Abstract
TET (Ten-Eleven-Translocation) proteins are Fe(II) and α-ketoglutarate-dependent dioxygenases1-3 that modify the methylation status of DNA by successively oxidizing 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC), 5-formylcytosine and 5-carboxycytosine1,3-5, potential intermediates in the active erasure of DNA methylation marks5,6. We show here that IDAX/ CXXC4, a player in the Wnt signaling pathway7 that has been implicated in malignant renal cell carcinoma8 and colonic villous adenoma9, functions as a negative regulator of TET2 protein expression. IDAX/ CXXC4 was originally encoded within an ancestral TET2 gene that underwent a chromosomal gene inversion during evolution, thus separating the TET2 CXXC domain from the catalytic domain. The Idax CXXC domain binds DNA sequences containing unmethylated CpGs, localises to promoters and CpG islands in genomic DNA, and interacts directly with the catalytic domain of Tet2. Unexpectedly, Idax expression resulted in caspase activation and Tet2 protein downregulation, in a manner that depended on DNA-binding through the Idax CXXC domain. Idax depletion prevented Tet2 downregulation in differentiating mouse embryonic stem (ES) cells, and shRNA against IDAX increased TET2 protein expression in the human monocytic cell line U937. Notably, we find that the expression and activity of TET3 are also regulated through its CXXC domain. Taken together, these results establish the separate and linked CXXC domains of TET2 and TET3 respectively as novel regulators of caspase activation and TET enzymatic activity.
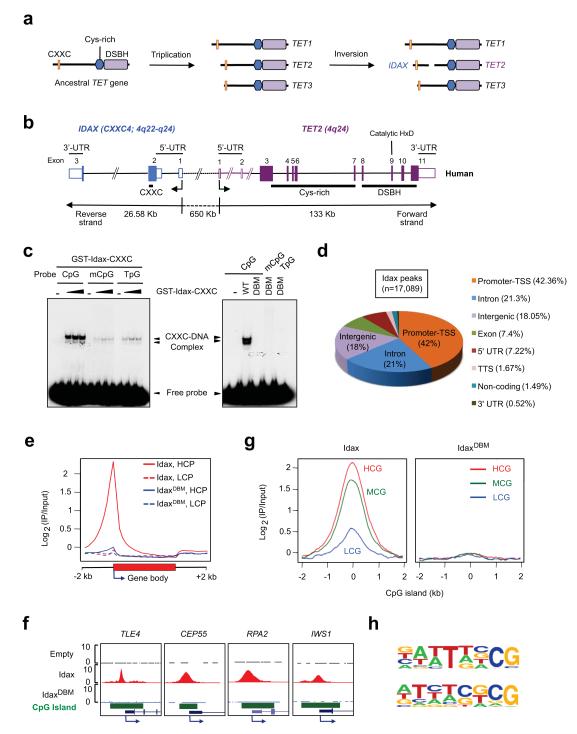
TET proteins are restricted to metazoa and their presence is strictly correlated with the presence of cytosine methylation2,10. Most animals have a single TET orthologue, characterized by an amino (N)-terminal CXXC-type zinc finger domain and a carboxy (C)-terminal catalytic Fe(II) and α-ketoglutarate-dependent dioxygenase domain with an inserted cysteine-rich domain2,10. In jawed vertebrates, the TET genes underwent triplication, and a subsequent chromosomal inversion split the TET2 gene into distinct segments encoding the catalytic and CXXC domains2,10 (Fig. 1a). The ancestral CXXC domain of TET2 is now encoded by a distinct gene, IDAX/ CXXC4, that is transcribed in the opposite direction (Fig. 1b, Supplementary Fig. 1a). Given the evolutionary relation between TET2 and IDAX, and the strong sequence conservation of IDAX/ CXX4 across species (Supplementary Fig. 1b), we asked whether IDAX could influence the nuclear function of TET2.
Fig. 1. IDAX/ CXXC4 preferentially binds CpG-rich DNA.
a. The evolutionary origin of IDAX/ CXXC4. Cys-rich, cysteine-rich; DSBH, double stranded β-helix.
b. The human IDAX and TET2 genes are transcribed in opposite directions.
c. Left, The GST-Idax-CXXC domain preferentially binds unmethylated CpGs in DNA. Right, The TGHQ > AGAA substitution abrogates the DNA-binding activity of the Idax-CXXC domain.
d. Genome-wide distribution of Idax binding sites in HEK293T cells.
e. Idax binds preferentially to TSS at high CpG density (HCP) but not low CpG density (LCP) promoters.
f. Examples of Idax peaks at CpG island (CGI) promoters.
g. Distribution of Idax across high (HCG), medium (MCG) and low (LCG) CpG density CGIs.
h. DNA motifs conserved in Idax-bound loci revealed by de-novo motif discovery analysis.
We assessed the DNA-binding specificity of the Idax CXXC domain. A GST-Idax CXXC domain fusion protein bound DNA oligonucleotides containing a single unmethylated CpG considerably more efficiently than oligonucleotides containing methylated CpG or no CpG (TpG), as also confirmed by competition with excess unlabeled oligonucleotides; the CFP1 CXXC domain, which preferentially binds DNA sequences containing unmethylated CpG11, was used as a control (Fig. 1c, left panel; Supplementary Fig. 2). The CXXC domain is found in several proteins involved in DNA methylation and chromatin modification10-16 (Supplementary Fig. 1c, d). In the structures of the Dnmt1-DNA complex and the MLL CXXC domain, short ‘RSKQ’ and ‘IKKQ’ loops mediate base-specific contacts with DNA containing unmethylated CpG dinucleotides12,13,15. These loops align with the ‘RMKQ’ motif of KDM2A/ CXXC8 required for DNA binding16, the IRQ sequence of the CFP1 CXXC domain that is critical for recognition of unmethylated CpG14, and the 162TGHQ165 sequence in the IDAX CXXC domain (Supplementary Fig. 1c). We generated a homology model of the IDAX CXXC domain and showed that a TGHQ > AGAA substitution abrogated DNA-binding (Fig. 1c, right panel; Supplementary Fig. 3). DNA tethering was required to maintain Idax in the nucleus, since epitope-tagged wild-type Idax and Tet2 were exclusively nuclear in HEK293T cells whereas the DNA-binding mutant, IdaxDBM, was partially present in the cytoplasm (Supplementary Fig. 4).
