Abstract
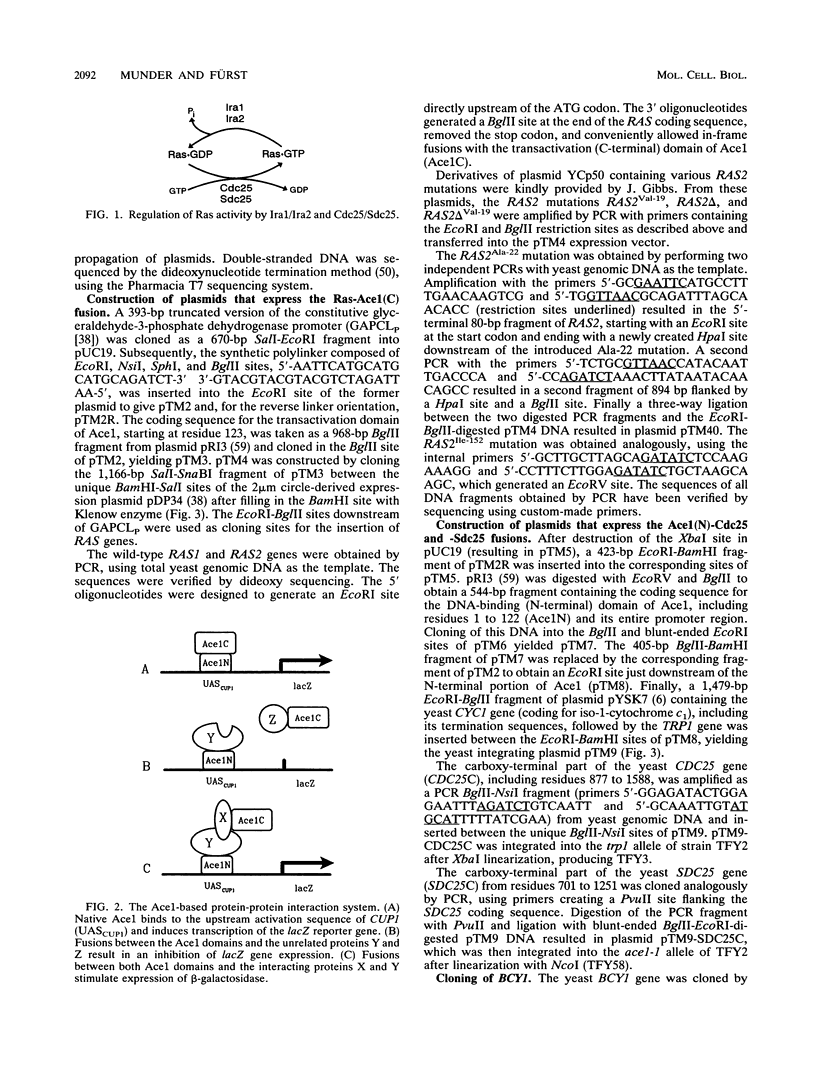
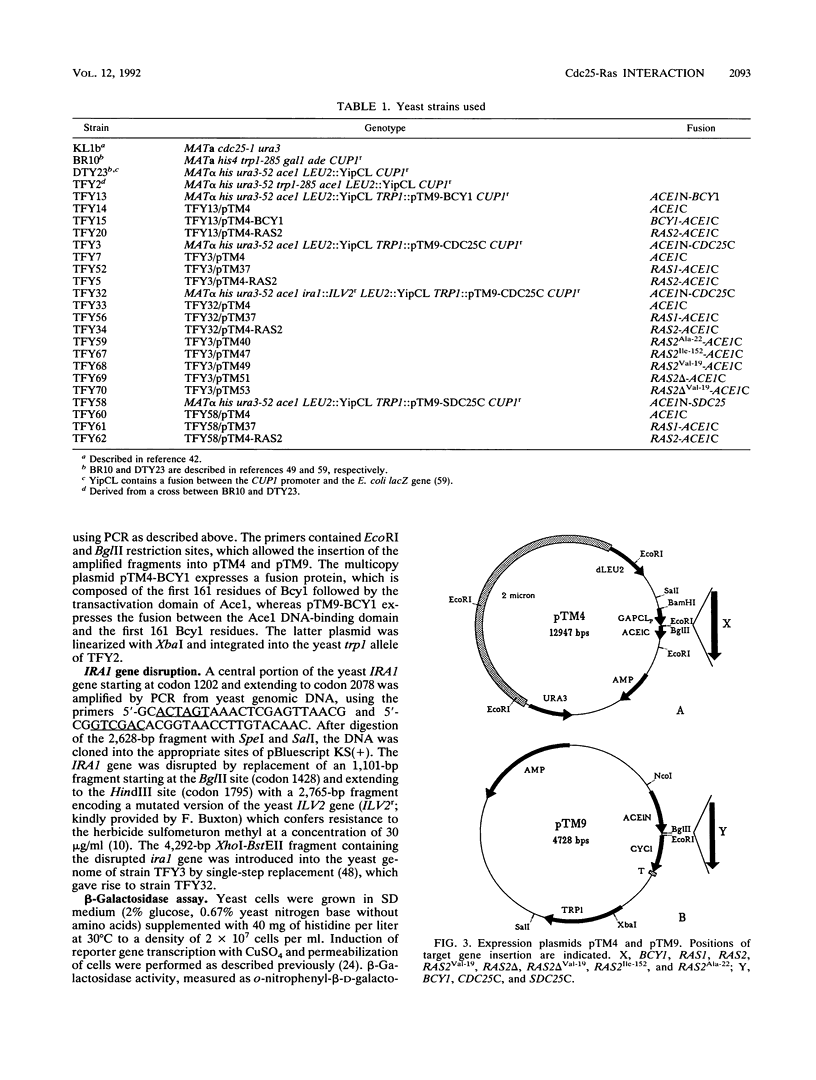
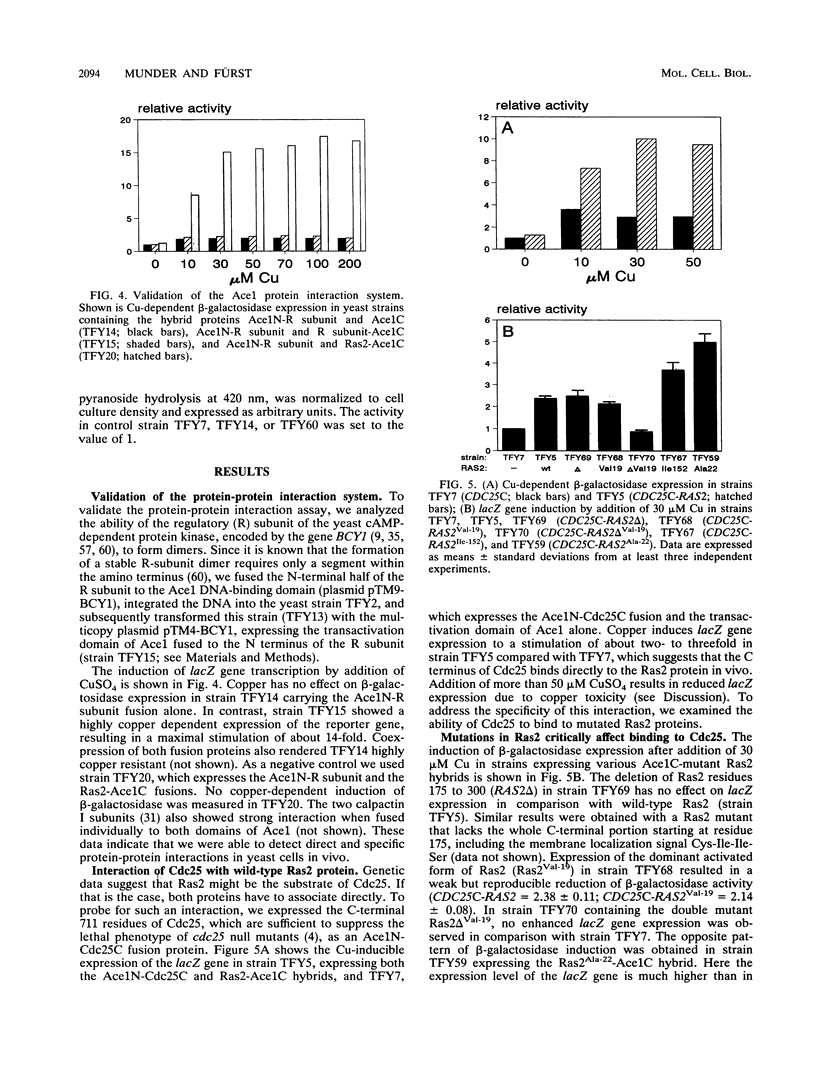
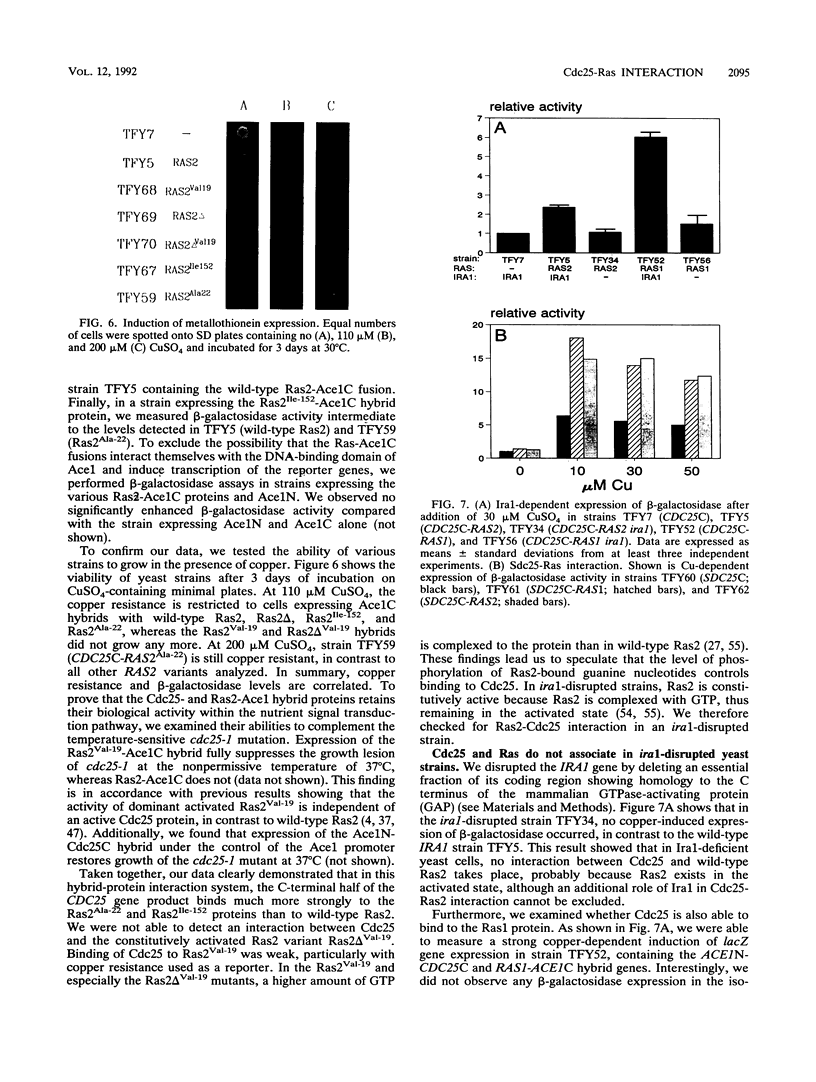
Genetic data suggest that the yeast cell cycle control gene CDC25 is an upstream regulator of RAS2. We have been able to show for the first time that the guanine nucleotide exchange proteins Cdc25 and Sdc25 from Saccharomyces cerevisiae bind directly to their targets Ras1 and Ras2 in vivo. Using the characteristics of the yeast Ace1 transcriptional activator to probe for protein-protein interaction, we found that the CDC25 gene product binds specifically to wild-type Ras2 but not to the mutated Ras2Val-19 and Ras2 delta Val-19 proteins. The binding properties of Cdc25 to Ras2 were strongly diminished in yeast cells expressing an inactive Ira1 protein, which normally acts as a negative regulator of Ras activity. On the basis of these data, we propose that the ability of Cdc25 to interact with Ras2 proteins is strongly dependent on the activation state of Ras2. Cdc25 binds predominantly to the catalytically inactive GDP-bound form of Ras2, whereas a conformational change of Ras2 to its activated GTP-bound state results in its loss of binding affinity to Cdc25.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Becker D. M., Guarente L. High-efficiency transformation of yeast by electroporation. Methods Enzymol. 1991;194:182–187. doi: 10.1016/0076-6879(91)94015-5. [DOI] [PubMed] [Google Scholar]
- Bourne H. R., Sanders D. A., McCormick F. The GTPase superfamily: conserved structure and molecular mechanism. Nature. 1991 Jan 10;349(6305):117–127. doi: 10.1038/349117a0. [DOI] [PubMed] [Google Scholar]
- Broach J. R., Deschenes R. J. The function of ras genes in Saccharomyces cerevisiae. Adv Cancer Res. 1990;54:79–139. doi: 10.1016/s0065-230x(08)60809-x. [DOI] [PubMed] [Google Scholar]
- Broek D., Toda T., Michaeli T., Levin L., Birchmeier C., Zoller M., Powers S., Wigler M. The S. cerevisiae CDC25 gene product regulates the RAS/adenylate cyclase pathway. Cell. 1987 Mar 13;48(5):789–799. doi: 10.1016/0092-8674(87)90076-6. [DOI] [PubMed] [Google Scholar]
