Abstract
The classic view of metastatic cancer progression is that it is a unidirectional process initiated at the primary tumor site, progressing to variably distant metastatic sites in a fairly predictable, though not perfectly understood, fashion. A Markov chain Monte Carlo mathematical approach can determine a pathway diagram that classifies metastatic tumors as ‘spreaders’ or ‘sponges’ and orders the timescales of progression from site to site. In light of recent experimental evidence highlighting the potential significance of self-seeding of primary tumors, we use a Markov chain Monte Carlo (MCMC) approach, based on large autopsy data sets, to quantify the stochastic, systemic, and often multi-directional aspects of cancer progression. We quantify three types of multi-directional mechanisms of progression: (i) self-seeding of the primary tumor; (ii) re-seeding of the primary tumor from a metastatic site (primary re-seeding); and (iii) re-seeding of metastatic tumors (metastasis re-seeding). The model shows that the combined characteristics of the primary and the first metastatic site to which it spreads largely determine the future pathways and timescales of systemic disease.
For lung cancer, the main ‘spreaders’ of systemic disease are the adrenal gland and kidney, whereas the main ‘sponges’ are regional lymph nodes, liver, and bone. Lung is a significant self-seeder, although it is a ‘sponge’ site with respect to progression characteristics.
1. Introduction
The classic view of metastatic progression, framed in part by the ‘seed-and-soil’ hypothesis of Paget (1), is that cancer spreads from the primary tumor site to distant metastatic locations in a unidirectional way. The ‘seeds’ responsible for the spread are circulating tumor cells (CTCs) (2-4) that detach from the primary tumor, enter the bloodstream and lymphatic system (3), and travel to new distant locations. If conditions are favorable, this initiates a complex (5-7) and not well understood metastatic cascade, ultimately leading to tumor growth at distant anatomic sites if their ‘soil’ is hospitable (1). The exclusively unidirectional nature of this process has been challenged recently in a series of papers (8-12, 28), which use mouse models to demonstrate a mechanism by which CTCs from the primary tumor can reenter the primary, a process called ‘self-seeding’ (12). These authors further comment that ‘it is tempting to speculate that self-seeding might occur not only at the primary tumor site, but also at distinct metastatic sites, … each site being a nesting ground’. The possibility of metastasis from metastases has also been discussed (23,29). While the underlying ‘agent’ responsible for the spread of cancer is the CTC, the disease progression pathways in different patients can be both predictable (from a statistical viewpoint), but often unpredictable and surprisingly distinct in patients with nominally the same disease (26,27), prompting the question ‘how can metastatic pathways be predictable and unpredictable at the same time’ (10)?
Motivated in part by these questions, we develop a Markov chain/Monte Carlo (MCMC) stochastic mathematical model for cancer progression to identify and quantify the multi-directional pathways and timescales associated with metastatic spread for primary lung cancer.
While stochastic in nature, our model shows that a defining aspect of both pathway selection and timescale determination is whether the disease spreads from the primary tumor to a metastatic site that is either a ‘spreader’ (adrenal gland and kidney), or a ‘sponge’ (regional lymph nodes, liver, bone). In contrast to the traditional view of cancer metastasis as a unidirectional process starting at the primary site and spreading to distant sites as time progresses, our model supports and quantifies the view that there are important multi-directional aspects to metastatic progression. These fall under three general classes: (i) self-seeding of the primary tumor; (ii) re-seeding of the primary tumor from a metastatic site (primary re-seeding); and (iii) re-seeding of metastatic tumors (metastasis re-seeding).
Using a discrete Markov chain (14) system of equations applied to a large autopsy data set of untreated cancer patients (15), we quantify the likelihoods of the top metastatic pathways in terms of probabilities and perform Monte Carlo computer simulations of cancer progression that statistically reflect the autopsy data regarding (non-Gaussian) distribution of disease. The stochastic Markov chain dynamical system takes place on a metastatic network based model of disease progression that we construct based on available autopsy data over large populations of patients. To obtain our baseline model, we use the data described in an autopsy analysis (15) in which metastatic tumor distributions in a population of 3827 untreated deceased cancer victims were recorded; 163 of these had primary lung cancer of some type, distributing a total of 619 metastatic tumors across 27 different sites. Information on lung cancer type in this data set is not possible to obtain as the samples were collected prior to the widespread use of immunohistochemistry (1914-1943), without which, the sub-categorization of non-small cell lung cancer is unreliable. However, it is probably safe to assume that the distribution of lung cancer type was not significantly different than current distributions, roughly 40% adenocarcinoma, 30% squamous-cell carcinoma, 9% large-cell carcinoma, and 21% small-cell carcinoma.
2. Materials and Methods
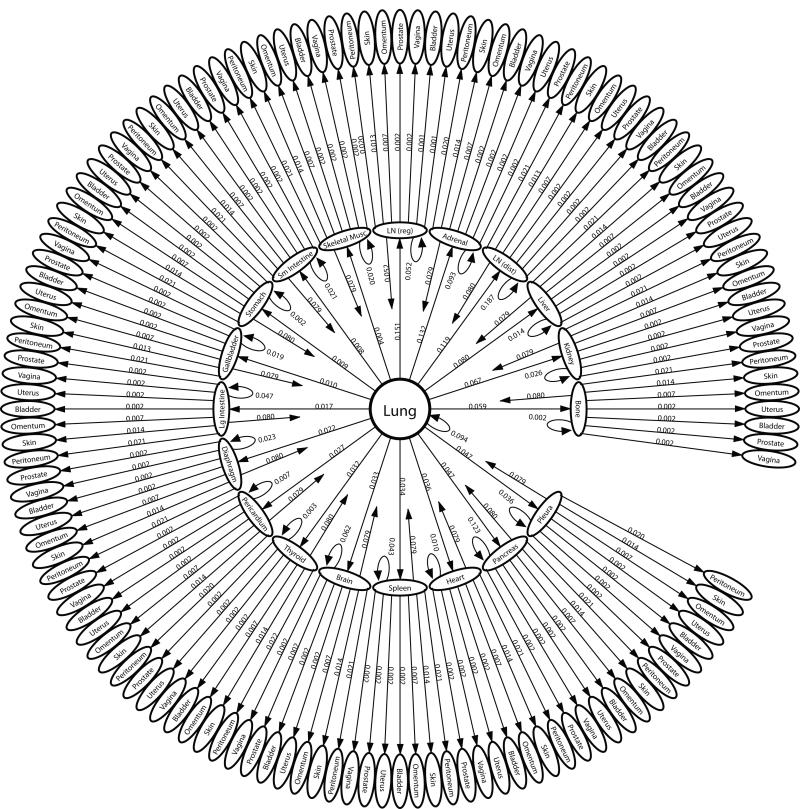
2.1 Structure of the lung cancer multi-step diagram
