Abstract
The bromodomain protein BRD4 links cell cycle and transcription, bookmarking active genes during mitosis and serving as a scaffold for transcription factors. Our recent discovery that BRD4 is a RNA Polymerase II CTD kinase identifies a novel transcriptional function. Here we discuss our model in the context of current knowledge.
Keywords: BRD4, BET protein, Atypical kinase, RNA Polymerase II, CTD kinase, P-TEFb
Introduction
The ubiquitously expressed BRD4, one of five mammalian bromodomain proteins, was first found associated with mitotic chromosomes and implicated in cell cycle control.1 BRD4 is now recognized as a critical mediator of normal and disease functions through its interactions with both cellular and viral factors.2 BRD4 plays a role in inflammatory diseases by binding NFkB and influencing immunological responses to infection.3 Regulation of HIV latency and transcription depend on the interaction between BRD4 and the transcription elongation factor, PTEFb.2,4 Similarly, HPV latency is regulated by a complex of BRD4 and the immediate early viral protein, E2, which represses transcription of the viral late proteins E6/E7.2 Importantly, BRD4 is associated with breast cancer survival and metastasis, acute myeloid leukemia, Burkitt's lymphoma and colon cancer.5-8 A BRD4 translocation fusion product, BRD4-NUT, is responsible for aggressive midline carcinomas.9 BRD4 is also critical in development: germline deletion of BRD4 results in early embryonic lethality.10
Despite its documented role in disease, an understanding of BRD4’s molecular functions is emerging only now. Indeed, the nature of BRD4 function differs throughout cell cycle. During mitosis, BRD4 remains bound to chromatin, identifying it as a bookmark for actively transcribed genes.1 During interphase BRD4 interacts with various cellular and viral transcription factors,2 which has led to the perspective that BRD4 is a passive scaffold for active factors in cellular functions. Challenging this notion, our recent discovery that BRD4 is a novel atypical kinase that phosphorylates Ser2 of the RNA Polymerase II (RNAP II) C-terminal domain (CTD) identifies BRD4 as an active transcription factor.11 Here, we review recent advances emphasizing BRD4’s novel kinase activity and present our working model that BRD4 links cell cycle and transcriptional regulation.
BRD4 structure and its functional implications
BRD4 belongs to the bromodomain and extra-terminal domain (BET) family of proteins that contain two tandem bromodomains (BD1 and BD2) and an extra-terminal domain (ET).1 The bromodomains bind acetylated histones and a wide range of non-histone proteins. The ET domain is thought to be a regulatory domain whose function is unclear. The mammalian members of the BET family, BRD2, BRD3, BRD4 and BRDT, exhibit similar domain organizations. BRD4 also shares strong structural similarity to the yeast BET proteins Bdf1 and Bdf2, kinases that are yeast homologs of TAF1, the kinase component of mammalian TFIID. There are two BRD4 isoforms. The predominant long isoform is responsible for almost all the biological functions attributed to BRD4. The function of the rare short form, also known as HUNK1, is unknown.2 The structures of the BRD4 BD1 and BD2 bromodomains have been solved and consist of 4 α-helices and 2 loops linking these α-helices. The BD1 domain primarily binds H3-K14ac, while the BD2 domain binds to H4-K5ac and H4-K16ac.12 It is these interactions that allow BRD4 to be retained on the chromatin during mitosis in a range of cell types. The solved ET domain structure consists of 3 α-helices and a loop.2
Interestingly, there may be multiple functionally distinct pools of BRD4: One pool remains associated with chromatin during mitosis, functioning as a mitotic bookmark responsible for accelerating post mitotic gene activation.13 Another BRD4 pool recruits PTEFb to sites of active transcription during interphase.14 A third pool of BRD4 is recruited by transcriptional Mediator complexes independent of PTEFb.2
BRD4 is a protein scaffold
BRD4 is a scaffold that recruits numerous proteins to chromatin in general and transcription sites in particular. In addition to the interactions noted above, BRD4 binds to the Kaposi's sarcoma herpes virus latency associated nuclear antigen 1 (LANA-1), recruiting it to chromatin thereby releasing a G1 cell cycle arrest.15 It also binds to and regulates the enzymatic activity of SIPA1, a GTPase activating protein that functions as a metastasis modifier.5 During myocyte differentiation, the interaction of BRD4 with myogenin induces chromatin remodeling and expression of muscle genes.16 Beyond these examples, BRD4 interacts broadly: A systematic large scale proteomic analysis of BRD4 interactions identified a variety of protein complexes that include the PAF (polymerase associated factor) complex, SEC (super elongation complex) and the Spliceosome assembly complex that play key roles in transcription.17 A large number of proteins, including those involved in heat shock responses and chromatin remodeling, as well as others with unknown functions were also identified by this study.
