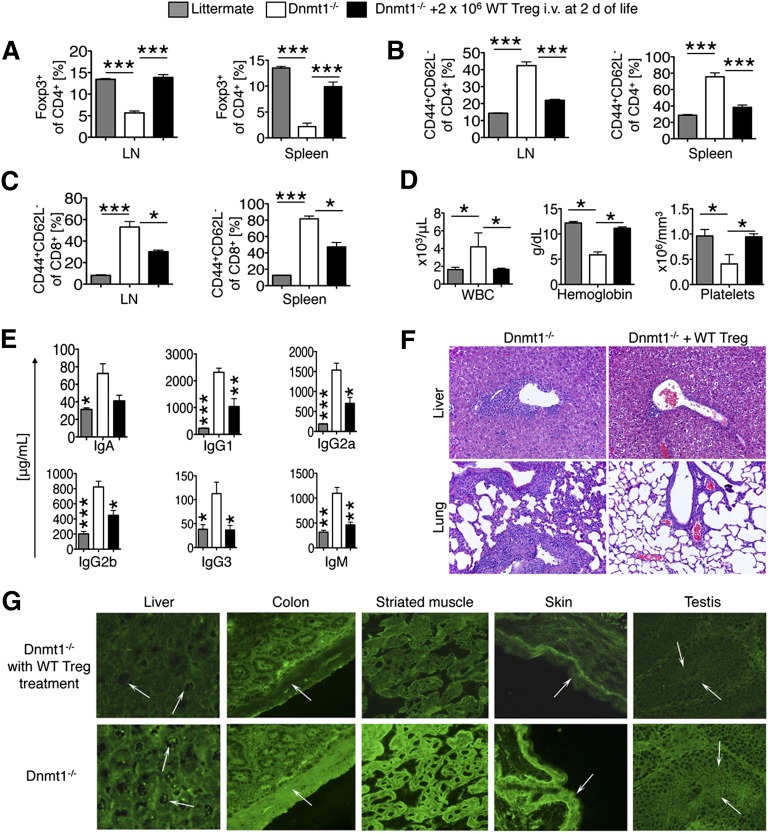
Given the immunopathology observed in mice lacking Dnmt1 within Foxp3+ Tregs (Figures 1-3), we tested whether adoptive transfer of WT (C57BL/6) Tregs with normal Dnmt1 expression could reverse the autoimmune phenotype. Intravenous injection of 2 × 106 WT Treg into 6 male fl-Dnmt1/Foxp3cre mice at 2 to 4 days of life resulted in reconstitution of their Treg population (Figure 5A). We followed three mice for survival, which was markedly extended compared with fl-Dnmt1/Foxp3cre mice without WT Treg injection (Figure 1C). The other 3 mice were sacrificed during their third week of life and analyzed for the same parameters that we had previously noted were abnormal in the fl-Dnmt1/Foxp3cre mice. Remarkably, CD4 and CD8 Tcon in treated fl-Dnmt1/Foxp3cre mice showed less cellular activation (CD44+CD62L-, Figure 5B-C). There was also normalization of hematologic parameters (Figure 5D) and immunoglobulin levels (Figure 5E) and markedly reduced infiltrates in liver and lung samples (Figure 5F). Correspondingly, adoptive transfer of WT Treg limited the amount of antibody formation in fl-Dnmt1/Foxp3cre mice (Figure 5G). Together, these data show that transfer of WT Treg can curtail development of autoimmunity in mice lacking Dnmt1 within Foxp3+ Treg.
Figure 5.