The 27 metastatic sites in the diagram shown in Figure 1 are organized in ring formation, with 23 sites surrounding lung on the inner ring, and the remaining 4 sites organized on the outmost ring, each connected to a site from the inner ring. The sites listed on the inner ring are called ‘first-order’ sites -they have direct edge connections from the lung, with edge probabilities decreasing from 12:00 clockwise around the ring. The most heavily weighted edge, hence the most likely first step of metastatic disease, is the transition from lung to regional lymph nodes (LN (reg)). The least heavily weighted, hence least likely first step is the transition from lung to skeletal muscle shown just to its left. The 4 sites making up the outermost ring are called ‘second-order’ sites, also organized with edge probabilities decreasing in clockwise order. These sites are classified as ‘second-order’ due to the fact that they have two-step probabilities via a first-order site that are equal or higher in probability than any direct one-step probability from the lung. In short, for disease to spread to a second-order site from lung, it most probably passes via a first-order.
Figure 1. The one-step pathways of metastatic lung cancer.
Ensemble averaged one-step pathway diagram. Primary lung tumor is at the center, next ring out are the 23 first-order sites showing their direct connection from the lung, with transition probabilities getting weaker in clockwise direction. Next ring out are the 4 second-order sites and their connections from the first-order sites. The three elements of multi-directional spread are highlighted in this diagram: (i) Self-seeding of the primary tumor (self-loop back to center); (ii) Re-seeding of the primary tumor from a first-order site (arrows back to center); (iii) Re-seeding of first-order sites (self-loops back to first-order site). Not shown in the diagram are the one-step paths from first order site to another first order site.
The general structure of the concentric diagram, with lung placed at the center, highlights the underlying classical unidirectional view of disease progression. However the diagram also highlights the three key mechanisms of multi-directional progression: (i) self-seeding of the primary lung tumor shown in the diagram as a self-loop in the 7th position, with an edge weight of 5.2%; (ii) re-seeding of the primary tumor from a first-order site, shown as arrows directed back to the center. Because we are using an ensemble average of 1000 trained lung cancer matrices to produce this diagram, the re-seeding edges are all roughly comparable in weight (8%); (iii) metastasis re-seeding of first-order sites shown as a self-loop back to each metastatic site. The strongest metastasis re-seeders are lymph nodes (regional and distant), followed by liver, adrenal, bone, and kidney.
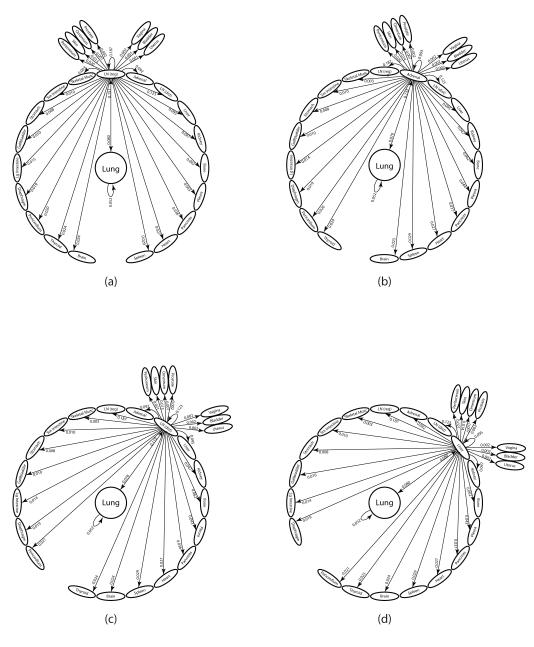
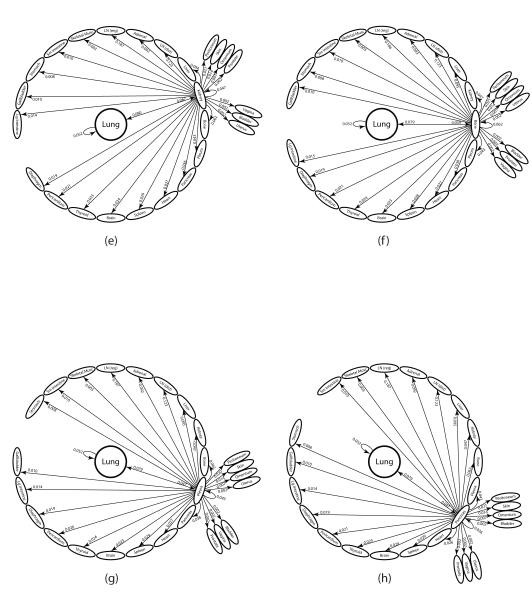
From this diagram, we can obtain all of the possible two-step pathway probabilities from the lung, by direct multiplication of the two edges making up any of the two-step paths starting from lung. The 756 distinct two-step paths from the lung, the top ones of which are shown in Figure 2, produce the statistical distribution produced by the Markov chain model. We calculate all of these and rank them in decreasing order in the next sub-sections. By comparing the probability distributions and (shown in Figure S2), we can see that after two steps, the distribution has nearly converged to the steady-state, so we expect our rankings of two-step pathways not to change much if we compare them to the top three-step and higher step pathways.
Figure 2. The two-step pathways through top 8 first-order sites.
Diagram of all 28 two-step pathways from lung to a tertiary site. (a) Lung through regional lymph nodes; (b) Lung through adrenal gland; (c) Lung through distant lymph nodes; (d) Lung through liver ; (e) Lung through kidney; (f) Lung through bone; (g) Lung through pleura; (h) Lung through pancreas.
Figure S1 shows a (ensemble) convergence and non-convergence plot associated with our search algorithm to calculate the Markov transition matrix based on the baseline data set (15). What is significant is the non-convergence of our algorithm when we constrain our searches to not allow for any multi-directional edges. In other words, when we forced our algorithm to not allow edges directly back to a site (no self-metastases nor primary reseeding), either separately or together, the algorithm would not converge to a solution. By contrast, the algorithm, in general, converged quickly to a solution when all connections were allowed, and produces a transition matrix with many multi-directional connections from site to site.
2.2 The autopsy data sets
The data in (15) compiles the metastatic tumor distributions in a population of 3827 deceased cancer victims, none of whom received chemotherapy or radiation, hence the model can be said to be based on the natural progression of the disease, although mastectomy for many breast cancer primaries was most likely performed at that time. In addition, brain metastases are likely under-represented by this data set since brain autopsies probably were not universally performed at that time. The autopsies were performed between 1914 and 1943 in 5 separate affiliated centers, with an ensemble distribution of 41 primary tumor types and 30 distinct metastatic locations. The total number of distinct primary and metastatic tumor locations is 50, which sets the size of our square Markov transition matrix (50 × 50) as well as the number of entries in the Markov state-vector . The data offers no direct information on the time history of the disease, either for individual patients comprising the ensemble, or in ensemble format. The data we use, therefore, only contains information on the ‘long-time’ distribution of metastatic tumors, where long-time is associated with end of life, a timescale that varies significantly from patient to patient. The model does, however, allow us to infer time histories from autopsy data based on the logic that if more metastatic tumors show up in a population of patients at a specific site, then on average, they would develop earlier in the progression history. Although this association is not perfect, if does allow us to extract meaningful temporal inferences from our Markov chain model. Details of how we infer the correct ensemble Markov transition matrix are described in (13).