To determine the genomic distribution of Idax, we used HEK293T cell lines stably expressing full-length Myc-Idax or Myc-IdaxDBM (Supplementary Fig. 5a). Chromatin immunoprecipitation followed by next-generation sequencing identified 17089 peaks for Idax but only 38 peaks for IdaxDBM (Supplementary Tables 1, 2). The majority of Idax peaks were located at promoters / transcription start sites (TSS) and CpG islands (CGIs) of high CpG content (Fig. 1d-g; Supplementary Tables 1, 2; Supplementary Fig. 5b, c). Idax-bound regions were enriched for sequences containing CG dinucleotides, tandem cytosines (as suggested previously for the TET3 CXXC domain17) or both (Fig. 1h, Supplementary Table 3), and were strongly associated with genes involved in mRNA splicing and transcriptional elongation (Supplementary Fig 5 d,e). Thus Idax preferentially associates with CpG-rich regions containing unmodified cytosines in vitro and in cells.
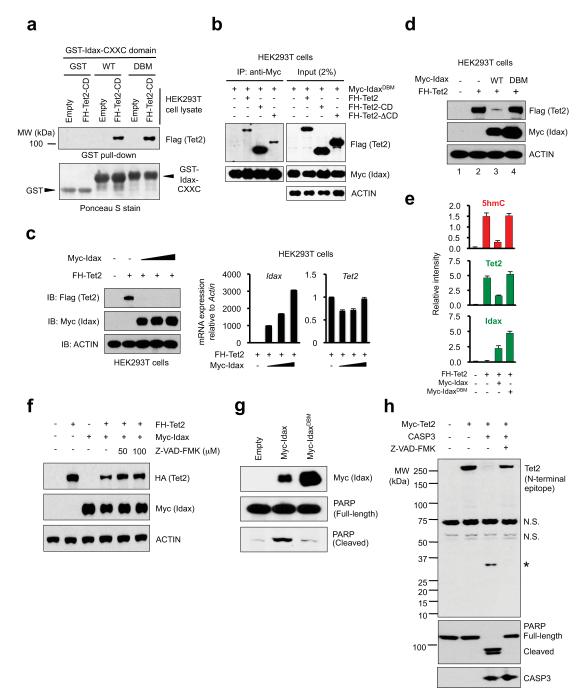
To ask whether Idax and Tet2 physically interact, we incubated GST-Idax-CXXC domain (Supplementary Fig. 2a) with lysates from HEK293T cells expressing Tet2 catalytic domain (Tet2-CD) or the N-terminal region lacking the catalytic domain (Tet2-ΔCD) (Supplementary Fig. 6a, b). Tet2-CD bound strongly to both wild-type Idax and IdaxDBM CXXC domain (Fig. 2a), whereas Tet2-ΔCD bound more weakly (Supplementary Fig. 6c). Co-immunoprecipitation assays confirmed a strong interaction between Myc-IdaxDBM (which does not downregulate Tet2 protein; see below) and full-length Tet2 or Tet2-CD in HEK293T cells (Fig. 2b). The interaction was direct, since Flag-tagged Tet2-CD expressed in insect cells bound GST-Idax-CXXC domain, but not GST-CFP1-CXXC domain, in pulldown assays (Supplementary Fig. 6d, e).
Fig. 2. Idax downregulates Tet2 protein through caspase activation.
a. Both WT and DBM mutant Idax-CXXC domain physically associate with Tet2.
b. Co-immunoprecipitation of Tet2 with IdaxDBM in HEK293T cell lysates.
c. Idax expression decreases the expression of Tet2 protein (left panel) but not Tet2 mRNA (right panel).
d. The DNA-binding activity of Idax is required to cause loss of Tet2 protein.
e. IdaxDBM does not affect Tet2 protein or 5hmC levels in HEK293T cells. Error bars indicate s.d. (n=24 wells).
f. Idax does not decrease Tet2 protein levels in the presence of pan-caspase inhibitor Z-VAD-FMK.
g. PARP cleavage induced by Idax but not IdaxDBM in transfected HEK293T cells.
h. Tet2 is a substrate for recombinant active caspase 3 in vitro.
Unexpectedly, transient co-expression of Myc-Idax and Flag-HA (FH)-tagged Tet2 in HEK293T cells led to disappearance of Tet2 protein and a decrease in 5hmC, with only a minor effect on Tet2 mRNA (Fig. 2c, Supplementary Fig. 7). Idax DNA-binding activity was required, since co-expressed Myc-IdaxDBM did not decrease Tet2 protein or 5hmC (Fig. 2d, e; Supplementary Fig. 8). Myc-IdaxDBM was expressed at considerably higher levels than WT Myc-Idax (Fig. 2d, e, g; Supplementary Fig. 8), suggesting that DNA-bound Idax recruits a degradation complex that targets both Idax and Tet2 (see below, Supplementary Fig. 16). Treatment of cells co-expressing Myc-Idax and Flag-HA-Tet2 with proteasome inhibitors variably rescued the loss of Tet2 protein, whereas treatment with lysosomal inhibitors had no effect (Supplementary Fig. 9a, b). However, Idax was unable to decrease Myc-Tet2 protein levels in cells treated with the pan-caspase inhibitor Z-VAD-FMK (Fig. 2f); moreover, Idax induced nuclear cleavage of PARP, a marker for caspase activation, whereas IdaxDBM did not (Fig. 2g, Supplementary Fig. 9c). Tet2 was a direct target for caspase cleavage, as shown by treatment of HEK293T cell lysates containing Myc-Tet2 with recombinant active human caspase 3 and caspase 8 (Fig. 2h, Supplementary Fig. 9d, e). Neither WT Idax nor IdaxDBM significantly influenced the enzymatic activity of Tet2 in vitro (Supplementary Fig. 10), indicating that the loss of genomic 5hmC in cells co-expressing Tet2 and Idax reflects the loss of Tet2 protein rather than any direct interference with Tet2 enzymatic activity.