Each of BRD4’s structural domains is involved in this multiplicity of interactions. Whereas binding to acetylated histones is mediated by its N-terminal bromodomains, BRD4’s interactions with key transcriptional regulators such as PTEFb, NFκb and HPV E2 occur through its C-terminal domain.1-3,18 A proteomic analysis using isolated domains/sections of BRD4 as bait identified several other epigenetic and transcriptional regulators interacting with specific domains on BRD4.19 Protein complexes interacting specifically with the BRD4 ET domain included those containing NSD3 (a histone methyl transferase), JMJD6 (a histone demethylase and splicing associated hydroxylase) and GLTSCR1 (unknown function). Importantly, depletion of NSD3, JMJD6 or GLTSCR1 resulted in the repression of BRD4-target genes, both viral and cellular.19 Together, these studies reinforce the conclusion that BRD4 plays a vital role as a scaffolding protein in physically linking chromatin remodeling and transcriptional regulation.
BRD4 is a mitotic bookmark
A mitotic bookmark identifies actively transcribed genes during mitosis by remaining associated chromatin when all other factors dissociate. The presence of a bookmark accelerates reinitiation of transcription after mitosis from active regions, many of which contain cell cycle genes, thus regulating cell cycle progression. In particular, in a model system with an MS2-YFP RNA reporter construct and fluorescently tagged BRD4, it was shown that BRD4 remains associated with H4K5ac histones on chromatin during mitosis, leading to rapid de-compaction of the surrounding chromatin and to transcription post-mitotically.14 BRD4 also marks the start sites of many M/G1 genes, accelerates expression of G1 genes and promotes cell cycle progression to S phase.13,20 BRD4 further regulates cell cycle progression through its transcriptional regulation of Aurora B kinase expression, essential for chromosome segregation and cytokinesis.21 In response to anti-mitotic agents, such as nocodozole, BRD4 is released from chromatin as part of the protective mechanism during mitotic arrest.22 This release is mediated through the JNK pathway and is correlated with the recovery of cells from drug-induced mitotic arrest. JNK inhibitors blocked BRD4 release and mitotic progression while JNK2−/− embryonic fibroblasts were defective in BRD4 release and cell growth after nocodozole treatment.22 A recent study also proposed a model for the transition of BRD4 from chromatin targeting to transcriptional regulation through a signal induced pathway possibly involving HDACs.23 Thus, BRD4 is a mitotic bookmark that plays a key role in maintaining epigenetic memory and chromatin structure, another mechanism by which it links cell cycle progression and gene expression.