We use the data set in two distinct ways to construct our model. First, we associate the distribution of metastatic tumors (after appropriate normalization) for primary lung cancer patients with the steady-state (long-time) probability distribution of our Markov chain (14). From this, we compute the ‘transition matrix’ for our Markov chain (ensemble averaged) that produces this steady-state. Since the problem is mathematically underdetermined, the calculation procedure requires an initial ‘candidate’ transition matrix obtained from the autopsy data and discussed in (13), which is then systematically iterated until a numerical convergence criterion is satisfied. Interestingly, we also show that when our search algorithm is constrained so as to not allow any multi-directional edges in the directed graph associated with the transition matrix, no self-consistent model can be produced (i.e. the search algorithm does not converge). Then, we update our baseline model with the more targeted data set described in (32) of 137 patients with adenocarcinoma of the lung (Stage I and II), all treated with complete lung resection, and show how the baseline model is able to adapt to this assimilated data set.
3. Results
3.1 Cancer metastasis as a stochastic multi-step process
The ensemble averaged lung cancer transition matrix associated with the Markov chain model (see Figure 1) depicts the complete metastatic pathway diagram (13). Each of the 2500 entries, aij, of the 50 × 50 transition matrix determines the probability of the disease (modeled as a random walker over the network) spreading from site ‘i’ to site ‘j’ in an effectively multi-step process before the statistical tumor distribution of the autopsy data set is filled out. The diagram rank orders (in decreasing clockwise order) all of the possible pathways emanating from the lung. One-step paths are defined by the edges leading directly out from the lung – the sum of these outgoing edges must be one. The single most likely one-step path of disease progression from the lung is to the regional lymph nodes, shown at the top of the diagram, with a probability of 15.1%, followed by the lung to adrenal gland path, with probability 13.2%. On the diagram ordering the first steps out of the lung, we also show the ‘self-seeding’ step directly back to the lung, represented by the edge from lung looping back to itself, with edge probability 5.2%. Two-step paths are made up of an edge from the lung to another site (or back to itself), followed by the edge from that site to a second site. There are 756 two-step paths emanating from the lung. The probability of taking a particular two-step path from the lung is obtained by multiplying the weights of the two edges making up the path. The sum of all of these two-step path probabilities must be one, and so on for three-step paths, four-step paths, etc. We focus on quantifying all of the two-step paths in this paper, because as shown in Figure S2 (See Supplementary Material), after two iterations of the Markov chain (k = 2), the state-vector has nearly converged to the steady-state target vector for metastatic tumors making metastatic progression for lung cancer effectively a two-step process. In Figure 2, we show all of the two-step paths emanating from the lung passing through each of the 8 most probable metastatic sites. To obtain the probability of cancer progression on one of these two-step paths, one multiplies the products of the two edges making up the two-step path.
3.2 Rank-ordering the two-step metastatic pathways towards the final state of the disease
We list the top multi-directional two-step pathways obtained from our model in Table 1. The first entries of Table 1 list the top 10 re-seeding pathways back to the lung from a first-order site, along with the running cumulative values. We highlight from this list several points. First, lymph nodes, adrenal gland, and liver are the most important intermediate sites that re-seed back to the lung. Their cumulative probability value (3.8 %) accounts for more than 1/2 of the total cumulative value from the entire list (6.2 %). This total cumulative value is slightly greater than, but roughly comparable in size to the lung to lung re-seeding path value of 5.2 %, indicating that cells that re-seed to the lung land there with roughly equal probabilities of having arrived via an intermediate site (see Table 1) vs. directly from the lung. The second half of Table 1 lists the top 10 two-step re-seeding pathways back to a metastatic site, a mechanism we call ‘metastasis re-seeding’. From this table, we can see that for lung cancer, lymph nodes and adrenal gland are the most active metastasis re-seeders, followed by liver, bone, and kidney.
Table 1.
Top two-step re-seeding pathways back to Lung: Primary  First-order site
First-order site  Primary. Top re-seeding pathways back to metastatic site: Primary
Primary. Top re-seeding pathways back to metastatic site: Primary  First-order site
First-order site  Back to first-order site. Cumulative values (obtained by adding the previous transition probabilities) are listed in third column.
Back to first-order site. Cumulative values (obtained by adding the previous transition probabilities) are listed in third column.
| Top re-seeding pathways back to Lung | Transition probability | Cum Values | ||||
|---|---|---|---|---|---|---|
| Lung |

