Abstract
Accurate segregation of the replicated genome requires chromosome biorientation on the spindle. Biorientation is ensured by Aurora B kinase, a member of the 4-subunit chromosomal passenger complex (CPC)1,2. Localization of the CPC to the inner centromere is central to the current model for how tension ensures chromosome biorientation—kinetochore-spindle attachments not under tension remain close to the inner centromere and are destabilized by Aurora B phosphorylation, whereas kinetochores under tension are pulled away from the influence of Aurora B, stabilizing their microtubule attachments3–5. Here we show that an engineered truncation of the INCENP/Sli15 subunit of budding yeast CPC that eliminates association with the inner centromere nevertheless supports proper chromosome segregation during both mitosis and meiosis. Truncated INCENP/Sli15 suppresses the deletion phenotypes of the inner centromere-targeting proteins Survivin/Bir1, Borealin/Nbl1, Bub1 and Sgo16. Unlike wildtype INCENP/Sli15, truncated INCENP/Sli15 localizes to pre-anaphase spindle microtubules. Premature targeting of full-length INCENP/Sli15 to microtubules by preventing Cdk1 phosphorylation also suppresses inviability of Survivin/Bir1 deletion. These results suggest that activation of Aurora B/Ipl1 by clustering either on chromatin or on microtubules is sufficient for chromosome biorientation.
All known mechanisms targeting the CPC to centromeric chromatin, in budding yeast and elsewhere, rely on the Survivin/Bir1 subunit. Budding yeast CPC (Fig. 1a) is targeted by two Bir1-dependent mechanisms: interaction of Bir1 with Sgo1, which recognizes histone H2a phosphorylated by the kinetochore-localized kinase Bub16, and direct binding of Bir1 to the Ndc10 subunit of the centromeric DNA-binding complex CBF37,8. In other species, Survivin binding to histone H3 phosphorylated on threonine 3 by Haspin kinase is also implicated in CPC centromere targeting9–11; however, deletion of the two haspin-like genes (Alk1 and Alk2) does not lead to a growth phenotype (see below) suggesting that this mechanism may not operate in budding yeast.
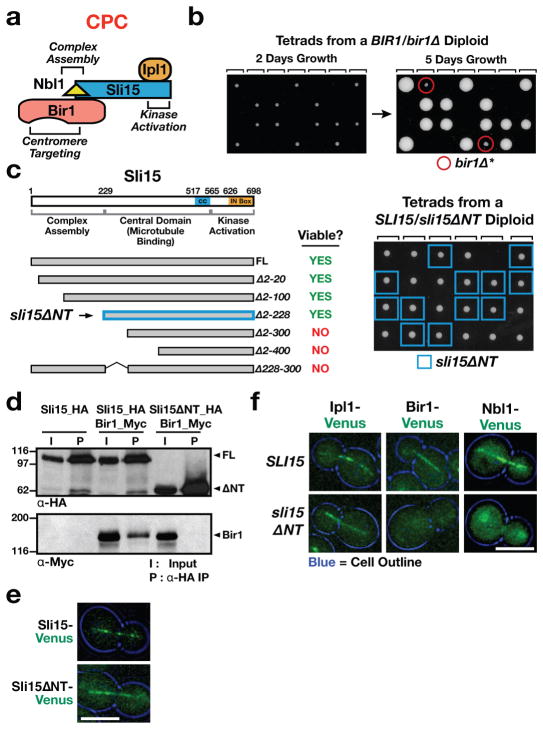
Figure 1. Deletion of the Sli15 N-terminus prevents association with Bir1 but does not affect cell viability or growth.
(a) Schematic of the CPC in budding yeast. Sli15=INCENP; Bir1=Survivin; Nbl1=Borealin/Dasra and Ipl1=Aurora B kinase.
(b) Phenotype of bir1Δ. Tetrad dissections from a bir1Δ heterozygote—the 4 spores from individual tetrads are arrayed in columns. Rare survivors (referred to as bir1Δ*) are observed after extended growth (right panel).
(c) Phenotype of Sli15 truncations. Tetrad dissections from a sli15ΔNT heterozygote are shown on the right.
(d) Co-immunoprecipitation analysis of full length (FL) Sli15 or Sli15ΔNT and Bir1. 9Myc and 6HA tags were inserted at endogenous loci to generate functional C-terminally tagged proteins. Bir1 coimmunoprecipitates with FL Sli15 but not with Sli15ΔNT.
(e) Localization of Sli15-Venus and Sli15ΔN-Venus to the anaphase spindle. Scale bar is 5 μm.