BRD4 is a transcription factor
In addition to its role as a mitotic bookmark, BRD4 is a critical regulator of transcription. Productive transcription depends on the phosphorylation of the C-terminal domain of RNAP II CTD, which consists of consecutive repeats of the heptad Y1S2P3T4S5P6S7. Phosphorylation of the CTD residues Serine 5 (Ser5) and Ser2 is necessary for the recruitment of RNA capping and splicing factors, respectively.24 The order, pattern and temporal separation of these phosphorylation events ensure an orderly transition from initiation to productive transcription elongation.25,26 CTD Ser5 residues are phosphorylated primarily by the CDK7 kinase component of TFIIH, while subsequent Ser2 phosphorylations release RNAP II from an early elongation block and are necessary during productive elongation.24 During elongation, Ser2 is phosphorylated by PTEFb, which depends on BRD4 for its nuclear localization and activation of its CDK9 kinase subunit.18,27
BRD4 recruits PTEFb to the transcription pre-initiation complex18 and initially travels with it in the transcription complex.4 Importantly, we have shown that BRD4 is also a kinase that phosphorylates the CTD Ser2 specifically both in vitro and in vivo.11 Although PTEFb was originally implicated as the predominant CTD Ser2 kinase for both release from an elongation block and productive elongation,27 we have proposed that BRD4 is the major RNAP II CTD Ser2 during transcription initiation and the transition to elongation, whereas PTEFb functions primarily during elongation.11 The evidence in support of this model is as follows. In somatic cells, overexpression of either wild type BRD4 or a mutant that does not bind PTEFb/CDK9 results in increased Ser2 phosphorylation. Furthermore, stem cell lines have negligible levels of PTEFb. Nevertheless, CTD Ser2 is phosphorylated to normal levels, indicating that PTEFb is not the primary Ser2 kinase. Instead, CTD Ser2 phosphorylation depends on BRD4.11
Based on our demonstration that BRD4 is a CTD Ser2 kinase, we propose a new model of transcription initiation in which the initial phosphorylation of the RNAP II CTD Ser2 is mediated by BRD4 during the transition from transcription initiation to elongation, and only subsequently by PTEFb during elongation. A number of additional lines of evidence support this view: i) BRD4 is part of the transcription pre-initiation complex and remains associated with the RNAP II transcription complex until productive elongation begins between +14 to +36 nt4; ii) BRD4 and PTEFb have distinct CTD substrate specificities. BRD4 directly phosphorylates Ser2 while PTEFb only phosphorylates Ser2 if Ser5 has not been previously phosphorylated, indicating that they play different roles during transcription. CTD Ser7, and not Ser5, phosphorylation by TFIIH primes the CTD for recognition by PTEFb;11,28 iii) BRD4 phosphorylation of CTD Ser2 is markedly enhanced by prior TFIIH phosphorylation at Ser5, suggesting that BRD4 functions immediately after TFIIH.11 Taken together, these findings argue that BRD4 is the initial CTD Ser2 kinase in the early steps of transcription until its release at the start of productive elongation. The functional role of BRD4 in vivo as the primary CTD Ser2 kinase was confirmed by our finding that preventing BRD4 from being recruited to the transcription site, or its siRNA knockdown, reduces Ser2 phosphorylation and nascent RNA levels whereas knockdown of PTEFb/CDK9 had only a minimal effect.11
Phosphorylations of RNAP II CTD are critical for the regulated maturation of nascent transcripts and coordination of transcription with other nuclear processes.24 Studies from our lab have begun to address the mechanisms that regulate the order and timing of those phosphorylations. We have recently identified a complex network of functional interactions among the CTD kinases that directly and indirectly govern their activities. Namely, TFIIH phosphorylates and inhibits the kinase activity of BRD4. BRD4 in turn phosphorylates PTEFb at two independent sites, one of which activates while the other inactivates the CDK9 CTD kinase activity. Conversely, BRD4 is phosphorylated and activated by PTEFb. BRD4, like TFIIH and PTEFb, also directly phosphorylates and modulates TAF7, a general transcription factor that regulates all three CTD kinases. These cross-phosphorylations have the potential to profoundly affect overall CTD phosphorylation and suggest a complex mechanism of modulating CTD phosphorylation during transcription initiation and early elongation.29 Together, these findings confirm that beyond its role in mitotic bookmarking, BRD4 functions as an essential transcription factor.