|
Lymph (reg) |

|
Lung | 0.01214 | |
| Lung |

|
Adrenal |

|
Lung | 0.01042 | 0.02256 |
| Lung |

|
Lymph (dist) |

|
Lung | 0.00952 | 0.03208 |
| Lung |

|
Liver |

|
Lung | 0.00645 | 0.03853 |
| Lung |

|
Kidney |

|
Lung | 0.00533 | 0.04386 |
| Lung |

|
Bone |

|
Lung | 0.00467 | 0.04853 |
| Lung |

|
Pleura |

|
Lung | 0.00375 | 0.05228 |
| Lung |

|
Pancreas |

|
Lung | 0.00367 | 0.05595 |
| Lung |

|
Heart |

|
Lung | 0.00288 | 0.05883 |
| Lung |

|
Lung |

|
Lung | 0.00273 | 0.06156 |
| Top metastasis re-seeders | Transition probability | Cum Values | ||||
| Lung |

|
Lymph (reg) |

|
Lymph (reg) | 0.02819 | |
| Lung |

|
Lymph (dist) |

|
Lymph (dist) | 0.01468 | 0.04287 |
| Lung |

|
Adrenal |

|
Adrenal | 0.01223 | 0.05510 |
| Lung |

|
Liver |

|
Liver | 0.00758 | 0.06268 |
| Lung |

|
Bone |

|
Bone | 0.00364 | 0.06632 |
| Lung |

|
Kidney |

|
Kidney | 0.00314 | 0.06946 |
| Lung |

|
Pleura |

|
Pleura | 0.00206 | 0.07152 |
| Lung |

|
Pancreas |

|
Pancreas | 0.00168 | 0.07320 |
| Lung |

|
Spleen |

|
Spleen | 0.00098 | 0.07418 |
| Lung |

|
Heart |

|
Heart | 0.00095 | 0.07513 |
3.3 Metastatic sites as spreaders or sponges
A careful analysis of all two-step pathways allows us to compute the key probabilistic quantity of interest associated with each two-step path which characterizes each site as a sponge or a spreader. The quantity is the ratio of Probability Out (Pout) over Probability In (Pin) to each of the sites. If Pout > Pin, the site is a spreader, whereas if Pin > Pout, we characterize it as a sponge. The ratio (Pout/Pin) of their exiting and incoming probabilities, in the case of a spreader, gives us what we call the amplification factor, since it is larger than one, while in the case of a sponge, we call the ratio the absorption factor, since it is less than one. Using these quantities, the top two spreaders are the adrenal gland and kidney, with amplification factors of 1.91 (adrenal gland) and 2.86 (kidney). The total number of two-step pathways into and out of the adrenal gland is 10, whereas the total into and out of kidney is only 3. For these reasons, we identify the adrenal gland as the key distant anatomical spreader of primary lung cancer.
The sponges associated with primary lung cancer are the regional lymph nodes, liver, and bone. Their respective absorption factors are 0.74 (regional lymph nodes), 0.87 (liver), and 0.75 (bone). The total number of two-step pathways into and out of the regional lymph nodes is 16, compared with 8 into and out of the liver, and 5 into and out of bone. For these reasons, we identify the regional lymph nodes as the key anatomical sponge associated with primary lung cancer, followed by both bone and liver.
3.4 The spatial pathways of lung cancer
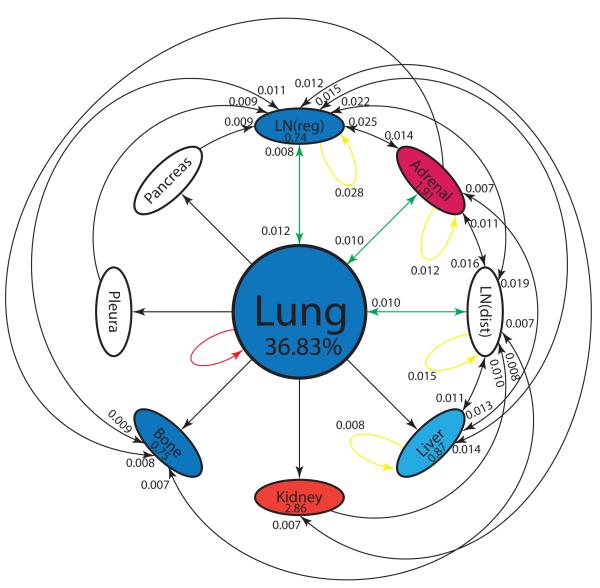
In order to compare the relative importance of two-step unidirectional pathways vs. two-step multi-directional pathways, we list the top 30 two-step pathways in decreasing order in Table S1. The top metastatic pathway (of any type) is the lung → lymph node (reg) → lymph node (reg) metastasis re-seeding pathway, while the top unidirectional pathway is the lung → adrenal → lymph node (reg) path. Looking at all of the multi-directional pathways, it is clear that the lymph nodes and adrenal gland are the key metastatic sites responsible for multi-directional spread, while lymph nodes, adrenal gland and liver are important sites responsible for unidirectional spread. In general terms, lymph nodes, adrenal gland, and liver feature very prominently as intermediate metastatic sites in many of the two-step pathways.
The information can then be combined into a reduced two-step diagram for progression, shown in Figure 3. The diagram visually demonstrates the centrality of lymph nodes and adrenal gland as key first metastatic sites, with many incoming and outgoing edges. The figure also captures all of the information regarding the spreader or sponge character of each site, with red indicating the color of the key spreaders (adrenal gland, kidney), and blue indicating the color of sponges (lung, regional lymph nodes, liver, bone). Amplification and absorption factors are shown in each of the ovals.
Figure 3. Reduced pathway diagram showing top 30 two-step paths.
Top 30 two-step pathways emanating from lung (representing 36.83% of the total pathway probabilities), obtained by multiplying the edges of the one-step edges comprising each two-step path. Edges without numbers are one-step paths emanating from lung. All other numbered edges mark the second edge in a two-step path, with numbers indicating the two-step probabilities. Colors indicate classification of each node as a ‘spreader’ (red), or ‘sponge’ (blue). Spreader amplification factor, and sponge absorption factor are listed in each oval. See text for more detailed descriptions.
3.5 Timescales of progression: Enhancing the Kaplan-Meier approach
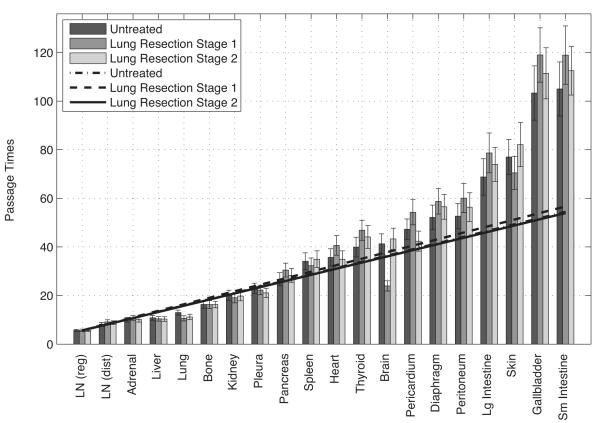