(f) Localization of CPC components during anaphase in cells with either wildtype Sli15 or Sli15ΔNT. Scale bar is 5 μm.
In agreement with the view that Bir1-directed targeting of the CPC to centromeres is critical for chromosome biorientation, the majority of bir1Δ spores fail to grow (Fig. 1b) and temperature-sensitive mutations in bir1 exhibit chromosome missegregation similar to that observed in ipl1 and sli15 mutants12,13. A low frequency (10%, n=60) of bir1Δ spore survival is observed after extended incubation (Fig. 1b: 14); these survivors, which we refer to as bir1Δ*, have high chromosome missegregation rates and harbor significant aneuploidy (Fig. S1a & not shown). To determine if the severe BIR1 deletion phenotype is due to inability of the CPC to target to the inner centromere, we generated truncations of the Sli15 N-terminus that are predicted to eliminate the interaction of Sli15-Ipl1 with Bir1-Nbl115. Surprisingly, truncations of up to 228 N-terminal amino acids of Sli15 (the longest viable truncation; referred to from now on as Sli15ΔNT; Fig. 1c) exhibited no lethality—cells harboring these truncations as the sole source of Sli15 grew indistinguishably from wildtype (Fig. 1c, 2b). Further truncations that encroached on the Sli15 central domain were lethal (Fig. 1c). Immunoprecipitation experiments indicated that deleting the Sli15 N-terminus eliminated the interaction with Bir1 (Fig. 1d). Analysis of CPC anaphase spindle localization, which is dependent on Sli15, confirmed this result. While Sli15ΔNT and Ipl1 localized on the anaphase spindle (Fig. 1e), Bir1 and Nbl1 were delocalized in sli15ΔNT mutant cells (Fig. 1f).
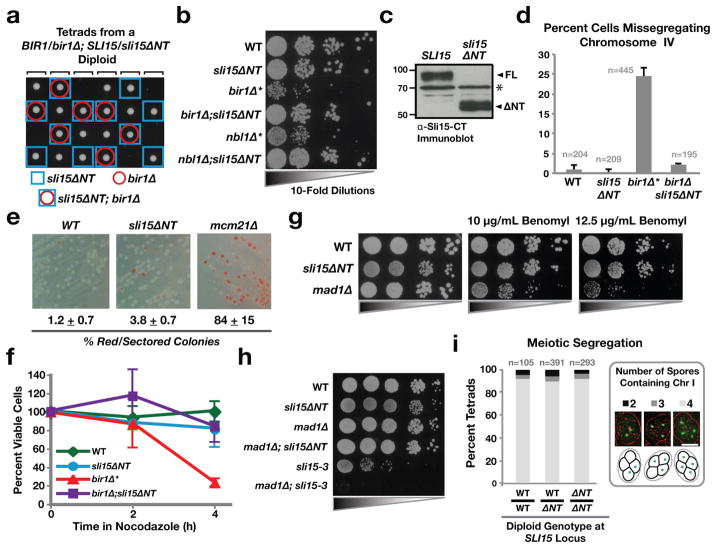
Figure 2. sli15ΔNT suppresses lethality of bir1Δ and nbl1Δ, and exhibits normal fidelity mitotic and meiotic chromosome segregation.
(a) Tetrad dissection showing viability of sli15ΔNT;bir1Δ double mutant spores.
(b) 10-fold serial dilutions of cells of indicated genotypes. bir1Δ* and nbl1Δ* represent rare survivors recovered as shown in Fig. 1B.
(c) Immunoblot of extracts prepared from strains expressing either wild type Sli15 or Sli15ΔNT and blotted using an antibody raised against the C-terminus of Sli15. The asterisk (*) indicates a non-specific band recognized by the primary antibody that serves as a loading control.
(d) Analysis of segregation fidelity of GFP-tagged chromosome IV in cells of the indicated genotypes. The average of 3 to 5 experiments is shown; error bars represent standard error.
(e) Minichromosome loss assay. The percentage and standard error of colonies that were either red or sectored is shown.
(f) Viability following transient nocodazole treatment of cells of the indicated genotypes. The average of 2 to 4 experiments is shown; error bars represent standard error.
(g) Serial dilutions of cells with the indicated genotypes spotted on plates with different concentrations of the microtubule-depolymerizing drug benomyl.
(h) Lack of synthetic lethality/sickness following checkpoint inhibition in sli15ΔNT cells. Plates were incubated at 37°C. sli15-3 is a temperature-sensitive mutant that compromises Ipl1 activation17.
(i) Meiotic segregation following sporulation of diploid cells. The presence or absence of Chromosome I was scored for each individual spore in a tetrad. Scale bar is 5 μm.