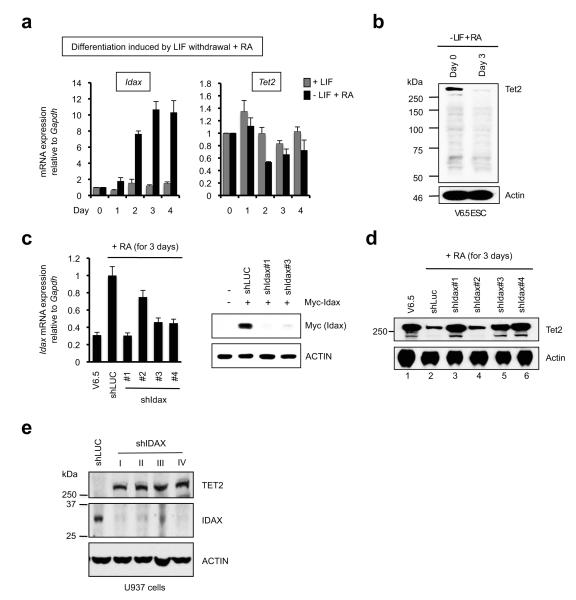
Regulation of Tet2 by Idax was observed in three independent systems. Idax mRNA levels were low in murine V6.5 ES cells, but increased progressively upon LIF withdrawal and supplementation of the culture medium with retinoic acid (RA) (Fig. 3a, left panel), concomitantly with decreased and increased expression of Tet1 and Tet3 respectively18 (Supplementary Fig. 11a). Under these conditions, Tet2 mRNA levels were only slightly altered (Fig. 3a, right panel), but Tet2 protein levels decreased precipitously over a 3-day period (Fig. 3b; for Tet2 antibody specificity, see Supplementary Fig. 11b-d). Lentiviral transduction of V6.5 ES cells with three effective shRNAs against Idax (shIdax #1, #3 and #4; Fig. 3c) substantially protected against the differentiation-induced downregulation of Tet2 protein, whereas transduction with an ineffective shRNA, shIdax #2, did not (Fig. 3d). Thus in differentiating murine ES cells, Tet2 protein downregulation can be directly attributed to Idax. Additionally, transduction of the human U937 monocytic cell line, which barely expresses TET2, with four separate lentiviral shRNAs against IDAX resulted in strong TET2 protein expression (Fig. 3e), suggesting that endogenous IDAX maintains low endogenous TET2 levels in U937 cells. Finally, Idax expression, like Tet2 deficiency3,19, skewed the differentiation of murine bone marrow haematopoietic stem/ progenitor cells toward the monocyte/ macrophage lineage (Supplementary Fig. 12).
Fig. 3. Reciprocal relation between Tet2 and Idax in ES cells (ESC) and U937 cells.
a. Tet2 and Idax mRNA levels in V6.5 ESC differentiated by LIF withdrawal plus 1 μM RA (− LIF + RA). Mean ± SEM of 4-6 independent experiments is shown.
b. Tet2 protein expression decreases within 3 d of ESC differentiation.
c. shIdax#1, #3 and #4 effectively deplete Idax mRNA in differentiating ESC (left panel), shRNAs #1 and #3 decrease Myc-Idax protein levels in HEK293T cells transfected with cDNA lacking the 3′ untranslated region (UTR). shIdax #4 was not tested in HEK293T cells as it is directed against the Idax 3′UTR. Error bars show the range of duplicates.
d. Idax depletion prevents the decrease in Tet2 protein expression in differentiating ESC. Undifferentiated ESC (lane 1) served as a control.
e. Depletion of endogenous IDAX in the human monocytic cell line U937 augments expression of endogenous TET2 protein.
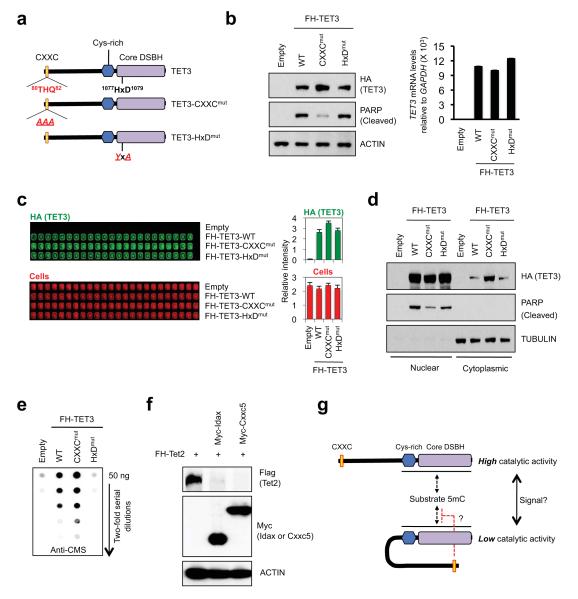
The direct binding of the CXXC domain of Idax to the catalytic domain of Tet2 suggested the possibility of corresponding interactions between the linked CXXC and catalytic domains of TET1 and TET3. Indeed, a CXXC domain mutant of TET3 (80THQ82 > AAA, TET3-CXXCmut, Fig. 4a) resembled IdaxDBM in several respects: it was expressed at higher levels in HEK293 cells compared with wild-type or catalytically-inactive TET3 (Fig. 4b, c); a fraction of TET3-CXXCmut was aberrantly present in the cytoplasm (Fig. 4d, top panel); and TET3-CXXCmut was weaker than wild-type TET3 at inducing PARP cleavage in the nucleus (Fig. 4d, middle panel). Cells expressing the TET3 CXXC mutant displayed higher levels of genomic 5hmC compared with cells expressing wild-type TET3 (Fig. 4e). This could not be attributed simply to increased protein levels of TET3 CXXC mutant versus wild-type TET3, since the nuclear levels of these two proteins were comparable (Fig. 4d, top panel). Together these results suggest that TET3 autoregulates its protein levels via caspase activation, through a mechanism that requires DNA binding by the CXXC domain. The catalytic activity of TET3 may also be controlled by an intramolecular interaction involving the CXXC domain (Fig. 4g); alternatively, the increased 5hmC in cells expressing TET3-CXXCmut could be due to loss of DNA tethering such that the mutant protein is more freely distributed than wild-type TET3 in the nucleus. In contrast, the expression and activity of TET1-CXXCmut were similar to those of wild-type TET1 (Supplementary Fig. 13), suggesting that TET1 and TET3 are regulated by different mechanisms.