Our model gives a useful measure of metastatic progression timescale, called first-passage time from lung to any given site, defined as the number of edges a ‘random walker’ leaving the lung must traverse in order to first arrive at that site. Monte Carlo simulations of random walk paths from the lung are performed computationally in order to obtain mean first-passage times (averages over 10,000 runs) to every other site in the model. The mean first-passage times (mfpts) act as a proxy timescale (model based) for metastatic progression. It is a model based relative measure of the time that it takes for a primary tumor to metastasize to a secondary site, or, roughly speaking, a model based measure of the timescale associated with successful extravasation and colonization (6). Timescales associated with metastatic disease are typically quantified by so-called Kaplan-Meier survival curves (30,31), which follow a cohort of patients from presentation until death, plotting the survival percentage associated with the cohort. Alternative methods have been proposed, but by and large, tracking survival of a cohort of patients remains the industry-standard way of tracking progression. There is very little in the literature that tracks the timescale of progression from metastatic site to metastatic site (24-27, 29).
Mean first-passage times from lung to each of the other sites are shown in Figure 4. The sites are ordered from shortest to longest mean first-passage time from lung. In drak, we show the baseline (untreated patients) model using the data set (13). The dashed-dot line is a linear curve fit to the first 9 sites, showing a clear linear increasing regime (roughly the top 16 sites), followed by a group of sites where mean first-passage times increase nonlinearly. The first 9 sites used in the reduced model set the basic linear timescales of progression for the high probability metastatic locations. Times increase following the general linear formula mfpt = a · t + b, where a = 2.56, b = 2.07 for the baseline (untreated) model, where ‘a’ is the slope, and ‘b’ is the y-intercept. In this formula, larger slopes indicate longer overall mean first-passage times from lung to metastatic sites. Spread to regional lymph nodes is fastest (with a normalized value of 1), followed by normalized times to distant lymph nodes (1.47), adrenal (1.72) and liver (1.75). One should interpret these timescales to indicate that it takes roughly 75% longer for cancer to metastasize to adrenal gland and liver than to regional lymph nodes. Self-seeding back to lung has a normalized mean first-passage time of 2.30, which is faster than to most of the first-order sites, but over twice the time as the lung to regional lymph node timescale.
Figure 4. Mean first-passage times from Lung to each of the metastatic sites.
Dark shows the baseline (untreated population) model, medium dark shows the baseline model with assimilated Stage I resections, light grey shows baseline model with assimilated Stage II resections. Lines are linear curve fits to first 9 entries. Error bars show one standard deviation from the mean. See text for details.
3.6 Assimilating new autopsy data of adenocarcinoma lung cancer patients undergoing complete resection
Figure 4 (more details are shown in Table S1) also shows metastatic pathways and mean first-passage times using the model with assimilated data from (32), an autopsy data set tracking a cohort of patients with adenocarcinoma of the lung (ACL) who underwent complete lung resection. Of these, 35 survived more than 30 days after resection, 22 were classified as Stage I, and 13 as Stage II. We assimilated their metastatic tumor distribution from an autopsy study into our baseline (untreated population) model, recalculated the Markov transition matrix and all mean first-passage times. The results are shown in Figure 4 (and the middle and right columns of Table S1). Stage I are shown in medium dark, Stage II in light grey.
Comparing the columns of Table S1, the main change in the spatial pathways shows up in the 5th entry down, where the Lung → Adrenal → LN (Dist) pathway drops in probability on the list of the Stage I treated patients, but not as much as for the Stage II treated patients. Lung resection seems to alter this important pathway, particularly for Stage I patients, making it less likely to occur, perhaps by disruption of lymphatic connections between the primary tumor and ipsilateral adrenal gland. The overall probabilities of each of the pathways in the treated population also decrease from the untreated population.