The above results show that Sli15ΔNT is viable even though it disrupts CPC formation and disconnects the kinase activity of the CPC from the Bir1 subunit that targets it to centromeres. Consistent with this finding, sli15ΔNT fully suppressed inviability of bir1Δ and nbl1Δ—double sli15ΔNT;bir1Δ or sli15ΔNT;nbl1Δ mutant spores formed colonies at the expected frequency with normal growth properties (Fig. 2a,b). Wild type Sli15 and Sli15ΔNT were expressed at similar levels indicating that suppression of bir1Δ and nbl1Δ lethality was not due to overexpression of mutant Sli15 (Fig. 2c). Sli15 was not detected in bir1Δ* and nbl1Δ* strains, suggesting that full length Sli15 is destabilized when complex formation is disrupted (Fig. S1b; see Suppl. Discussion). We conclude that Sli15ΔNT-Ipl1 is sufficient to perform the CPC’s essential function(s) in the complete absence of Bir1 or Nbl1.
To assess the degree to which Sli15ΔNT-Ipl1 substitutes for the full CPC, we analyzed chromosome segregation and biorientation using multiple assays. First, we monitored a single tagged chromosome, which revealed high fidelity segregation for both the single sli15ΔNT and double sli15ΔNT;bir1Δ mutants (Fig. 2d); in contrast, extensive missegregation was observed in the rare bir1Δ* survivors (Fig. 2d; Fig. S1a) and has been previously reported for ipl1 kinase activity-defective mutants3,16,17. Second, we monitored segregation of a minichromosome (non-essential chromosome III fragment) in a sensitive colony color assay, which revealed near-normal fidelity of segregation in sli15ΔNT cells (Fig. 2e); mcm21Δ cells, which are also viable and do not exhibit significant growth defects but reduce chromosome segregation fidelity, are shown for comparison (Fig. 2e). Third, modest defects in chromosome biorientation are enhanced by transient nocodazole treatment and removal, due to formation of multiple incorrect attachments during the recovery of collapsed spindles18. We therefore tested cell viability following transient nocodazole treatment. bir1Δ* spores exhibited rapid death following transient nocodazole treatment; in contrast, sli15ΔNT and sli15ΔNT;bir1Δ cells behaved similarly to wild-type cells (Fig. 2f). Fourth, mutations that increase chromosome segregation errors exhibit sensitivity to microtubule depolymerizing drugs such as benomyl18,19. sli15ΔNT growth rates were similar to wildtype in the presence of benomyl (Fig. 2g). Fifth, mild defects in CPC activity, as well as deletion mutants of non-essential chromosome segregation proteins, show strong synthetic lethal/sick interactions with mutations in the spindle checkpoint (such as mad1Δ or mad2Δ) (e.g. 20). sli15ΔNT mutant cells did not exhibit a synthetic lethal/sick interaction with mad1Δ (Fig. 2h). Sixth, sli15ΔNT mutant cells delayed cell cycle progression in response to lack of sister chromatid cohesion, indicating that they are competent to communicate absence of tension to the spindle checkpoint (Fig. S2a). Seventh, subtle defects in chromosome segregation are often magnified during meiosis21, but sli15ΔNT cells did not exhibit increased meiotic missegregation relative to control cells (Fig. 2i). Thus, sli15ΔNT cells exhibit remarkably normal chromosome segregation fidelity during mitosis and meiosis in budding yeast.
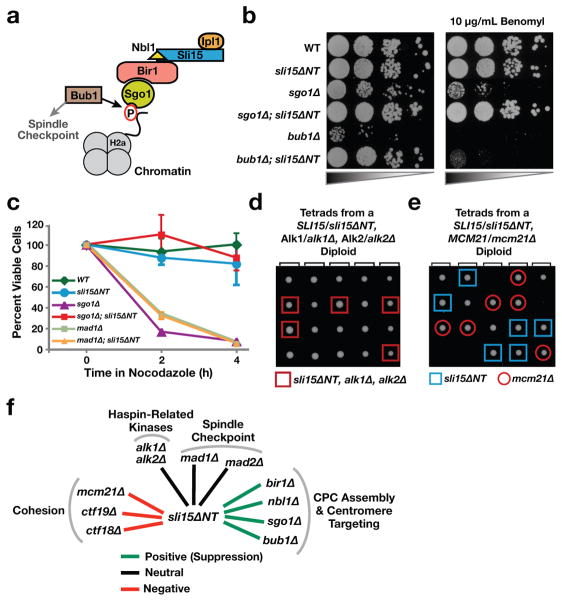
We next tested if Sli15ΔNT also bypasses mutations in the Bir1 localization pathway. Bir1-dependent targeting of the CPC to the inner centromere involves recognition of phosphorylated histone H2a by Sgo1 (Fig. 3a). Chromosome biorientation errors in bub1Δ and sgo1Δ mutants lead to severe growth defects, sensitivity to benomyl, and difficulty correcting defective attachments following nocodazole washout18. sli15ΔNT suppressed the severe growth defect of both bub1Δ and sgo1Δ cells (Fig. 3b); in addition, the benomyl sensitivity, the rapid loss of viability following transient nocodazole treatment, and the chromosome missegregation of sgo1Δ cells were also suppressed (Fig. 3b,c; Fig. S1c). The benomyl sensitivity of bub1Δ was not suppressed as the spindle checkpoint function of Bub1 is independent of its role in CPC localization. While Haspin kinases contribute to CPC targeting in other organisms via creating a binding site on centromeric chromatin for Survivin, deletion of the two Haspin homologues in budding yeast had no growth phenotype on their own or in combination with sli15ΔNT (Fig. 3d).