- Brünger A. T., Milburn M. V., Tong L., deVos A. M., Jancarik J., Yamaizumi Z., Nishimura S., Ohtsuka E., Kim S. H. Crystal structure of an active form of RAS protein, a complex of a GTP analog and the HRAS p21 catalytic domain. Proc Natl Acad Sci U S A. 1990 Jun;87(12):4849–4853. doi: 10.1073/pnas.87.12.4849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler G., Thiele D. J. ACE2, an activator of yeast metallothionein expression which is homologous to SWI5. Mol Cell Biol. 1991 Jan;11(1):476–485. doi: 10.1128/mcb.11.1.476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butt T. R., Sternberg E. J., Gorman J. A., Clark P., Hamer D., Rosenberg M., Crooke S. T. Copper metallothionein of yeast, structure of the gene, and regulation of expression. Proc Natl Acad Sci U S A. 1984 Jun;81(11):3332–3336. doi: 10.1073/pnas.81.11.3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camonis J. H., Jacquet M. A new RAS mutation that suppresses the CDC25 gene requirement for growth of Saccharomyces cerevisiae. Mol Cell Biol. 1988 Jul;8(7):2980–2983. doi: 10.1128/mcb.8.7.2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon J. F., Tatchell K. Characterization of Saccharomyces cerevisiae genes encoding subunits of cyclic AMP-dependent protein kinase. Mol Cell Biol. 1987 Aug;7(8):2653–2663. doi: 10.1128/mcb.7.8.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Créchet J. B., Poullet P., Camonis J., Jacquet M., Parmeggiani A. Different kinetic properties of the two mutants, RAS2Ile152 and RAS2Val19, that suppress the CDC25 requirement in RAS/adenylate cyclase pathway in Saccharomyces cerevisiae. J Biol Chem. 1990 Jan 25;265(3):1563–1568. [PubMed] [Google Scholar]
- Créchet J. B., Poullet P., Mistou M. Y., Parmeggiani A., Camonis J., Boy-Marcotte E., Damak F., Jacquet M. Enhancement of the GDP-GTP exchange of RAS proteins by the carboxyl-terminal domain of SCD25. Science. 1990 May 18;248(4957):866–868. doi: 10.1126/science.2188363. [DOI] [PubMed] [Google Scholar]
- Culotta V. C., Hsu T., Hu S., Fürst P., Hamer D. Copper and the ACE1 regulatory protein reversibly induce yeast metallothionein gene transcription in a mouse extract. Proc Natl Acad Sci U S A. 1989 Nov;86(21):8377–8381. doi: 10.1073/pnas.86.21.8377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damak F., Boy-Marcotte E., Le-Roscouet D., Guilbaud R., Jacquet M. SDC25, a CDC25-like gene which contains a RAS-activating domain and is a dispensable gene of Saccharomyces cerevisiae. Mol Cell Biol. 1991 Jan;11(1):202–212. doi: 10.1128/mcb.11.1.