Abstract
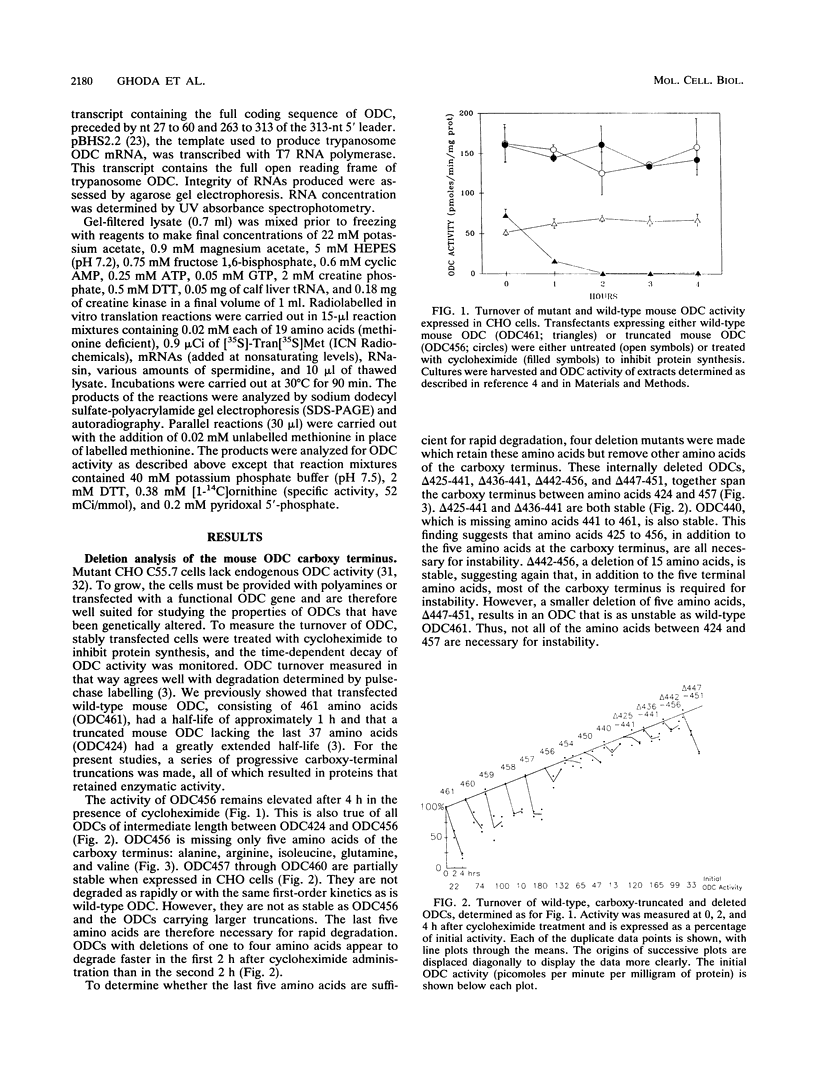
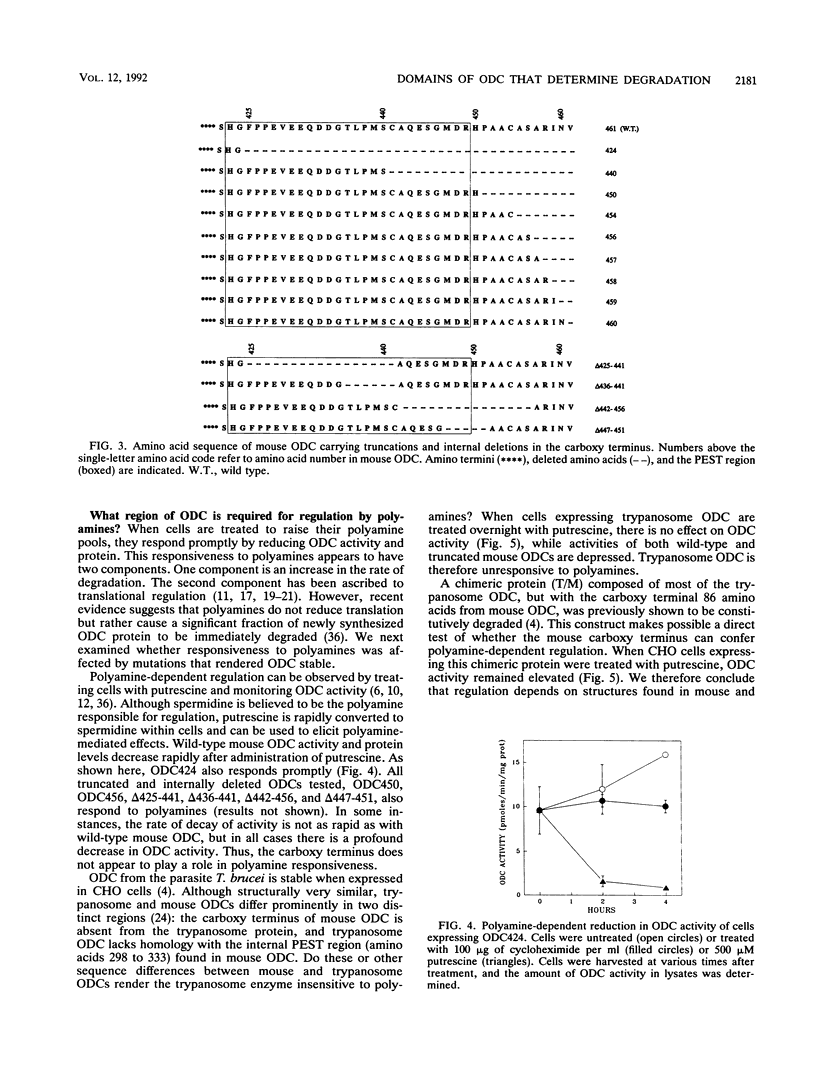
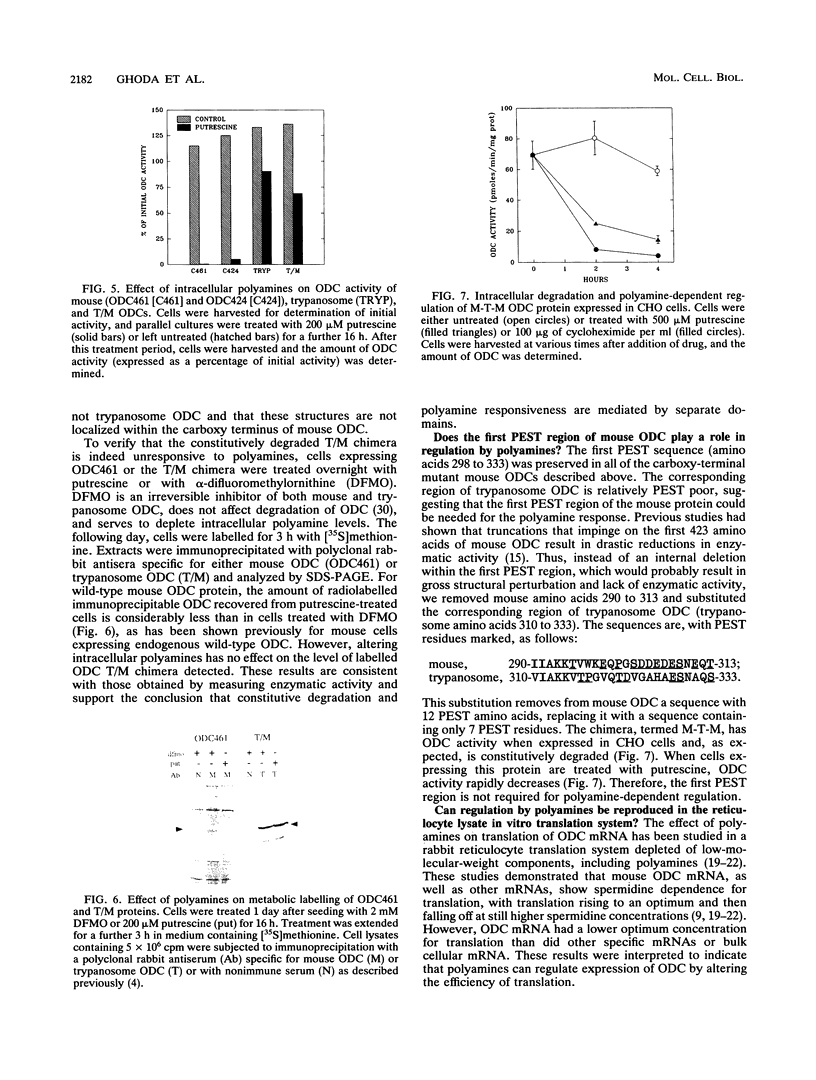
Mammalian ornithine decarboxylase (ODC), a key enzyme in polyamine biosynthesis, is rapidly degraded in cells, an attribute important to the regulation of its activity. Mutant and chimeric ODCs were created to determine the structural requirements for two modes of proteolysis. Constitutive degradation requires the carboxy terminus and is independent of intracellular polyamines. Truncation of five or more carboxy-terminal amino acids prevents this mode of degradation, as do several internal deletions within the 37 carboxy-most amino acids that spare the last five residues. Polyamine-dependent degradation of ODC requires a distinct region outside the carboxy terminus. The ODC of a parasite, Trypanosoma brucei, is structurally very similar to mouse ODC but lacks the carboxy-terminal domain; it is not a substrate for either pathway. The regulatory properties of enzymatically active chimeric proteins incorporating regions of the two ODCs support the conclusion that distinct domains of mouse ODC confer constitutive degradation and polyamine-mediated regulation. Mouse ODC contains two PEST regions. The first was not required for either form of degradation; major deletions within the second ablated constitutive degradation. When mouse and T. brucei ODC RNAs were translated in vitro in a reticulocyte lysate system, the effects of polyamine concentration on ODC protein production and activity were similar for the two mRNAs, which contradicts claims that this system accurately reflects the in vivo effects of polyamines on responsive ODCs.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Clark J. L. Specific induction of ornithine decarboxylase in 3T3 mouse fibroblasts by pituitary growth factors: cell density-dependent biphasic response and alteration of half-life. Biochemistry. 1974 Oct 22;13(22):4668–4674. doi: 10.1021/bi00719a031. [DOI] [PubMed] [Google Scholar]