Fig. 4. Negative regulation of TET3 by its CXXC domain.
a. Schematic representation of TET3, TET3-CXXCmut and catalytically-inactive TET3-HxDmut.
b. Increased protein expression of TET3-CXXCmut relative to WT TET3 or TET3-HxDmut (left panel), without a change in TET3 mRNA levels (right panel).
c. In-cell western assays confirm that TET3-CXXCmut is expressed at higher levels than WT TET3 or TET3-HxDmut.
d. WT TET3 and TET3-HxDmut are mainly nuclear, whereas TET3-CXXCmut is found in both cytoplasmic and nuclear fractions and is less effective at inducing PARP cleavage than WT TET3.
e. Cells expressing TET3-CXXCmut show higher genomic 5hmC than cells expressing WT TET3 or TET3-HxDmut (anti-CMS dot blot3).
f. Cxxc5 expression results in a decrease in protein levels of co-expressed Tet2 in HEK293T cells.
g. Potential intramolecular (auto-inhibitory) interaction between the linked CXXC and catalytic domains of TET3.
The IDAX-related protein, CXXC5/ RINF, is overexpressed in solid cancers and its high expression correlates with poor prognosis in breast cancer20. Notably, CXXC5 resembles Idax/ CXXC4 in being a negative regulator of Wnt signalling21; moreover, like Tet2, CXXC5 has a role in normal myelopoiesis as well as in myeloid cancers and the CXXC5 gene is often deleted in myeloid leukemias22. Like Idax/ Cxxc4, mouse Cxxc5 downregulated Tet2 protein expression in a dose-dependent manner (Fig. 4f, Supplementary Fig. 14), suggesting that Cxxc5 might also regulate the genomic localisation, expression and enzymatic function of Tet2.
We present a striking example of how a gene fission event, followed by a chromosomal inversion, facilitated the emergence of a novel regulatory mechanism for DNA cytosine modification in vertebrates. The Idax CXXC domain, which separated from the Tet2 catalytic domain during vertebrate evolution2,10, binds Tet2 and potentially recruits it to promoters and CpG islands in genomic DNA. We propose that DNA-bound Idax activates one or more caspases, perhaps at a transcriptional level through promoter/TSS binding (Supplementary Fig. 15). Indeed, caspase 3 activation and Idax upregulation occur with similar kinetics during ES cell differentiation23. As proposed for ubiquitylation24, caspase-mediated cleavage could be involved in the initial activation as well as the subsequent degradation of Tet2 and Idax (Supplementary Fig. 16).
The cellular and subcellular distribution of Idax, and its sensitivity to local environmental signals, may contribute to differences in 5hmC and TET2 levels in different cell types and organs3,25-27. The DNA-binding mutant of Idax is partly in the cytoplasm, and Idax is reported to bind Dishevelled in the cytoplasm in the absence of Wnt signalling7. Since Idax is expressed at very low levels in ES cells but becomes upregulated upon differentiation, the genomic distribution of Tet2 and oxidised methylcytosines in ES cells would be predicted to differ from those in more differentiated tissues, such as the primitive streak of the mouse embryo where Wnt signalling is prominent28. Further analyses will resolve these questions.
Many malignant tissues have been reported to contain low levels of 5hmC compared with normal tissues29,30, and TET2 loss-of-function correlates with decreased 5hmC levels in patients with myeloid malignancies3. However, an appreciable proportion of patients with wild-type TET2 show low 5hmC3, suggesting the existence of additional factors that impair TET2 expression at the protein level and/or TET2 enzymatic activity. For instance, IDAX overexpression, reported in villous adenomas of the colon9, might promote TET2 degradation, leading to depletion of 5hmC in TET2 target genes. Conversely, the homozygous deletion of IDAX/ CXXC4 reported in an aggressive renal cell carcinoma8 could result in TET2 overexpression and its aberrant recruitment to genomic regions where it is not normally found. It will be important in future studies to explore the genomic targets of TET2, IDAX and RINF/CXXC5 in normal development and cancer, and define the effects of cancer-associated mutations in these proteins on patterns of cytosine modification in DNA.
Methods
Recombinant protein expression and purification
Competent BL21 cells were transformed with appropriate pGEX constructs, then induced to produce proteins using 100 ml Overnight Express Autoinduction System 2 (EMD Biosciences). Induced bacteria were harvested and washed with 1X PBS. Then, cells were resuspended in 15 ml PBS containing protease inhibitors (Roche) and lysed by sonication. The lysate was incubated in the presence of lysozyme (1 mg/ml) and Triton X-100 (1%) with gentle rotation for 30 min at 4°C and hard spun at 20,000 g for 30 min at 4°C. The soluble fraction was incubated with 2 ml Glutathione Sepharose 4B slurry (HE Healthcare) with gentle rotation for 30 min at 4°C and washed with cold 1X PBS three-four times. The recombinant GST fusion proteins were incubated in elution buffer (50 mM Tris-HCl, pH 8.0, 10 mM reduced glutathoine) for 10 min at 4°C and the eluted proteins were collected by centrifugation. The elution and incubation steps were repeated three times to collect all proteins. Subsequently, the eluted protein was dialyzed overnight in cold 1X PBS, 10% glycerol, 10 μM ZnCl2, 10 mM β-mercaptoethanol and flash frozen. Protein was concentrated by Amicon Ultra Spin columns 10,000 molecular weight cut-off. Protein expression and purification in Sf9 cells were described previously1. Integrity and purity of proteins were verified by SDS-PAGE and Coomassie Brilliant Blue staining, and purified protein was quantified using a Bradford Assay.