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeFeo-Jones D., Tatchell K., Robinson L. C., Sigal I. S., Vass W. C., Lowy D. R., Scolnick E. M. Mammalian and yeast ras gene products: biological function in their heterologous systems. Science. 1985 Apr 12;228(4696):179–184. doi: 10.1126/science.3883495. [DOI] [PubMed] [Google Scholar]
- Engelberg D., Simchen G., Levitzki A. In vitro reconstitution of cdc25 regulated S. cerevisiae adenylyl cyclase and its kinetic properties. EMBO J. 1990 Mar;9(3):641–651. doi: 10.1002/j.1460-2075.1990.tb08156.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedor-Chaiken M., Deschenes R. J., Broach J. R. SRV2, a gene required for RAS activation of adenylate cyclase in yeast. Cell. 1990 Apr 20;61(2):329–340. doi: 10.1016/0092-8674(90)90813-t. [DOI] [PubMed] [Google Scholar]
- Feig L. A., Cooper G. M. Inhibition of NIH 3T3 cell proliferation by a mutant ras protein with preferential affinity for GDP. Mol Cell Biol. 1988 Aug;8(8):3235–3243. doi: 10.1128/mcb.8.8.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feig L. A., Pan B. T., Roberts T. M., Cooper G. M. Isolation of ras GTP-binding mutants using an in situ colony-binding assay. Proc Natl Acad Sci U S A. 1986 Jul;83(13):4607–4611. doi: 10.1073/pnas.83.13.4607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field J., Vojtek A., Ballester R., Bolger G., Colicelli J., Ferguson K., Gerst J., Kataoka T., Michaeli T., Powers S. Cloning and characterization of CAP, the S. cerevisiae gene encoding the 70 kd adenylyl cyclase-associated protein. Cell. 1990 Apr 20;61(2):319–327. doi: 10.1016/0092-8674(90)90812-s. [DOI] [PubMed] [Google Scholar]
- Field J., Xu H. P., Michaeli T., Ballester R., Sass P., Wigler M., Colicelli J. Mutations of the adenylyl cyclase gene that block RAS function in Saccharomyces cerevisiae. Science. 1990 Jan 26;247(4941):464–467. doi: 10.1126/science.2405488. [DOI] [PubMed] [Google Scholar]