- Dasso M. C., Jackson R. J. On the fidelity of mRNA translation in the nuclease-treated rabbit reticulocyte lysate system. Nucleic Acids Res. 1989 Apr 25;17(8):3129–3144. doi: 10.1093/nar/17.8.3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghoda L., Phillips M. A., Bass K. E., Wang C. C., Coffino P. Trypanosome ornithine decarboxylase is stable because it lacks sequences found in the carboxyl terminus of the mouse enzyme which target the latter for intracellular degradation. J Biol Chem. 1990 Jul 15;265(20):11823–11826. [PubMed] [Google Scholar]
- Ghoda L., van Daalen Wetters T., Macrae M., Ascherman D., Coffino P. Prevention of rapid intracellular degradation of ODC by a carboxyl-terminal truncation. Science. 1989 Mar 17;243(4897):1493–1495. doi: 10.1126/science.2928784. [DOI] [PubMed] [Google Scholar]
- Gupta M., Coffino P. Mouse ornithine decarboxylase. Complete amino acid sequence deduced from cDNA. J Biol Chem. 1985 Mar 10;260(5):2941–2944. [PubMed] [Google Scholar]
- Holm I., Persson L., Stjernborg L., Thorsson L., Heby O. Feedback control of ornithine decarboxylase expression by polyamines. Analysis of ornithine decarboxylase mRNA distribution in polysome profiles and of translation of this mRNA in vitro. Biochem J. 1989 Mar 1;258(2):343–350. doi: 10.1042/bj2580343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hölttä E., Pohjanpelto P. Control of ornithine decarboxylase in Chinese hamster ovary cells by polyamines. Translational inhibition of synthesis and acceleration of degradation of the enzyme by putrescine, spermidine, and spermine. J Biol Chem. 1986 Jul 15;261(20):9502–9508. [PubMed] [Google Scholar]
- Isomaa V. V., Pajunen A. E., Bardin C. W., Jänne O. A. Ornithine decarboxylase in mouse kidney. Purification, characterization, and radioimmunological determination of the enzyme protein. J Biol Chem. 1983 Jun 10;258(11):6735–6740. [PubMed] [Google Scholar]
- Isomaa V. V., Pajunen A. E., Bardin C. W., Jänne O. A. Ornithine decarboxylase in mouse kidney. Purification, characterization, and radioimmunological determination of the enzyme protein. J Biol Chem. 1983 Jun 10;258(11):6735–6740. [PubMed] [Google Scholar]
- Jackson R. J., Hunt T. Preparation and use of nuclease-treated rabbit reticulocyte lysates for the translation of eukaryotic messenger RNA. Methods Enzymol. 1983;96:50–74. doi: 10.1016/s0076-6879(83)96008-1. [DOI] [PubMed] [Google Scholar]