Figure 3. sli15ΔNT suppresses mutants in the Bir1-dependent CPC targeting pathway but is synthetically lethal with genes implicated in centromere cohesion.
(a) Schematic of the CPC targeting mechanism involving Bub1 kinase and Sgo1.
(b) Suppression of sgo1Δ and bub1Δ growth phenotypes by sli15ΔNT. Compromised growth of sgo1Δ on benomyl is also suppressed by sli15ΔNT.
(c) Viability following transient nocodazole treatment of cells of the indicated genotypes. The average of 2 to 4 experiments is shown; error bars represent standard error. WT and sli15ΔNT measurements are the same as in Fig. 2f.
(d) Tetrad analysis showing no synthetic defect for cells that are triple mutants for sli15ΔNT alk1Δ and alk1Δ.
(e) Tetrad analysis showing synthetic lethality of sli15ΔNT and mcm21Δ. Similar results were observed for ctf19Δ (Fig. S2b).
(f) Summary of genetic interactions exhibited by sli15ΔNT. A positive genetic interaction (green) indicates suppression; negative genetic interaction (red) indicates synthetic lethality/sickness and neutral interaction (black) indicates lack of a synthetic phenotype.
An ipl1 temperature-sensitive mutant is synthetic lethal with deletion mutants of the Ctf19 and Mcm21 subunits of the Ctf19/COMA kinetochore complex, which provides a non-essential function in centromeric cohesion20. In contrast to the bir1Δ, nbl1Δ, sgo1Δ, and bub1Δ mutants, whose lethality/severe growth defects were suppressed by sli15ΔNT, ctf19Δ and mcm21Δ mutants exhibited synthetic lethality/sickness with sli15ΔNT (Fig. 3e,f; Fig. S2b,c). Combining sli15ΔNT with a deletion of CTF18, a separate non-essential mutant affecting cohesion establishment, also led to a synthetic sick phenotype (Fig. S2b,c). Thus, while Bir1-dependent CPC targeting is dispensable for chromosome biorientation and segregation, this targeted CPC pool exhibits a functional connection with cohesion (Fig. 3f; see Suppl. Discussion).
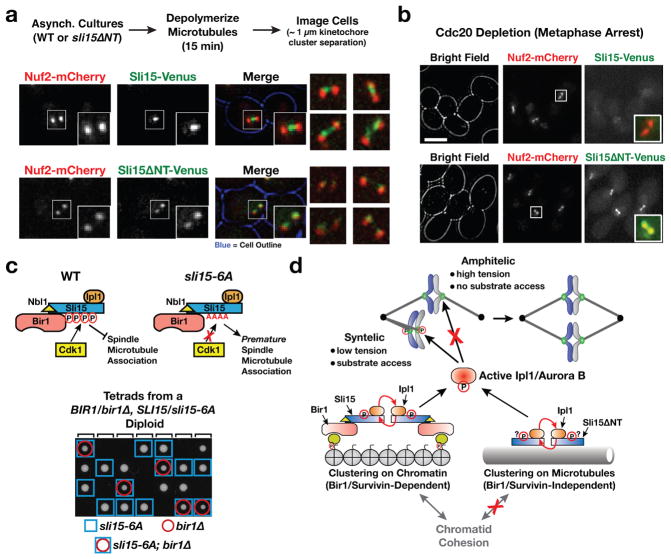
We next examined the localization of CPC subunits in wildtype cells and in cells expressing Sli15ΔNT. In cells arrested in metaphase by depletion of the anaphase activator Cdc20, where the chromosomes are already bioriented, significant localization of Sli15 or Ipl1 on chromatin was not detected (Fig. 4b top row; Fig. S3b). However, localization of Sli15 and Ipl1 between sister kinetochore clusters (analogous to the inner centromere localization in other species) was observed following brief microtubule depolymerization in asynchronously growing cells (Fig. 4a; Fig. S3a). In sli15ΔNT cells, localization between sister kinetochore clusters was lost for both Sli15ΔNT and Ipl1 (Fig. 4a; Fig. S3a); instead weak localization was observed coincident with kinetochore clusters (Fig. 4a; see Suppl. Discussion). Thus, the Sli15ΔNT-Ipl1 complex supports accurate chromosome segregation without enriching between sister kinetochores in vivo.