Adoptive transfer of WT Treg prevents autoimmunity in mice with Dnmt1−/− Foxp3+ Tregs. We adoptively transferred 2 × 106 WT Treg intravenously to fl-Dnmt1/Foxp3cre (Dnmt1−/−) mice at 2 days of life and assessed immunopathology at 3 weeks compared with fl-Dnmt1/Foxp3cre without treatment and littermate control (Foxp3cre). (A) CD4+Foxp3+ Treg population is normalized in Dnmt1−/− mice adoptively transferred with WT Treg. (B) CD4 and (C) CD8 Tcon showed a reduced level of activation in fl-Dnmt1/Foxp3cre mice treated with WT Treg. (D) Normalization of key hematologic parameters and (E) reduction of increased immunoglobulin levels in Dnmt1−/− mice after WT Treg administration. (F) Rescue of Dnmt1−/− mice with WT Treg alleviates autoimmune pathology of the lung and liver; hematoxylin and eosin staining, ×200 magnification. (G) Sera from Dnmt1−/− mice without WT Treg treatment (lower row) contained antibodies against nuclei (3 of 3 mice, nucleolar pattern in, eg, liver, 1/40 to 1/80 titers), smooth muscle (3 of 3 mice, eg, colon, 1/10 to 1/40 titers), striated muscle (3 of 3 mice, 1/20 to 1/80 titers), stratified squamous keratinizing epithelium in the skin (2 of 3 mice, ear section, 1/20 to 1/80 titers), and against sperm cells (1 of 3 mice, 1/10 to 1/20 titers). Sera from Dnmt1−/− mice with WT Treg treatment (upper row) had no autoantibodies in corresponding tissues and were indistinguishable from WT controls, with the exception of low smooth muscle antibody titers (1 of 3 mice, 1/10) and striated muscle antibodies (1 of 3 mice, 1/10 titer). All data are shown as mean ± SEM with n = 3 to 4 mice per group. *P < .05, **P < .01, ***P < .001 (analysis of variance with Tukey’s multiple comparison test).
Loss of Dnmt1 does not affect methylation of foxp3 TSDR or promoter regions
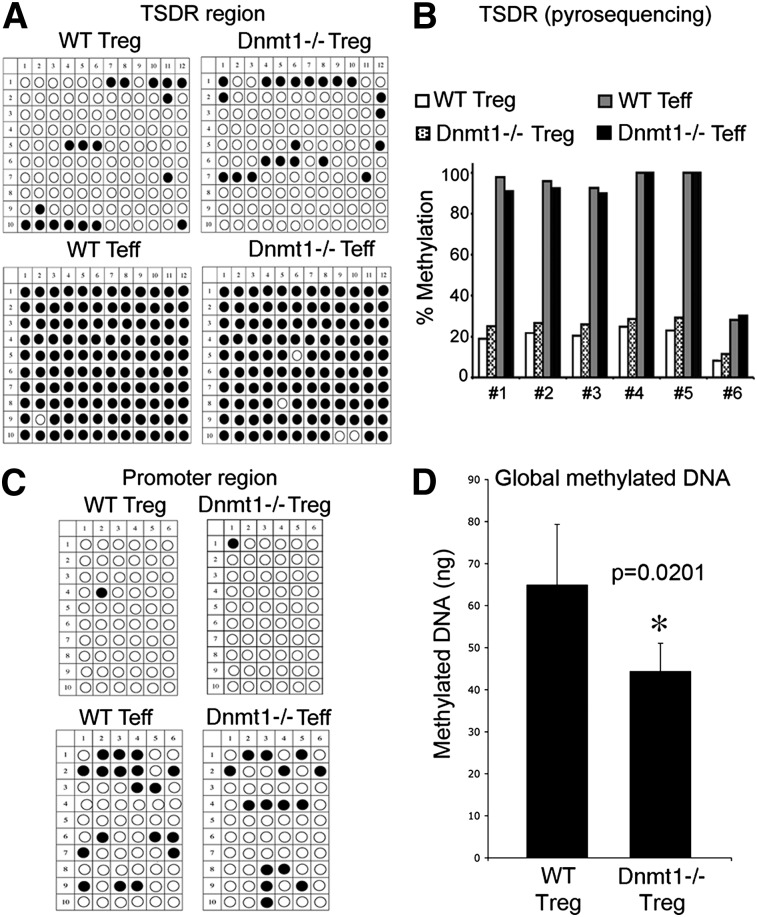
Given the marked effects arising from targeted deletion of Dnmt1 in Tregs, we analyzed DNA methylation in Dnmt1−/− Foxp3+ Treg cells. CpG islands of the foxp3 intronic site known as the Treg-specific demethylation region (TSDR) are normally fully demethylated in Treg cells, but are fully methylated in Tcon cells.20 We first assessed TSDR methylation by bisulphite conversion, cloning, and sequencing.31 We found that the TSDR site was still demethylated in Dnmt1−/− Treg cells and methylated in Tcon cells (Figure 6A), and our results were confirmed by pyrosequencing (Figure 6B).32 In addition to TSDR demethylation, the CpG islands of the foxp3 promoter are normally fully demethylated in Treg cells but partially methylated in Tcon cells.20 Again, we found that the normal patterns of demethylation of the foxp3 promoter were unchanged in Treg and Tcon cells of Dnmt1−/− versus WT mice (Figure 6C), and demethylation of the foxp3 promoter was unaffected by CD3/CD28 mAb stimulation (supplemental Figure 11). By contrast, global DNA methylation was significantly decreased in Tregs of Dnmt1−/− versus WT mice (Figure 6D). Hence, loss of Treg expression of Dnmt1 did not affect methylation at key foxp3 regulatory sites but did broadly reduce DNA methylation in Treg cells.