- Jänne J., Hölttä E. Regulation of ornithine decarboxylase activity by putrescine and spermidine in rat liver. Biochem Biophys Res Commun. 1974 Nov 27;61(2):449–456. doi: 10.1016/0006-291x(74)90977-2. [DOI] [PubMed] [Google Scholar]
- Kahana C., Nathans D. Translational regulation of mammalian ornithine decarboxylase by polyamines. J Biol Chem. 1985 Dec 15;260(29):15390–15393. [PubMed] [Google Scholar]
- Kay J. E., Lindsay V. J. Control of ornithine decarboxylase activity in stimulated human lymphocytes by putrescine and spermidine. Biochem J. 1973 Apr;132(4):791–796. doi: 10.1042/bj1320791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U S A. 1985 Jan;82(2):488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loetscher P., Pratt G., Rechsteiner M. The C terminus of mouse ornithine decarboxylase confers rapid degradation on dihydrofolate reductase. Support for the pest hypothesis. J Biol Chem. 1991 Jun 15;266(17):11213–11220. [PubMed] [Google Scholar]
- Macrae M., Coffino P. Complementation of a polyamine-deficient Escherichia coli mutant by expression of mouse ornithine decarboxylase. Mol Cell Biol. 1987 Jan;7(1):564–567. doi: 10.1128/mcb.7.1.564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConlogue L., Dana S. L., Coffino P. Multiple mechanisms are responsible for altered expression of ornithine decarboxylase in overproducing variant cells. Mol Cell Biol. 1986 Aug;6(8):2865–2871. doi: 10.1128/mcb.6.8.2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsell D. A., Silber K. R., Sauer R. T. Carboxy-terminal determinants of intracellular protein degradation. Genes Dev. 1990 Feb;4(2):277–286. doi: 10.1101/gad.4.2.277. [DOI] [PubMed] [Google Scholar]
- Pegg A. E., Madhubala R., Kameji T., Bergeron R. J. Control of ornithine decarboxylase activity in alpha-difluoromethylornithine-resistant L1210 cells by polyamines and synthetic analogues. J Biol Chem. 1988 Aug 5;263(22):11008–11014. [PubMed] [Google Scholar]