Figure 4. Localization of Sli15ΔNT and relationship between Sli15 microtubule localization and suppression of bir1Δ.
(a) Images of cells expressing Nuf2-mCherry and either Sli15-Venus or Sli15ΔNT-Venus after brief microtubule depolymerization. A cell with ~1 μm separation of Nuf2-mCherry clusters is shown for each. Boxes are 2.1 μm square. 4 additional examples are shown on the right. See Fig. S3A for similar analysis of Ipl1 localization.
(b) Images of cells arrested in metaphase by Cdc20 depletion expressing Nuf2-mCherry and either Sli15-Venus or Sli15ΔNT-Venus. Scale bar is 5 μm; merged insets are magnified 2.5-fold. See Fig. S3B for similar analysis of of Ipl1 localization.
(c) Schematic summarizing prior work on Cdk1 regulation of Sli15 spindle localization22 and tetrad analysis showing growth of sli15-6A;bir1Δ double mutant cells.
(d) Model for mechanism of chromosome biorientation. Bir1-dependent chromatin clustering of the CPC or Bir1-independent clustering on microtubules of Sli15ΔNT-Ipl1 generates active Ipl1 kinase, which is capable of discriminating between correct and incorrect attachments.
In wildtype cells, the CPC is prevented from localizing to the spindle by Cdk1 phosphorylation until anaphase onset, when it is recruited to spindle microtubules and functions in spindle elongation22–24. However, the Sli15ΔNT-Ipl1 complex exhibited robust accumulation on the spindle in cells held in metaphase by depletion of the anaphase activator Cdc20 (Fig. 4b; Fig. S3b). The central domain of Sli15, which harbors microtubule-binding activity, is required for chromosome biorientation (14; Fig. 1c) and INCENP microtubule binding activates Aurora B kinase through local clustering25–27. We therefore hypothesized that the Sli15ΔNT mutant may rely on clustering mediated by microtubule binding to activate Ipl1, which in turn detects and corrects defective kinetochore-spindle attachments. To test this idea, we wanted to determine if prematurely clustering Sli15 on microtubules by another means bypassed the requirement for Bir1 for viability. For this purpose, we used the previously described sli15-6A mutant that prevents Cdk1 phosphorylation in the central domain of Sli15 and prematurely targets it to preanaphase spindle microtubules22. Consistent with our hypothesis, sli15-6A suppressed the inviability of bir1Δ—in contrast to bir1Δ alone (Fig. 1b) all double mutant sli15-6A;bir1Δ spores formed colonies (Fig. 4c). However, the sli15-6A;bir1Δ double mutant cells grew slowly and exhibited benomyl sensitivity indicating that the suppression was partial, unlike the case for sli15ΔNT (not shown). As with sli15ΔNT, the suppression of BIR1 deletion by sli15-6A was not due to overexpression of the mutant protein (Fig. S3d). A Sli15 mutant that cannot be phosphorylated by Ipl1 and displays premature localization to the metaphase spindle24 also partially rescued BIR1 deletion (Fig. S3c).
Our finding that biorientation occurs normally in the absence of Bir1-dependent targeting of the CPC has significant implications for how the discrimination of correct (amphitelic; with tension) and incorrect (syntelic; lacking tension) attachments is achieved (Fig. 4d; see Suppl. Discussion). Current models for biorientation emphasize the distance between inner centromere-localized Aurora B kinase and outer kinetochore-localized phosphatase activity as being critical for this discrimination3–5. Our findings instead suggest that active Aurora B, generated by clustering on either microtubules or centromeric chromatin, is capable of discriminating between correct and incorrect attachments and that this discrimination is intrinsic to the kinetochore (Fig. 4d). A parsimonious explanation for how this discrimination is achieved is substrate access, with correct, tense attachments becoming less sensitive to Aurora B activity. Super-resolution imaging studies have documented structural changes in microtubule-attached kinetochores under tension versus lacking tension28, which may lead to changes in susceptibility to Aurora B phosphorylation. Defining the property that is detected by Aurora B to discriminate correct versus incorrect attachments should be facilitated by the finding that Survivin-mediated centromere targeting of the CPC is not necessary for tension-sensing and chromosome biorientation in budding yeast.