Model for BRD4 function
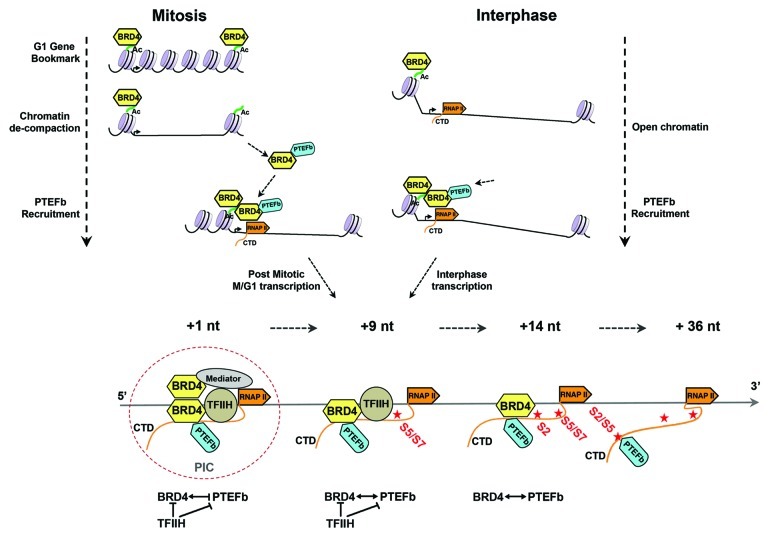
The multiplicity of BRD4 functions—as a mitotic bookmark, a scaffold that recruits transcription factors to promoters and a CTD kinase that regulates RNAP II CTD phosphorylation—suggest a model in which BRD4 regulates transcription, coordinating it with cell cycle, as summarized in Figure 1. Briefly, during mitosis BRD4 is bound to the condensed chromatin, marking immediate-early post-mitotic (M/G1) genes. At late anaphase, a portion of the bound BRD4 is released to recruit dephosphorylated, inactive PTEFb from the HEXIM complex in the cytoplasm.18,20 Although the JNK pathway is conditionally implicated,22 the general factors responsible for the release of a subset of BRD4 remains to be determined, as does the mechanism by which condensed chromatin is de-compacted by the remaining BRD4 bound to it.14 The chromatin-bound BRD4 bookmark may serve as an anchor together with acetylated histones, recruiting BRD4/PTEFb complexes to sites of transcription through transient dimerization with the BRD4.30 BRD4/PTEFb complexes are also recruited to active transcription sites during interphase.14 The interactions between PTEFb and BRD4 results in their mutual phosphorylation, the net effect of which is to activate the BRD4 kinase while conditionally repressing PTEFb kinase.29 In addition to its role in recruiting and regulating PTEFb, BRD4 also has a critical role in transcription through its own CTD kinase activity, which is regulated by TFIIH. In addition, BRD4 recruits other transcription factors (e.g., NFkB and HPV E2) to sites of active transcription through its interaction with the Mediator.2
Figure 1. Model of BRD4 function in cell cycle and gene transcription. During mitosis (upper left), BRD4 functions as a G1 gene bookmark by binding to acetylated chromatin, as a PTEFb recruiter and chromatin de-compacter. During interphase (upper right), it recruits PTEFb to RNAP II, either directly binding chromatin or dimerizing with chromatin-bound BRD4, thereby activating both post mitotic and interphase transcription. During transcription initiation (lower panel), direct and indirect interactions among the CTD kinases, including BRD4, initially keeps their activity in check, with the release of their activities as the transcription complexes proceeds. BRD4 plays a central role in this cross-talk of the CTD kinases.