- Pegg A. E. Recent advances in the biochemistry of polyamines in eukaryotes. Biochem J. 1986 Mar 1;234(2):249–262. doi: 10.1042/bj2340249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson L., Holm I., Heby O. Regulation of ornithine decarboxylase mRNA translation by polyamines. Studies using a cell-free system and a cell line with an amplified ornithine decarboxylase gene. J Biol Chem. 1988 Mar 5;263(7):3528–3533. [PubMed] [Google Scholar]
- Persson L., Holm I., Heby O. Translational regulation of ornithine decarboxylase by polyamines. FEBS Lett. 1986 Sep 15;205(2):175–178. doi: 10.1016/0014-5793(86)80892-4. [DOI] [PubMed] [Google Scholar]
- Persson L., Oredsson S. M., Anehus S., Heby O. Ornithine decarboxylase inhibitors increase the cellular content of the enzyme: implications for translational regulation. Biochem Biophys Res Commun. 1985 Aug 30;131(1):239–245. doi: 10.1016/0006-291x(85)91794-2. [DOI] [PubMed] [Google Scholar]
- Phillips M. A., Coffino P., Wang C. C. Cloning and sequencing of the ornithine decarboxylase gene from Trypanosoma brucei. Implications for enzyme turnover and selective difluoromethylornithine inhibition. J Biol Chem. 1987 Jun 25;262(18):8721–8727. [PubMed] [Google Scholar]
- Phillips M. A., Coffino P., Wang C. C. Trypanosoma brucei ornithine decarboxylase: enzyme purification, characterization, and expression in Escherichia coli. J Biol Chem. 1988 Dec 5;263(34):17933–17941. [PubMed] [Google Scholar]
- Rogers S., Wells R., Rechsteiner M. Amino acid sequences common to rapidly degraded proteins: the PEST hypothesis. Science. 1986 Oct 17;234(4774):364–368. doi: 10.1126/science.2876518. [DOI] [PubMed] [Google Scholar]
- Rosenberg-Hasson Y., Bercovich Z., Kahana C. Characterization of sequences involved in mediating degradation of ornithine decarboxylase in cells and in reticulocyte lysate. Eur J Biochem. 1991 Mar 28;196(3):647–651. doi: 10.1111/j.1432-1033.1991.tb15861.x. [DOI] [PubMed] [Google Scholar]
- Rosenberg-Hasson Y., Strumpf D., Kahana C. Mouse ornithine decarboxylase is phosphorylated by casein kinase-II at a predominant single location (serine 303). Eur J Biochem. 1991 Apr 23;197(2):419–424. doi: 10.1111/j.1432-1033.1991.tb15927.x. [DOI] [PubMed] [Google Scholar]
- Russell D. H., Snyder S. H. Amine synthesis in regenerating rat liver: extremely rapid turnover of ornithine decarboxylase. Mol Pharmacol. 1969 May;5(3):253–262. [PubMed] [Google Scholar]