The effect of treatment on the overall mean first-passage times is shown in Figure 4. The corresponding curve fit to the first 9 sites follow the same general linear trend as in the untreated population, mfpt = a · t + b, but with a = 2.68, b=1.55 (Stage I, medium dark); a = 2.54, b=1.91 (Stage II, light grey). The conclusions we can draw are clear: mean first-passage times increase overall with the Stage I treated cohort, shown by the increase in slope over the untreated slope, but not with the Stage II treated cohort. Interestingly, the mfpt back to lung in the treated cohort actually decreases with treatment. Since lung is classified as a sponge in our model, this does not seem to have a negative overall effect on the general trend of increasing passage times with treatment. By contrast, the mfpt back to adrenal gland (the key spreader) with the treated cohort increases. This enhances the overall increase in mfpts for the treated cohort. The mean first-passage times increase most in the sub-group of Stage I patients, indicating that complete lung resection is more effective in this group compared with the Stage II sub-group. To summarize, our model shows that lung resection for ACL patients seems to generally increase overall mfpts of metastases for Stage I patients, and it does this by (i) altering a key pathway from lung to adrenal gland to lymph nodes (distal); (ii) increasing mean first-passage times to the adrenal gland (spreader), (iii) decreasing mean first-passage times back to the lung (sponge); (iv) reducing the overall top pathway probabilities. Lung resection seems to have very little impact on Stage II patients. The failure of resection to improve metastasis free survival in stage II lung cancer patients may occur because the regional lymph nodes act as a sponge (figure 3), potentially suppressing early metastasis when not removed. However, because of the risk of local disease is high in lung cancer, surgery remains the preferred treatment in stage II disease.
4. Discussion
Our model depicts cancer progression as effectively a multi-step (two-step), multi-directional, stochastic process, spreading probabilistically from site to site in individual patients, but filling out a well-defined and predictable metastatic tumor distribution for large ensembles of patients. This stable, robust, and predictable ensemble tumor distribution available over large autopsy data sets is exploited to build a Markov transition matrix for lung cancer progression. We identify the top unidirectional and multi-directional metastatic pathways of primary lung cancer by means of a probabilistic comparison of all two-step paths emanating from the lung. The results support the view that multi-directional pathways play an important role in cancer progression. We identify three main mechanisms of multi-directionality needed to obtain consistency with ensemble autopsy data: (i) primary tumor self-seeding; (ii) re-seeding of the primary tumor from a metastatic tumor; (iii) metastasis re-seeding. Of these, the most important are metastasis re-seeding of the lymph nodes (both regional and distant) and adrenal gland, and primary lung re-seeding via the regional lymph nodes. Also significant is metastasis re-seeding of the liver, and primary self-seeding of the lung, but neither seem to be as significant as passage of the disease through the regional lymph nodes.
While very few patients die from their first metastasis, the characterization of the first metastatic site as a spreader or sponge yields important insights into metastatic pathway selection and the determination of progression timescales for patients. The model may have implications for decisions surrounding surgical resection of oligometastatic disease (33) as one might predict different outcomes for patients whose solitary site of disease is a sponge or spreader. Historically, resection of isolated adrenal metastasis has entered clinical practice in lung cancer, and removal of this spreader site, has benefited patients (34). Conversely, there has never been an established role for resection of isolated liver metastasis, a sponge site, despite there being a track record of success doing this in colon cancer (35-39).
A careful inspection of the top two-step pathways supports the dominance of unidirectional metastatic spread over multi-directional processes, which perhaps explains why the prevailing historical view is one of unidirectional spread (5). However we should emphasize that our search algorithm for a Markov transition matrix could not converge to any solution when we constrained it so that multi-directional edges were ruled out, but did converge consistently to an ensemble of transition matrices when unconstrained so that all possible paths were allowed (See Supplementary Material). In other words, we were not able to find a Markov transition matrix that produced a steady-state consistent with the autopsy data unless multi-directional edge connections were allowed. Therefore we stress the viewpoint that multi-directional processes play a key role in pathway selection and timescale determination for metastatic lung cancer.
Supplementary Material
Quick guide to equations and assumptions.
Assumptions of the model
The disease progression starts with an isolated tumor in the lung position.