This aggregation of interactions results in a cascade of phosphorylations that serve to prevent premature transcription initiation: Mediator-associated CDK8 phosphorylation of TFIIH/cyclin H has been reported to repress its CTD kinase activity.31 TFIIH phosphorylation of BRD4 represses its CTD kinase activity. BRD4 phosphorylation of PTEFb/CDK9 initially represses its activity by phosphorylating its inactivating site. When PIC assembly is completed, transcription initiates, possibly involving the release of each inhibited kinase by phosphatases that remain to be identified. After transcription initiates, CDK7 CTD kinase activity is released from inhibition by Mediator/CDK8, allowing it to phosphorylate the CTD Ser5/7.31 Importantly, TFIIH/CDK7 also phosphorylates the BRD4 complexed with PTEFb, thereby repressing BRD4 CTD kinase activity. This ensures that the first CTD phosphorylation is that of Ser5/7 by CDK7. Where TFIIH is left behind by the transcriptional complex between + 14 nt and +36 nt, BRD4 kinase activity would be regained, allowing it to phosphorylate the Ser2 sites of CTD pre-phosphorylated at Ser5, a preferred substrate for BRD4 but not PTEFb. BRD4 is released from the PIC around +36 nt. At this point, PTEFb/CDK9 CTD kinase activity is restored by PP2A phosphatase, which specifically dephosphorylates the inactivating mark placed by BRD4, leaving only the activating phosphorylation mark.4 As noted earlier, although PTEFb is a Ser2 CTD kinase, it requires CTD dephosphorylated of prior TFIIH-mediated Ser5P.28 This is accomplished by the RPAP2 phosphatase,25 allowing PTEFb to continue phosphorylating CTD Ser2/Ser5 sites during productive elongation.
As evident by its overarching role in disease and normal cell biology, BRD4 is a central coordinator of transcription, linking cell cycle with transcription through its functions as a mitotic bookmark, scaffold and RNAP II CTD kinase. The discovery of its intrinsic kinase activity extends the function of BRD4 from its previously described function as a chromatin modifier to transcriptional regulation. In addition, its kinase activity suggests a possible mechanism by which it regulates its numerous interacting partners. The identification of the JQ1 inhibitor that prevents BRD4 from binding chromatin was an important advance for targeting BRD4 in therapeutic strategies.32 Efforts are underway in our lab to identify a BRD4 specific kinase inhibitor, which may be similarly useful in treating diseases where BRD4 kinase activity plays a role in regulating transcription. Much remains to be learned about BRD4. It remains to be determined whether BRD4’s function as a transcription factor is global or restricted to only those genes it bookmarks during mitosis. Similarly, although BRD4 is required during embryonic development, its role in differentiated cells and their functions has not been established. Future studies will be needed to address these questions.
Acknowledgments
We thank members of our lab for thoughtful discussions and apologize to the researchers whose work could not be referenced due to space limitations. This research was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
Footnotes
Previously published online: www.landesbioscience.com/journals/transcription/article/22542
References
- 1.Dey A, Ellenberg J, Farina A, Coleman AE, Maruyama T, Sciortino S, et al. A bromodomain protein, MCAP, associates with mitotic chromosomes and affects G(2)-to-M transition. Mol Cell Biol. 2000;20:6537–49. doi: 10.1128/MCB.20.17.6537-6549.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu SY, Chiang CM. The double bromodomain-containing chromatin adaptor Brd4 and transcriptional regulation. J Biol Chem. 2007;282:13141–5. doi: 10.1074/jbc.R700001200. [DOI] [PubMed] [Google Scholar]