Figure 6.
Demethylation at the Foxp3 intronic TSDR region in WT versus Dnmt1−/− Tregs and Tcon. Demethylation was determined by (A) bisulphite conversion, cloning, and sequencing and was (B) confirmed by pyrosequencing. (C) Assessment of demethylation of the Foxp3 promoter region in WT versus Dnmt1−/− Tregs and Tcon was determined by bisulphite conversion. Open circles indicate demethylated CpG and closed circles indicate methylated CpG. (D) Measurement of total DNA methylation in WT versus Dnmt1−/− Treg cells (mean ± SD, n = 3 per group).
Loss of Dnmt1 affects genome-wide Treg methylation and gene expression
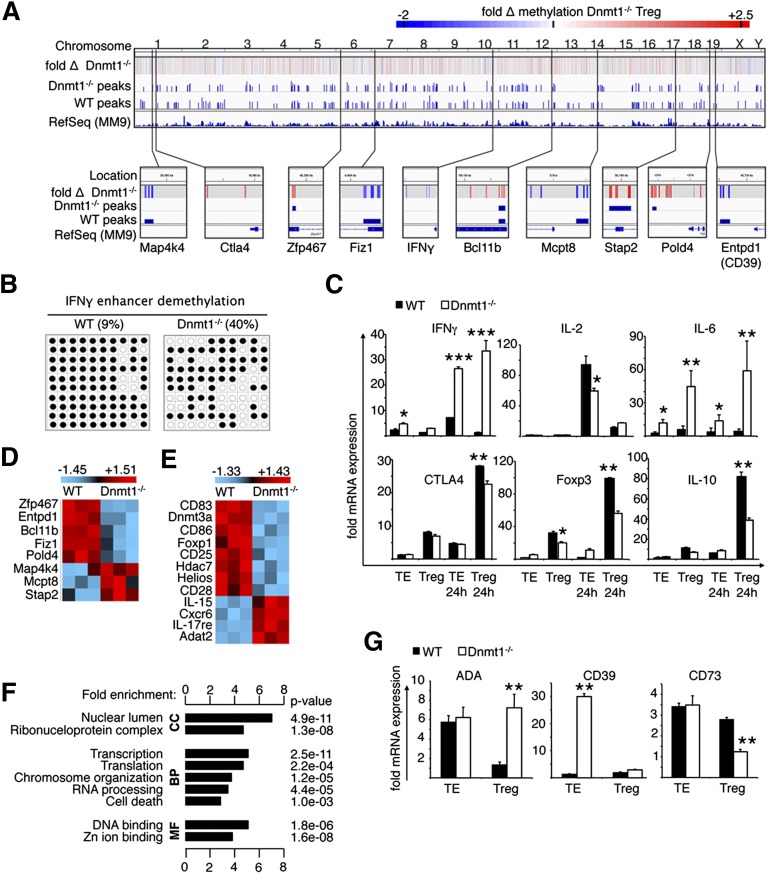
Given the overall importance of Dnmt1 in transcriptional gene regulation, we assessed genome wide DNA methylation using methylated DNA immunoprecipitation (Figure 7A and supplemental Figure 12). Deletion of Dnmt1 in Treg led to significant differential methylation in 6.9% of the genome. Further detailed analysis of mapped peaks indicated enhancer and promoter hypermethylation of genes essential to Treg function, such as CTLA4 and Bcl11b.33 On the other hand, we noted demethylation of genes normally suppressed in Treg, such as Mcpt8 (protease-8), a highly basophil-specific marker,34 as well as the enhancer-specific region of interferon (IFN)-γ (Figure 7B). The DNA methylation findings correlated well with gene expression data, showing prominent increase in IFN-γ and other transcripts of identified genes with altered methylation (Figure 7C-D and supplemental Figure 13), as well as genes of relevance to Treg function.
Figure 7.