Electrophoretic mobility shift assay (EMSA)
Single-stranded oligonucleotides used for substrate preparation were synthesized at IDT (Sequences in Supplementary Fig. 2b). One nmol of complementary single stranded oligonucleotides was mixed in annealing buffer (10 mM Tris, pH8.0, 50 mM NaCl and 1 mM EDTA), boiled for 4 min and gradually cooled overnight by transferring to a water bath pre-heated to 90°C and turned off. The annealed oligonucleotides and single-stranded oligos as controls were separated by running on a 15% acrylamide gel in 0.5X TBE gel. The gel was placed on an X-ray intensifying screen and irradiated with short wave UV to visualize DNA and excise the region of the gel containing the correct sized double-stranded oligonucleotides. The excised gel slice was crushed and incubated with 10 mM Tris pH 8.0, 0.1 mM EDTA, 300 mM sodium acetate, pH 7.4 overnight at 37°C with agitation. Supernatant was extracted, spun for 5 min at 14,000 rpm to remove undissolved acrylamide, and the eluted DNA was purified by ethanol precipitation. 100 ng double-stranded DNA was radiolabeled using T4 PNK enzyme (NEB) according to the manufacturer’s protocols. Unincorporated ATP was removed using an Illustra G-25 Micro Column (GE Healthcare). For the experiment shown in the left panel of Fig. 1c, 0, 100, 200, 400 ng of GST-Idax CXXC domain was used. Except where otherwise noted, 500 ng of purified GST-tagged wild-type or DNA-binding mutant of Idax CXXC domain was incubated in a total of 20μl reaction mixture containing 1 ng of radiolabelled oligonucleotide and 2 μg of poly (dA-dT), 20 mM HEPES (pH 7.9), 40 mM KCl, 2.5 mM MgCl2, 1 mM dithiothreitol (DTT) and 5% glycerol for 10 min on ice, then an additional 30 min at room temperature (RT). For competition assays, the extracts were pre-incubated with unlabeled DNA competitors before addition of the reaction mixture. The resulting protein-DNA complexes were resolved in a non-denaturating 5% acrylamide gel in 0.5X TBE buffer. Gels were dried and visualized by autoradiography.
Chromatin immunoprecipitation followed by next-generation sequencing (ChIP-seq)
Chromatin was sheared using truChIP™ High Cell Chromatin Shearing Kit with Non-ionic Shearing Buffer (Covaris). Briefly, cells were cross-linked with 1X Covaris fixing buffer at RT for 5 min with rotation, followed by addition of 1X Covaris quenching buffer. After further incubation at RT for 5 min with rotation, cells were washed with PBS two times at 4°C and nuclei were prepared as per manufacturer’s instructions. The nuclei pellet was resuspended in 1X Covaris shearing buffer and chromatin was sheared into 200-300 bp fragments using a Covaris S2 instrument (Duty Cycle: 5%, Intenstiy 4, 200 Cycles per burst, 20 min). Chromatin fragments from two biologically independent cells expressing empty vector, Myc-Idax or Myc-IdaxDBM were immunoprecipitated using anti-Myc antibody conjugated to agarose (Sigma). ChIP-seq library was constructed using TruSeq Sample Prep kit (Illumina) and Illumina HiSeq sequencing was performed as per manufacturer’s instructions.
Data analyses
Sequencing reads were mapped against hg19 using Bowtie31 (bowtie-0.12.7 -n 2 -l 28 -m 3 -k 1 --best -- chunkmbs 512 -p 8 -S -q) by suppressing all alignments for a particular read, if more than three reportable alignments exist, and by reporting only the best valid alignment per read. Mapping results from both ChIP-seq experiments per condition were combined and clonal reads (i.e. reads mapped to the same genomic position) were replaced by one representative, resulting in 52.4 Mio, 43 Mio, and 29.9 Mio reported reads for Myc-Idax, Myc-IdaxDBM, and cells expressing empty vector, respectively. Genomic regions enriched for short reads obtained from Myc-Idax (or Myc-IdaxDBM, respectively) were identified by using a local background estimated from short reads obtained from cells expressing empty vector employing the statistical software MACS32. Annotation (hg19) of the obtained peaks has been performed by using the annotatePeaks.pl function of the Homer software package33. The promoter region was defined as −1 kb to +0.1 kb relative to transcription start site (TSS). In addition, we created Myc-Idax and Myc-IdaxDBM specific peak sets by excluding all overlapping peaks. For the Myc-Idax specific peaks, we performed de-novo motif discovery by using the findMotifsGenome.pl function of the Homer software package33 using default parameter settings. Moreover, we calculated the CpG densities of the DNA sequences underlying the identified peaks, by the CpG observed/expected ratio ((number of CpG / (number of C × number of G)) × total number of nucleotides in the sequence)34.
In order to calculate IP enrichment at gene regions and at CpG islands, we accessed hg19 RefSeq gene annotations and CpG island annotations provided by the UCSC genome browser database (11/19/2012)35. According to their promoter CpG density, we divided all genes into low CpG density promoter (LCP) and high CpG density promoter (HCP) genes. For this, we calculated the CpG observed/ expected ratio34 of the sequences underlying the −1 kb to +0.5 kb region around their transcription start sites. By visual inspection of the CpG density distribution, we decided on a threshold of 0.44 for dividing the genes into LCP and HCP. In order to divide the CpG islands into high CpG (HCG), intermediate CpG (ICG), and low CpG (LCG) density islands, we calculated the CpG observed/ expected ratio34 of the underlying sequences. Subsequently, we sorted all CpG islands into five equally sized groups according to their descending CpG observed/ expected ratio. We used the first, third and fifth group for defining HCG, ICG, and LCG islands.