- Fields S., Song O. A novel genetic system to detect protein-protein interactions. Nature. 1989 Jul 20;340(6230):245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- Fogel S., Welch J. W. Tandem gene amplification mediates copper resistance in yeast. Proc Natl Acad Sci U S A. 1982 Sep;79(17):5342–5346. doi: 10.1073/pnas.79.17.5342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fürst P., Hu S., Hackett R., Hamer D. Copper activates metallothionein gene transcription by altering the conformation of a specific DNA binding protein. Cell. 1988 Nov 18;55(4):705–717. doi: 10.1016/0092-8674(88)90229-2. [DOI] [PubMed] [Google Scholar]
- Gerst J. E., Ferguson K., Vojtek A., Wigler M., Field J. CAP is a bifunctional component of the Saccharomyces cerevisiae adenylyl cyclase complex. Mol Cell Biol. 1991 Mar;11(3):1248–1257. doi: 10.1128/mcb.11.3.1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs J. B., Marshall M. S. The ras oncogene--an important regulatory element in lower eucaryotic organisms. Microbiol Rev. 1989 Jun;53(2):171–185. doi: 10.1128/mr.53.2.171-185.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs J. B., Schaber M. D., Marshall M. S., Scolnick E. M., Sigal I. S. Identification of guanine nucleotides bound to ras-encoded proteins in growing yeast cells. J Biol Chem. 1987 Aug 5;262(22):10426–10429. [PubMed] [Google Scholar]
- Goody R. S., Frech M., Wittinghofer A. Affinity of guanine nucleotide binding proteins for their ligands: facts and artefacts. Trends Biochem Sci. 1991 Sep;16(9):327–328. doi: 10.1016/0968-0004(91)90134-h. [DOI] [PubMed] [Google Scholar]
- Hamer D. H. Metallothionein. Annu Rev Biochem. 1986;55:913–951. doi: 10.1146/annurev.bi.55.070186.004405. [DOI] [PubMed] [Google Scholar]
- Herskowitz I. Functional inactivation of genes by dominant negative mutations. Nature. 1987 Sep 17;329(6136):219–222. doi: 10.1038/329219a0. [DOI] [PubMed] [Google Scholar]
- Johnsson N., Marriott G., Weber K. p36, the major cytoplasmic substrate of src tyrosine protein kinase, binds to its p11 regulatory subunit via a short amino-terminal amphiphatic helix. EMBO J. 1988 Aug;7(8):2435–2442. doi: 10.1002/j.1460-2075.1988.tb03089.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones S., Vignais M. L., Broach J. R. The CDC25 protein of Saccharomyces cerevisiae promotes exchange of guanine nucleotides bound to ras. Mol Cell Biol. 1991 May;11(5):2641–2646. doi: 10.1128/mcb.11.5.2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataoka T., Powers S., Cameron S., Fasano O., Goldfarb M., Broach J., Wigler M. Functional homology of mammalian and yeast RAS genes. Cell. 1985 Jan;40(1):19–26. doi: 10.1016/0092-8674(85)90304-6. [DOI] [PubMed] [Google Scholar]