Dnmt1 affects genome-wide methylation, leading to loss of the Treg phenotype. (A) Genome-wide overview of fold change in scaled log2 ratios and peak scores of methylated genomic DNA from Dnmt1−/− and WT Treg. Heatmap scale optimized for visualization of −2 to +2.5–fold changes (>96.6% of data; range, −5.68 to +9.28). Hypomethylation shown indicated in blue, and hypermethylation is indicated in red. Results pooled from 3 independent experiments and visualized with the Broad Institute’s Integrative Genomics Viewer. (B) Methylation assessment of the IFN-γ enhancer region, as in Figure 5, confirmed greater than 4× increase in demethylation in Dnmt1−/− Treg. (C) Real-time PCR analysis (mean ± SD, n = 4 per group) of the effects of Dnmt1 loss on Treg expression of various cytokines, and signature Treg genes showed increased IFN-γ expression, consistent with enhancer demethylation. (D-E) Selection transcripts with greater than 2× differential expression; microarray data are shown after z-score transformation (n = 3 per group), data shown after z-score transformation. (D) Transcripts identified as differentially methylated in (A). (E) Treg-relevant genes. (F) Functional annotation enrichment of probes with greater than 5 higher raw scores in Dnmt1−/− versus WT Treg (P < .05). Gene ontology (GO), cellular component (CC), biological process (BP), molecular function (MF). Functional Annotation Clustering calculated using DAVID.50 (G) Loss of Dnmt1 disrupted the normal expression of genes associated with adenosine production (real-time PCR, mean ± SD, n = 4 per group).
Next, we investigated global transcription by microarray analysis to assess differences in gene expression by Dnmt1−/− versus WT Treg cells. We observed altered expression of many Treg-associated genes (Figure 7E), with downregulation of genes encoding surface proteins important to Treg development and maintenance in the periphery (eg, transforming growth factor–β receptor, CD25, CD28, CD83, CD86); proteins that complex with Foxp3 and are important to Treg function (eg, Foxp1, HDAC7); additional transcription factors implicated in Treg biology (eg, Ikzf2/Helios, Egr2); and Dnmt3a. Loss of Dnmt1 led to upregulated expression in Tregs of genes encoding cytokines (IL-6, IL-15, IL-17, and IFN-γ), chemokine receptors (eg, CCR1 and CXCR6), and surface molecules characteristic of other lineages, including genes typical of developing Th17 cells, such as IL-17, IL-17R, and STAT3. Remarkably, similarly to DNA methylation, we found ∼4% of the entire array to be differentially expressed, indicating the manifold actions of Dnmt1 in Foxp3+ Treg cells (supplemental Figure 14).
Previous studies indicated that loss of Dnmt1 caused cell type–specific changes in gene expression, including expression of imprinted genes, cell-cycle control, and growth factor/receptor signal transduction genes.22,24 Consistent with this, functional annotation clustering showed that Dnmt1 deletion upregulated genes associated with diverse functions ranging from control of transcription and translation to nuclear organization (Figure 7F). Likewise, Dnmt1 targeting disrupted the functionally important pathway regulating extracellular adenosine production (Figure 7G).35-37 Loss of Dnmt1 in Foxp3+ Tregs led to downregulation of CD73 expression and an increase in their expression of adenosine deaminase, suggesting that Dnmt1−/− Tregs actively metabolize adenosine rather than using its production to regulate the cAMP levels and associated threshold for cellular activation of Tcon cells. Hence, Dnmt1 deletion in Tregs has effects on expression of a broad range of genes that are well-established in Treg biology but, remarkably, are also profoundly altered expression of many other genes typically associated with core cellular functions.
Discussion
The autoimmunity developing in mice whose Foxp3+ Tregs lack Dnmt1 is notable for its rapidity and severity, and for its remarkable similarities to the lethal X-linked autoimmunity arising in Scurfy mice, which is regarded as the most severe form of autoimmunity arising in mammals.30,38,39 However, although the autoimmunity in both cases arises from almost complete loss of Treg suppressive function, Scurfy mice and patients with IPEX syndrome produce an abnormal Foxp3 protein that lacks a DNA-binding forkhead domain, whereas in the current study, a normal Foxp3 protein is expressed but a global reduction in methylation leads to altered expression of many genes in Treg cells. Thus, despite persistence of Foxp3 expression, Dnmt1−/− Tregs produce a range of cytokines (eg, IFN-γ, IL-6, IL-12, IL-17, IL-22) and transcription factors (eg, Irf7, Klf1, Runx2, Traf6) characteristic of other T-cell subsets. DNA methylation represses gene expression through both direct and indirect mechanisms. The direct mechanism entails blocking the binding of transcription factors, whereas the indirect mechanisms include recruitment of methyl-binding domain proteins, histone deacetylases, and histone methyltransferases that promote chromatin compaction.40 Global reductions in T-cell DNA methylation are correlated with the development of autoimmunity, including systemic lupus erythematosus, rheumatoid arthritis, systemic sclerosis, and dermatomyositis.41,42 There are also murine models of systemic lupus erythematosus that are induced by use of Dnmt1 inhibitors,43 but the current model is the first known example of aggressive autoimmunity arising from precise genetic targeting of a single Dnmt enzyme.