For the gene-centric enrichment analysis, each gene was divided into 20 bins. In addition, the 2 kb region upstream of the transcription start site, as well as the 2 kb region downstream of the transcription end site, were divided into 10 bins each. For the CpG island and transcription start site-centric analyses, we divided the −2 kb to +2 kb range around the midpoint of each island (or the transcription start site, respectively) into 80 bins. Short reads were extended to a length of 200 bp along their sequencing direction and bin-wise coverage was calculated using the Bioconductor36 environment. In order to avoid division by zero, we added one to each bin of the IP and of the control sample. For each IP sample, a correction value for the library size was estimated by calculating the mean over the ratios of IP over control short read coverage at genome wide 100 bp windows. Subsequently, IP signals at the tested bins were divided by the correction value. Enrichment is defined by the log2 of corrected IP signals divided by the control signals, where Myc-Idax (or Myc-IdaxDBM, respectively) is considered as IP and cells expressing empty vector is considered as control. For the average enrichment profiles, we calculate the log2 of the mean over all corrected IP over control ratios at every tested position of stacked annotations.
In-cell western blot
In-cell western was performed using standard immunocytochemistry procedures. Diluted DNA was mixed with 0.25 μl of lipofectamine 2000 (Invitrogen) diluted in Opti-MEM reduced serum medium (Invitrogen) and incubated at RT for 20 min. HEK239T cells were trypsinized, washed and counted and 2X104 cells were plated in each well of amine-coated BD PureCoat™ 384-well plate (BD Bioscience), followed by addition of the DNA-lipofectamine mixture. Three days later, cells were fixed with 4% paraformaldehyde in PBS for 15 min and permeabilized with 0.2% Triton X-100 in PBS for 15 min at RT. Subsequently, DNAs were denatured with 2N HCl at RT for 30 min and neutralized with 100 mM Tris-HCl buffer (pH 8.5) for 10 min. Cells were incubated in Odyssey Blocking Buffer (Li-COR Biosciences) diluted 1:1 with PBS at RT for 1 hr. Then, rabbit anti-5hmC polyclonal antibody (produced in-house; diluted to 1:2,500) for 5hmC staining or mouse anti-HA antibody (Covance, HA.11 clone 16B12, diluted to 1:1,000) and mouse anti-Myc antibody (Sigma, diluted to 1:2,000) for HA-Tet2 and Myc-Idax staining, respectively, were added into the blocking buffer for 3 hr at RT. The cells were rinsed 3 times with 0.2% Triton X-100 in PBS and incubated at RT for 1 hr with an IRDye 800CW-conjugated donkey anti-mouse IgG (1:4,000) or IRDye 680LT-conjugated donkey anti-rabbit IgG secondary antibody (1:1,000) (LI-COR Biosciences) in blocking buffer, and again washed 4 times with wash buffer, followed by addition of 50 μl of PBS. Plates were scanned on the OdysseyR Sa infrared imaging system and staining was quantified using the scanner software. To quantify cell density, cells were stained with IR Dye 680 LT maleimide (diluted into 10 ng/ml in PBS, Li-COR Biosciences) at RT for 15 min. After extensive washing with PBS three times, and the plate was scanned again and staining was quantified using the scanner software.
Western blotting and caspase-mediated Tet2 cleavage in vitro
Cells were lysed with RIPA buffer (150 mM NaCl, 50 mM Tris-HCl, pH 8.0, 1% Triton X-100, 0.5% sodium deoxycholate and 0.1% SDS) supplemented with protease inhibitor cocktail (Roche) or mixture of protease/ phosphatase inhibitors (20 mM β-glycerophosphate, 10 mM sodium pyrophosphate, 1 mM sodium o-vanadate, 10 μM leupeptin, 10 μg/ml aprotinin, 1 mM freshly prepared PMSF) and incubated on ice for 20 min. Cell debris was removed by centrifuging at 12,000 rpm for 15 min at 4°C. The protein concentration was measured by Bradford protein assay. Samples were mixed with SDS sample buffer and boiled for 4 min. Whole cell lysates were separated by 7.5% or 10% SDS-PAGE and transferred onto nitrocellulose membranes. Proteins were detected by immunoblotting in TBST (150 mM NaCl, 10 mM Tris-Cl, pH 8.0, 0.5% Tween-20) containing 5% low-fat milk and antibodies against Tet2 (Abcam, ab94580 or Abiocode, R1086-vp), DDK (Flag) epitope (Origene, TA50011-100), Myc epitope (clone 9E10 or hybridoma supernatant prepared in-house), HA epitope (Covance, HA.11 clone 16B12), Actin (Sigma A5441), CASP3, CASP8, PARP, cleaved PARP (Cell Signaling) proteins, followed by incubation with HRP-conjugated secondary antibodies and enhanced chemiluminescence. Except where otherwise noted, immunoblot analyses were performed 48 h after transfection. For in vitro Tet2 cleavage assay using recombinant caspases, lysates (~ 20 μg) from HEK293T cells expressing Myc-Tet2 proteins were incubated with ~ 0.4 μg of recombinant active human caspases 3 or 8 (BD Biosciences) in 1X caspase 3 (20 mM HEPES, pH 7.4, 0.1% CHAPS, 5 mM DTT and 2 mM EDTA) or caspase 8 (20 mM HEPES, pH 7.4, 0.1% CHAPS, 5 mM DTT and 2 mM EDTA, 5% sucrose) assay buffer supplemented with protease/ phosphatase inhibitors, respectively at 37°C for 3 hr with gentle shaking. As a control, Z-VAD-FMK (100 μM) was added to the reaction mixture before addition of caspases. Then, lysates were separated by SDS-PAGE.