METHODS
Yeast strains and media
All yeast strains and plasmids used in this study are listed in Supplementary Table 1. Strains were grown in either yeast extract/peptone or synthetic media at 30°C unless otherwise indicated. Epitope and Fluorescent tags were inserted at the C-terminus of genes at their native loci as previously described29. Gene truncations were made with the QuikChange Mutagenesis Kit (Agilent Technologies). Truncations and point mutants were integrated either at their native loci (by digesting the plasmid with the unique NruI site in the Sli15 promoter), or the URA3 locus (by digesting with the StuI site in the Ura3 gene). For native locus integration, the wild-type copies of the gene were then excised by growing overnight in YPD and selecting for growth on 5-FOA plates. All integrations were then checked by PCR and sequencing.
Immunoprecipitation and Western Blotting
Cells in exponential growth were pelleted and resuspended in 600μL IP buffer with protease inhibitors [50mM Tris pH7.6, 150mM NaCl, 1% Triton X-100, 1mM EDTA, 2mM PMSF, 4mM Benzamidine, cOmplete EDTA-free protease inhibitor cocktail (Roche)] and vortexed for 45 min with 400μL glass beads. Lysates were cleared at 18,000g for 10 minutes, transferred to a new tube and centrifuged again. 25 μL of antibody-bead slurry (Anti-HA rat monoclonal clone 3F10; Roche) was combined with 500 μL of cleared lysate and rotated at 4°C for one hour. Beads were washed once with IP buffer and 3 times with TBS (Tris buffer saline, pH 7.4) and then resuspended in protein sample buffer. Samples were analyzed by 8 or 10% SDS-PAGE and immunoblotted with anti-HA clone 3F10 (Roche) and anti-myc mouse monoclonal 4A6 (Millipore), followed by HRP-conjugated secondary antibodies.
Nocodazole Recovery Assay
Yeast cultures were diluted to an OD600 of 0.1 in YPD and incubated at 30°C for 1.5 hours. 10 μM α-factor was added for 2.5 hours. Cells were washed 5 times with YPD, resuspended in YPD plus 10 μg/mL benomyl and 15 μg/mL nocodazole, and incubated at 23°C. Samples were collected every 2 hours and 100 μL of a 1:10,000 dilution was plated on YPD agar plates. The percentage of colonies formed was determined by dividing the number of colonies at the indicated time-point to the number at time zero after 3 days growth on plates. A minimum of 200 colonies were counted for each mutant at time zero.
Analysis of Cell Cycle Progression Following Cohesin Depletion
Overnight cultures of GAL-MCD1 strains were diluted into fresh YPG media and arrested in G1 with 1μM α-factor. Cells were then washed five times and released into fresh medium. 1μM α-factor was added again when the cells had small buds to prevent entry into the next cell cycle. Samples were taken at the indicated time points and lysed by vortexing for two minutes with glass beads in sample buffer. The samples were then analyzed by SDS-PAGE and western blot.
Microscopy
Overnight cultures were diluted ~ 100-fold and grown for 5 hours. Cells were pelleted, washed once and resuspended in water, placed on 1% agar pads supplemented with complete synthetic media, covered with a cover-slip and sealed around the edges with VALAP (a 1:1:1 mixture of Vaseline®, lanolin (Fisher) and paraffin (Fisher) by weight). Images were collected on a Deltavision microscopy system (Applied Precision) using a 100x, 1.3 NA Olympus U-PlanApo objective. 14 z-sections were taken with 0.5 μm steps and deconvolved with SoftWoRx software. Further image analyses, including maximum intensity projections and contrast adjustments, were performed in ImageJ (NIH). Images within each figure were all collected under the same conditions and contrast adjusted identically. For metaphase arrest with Cdc20 depletions, asynchronous cultures in rich media with 1% galactose and 1% raffinose were washed 3 times and switched to media containing 2% glucose for 2.5 hours before imaging. For mitotic chromosome segregation assays, cells were grown overnight in media selective for the lacO cassette and LacI-GFP and then switched to rich media for 5 hours. The cells were then fixed with 4% formaldehyde, washed once, stored in storage solution (100mM potassium phosphate pH7.5, 1M Sorbitol) at 4°C, and imaged no more than 2 days later. For meiosis microscopy, saturated cultures in YPD were pelleted, resuspended in 1% potassium acetate, and incubated with rotation for 24 hours at 23°C. For imaging following microtubule depolymerization, cultures undergoing exponential growth were treated with 10 μg/mL benomyl and 15 μg/mL nocodazole for 15 minutes, washed once with water, put on agar pads with complete synthetic media and 10 μg/mL benomyl, and immediately imaged.