As a result of defective Treg function, mice in the current study showed a complex and systemic autoimmunity involving both T- and B-cell lineages. Affected mice showed progressive mononuclear cell infiltration of the lung, liver, skin, bone marrow, and other sites, leading to disorganized lymphoid architecture, as well as anemia, thrombocytopenia, and neutropenia, but a lack of colitis or hyperglycemia; these are all findings very typical of the autoimmunity seen in Scurfy mice.30,38,39 Although the precise mechanisms can vary by context, Foxp3+ Tregs usually suppress the activation of T, DC, and NK cells.44 Treg cells can also induce B-cell death and suppress antibody production, and patients with IPEX syndrome and Scurfy mice display a loss of B-cell anergy and produce a diverse array of autoantibodies,45,46 as found in the current study.
In addition to loss of Treg function at the single-cell level, mice with conditional deletion of Dnmt1 continued to produce Foxp3+ Tregs in the thymus but had fewer than normal Treg cells in the periphery. This reduction likely has several components, including a shortened lifespan of nTregs in the periphery, reduced capacity for their supplementation via peripheral conversion in the periphery (iTreg formation), and a loss of Treg lineage commitment, as suggested by our microarray data.
Finally, a potentially important translational aspect of this work relates to certain proposed strategies for clinical cell therapy using Tregs. Administration of Dnmt inhibitors has been suggested as a way to enhance nTreg cell function and fidelity in vivo,47,48 consistent with the initial clinical data from bone marrow or stem-cell transplant recipients that co-infused Foxp3+ Tregs became less effective the longer they were cultured before use.49 Indeed, in protocols to develop such cells for clinical infusion, it was reported that expanded Tregs, regardless of their initial naïve or memory phenotype, underwent homeostatic proliferation and transformed into effector memorylike cells, which produced IL-10 but also IL-6, IL-17, and IFN-γ.49 Given the current data, it is possible that the sustained use of Dnmt inhibitors, in efforts to promote and stabilize the Foxp3+ Treg lineage, may actually be counterproductive unless they are carefully monitored. Thus, reduction of Dnmt1 activity, below a basal level required for maintenance of a classical Treg lineage and associated optimal Treg cell function, may provoke significant harm.
Supplementary Material
Acknowledgments
The authors gratefully acknowledge the input and comments of Drs. Talal Chatila and Magali Rivas of the Children’s Hospital of Boston and Harvard Medical School.
This work was supported by grants from the National Institutes of Health (K08AI095353 to U.H.B., and P01AI073489 and 1R01CA158941 to W.W.H.)
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: L.W. performed most of the experiments and analyzed the data. Additional studies were contributed by Y.L., R.H., T.R.B., and T.A. U.H.B analyzed the microarray and methylated DNA immunoprecipitation data. W.W.H. devised the project. L.W., U.H.B., and W.W.H. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Wayne W. Hancock, Division of Transplant Immunology, Pathology and Laboratory Medicine, 916B Abramson Research Center, The Children's Hospital of Philadelphia, 3615 Civic Center Blvd., Philadelphia, PA 19104-4318; e-mail: whancock@mail.med.upenn.edu.