ES cell culture and transduction
V6.5 mouse ES cells were maintained in culture as previously described18. Briefly, V6.5 mouse ES cells were cultured on primary mouse embryonic fibroblasts that were pre-treated with Mitomycin C (Sigma). The culture medium contains DMEM knockout (Invitrogen), 15% Stasis™ Stem Cell FBS (Gemini Bio-Products), 0.1 mM non-essential amino acids (Invitrogen), 2 mM L-glutamine (Invitrogen), 0.1 mM β-mercaptoethanol (Invitrogen), 50 units/ml penicillin/streptomycin (Invitrogen) and 0.2% LIF-conditioned medium (produced in-house). Culture media were changed every day with fresh complete ES cell media. In experiments to induce differentiation, cells were first trypsinized (or split using TrypLE Express) and plated onto gelatin-coated plates for 45 min two times to remove feeder cells. The floating feeder-depleted V6.5 cells were collected and washed with 1X PBS two times. Then, cells were resuspended in ES cell media lacking LIF and re-plated on gelatin-coated plates or dishes. 1 μM of all-trans retinoic acid (RA) was added to induce differentiation. Culture media were changed every day with fresh RA-supplemented media without LIF. As controls, cells were cultured on gelatin-coated plates in the presence of LIF. For experiments in Fig. 3 b-d, V6.5 cells were differentiated for 3 days.
To produce lentivirus, HEK293T cells were transfected with lentiviral vectors and packaging constructs (pLP1, pLP2 and pLP/VSV-G from Invitrogen) using Lipofectamine 2000. After 48 h, viral supernatant was collected and passed through 0.45 μm low-protein binding filters. To transduce V6.5, cells were plated on gelatin-coated plates in complete ES cell media and incubated overnight at 37°C, then viral supernatant was added together with 8 μg/ml polybrene. Six hours later, media was replaced with fresh ES cell media. Transduction was repeated two times, followed by transferring cells to the plates containing mitomycin C-treated puromycin-resistant MEFs. Cells were selected on puromycin-resistant feeders with 2 μg/ml puromycin for at least 1 week.
Homology modeling of IDAX
Homology modeling of human IDAX/ CXXC4 protein was done on the Protein Model Portal server37 (http://www.proteinmodelportal.org), which gives access to various comparative modeling methods provided by partner sites. Briefly, IDAX (UniPort ID: Q9H2H0) was entered as a query, and a list of models generated from their respective template structures ranked by sequence homology was returned. Among the top two solutions, #1 is a ModBase model38, whose template is the solution structure of the nonmethyl-CpG-binding CXXC domain of the leukaemia-associated MLL histone methyltransferase (PDB ID 2J2S). The modeled region shares 40% sequence identity when aligned with residues 127 to 179 of the target protein. #2 is generated by Swiss-Model39, and it is based on the crystal structure of Dnmt1-DNA complex (PDB ID 3PT6), which shares 35% sequence identity from residues 135 to 176 of the target protein. The alignment of the target sequence with its templates suggests that the TGHQ sequence in IDAX is part of a DNA-binding loop, because it is aligned with the RSKQ DNA binding loop from Dnmt1, and the IKKQ DNA binding sequence in MLL. The superposition of the model with their respective templates was made using Coot40, and the figure was created using Chimera41.
Supplementary Material
Acknowledgements
We thank Gregory Seumois, Manching Ku and Jeremy Day for help with library preparation, Dr. Bing Ren (UCSD) for use of his Illumina Hi-Seq 2000, Jorge A. Zepeda-Martínez for the recombinant Flag-Tet2-CD, and members of the Rao lab for productive discussions. This work was supported by NIH R01 grants HD065812 and CA151535, grant RM-01729 from the California Institute of Regenerative Medicine and Translational Research grant TRP 6187-12 from the Leukemia and Lymphoma Society (to A.R), and NIH R01 grant AI40127 (to P.G.H. and A.R). We also gratefully acknowledge a Special Fellow Award from the Leukemia and Lymphoma Society (to M.K.), postdoctoral fellowships from the Lady Tata Memorial Trust and from the GlaxoSmithKline-Immune Disease Institute Alliance (to H.S.B.), and a predoctoral graduate research fellowship from the National Science Foundation (to W.A.P.).
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions. L.A., P.G.H. and A.R. conceived the project and supervised project planning and execution. M.K. and J.A. performed cellular and molecular experiments including ChIP-Seq, gene knockdown, establishment of stable cell lines, site-directed mutagenesis, dot blot, immunocytochemistry, in vitro caspase and TET assays, and in vitro differentiation studies. J. A. performed the in-cell western blots. H.S.B. obtained the initial data showing downregulation of Tet2 protein by Idax. M. K. conducted the electrophoretic mobility shift assays with help from W.A.P. and M.F.S. H. Li and P.G.H. generated the homology model of the Idax CXXC domain. K.P.K. provided mRNAs from ES cell samples. L.C., T.A. and H. Lähdesmäki performed the bioinformatic analyses of ChIP-seq data. M.K. and A.R. wrote the manuscript with input from other authors.
Author Information The ChIP-Seq data have been deposited in the Gene Expression Omnibus under accession number GSE42958. Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests. Readers are welcome to comment on the online version of the paper.