- Schimke R. T. Control of enzyme levels in mammalian tissues. Adv Enzymol Relat Areas Mol Biol. 1973;37:135–187. doi: 10.1002/9780470122822.ch3. [DOI] [PubMed] [Google Scholar]
- Seely J. E., Pegg A. E. Changes in mouse kidney ornithine decarboxylase activity are brought about by changes in the amount of enzyme protein as measured by radioimmunoassay. J Biol Chem. 1983 Feb 25;258(4):2496–2500. [PubMed] [Google Scholar]
- Seely J. E., Pösö H., Pegg A. E. Effect of androgens on turnover of ornithine decarboxylase in mouse kidney. Studies using labeling of the enzyme by reaction with [14C] alpha-difluoromethylornithine. J Biol Chem. 1982 Jul 10;257(13):7549–7553. [PubMed] [Google Scholar]
- Steglich C., Grens A., Scheffler I. E. Chinese hamster cells deficient in ornithine decarboxylase activity: reversion by gene amplification and by azacytidine treatment. Somat Cell Mol Genet. 1985 Jan;11(1):11–23. doi: 10.1007/BF01534730. [DOI] [PubMed] [Google Scholar]
- Steglich C., Scheffler I. E. An ornithine decarboxylase-deficient mutant of Chinese hamster ovary cells. J Biol Chem. 1982 Apr 25;257(8):4603–4609. [PubMed] [Google Scholar]
- Tabor C. W., Tabor H. Polyamines. Annu Rev Biochem. 1984;53:749–790. doi: 10.1146/annurev.bi.53.070184.003533. [DOI] [PubMed] [Google Scholar]
- Thomas K. R., Capecchi M. R. Site-directed mutagenesis by gene targeting in mouse embryo-derived stem cells. Cell. 1987 Nov 6;51(3):503–512. doi: 10.1016/0092-8674(87)90646-5. [DOI] [PubMed] [Google Scholar]
- van Daalen Wetters T., Brabant M., Coffino P. Regulation of mouse ornithine decarboxylase activity by cell growth, serum and tetradecanoyl phorbol acetate is governed primarily by sequences within the coding region of the gene. Nucleic Acids Res. 1989 Dec 11;17(23):9843–9860. doi: 10.1093/nar/17.23.9843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Daalen Wetters T., Macrae M., Brabant M., Sittler A., Coffino P. Polyamine-mediated regulation of mouse ornithine decarboxylase is posttranslational. Mol Cell Biol. 1989 Dec;9(12):5484–5490. doi: 10.1128/mcb.9.12.5484. [DOI] [PMC free article] [PubMed] [Google Scholar]