The progression dynamics follows a Markov stochastic process (14), moving from site ‘i’ to site ‘j’ according to a transition probability Pij that depends only on those two sites, not on the past history of how it arrived at site ‘i’.
The Markov transition matrix of our model is calculated so that the steady-state vector of the Markov chain model corresponds to the metastatic distribution of tumors found from the autopsy data set described in reference (15).
Key equations
- A Markov chain dynamical system (13,14) is defined by the equation:
, where A is called the Markov transition matrix, and is called the state-vector. The entries of the state-vector give the probabilities of metastatic tumors developing at the 50 different sites in our model. The sum of the entries must be 1. The entries of the Markov transition matrix are the transition probabilities Pij from site ‘i’ to site ‘j’. In our model, A is a 50 × 50 square matrix, and is a vector in . is our initial state vector, which has a 1 placed in the 23rd position, corresponding to the ‘lung’ entry.
is called the steady-state vector associated with the Markov chain. It can be obtained by solving the eigenvalue problem: . Therefore the steady-state vector is a left-eigenvector of the Markov transition matrix.
Acknowledgments
The project described was supported by Award Number U54CA143906 from the National Cancer Institute and the Bill and Melinda Gates Foundation through the Gates Millennium Fellowship Program. The content is solely the responsibility of the authors and does not necessarily represent the official view of the National Cancer Institute or the National Institutes of Health.
Footnotes
Conflict of interest statement: The authors have no conflicts of interest to report.
References
- 1.Paget S. The distribution of secondary growths in cancer of the breast. Lancet. 1889;1:571–573. [PubMed] [Google Scholar]
- 2.Weinberg RA. The Biology of Cancer. Garland Science. 2006 [Google Scholar]
- 3.Weiss L, Ward PM. Cell detachment and metastasis. Cancer Metastasis Rev. 1983;2:111–127. doi: 10.1007/BF00048965. [DOI] [PubMed] [Google Scholar]
- 4.Nieva J, Wendel M, Luttgen MS, Marrinucci D, Bazhenova L, Kolatkar A, et al. High-definition imaging of circulating tumor cells and associated cellular events in non-small cell lung cancer patients: a longitudinal analysis. Physical Biology. 2012;9:1. doi: 10.1088/1478-3975/9/1/016004. doi:10.1088/1478-3975/9/1/016004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weiss L. Metastasis of cancer: a conceptual history from antiquity to the 1990’s. Cancer Metastasis Rev. 2000;19:193–204. [PubMed] [Google Scholar]
- 6.Fidler IJ. Timeline: The pathogenesis of cancer metastasis: the ‘seed and soil’ hypothesis revisited. Nat. Rev. Cancer. 2003;3:453–458. doi: 10.1038/nrc1098. [DOI] [PubMed] [Google Scholar]
- 7.Chambers AF, Groom AC, MacDonald IC. Dissemination and growth of cancer cells in metastatic sites. Nature Reviews Cancer. 2002 Aug.2:563–573. doi: 10.1038/nrc865. [DOI] [PubMed] [Google Scholar]
- 8.Leung CT, Oskarsson T, Acharyya S, Nguyen DX, Zhang XH-F, Norton L, et al. Tumor self-seeding by circulating cancer cells. Cell. 2009;139:1315–1326. doi: 10.1016/j.cell.2009.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reynolds S. Coming home to roost: The self-seeding hypothesis of tumor growth. NCI Cancer Bulletin. 2011 Feb. [Google Scholar]
- 10.Comen E, Norton L, Massagué J. Clinical implications of cancer self-seeding. Nature Reviews Clinical Oncology. 2011;8:369–377. doi: 10.1038/nrclinonc.2011.64. [DOI] [PubMed] [Google Scholar]
- 11.Aguirre-Ghiso JA. On the theory of tumor self-seeding: implications for metastatic progression in humans. Breast Cancer Research. 2010;12:304. doi: 10.1186/bcr2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Norton L, Massagué J. Is cancer a disease of self-seeding? Nature Med. 2006;12:875–878. doi: 10.1038/nm0806-875. [DOI] [PubMed] [Google Scholar]
- 13.Newton PK, Mason J, Nieva J, Bethel K, Bazhenova LA, Kuhn P. A stochastic Markov chain model to describe lung cancer growth and metastasis. PLoS ONE. 2012 Apr;7(4):e34637. doi: 10.1371/journal.pone.0034637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Norris JR. Markov Chains. Cambridge University Press; 1997. [Google Scholar]
- 15.DiSibio G, French SW. Metastatic patterns of cancers: Results from a large autopsy study. Arch Pathol Lab Med. 2008 Jun;132:931–939. doi: 10.5858/2008-132-931-MPOCRF. [DOI] [PubMed] [Google Scholar]
- 16.Norton L. A Gompertzian growth model of human breast cancer growth. Cancer Research. 1988;48:7067–7071. [PubMed] [Google Scholar]
- 17.Norton L, Simon R, Brereton HD, Bogden AE. Predicting the course of Gompertzian growth. Nature. 1976 Dec.264:542–545. doi: 10.1038/264542a0. [DOI] [PubMed] [Google Scholar]