References
- 1.Allis CD, Muir TW. Spreading chromatin into chemical biology. ChemBioChem. 2011;12(2):264–279. doi: 10.1002/cbic.201000761. [DOI] [PubMed] [Google Scholar]
- 2.Chen ZX, Riggs AD. DNA methylation and demethylation in mammals. J Biol Chem. 2011;286(21):18347–18353. doi: 10.1074/jbc.R110.205286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lan J, Hua S, He X, Zhang Y. DNA methyltransferases and methyl-binding proteins of mammals. Acta Biochim Biophys Sin (Shanghai) 2010;42(4):243–252. doi: 10.1093/abbs/gmq015. [DOI] [PubMed] [Google Scholar]
- 4.He XJ, Chen T, Zhu JK. Regulation and function of DNA methylation in plants and animals. Cell Res. 2011;21(3):442–465. doi: 10.1038/cr.2011.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Felle M, Joppien S, Németh A, et al. The USP7/Dnmt1 complex stimulates the DNA methylation activity of Dnmt1 and regulates the stability of UHRF1. Nucleic Acids Res. 2011;39(19):8355–8365. doi: 10.1093/nar/gkr528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qin W, Leonhardt H, Spada F. Usp7 and Uhrf1 control ubiquitination and stability of the maintenance DNA methyltransferase Dnmt1. J Cell Biochem. 2011;112(2):439–444. doi: 10.1002/jcb.22998. [DOI] [PubMed] [Google Scholar]
- 7.Jurkowska RZ, Jurkowski TP, Jeltsch A. Structure and function of mammalian DNA methyltransferases. ChemBioChem. 2011;12(2):206–222. doi: 10.1002/cbic.201000195. [DOI] [PubMed] [Google Scholar]
- 8.Bronner C. Control of DNMT1 abundance in epigenetic inheritance by acetylation, ubiquitylation, and the histone code. Sci Signal. 2011;4(157):pe3. doi: 10.1126/scisignal.2001764. [DOI] [PubMed] [Google Scholar]
- 9.Estève PO, Chang Y, Samaranayake M, et al. A methylation and phosphorylation switch between an adjacent lysine and serine determines human DNMT1 stability. Nat Struct Mol Biol. 2011;18(1):42–48. doi: 10.1038/nsmb.1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brunkow ME, Jeffery EW, Hjerrild KA, et al. Disruption of a new forkhead/winged-helix protein, scurfin, results in the fatal lymphoproliferative disorder of the scurfy mouse. Nat Genet. 2001;27(1):68–73. doi: 10.1038/83784. [DOI] [PubMed] [Google Scholar]
- 11.Kim J, Lahl K, Hori S, et al. Cutting edge: depletion of Foxp3+ cells leads to induction of autoimmunity by specific ablation of regulatory T cells in genetically targeted mice. J Immunol. 2009;183(12):7631–7634. doi: 10.4049/jimmunol.0804308. [DOI] [PubMed] [Google Scholar]
- 12.Fontenot JD, Gavin MA, Rudensky AY. Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat Immunol. 2003;4(4):330–336. doi: 10.1038/ni904. [DOI] [PubMed] [Google Scholar]
- 13.Setoguchi R, Hori S, Takahashi T, Sakaguchi S. Homeostatic maintenance of natural Foxp3(+) CD25(+) CD4(+) regulatory T cells by interleukin (IL)-2 and induction of autoimmune disease by IL-2 neutralization. J Exp Med. 2005;201(5):723–735. doi: 10.1084/jem.20041982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang L, de Zoeten EF, Greene MI, Hancock WW. Immunomodulatory effects of deacetylase inhibitors: therapeutic targeting of FOXP3+ regulatory T cells. Nat Rev Drug Discov. 2009;8(12):969–981. doi: 10.1038/nrd3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang L, Tao R, Hancock WW. Using histone deacetylase inhibitors to enhance Foxp3(+) regulatory T-cell function and induce allograft tolerance. Immunol Cell Biol. 2009;87(3):195–202. doi: 10.1038/icb.2008.106. [DOI] [PubMed] [Google Scholar]
- 16.Tao R, de Zoeten EF, Ozkaynak E, et al. Deacetylase inhibition promotes the generation and function of regulatory T cells. Nat Med. 2007;13(11):1299–1307. doi: 10.1038/nm1652. [DOI] [PubMed] [Google Scholar]