References
- 1.Tahiliani M, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Iyer LM, Tahiliani M, Rao A, Aravind L. Prediction of novel families of enzymes involved in oxidative and other complex modifications of bases in nucleic acids. Cell Cycle. 2009;8:1698–1710. doi: 10.4161/cc.8.11.8580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ko M, et al. Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature. 2010;468:839–843. doi: 10.1038/nature09586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ito S, et al. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.He YF, et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333:1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maiti A, Drohat AC. Dependence of substrate binding and catalysis on pH, ionic strength, and temperature for thymine DNA glycosylase: Insights into recognition and processing of G.T mispairs. DNA repair. 2011;10:545–553. doi: 10.1016/j.dnarep.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hino S, et al. Inhibition of the Wnt signaling pathway by Idax, a novel Dvl-binding protein. Mol Cell Biol. 2001;21:330–342. doi: 10.1128/MCB.21.1.330-342.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kojima T, et al. Decreased expression of CXXC4 promotes a malignant phenotype in renal cell carcinoma by activating Wnt signaling. Oncogene. 2009;28:297–305. doi: 10.1038/onc.2008.391. [DOI] [PubMed] [Google Scholar]
- 9.Nguyen AV, Albers CG, Holcombe RF. Differentiation of tubular and villous adenomas based on Wnt pathway-related gene expression profiles. Int J Mol Med. 2010;26:121–125. doi: 10.3892/ijmm_00000443. [DOI] [PubMed] [Google Scholar]
- 10.Iyer LM, Abhiman S, Aravind L. Natural history of eukaryotic DNA methylation systems. Prog Mol Biol Transl Sci. 2011;101:25–104. doi: 10.1016/B978-0-12-387685-0.00002-0. [DOI] [PubMed] [Google Scholar]
- 11.Lee JH, Voo KS, Skalnik DG. Identification and characterization of the DNA binding domain of CpG-binding protein. J Biol Chem. 2001;276:44669–44676. doi: 10.1074/jbc.M107179200. [DOI] [PubMed] [Google Scholar]
- 12.Cierpicki T, et al. Structure of the MLL CXXC domain-DNA complex and its functional role in MLL-AF9 leukemia. Nature structural & molecular biology. 2010;17:62–68. doi: 10.1038/nsmb.1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Allen MD, et al. Solution structure of the nonmethyl-CpG-binding CXXC domain of the leukaemia-associated MLL histone methyltransferase. Embo J. 2006;25:4503–4512. doi: 10.1038/sj.emboj.7601340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu C, Bian C, Lam R, Dong A, Min J. The structural basis for selective binding of non-methylated CpG islands by the CFP1 CXXC domain. Nature communications. 2011;2:227. doi: 10.1038/ncomms1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Song J, Rechkoblit O, Bestor TH, Patel DJ. Structure of DNMT1-DNA complex reveals a role for autoinhibition in maintenance DNA methylation. Science. 2011;331:1036–1040. doi: 10.1126/science.1195380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blackledge NP, et al. CpG islands recruit a histone H3 lysine 36 demethylase. Mol Cell. 38:179–190. doi: 10.1016/j.molcel.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu Y, et al. Tet3 CXXC domain and dioxygenase activity cooperatively regulate key genes for Xenopus eye and neural development. Cell. 2012;151:1200–1213. doi: 10.1016/j.cell.2012.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koh KP, et al. Tet1 and Tet2 regulate 5-hydroxymethylcytosine production and cell lineage specification in mouse embryonic stem cells. Cell Stem Cell. 2011;8:200–213. doi: 10.1016/j.stem.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ko M, et al. Ten-Eleven-Translocation 2 (TET2) negatively regulates homeostasis and differentiation of hematopoietic stem cells in mice. Proc Natl Acad Sci U S A. 2011;108:14566–14571. doi: 10.1073/pnas.1112317108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Knappskog S, et al. RINF (CXXC5) is overexpressed in solid tumors and is an unfavorable prognostic factor in breast cancer. Ann Oncol. 2011;22:2208–2215. doi: 10.1093/annonc/mdq737. [DOI] [PubMed] [Google Scholar]
- 21.Kim MS, et al. A novel Wilms tumor 1 (WT1) target gene negatively regulates the WNT signaling pathway. J Biol Chem. 2010;285:14585–14593. doi: 10.1074/jbc.M109.094334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pendino F, et al. Functional involvement of RINF, retinoid-inducible nuclear factor (CXXC5), in normal and tumoral human myelopoiesis. Blood. 2009;113:3172–3181. doi: 10.1182/blood-2008-07-170035. [DOI] [PubMed] [Google Scholar]
- 23.Fujita J, et al. Caspase activity mediates the differentiation of embryonic stem cells. Cell Stem Cell. 2008;2:595–601. doi: 10.1016/j.stem.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Geng F, Wenzel S, Tansey WP. Ubiquitin and proteasomes in transcription. Annu Rev Biochem. 2012;81:177–201. doi: 10.1146/annurev-biochem-052110-120012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nestor CE, et al. Tissue type is a major modifier of the 5-hydroxymethylcytosine content of human genes. Genome Res. 2011 doi: 10.1101/gr.126417.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Globisch D, et al. Tissue distribution of 5-hydroxymethylcytosine and search for active demethylation intermediates. PLoS One. 5:e15367. doi: 10.1371/journal.pone.0015367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haffner MC, et al. Global 5-hydroxymethylcytosine content is significantly reduced in tissue stem/progenitor cell compartments and in human cancers. Oncotarget. 2:627–637. doi: 10.18632/oncotarget.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sokol SY. Maintaining embryonic stem cell pluripotency with Wnt signaling. Development. 2011;138:4341–4350. doi: 10.1242/dev.066209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang H, et al. Tumor development is associated with decrease of TET gene expression and 5-methylcytosine hydroxylation. Oncogene. 2012 doi: 10.1038/onc.2012.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lian CG, et al. Loss of 5-hydroxymethylcytosine is an epigenetic hallmark of melanoma. Cell. 2012;150:1135–1146. doi: 10.1016/j.cell.2012.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
References
- 31.Langemead B, et al. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heinz S, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010;38:576–89. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gardiner-Garden M, et al. CpG islands in vertebrate genomes. J. Mol. Biol. 1987;196:261–82. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- 35.Dreszer TR, et al. The UCSC Genome Browser database: extensions and updates 2011. Nucleic Acids Res. 2012;40(Database issue):D918–23. doi: 10.1093/nar/gkr1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gentleman RC, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Arnold K, et al. The Protein Model Portal. J Struct Funct Genomics. 2009;10:1–8. doi: 10.1007/s10969-008-9048-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pieper, et al. MODBASE, a database of annotated comparative protein structure models and associated resources. Nucleic Acids Res. 2004;32:D217–22. doi: 10.1093/nar/gkh095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.The SWISS-MODEL Repository. http://swissmodel.expasy.org/repository/
- 40.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Cryst D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 41.Huang, et al. Chimera: An Extensible Molecular Modeling Application Constructed Using Standard Components. Pacific Symposium on Biocomputing. 1996;1:724. URL: http://www.cgl.ucsf.edu/chimera. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.