- 18.Chen LL, Blumm N, Christakis NA, Barabasi AL, Deisboeck TS. Cancer metastasis networks and the prediction of progression patterns. British J. of Cancer. 2009;101:749–758. doi: 10.1038/sj.bjc.6605214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goh KI, Cusick ME, Valle D, Childs B, Vidal M, Barabasi AL. The human disease network. Proc. Nat’l Acad. Sci. 2007;104:8685–8690. doi: 10.1073/pnas.0701361104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Girvan M, Newman MEJ. Community structure in social and biological networks. Proc. Natl. Acad. Sci. 2002;99:12, 7821–7826. doi: 10.1073/pnas.122653799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Salsbury AJ. The significance of the circulating cancer cell. Cancer Treat. Rev. 1975 Mar;2(1):55–72. doi: 10.1016/s0305-7372(75)80015-6. [DOI] [PubMed] [Google Scholar]
- 22.Zhang J, Zhou CS, Xu XK. Mapping from structure to dynamics: A unified view of dynamical processes on networks. Phys. Rev. E. 2010;82:2, 026116. doi: 10.1103/PhysRevE.82.026116. [DOI] [PubMed] [Google Scholar]
- 23.Hoover HC, Ketcham AS. Metastasis of metastases. The American Journal of Surgery. 1975 Oct.130:405–411. doi: 10.1016/0002-9610(75)90473-0. [DOI] [PubMed] [Google Scholar]
- 24.Iwata K, Kawasaki K, Shigesada N. A dynamical model for the growth and size distribution of multiple metastatic tumors. J. Theor. Biol. 2000;203:177–186. doi: 10.1006/jtbi.2000.1075. [DOI] [PubMed] [Google Scholar]
- 25.Haustein V, Schumacher U. A dynamic model for tumor growth and metastasis formation. J. Clinical Bioinformatics. 2012 May 1;2:11. doi: 10.1186/2043-9113-2-11. doi:10.1186/2043-9113-2-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yokota J. Tumor progression and metastasis. Carcinogenisis. 2000;21(3):497–503. doi: 10.1093/carcin/21.3.497. [DOI] [PubMed] [Google Scholar]
- 27.Edelman EJ, Guinney J, Jen-Tsan C, Phillip G, Febbo PG, Mukherjee S. Modeling cancer progression via pathway dependencies. PLoS Comp. Bio. 2008 Feb.4(2):e28. doi: 10.1371/journal.pcbi.0040028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klein CA. Parallel progression of primary tumours and metastases. Nature Perspectives. 2009 Apr;9:302–312. doi: 10.1038/nrc2627. [DOI] [PubMed] [Google Scholar]
- 29.Bethge A, Schumacher U, Wree A, Wedemann G. Are metastases from metastases clinically relevant? Computer modeling of cancer spread in a case of Hepatocellular Carcinoma. PLoS ONE. 2012 Apr;12(4):e35689. doi: 10.1371/journal.pone.0035689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lamelas IP. Long-term survival of lung cancer in a province of Spain. J Pulmonar Respirat Med. 2011;S:5 [Google Scholar]
- 31.Nordquist LT, Simon GR, Cantor A, Alberts WM, Bepler G. Improved Survival in Never-Smokers vs Current Smokers With Primary Adenocarcinoma of the Lung. Chest. 2004;126:347–351. doi: 10.1378/chest.126.2.347. DOI 10.1378/chest.126.2.347. [DOI] [PubMed] [Google Scholar]
- 32.Stenbygaard LE, Sorensen JB, Olsen JE. Metastatic pattern in adenocarcinoma of the lung: An autopsy study from a cohort of 137 consecutive patients with complete resection. Journal of Thoracic and Cardiovascular Surgery. 1995;110(4)(Part 1):1130–1135. doi: 10.1016/s0022-5223(05)80183-7. [DOI] [PubMed] [Google Scholar]
- 33.Weichselbaum RR, Hellman S. Oligometastases revisited. Nature Perspectives. 2011 Jun;8:378–382. doi: 10.1038/nrclinonc.2011.44. [DOI] [PubMed] [Google Scholar]
- 34.Bretcha-Boix P, Rami-Porta R, Mateu-Navarro M, Hoyvela-Alonso C, Marco-Molina C. Surgical treatment of lung cancer with adrenal metastases. Lung Cancer. 2000;27:101–105. doi: 10.1016/s0169-5002(99)00097-5. [DOI] [PubMed] [Google Scholar]
- 35.Garden OJ, Rees M, Poston GJ, Mirza D, Saunders M, Lederman J, et al. Guidelines for resection of colorectal cancer liver metastases. GUT. 2002;55(Supp III) doi: 10.1136/gut.2006.098053. doi 10.1136/gut.2006.098053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fong Y, Cohen AM, Fortner JG, Enker WE, Turnbull AD, Coit DG, et al. Liver resection for colorectal metastases. J. Clin. Onc. 1997;15:938–946. doi: 10.1200/JCO.1997.15.3.938. [DOI] [PubMed] [Google Scholar]
- 37.Hughes KS, Simon R, Songhorabodi S, Adson MA, Ilstrup DM, Former JG, et al. Resection of the liver for colorectal carcinoma metastases: A multi-institutional study of indications for resection. Surgery. 1988 Mar;103(3):278–288. [PubMed] [Google Scholar]
- 38.Abdalla EK, Vauthey J-N, Ellis LM, Ellis V, Pollock R, Broglio KR, et al. Recurrence and outcomes following hepatic resection, radiofrequency ablation, and combined resection/ablation for colorectal liver metastases. Annals of Surgery. 2004;Vol. 239(No. 6):818–827. doi: 10.1097/01.sla.0000128305.90650.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pawlick TM, Abdalla EK, Ellis LM, Vauthey J-N, Curley SA. Debunking dogma: Surgery for four or more colorectal liver metastases is justified. The Society for Surg. of the Aliment. Tract. 2006;Vol. 10(No. 2):240–248. doi: 10.1016/j.gassur.2005.07.027. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.