Tetrad Dissection
Diploids were sporulated by transferring patches of yeast from YPD plates grown overnight at 30°C to sporulation plates (8.2 mg/mL Sodium Acetate, 0.35 mg/mL Magnesium Sulfate, 1.9 mg/mL Potassium Chloride, 1.2 mg/mL Sodium Chloride, 16 mg/mL agar) and incubated at 23°C for 2–3 days. Spores were then washed with water and digested with 1 mg/mL zymolyase for 5 minutes at 30°C and then dissected onto YPD plates. Genotyping was performed by replica plating colonies onto selective media. For Sli15 constructs integrated at their endogenous loci, a G418 resistance cassette was integrated 1.5 kilobases upstream of either the wild-type or mutant protein for genotyping purposes.
Minichromosome Loss Assay
Cultures were started overnight in selective (-HIS) media and then diluted into YPD and grown without selection for six hours. The cultures were then plated on synthetic media with low (6μg/mL) adenine to enhance the color change30 and grown for three days at 23°C. The percent of red or sectored versus completely white colonies was then counted.
Supplementary Material
Acknowledgments
The authors would like to thank the Desai and Oegema labs for discussions; S. Sandall, H. Hu and E. Manrriquez for assistance; B. Ren’s lab for help with ChIP experiments, S. Biggins, D. Dawson, G. Pereira, G. Barnes, Phil Hieter and the Yeast Resource Center for strains and plasmids; K. Oegema, R. Green and J. DeLuca for comments on the manuscript. This work was supported by a NIH grant (GM074215) to A.D. and a Damon Runyon Cancer Research Foundation Fellowship (DRG 2007-09) to C.C. A.D. receives salary and other support from the Ludwig Institute for Cancer Research.
Footnotes
AUTHOR CONTRIBUTIONS
C.C. and A.D. designed experiments and wrote the manuscript. C.C. performed the experiments and analyzed the data.
References
- 1.Ruchaud S, Carmena M, Earnshaw WC. Chromosomal passengers: conducting cell division. Nat Rev Mol Cell Biol. 2007;8:798–812. doi: 10.1038/nrm2257. [DOI] [PubMed] [Google Scholar]
- 2.Pinsky BA, Biggins S. The spindle checkpoint: tension versus attachment. Trends Cell Biol. 2005;15:486–493. doi: 10.1016/j.tcb.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 3.Tanaka TU, et al. Evidence that the Ipl1-Sli15 (Aurora kinase-INCENP) complex promotes chromosome bi-orientation by altering kinetochore-spindle pole connections. Cell. 2002;108:317–329. doi: 10.1016/s0092-8674(02)00633-5. [DOI] [PubMed] [Google Scholar]
- 4.Liu D, Vader G, Vromans MJM, Lampson MA, Lens SMA. Sensing Chromosome Bi-Orientation by Spatial Separation of Aurora B Kinase from Kinetochore Substrates. Science. 2009;323:1350–1353. doi: 10.1126/science.1167000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lampson MA, Cheeseman IM. Sensing centromere tension: Aurora B and the regulation of kinetochore function. Trends Cell Biol. 2011;21:133–140. doi: 10.1016/j.tcb.2010.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kawashima SA, Yamagishi Y, Honda T, Ishiguro KI, Watanabe Y. Phosphorylation of H2A by Bub1 prevents chromosomal instability through localizing shugoshin. Science. 2010;327:172–177. doi: 10.1126/science.1180189. [DOI] [PubMed] [Google Scholar]
- 7.Yoon HJ, Carbon J. Participation of Bir1p, a member of the inhibitor of apoptosis family, in yeast chromosome segregation events. Proc Natl Acad Sci USA. 1999;96:13208–13213. doi: 10.1073/pnas.96.23.13208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cho U-S, Harrison SC. Ndc10 is a platform for inner kinetochore assembly in budding yeast. Nat Struct Mol Biol. 2011 doi: 10.1038/nsmb.2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yamagishi Y, Honda T, Tanno Y, Watanabe Y. Two histone marks establish the inner centromere and chromosome bi-orientation. Science. 2010;330:239–243. doi: 10.1126/science.1194498. [DOI] [PubMed] [Google Scholar]