- 17.Lal G, Bromberg JS. Epigenetic mechanisms of regulation of Foxp3 expression. Blood. 2009;114(18):3727–3735. doi: 10.1182/blood-2009-05-219584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lal G, Zhang N, van der Touw W, et al. Epigenetic regulation of Foxp3 expression in regulatory T cells by DNA methylation. J Immunol. 2009;182(1):259–273. doi: 10.4049/jimmunol.182.1.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zheng Y, Josefowicz S, Chaudhry A, Peng XP, Forbush K, Rudensky AY. Role of conserved non-coding DNA elements in the Foxp3 gene in regulatory T-cell fate. Nature. 2010;463(7282):808–812. doi: 10.1038/nature08750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim HP, Leonard WJ. CREB/ATF-dependent T cell receptor-induced FoxP3 gene expression: a role for DNA methylation. J Exp Med. 2007;204(7):1543–1551. doi: 10.1084/jem.20070109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Josefowicz SZ, Wilson CB, Rudensky AY. Cutting edge: TCR stimulation is sufficient for induction of Foxp3 expression in the absence of DNA methyltransferase 1. J Immunol. 2009;182(11):6648–6652. doi: 10.4049/jimmunol.0803320. [DOI] [PubMed] [Google Scholar]
- 22.Jackson-Grusby L, Beard C, Possemato R, et al. Loss of genomic methylation causes p53-dependent apoptosis and epigenetic deregulation. Nat Genet. 2001;27(1):31–39. doi: 10.1038/83730. [DOI] [PubMed] [Google Scholar]
- 23.Kaneda M, Okano M, Hata K, Sado T, Tsujimoto N, Li E, Sasaki H. Essential role for de novo DNA methyltransferase Dnmt3a in paternal and maternal imprinting. Nature. 2004;429(6994):900–903. doi: 10.1038/nature02633. [DOI] [PubMed] [Google Scholar]
- 24.Lee PP, Fitzpatrick DR, Beard C, et al. A critical role for Dnmt1 and DNA methylation in T cell development, function, and survival. Immunity. 2001;15(5):763–774. doi: 10.1016/s1074-7613(01)00227-8. [DOI] [PubMed] [Google Scholar]
- 25.Rubtsov YP, Rasmussen JP, Chi EY, et al. Regulatory T cell-derived interleukin-10 limits inflammation at environmental interfaces. Immunity. 2008;28(4):546–558. doi: 10.1016/j.immuni.2008.02.017. [DOI] [PubMed] [Google Scholar]
- 26.Northrop JK, Thomas RM, Wells AD, Shen H. Epigenetic remodeling of the IL-2 and IFN-gamma loci in memory CD8 T cells is influenced by CD4 T cells. J Immunol. 2006;177(2):1062–1069. doi: 10.4049/jimmunol.177.2.1062. [DOI] [PubMed] [Google Scholar]
- 27.Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. Integrative genomics viewer. Nat Biotechnol. 2011;29(1):24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Battke F, Symons S, Nieselt K. Mayday—integrative analytics for expression data. BMC Bioinformatics. 2010;11:121. doi: 10.1186/1471-2105-11-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Datta J, Ghoshal K, Sharma SM, Tajima S, Jacob ST. Biochemical fractionation reveals association of DNA methyltransferase (Dnmt) 3b with Dnmt1 and that of Dnmt 3a with a histone H3 methyltransferase and Hdac1. J Cell Biochem. 2003;88(5):855–864. doi: 10.1002/jcb.10457. [DOI] [PubMed] [Google Scholar]
- 30.Lyon MF, Peters J, Glenister PH, Ball S, Wright E. The scurfy mouse mutant has previously unrecognized hematological abnormalities and resembles Wiskott-Aldrich syndrome. Proc Natl Acad Sci USA. 1990;87(7):2433–2437. doi: 10.1073/pnas.87.7.2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thomas RM, Gao L, Wells AD. Signals from CD28 induce stable epigenetic modification of the IL-2 promoter. J Immunol. 2005;174(8):4639–4646. doi: 10.4049/jimmunol.174.8.4639. [DOI] [PubMed] [Google Scholar]
- 32.Ronaghi M. Pyrosequencing sheds light on DNA sequencing. Genome Res. 2001;11(1):3–11. doi: 10.1101/gr.11.1.3. [DOI] [PubMed] [Google Scholar]
- 33.Vanvalkenburgh J, Albu DI, Bapanpally C, et al. Critical role of Bcl11b in suppressor function of T regulatory cells and prevention of inflammatory bowel disease. J Exp Med. 2011;208(10):2069–2081. doi: 10.1084/jem.20102683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sullivan BM, Liang HE, Bando JK, et al. Genetic analysis of basophil function in vivo. Nat Immunol. 2011;12(6):527–535. doi: 10.1038/ni.2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kobie JJ, Shah PR, Yang L, Rebhahn JA, Fowell DJ, Mosmann TR. T regulatory and primed uncommitted CD4 T cells express CD73, which suppresses effector CD4 T cells by converting 5′-adenosine monophosphate to adenosine. J Immunol. 2006;177(10):6780–6786. doi: 10.4049/jimmunol.177.10.6780. [DOI] [PubMed] [Google Scholar]