- Krengel U., Schlichting I., Scherer A., Schumann R., Frech M., John J., Kabsch W., Pai E. F., Wittinghofer A. Three-dimensional structures of H-ras p21 mutants: molecular basis for their inability to function as signal switch molecules. Cell. 1990 Aug 10;62(3):539–548. doi: 10.1016/0092-8674(90)90018-a. [DOI] [PubMed] [Google Scholar]
- Kunisawa R., Davis T. N., Urdea M. S., Thorner J. Complete nucleotide sequence of the gene encoding the regulatory subunit of 3',5'-cyclic AMP-dependent protein kinase from the yeast Saccharomyces cerevisiae. Nucleic Acids Res. 1987 Jan 12;15(1):368–369. doi: 10.1093/nar/15.1.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall M. S., Gibbs J. B., Scolnick E. M., Sigal I. S. Regulatory function of the Saccharomyces cerevisiae RAS C-terminus. Mol Cell Biol. 1987 Jul;7(7):2309–2315. doi: 10.1128/mcb.7.7.2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milburn M. V., Tong L., deVos A. M., Brünger A., Yamaizumi Z., Nishimura S., Kim S. H. Molecular switch for signal transduction: structural differences between active and inactive forms of protooncogenic ras proteins. Science. 1990 Feb 23;247(4945):939–945. doi: 10.1126/science.2406906. [DOI] [PubMed] [Google Scholar]
- Mitts M. R., Bradshaw-Rouse J., Heideman W. Interactions between adenylate cyclase and the yeast GTPase-activating protein IRA1. Mol Cell Biol. 1991 Sep;11(9):4591–4598. doi: 10.1128/mcb.11.9.4591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munder T., Küntzel H. Glucose-induced cAMP signaling in Saccharomyces cerevisiae is mediated by the CDC25 protein. FEBS Lett. 1989 Jan 2;242(2):341–345. doi: 10.1016/0014-5793(89)80498-3. [DOI] [PubMed] [Google Scholar]
- Munder T., Mink M., Küntzel H. Domains of the Saccharomyces cerevisiae CDC25 gene controlling mitosis and meiosis. Mol Gen Genet. 1988 Oct;214(2):271–277. doi: 10.1007/BF00337721. [DOI] [PubMed] [Google Scholar]