- 10.Kelly AE, et al. Survivin reads phosphorylated histone H3 threonine 3 to activate the mitotic kinase Aurora B. Science. 2010;330:235–239. doi: 10.1126/science.1189505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang F, et al. Histone H3 Thr-3 phosphorylation by Haspin positions Aurora B at centromeres in mitosis. Science. 2010;330:231–235. doi: 10.1126/science.1189435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shimogawa MM, Widlund PO, Riffle M, Ess M, Davis TN. Bir1 is required for the tension checkpoint. Mol Biol Cell. 2009;20:915–923. doi: 10.1091/mbc.E08-07-0723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Makrantoni V, Stark M. Efficient chromosome bi-orientation and the tension checkpoint in Saccharomyces cerevisiae both require Bir1. Mol Cell Biol. 2009;29:4552–4562. doi: 10.1128/MCB.01911-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sandall S, et al. A Bir1-Sli15 complex connects centromeres to microtubules and is required to sense kinetochore tension. Cell. 2006;127:1179–1191. doi: 10.1016/j.cell.2006.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jeyaprakash AA, et al. Structure of a Survivin-Borealin-INCENP core complex reveals how chromosomal passengers travel together. Cell. 2007;131:271–285. doi: 10.1016/j.cell.2007.07.045. [DOI] [PubMed] [Google Scholar]
- 16.Biggins S, Murray AW. The budding yeast protein kinase Ipl1/Aurora allows the absence of tension to activate the spindle checkpoint. Genes Dev. 2001;15:3118–3129. doi: 10.1101/gad.934801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim JH, Kang JS, Chan CS. Sli15 associates with the ipl1 protein kinase to promote proper chromosome segregation in Saccharomyces cerevisiae. J Cell Biol. 1999;145:1381–1394. doi: 10.1083/jcb.145.7.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Indjeian VB. The Centromeric Protein Sgo1 Is Required to Sense Lack of Tension on Mitotic Chromosomes. Science. 2005;307:130–133. doi: 10.1126/science.1101366. [DOI] [PubMed] [Google Scholar]
- 19.Li R, Murray AW. Feedback control of mitosis in budding yeast. Cell. 1991;66:519–531. doi: 10.1016/0092-8674(81)90015-5. [DOI] [PubMed] [Google Scholar]
- 20.Ng TM, Waples WG, Lavoie BD, Biggins S. Pericentromeric sister chromatid cohesion promotes kinetochore biorientation. Mol Biol Cell. 2009;20:3818–3827. doi: 10.1091/mbc.E09-04-0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shonn MA, McCarroll R, Murray AW. Requirement of the spindle checkpoint for proper chromosome segregation in budding yeast meiosis. Science. 2000;289:300–303. doi: 10.1126/science.289.5477.300. [DOI] [PubMed] [Google Scholar]
- 22.Pereira G. Separase Regulates INCENP-Aurora B Anaphase Spindle Function Through Cdc14. Science. 2003;302:2120–2124. doi: 10.1126/science.1091936. [DOI] [PubMed] [Google Scholar]
- 23.Rozelle DK, Hansen SD, Kaplan KB. Chromosome passenger complexes control anaphase duration and spindle elongation via a kinesin-5 brake. J Cell Biol. 2011;193:285–294. doi: 10.1083/jcb.201011002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nakajima Y, et al. Ipl1/Aurora-dependent phosphorylation of Sli15/INCENP regulates CPC-spindle interaction to ensure proper microtubule dynamics. J Cell Biol. 2011;194:137–153. doi: 10.1083/jcb.201009137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sessa F, et al. Mechanism of Aurora B activation by INCENP and inhibition by hesperadin. Mol Cell. 2005;18:379–391. doi: 10.1016/j.molcel.2005.03.031. [DOI] [PubMed] [Google Scholar]
- 26.Kelly AE, et al. Chromosomal enrichment and activation of the aurora B pathway are coupled to spatially regulate spindle assembly. Dev Cell. 2007;12:31–43. doi: 10.1016/j.devcel.2006.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tseng BS, Tan L, Kapoor TM, Funabiki H. Dual detection of chromosomes and microtubules by the chromosomal passenger complex drives spindle assembly. Dev Cell. 2010;18:903–912. doi: 10.1016/j.devcel.2010.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wan X, et al. Protein architecture of the human kinetochore microtubule attachment site. Cell. 2009;137:672–684. doi: 10.1016/j.cell.2009.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Longtine MS, et al. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 30.Hieter P, Mann C, Snyder M, Davis RW. Mitotic stability of yeast chromosomes: a colony color assay that measures nondisjunction and chromosome loss. Cell. 1985;40:381–392. doi: 10.1016/0092-8674(85)90152-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.