- 36.Deaglio S, Dwyer KM, Gao W, et al. Adenosine generation catalyzed by CD39 and CD73 expressed on regulatory T cells mediates immune suppression. J Exp Med. 2007;204(6):1257–1265. doi: 10.1084/jem.20062512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mandapathil M, Hilldorfer B, Szczepanski MJ, et al. Generation and accumulation of immunosuppressive adenosine by human CD4+CD25highFOXP3+ regulatory T cells. J Biol Chem. 2010;285(10):7176–7186. doi: 10.1074/jbc.M109.047423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Godfrey VL, Wilkinson JE, Russell LB. X-linked lymphoreticular disease in the scurfy (sf) mutant mouse. Am J Pathol. 1991;138(6):1379–1387. [PMC free article] [PubMed] [Google Scholar]
- 39.Zheng L, Sharma R, Gaskin F, Fu SM, Ju ST. A novel role of IL-2 in organ-specific autoimmune inflammation beyond regulatory T cell checkpoint: both IL-2 knockout and Fas mutation prolong lifespan of Scurfy mice but by different mechanisms. J Immunol. 2007;179(12):8035–8041. doi: 10.4049/jimmunol.179.12.8035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wilson CB, Makar KW, Shnyreva M, Fitzpatrick DR. DNA methylation and the expanding epigenetics of T cell lineage commitment. Semin Immunol. 2005;17(2):105–119. doi: 10.1016/j.smim.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 41.Richardson B, Scheinbart L, Strahler J, Gross L, Hanash S, Johnson M. Evidence for impaired T cell DNA methylation in systemic lupus erythematosus and rheumatoid arthritis. Arthritis Rheum. 1990;33(11):1665–1673. doi: 10.1002/art.1780331109. [DOI] [PubMed] [Google Scholar]
- 42.Lei W, Luo Y, Lei W, et al. Abnormal DNA methylation in CD4+ T cells from patients with systemic lupus erythematosus, systemic sclerosis, and dermatomyositis. Scand J Rheumatol. 2009;38(5):369–374. doi: 10.1080/03009740902758875. [DOI] [PubMed] [Google Scholar]
- 43.Quddus J, Johnson KJ, Gavalchin J, Amento EP, Chrisp CE, Yung RL, Richardson BC. Treating activated CD4+ T cells with either of two distinct DNA methyltransferase inhibitors, 5-azacytidine or procainamide, is sufficient to cause a lupus-like disease in syngeneic mice. J Clin Invest. 1993;92(1):38–53. doi: 10.1172/JCI116576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tang Q, Bluestone JA. The Foxp3+ regulatory T cell: a jack of all trades, master of regulation. Nat Immunol. 2008;9(3):239–244. doi: 10.1038/ni1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Crispín JC. Regulatory T cells as modulators of B cell antibody production. Clin Immunol. 2011;140(3):216–217. doi: 10.1016/j.clim.2011.05.006. [DOI] [PubMed] [Google Scholar]
- 46.Leonardo SM, Josephson JA, Hartog NL, Gauld SB. Altered B cell development and anergy in the absence of Foxp3. J Immunol. 2010;185(4):2147–2156. doi: 10.4049/jimmunol.1000136. [DOI] [PubMed] [Google Scholar]
- 47.Riley JL, June CH, Blazar BR. Human T regulatory cell therapy: take a billion or so and call me in the morning. Immunity. 2009;30(5):656–665. doi: 10.1016/j.immuni.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hippen KL, Riley JL, June CH, Blazar BR. Clinical perspectives for regulatory T cells in transplantation tolerance. Semin Immunol. 2011;23(6):462–468. doi: 10.1016/j.smim.2011.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Trzonkowski P, Bieniaszewska M, Juścińska J, et al. First-in-man clinical results of the treatment of patients with graft versus host disease with human ex vivo expanded CD4+CD25+CD127- T regulatory cells. Clin Immunol. 2009;133(1):22–26. doi: 10.1016/j.clim.2009.06.001. [DOI] [PubMed] [Google Scholar]
- 50.Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.