- Pai E. F., Krengel U., Petsko G. A., Goody R. S., Kabsch W., Wittinghofer A. Refined crystal structure of the triphosphate conformation of H-ras p21 at 1.35 A resolution: implications for the mechanism of GTP hydrolysis. EMBO J. 1990 Aug;9(8):2351–2359. doi: 10.1002/j.1460-2075.1990.tb07409.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers S., O'Neill K., Wigler M. Dominant yeast and mammalian RAS mutants that interfere with the CDC25-dependent activation of wild-type RAS in Saccharomyces cerevisiae. Mol Cell Biol. 1989 Feb;9(2):390–395. doi: 10.1128/mcb.9.2.390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson L. C., Gibbs J. B., Marshall M. S., Sigal I. S., Tatchell K. CDC25: a component of the RAS-adenylate cyclase pathway in Saccharomyces cerevisiae. Science. 1987 Mar 6;235(4793):1218–1221. doi: 10.1126/science.3547648. [DOI] [PubMed] [Google Scholar]
- Rothstein R. J. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- Rymond B. C., Zitomer R. S., Schümperli D., Rosenberg M. The expression in yeast of the Escherichia coli galK gene on CYC1::galK fusion plasmids. Gene. 1983 Nov;25(2-3):249–262. doi: 10.1016/0378-1119(83)90229-9. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigal I. S., Gibbs J. B., D'Alonzo J. S., Temeles G. L., Wolanski B. S., Socher S. H., Scolnick E. M. Mutant ras-encoded proteins with altered nucleotide binding exert dominant biological effects. Proc Natl Acad Sci U S A. 1986 Feb;83(4):952–956. doi: 10.1073/pnas.83.4.952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stacey D. W., Feig L. A., Gibbs J. B. Dominant inhibitory Ras mutants selectively inhibit the activity of either cellular or oncogenic Ras. Mol Cell Biol. 1991 Aug;11(8):4053–4064. doi: 10.1128/mcb.11.8.4053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K., Lin B. K., Wood D. R., Tamanoi F. IRA2, an upstream negative regulator of RAS in yeast, is a RAS GTPase-activating protein. Proc Natl Acad Sci U S A. 1991 Jan 15;88(2):468–472. doi: 10.1073/pnas.88.2.468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K., Matsumoto K., Toh-E A. IRA1, an inhibitory regulator of the RAS-cyclic AMP pathway in Saccharomyces cerevisiae. Mol Cell Biol. 1989 Feb;9(2):757–768. doi: 10.1128/mcb.9.2.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K., Nakafuku M., Satoh T., Marshall M. S., Gibbs J. B., Matsumoto K., Kaziro Y., Toh-e A. S. cerevisiae genes IRA1 and IRA2 encode proteins that may be functionally equivalent to mammalian ras GTPase activating protein. Cell. 1990 Mar 9;60(5):803–807. doi: 10.1016/0092-8674(90)90094-u. [DOI] [PubMed] [Google Scholar]
- Tanaka K., Nakafuku M., Tamanoi F., Kaziro Y., Matsumoto K., Toh-e A. IRA2, a second gene of Saccharomyces cerevisiae that encodes a protein with a domain homologous to mammalian ras GTPase-activating protein. Mol Cell Biol. 1990 Aug;10(8):4303–4313. doi: 10.1128/mcb.10.8.4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor S. S., Buechler J. A., Yonemoto W. cAMP-dependent protein kinase: framework for a diverse family of regulatory enzymes. Annu Rev Biochem. 1990;59:971–1005. doi: 10.1146/annurev.bi.59.070190.004543. [DOI] [PubMed] [Google Scholar]
- Thevelein J. M. Fermentable sugars and intracellular acidification as specific activators of the RAS-adenylate cyclase signalling pathway in yeast: the relationship to nutrient-induced cell cycle control. Mol Microbiol. 1991 Jun;5(6):1301–1307. doi: 10.1111/j.1365-2958.1991.tb00776.x. [DOI] [PubMed] [Google Scholar]
- Thiele D. J. ACE1 regulates expression of the Saccharomyces cerevisiae metallothionein gene. Mol Cell Biol. 1988 Jul;8(7):2745–2752. doi: 10.1128/mcb.8.7.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toda T., Cameron S., Sass P., Zoller M., Scott J. D., McMullen B., Hurwitz M., Krebs E. G., Wigler M. Cloning and characterization of BCY1, a locus encoding a regulatory subunit of the cyclic AMP-dependent protein kinase in Saccharomyces cerevisiae. Mol Cell Biol. 1987 Apr;7(4):1371–1377. doi: 10.1128/mcb.7.4.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch J., Fogel S., Buchman C., Karin M. The CUP2 gene product regulates the expression of the CUP1 gene, coding for yeast metallothionein. EMBO J. 1989 Jan;8(1):255–260. doi: 10.1002/j.1460-2075.1989.tb03371.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigler M., Field J., Powers S., Broek D., Toda T., Cameron S., Nikawa J., Michaeli T., Colicelli J., Ferguson K. Studies of RAS function in the yeast Saccharomyces cerevisiae. Cold Spring Harb Symp Quant Biol. 1988;53(Pt 2):649–655. doi: 10.1101/sqb.1988.053.01.074. [DOI] [PubMed] [Google Scholar]
- Yang W. M., Gahl W., Hamer D. Role of heat shock transcription factor in yeast metallothionein gene expression. Mol Cell Biol. 1991 Jul;11(7):3676–3681. doi: 10.1128/mcb.11.7.3676. [DOI] [PMC free article] [PubMed] [Google Scholar]