- 3.Nicodeme E, Jeffrey KL, Schaefer U, Beinke S, Dewell S, Chung CW, et al. Suppression of inflammation by a synthetic histone mimic. Nature. 2010;468:1119–23. doi: 10.1038/nature09589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou M, Huang K, Jung KJ, Cho WK, Klase Z, Kashanchi F, et al. Bromodomain protein Brd4 regulates human immunodeficiency virus transcription through phosphorylation of CDK9 at threonine 29. J Virol. 2009;83:1036–44. doi: 10.1128/JVI.01316-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crawford NP, Alsarraj J, Lukes L, Walker RC, Officewala JS, Yang HH, et al. Bromodomain 4 activation predicts breast cancer survival. Proc Natl Acad Sci U S A. 2008;105:6380–5. doi: 10.1073/pnas.0710331105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zuber J, Shi J, Wang E, Rappaport AR, Herrmann H, Sison EA, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011;478:524–8. doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mertz JA, Conery AR, Bryant BM, Sandy P, Balasubramanian S, Mele DA, et al. Targeting MYC dependence in cancer by inhibiting BET bromodomains. Proc Natl Acad Sci U S A. 2011;108:16669–74. doi: 10.1073/pnas.1108190108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rodriguez RM, Huidobro C, Urdinguio RG, Mangas C, Soldevilla B, Domínguez G, et al. Aberrant epigenetic regulation of bromodomain BRD4 in human colon cancer. J Mol Med (Berl) 2012;90:587–95. doi: 10.1007/s00109-011-0837-0. [DOI] [PubMed] [Google Scholar]
- 9.French CA, Miyoshi I, Kubonishi I, Grier HE, Perez-Atayde AR, Fletcher JA. BRD4-NUT fusion oncogene: a novel mechanism in aggressive carcinoma. Cancer Res. 2003;63:304–7. [PubMed] [Google Scholar]
- 10.Houzelstein D, Bullock SL, Lynch DE, Grigorieva EF, Wilson VA, Beddington RS. Growth and early postimplantation defects in mice deficient for the bromodomain-containing protein Brd4. Mol Cell Biol. 2002;22:3794–802. doi: 10.1128/MCB.22.11.3794-3802.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Devaiah BN, Lewis BA, Cherman N, Hewitt MC, Albrecht BK, Robey PG, et al. BRD4 is an atypical kinase that phosphorylates serine2 of the RNA polymerase II carboxy-terminal domain. Proc Natl Acad Sci U S A. 2012;109:6927–32. doi: 10.1073/pnas.1120422109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vollmuth F, Blankenfeldt W, Geyer M. Structures of the dual bromodomains of the P-TEFb-activating protein Brd4 at atomic resolution. J Biol Chem. 2009;284:36547–56. doi: 10.1074/jbc.M109.033712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dey A, Nishiyama A, Karpova T, McNally J, Ozato K. Brd4 marks select genes on mitotic chromatin and directs postmitotic transcription. Mol Biol Cell. 2009;20:4899–909. doi: 10.1091/mbc.E09-05-0380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao R, Nakamura T, Fu Y, Lazar Z, Spector DL. Gene bookmarking accelerates the kinetics of post-mitotic transcriptional re-activation. Nat Cell Biol. 2011;13:1295–304. doi: 10.1038/ncb2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ottinger M, Christalla T, Nathan K, Brinkmann MM, Viejo-Borbolla A, Schulz TF. Kaposi’s sarcoma-associated herpesvirus LANA-1 interacts with the short variant of BRD4 and releases cells from a BRD4- and BRD2/RING3-induced G1 cell cycle arrest. J Virol. 2006;80:10772–86. doi: 10.1128/JVI.00804-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Du C, Jin YQ, Qi JJ, Ji ZX, Li SY, An GS, et al. Effects of myogenin on expression of late muscle genes through MyoD-dependent chromatin remodeling ability of myogenin. Mol Cells. 2012;34:133–42. doi: 10.1007/s10059-012-2286-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dawson MA, Prinjha RK, Dittmann A, Giotopoulos G, Bantscheff M, Chan WI, et al. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature. 2011;478:529–33. doi: 10.1038/nature10509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jang MK, Mochizuki K, Zhou M, Jeong HS, Brady JN, Ozato K. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol Cell. 2005;19:523–34. doi: 10.1016/j.molcel.2005.06.027. [DOI] [PubMed] [Google Scholar]
- 19.Rahman S, Sowa ME, Ottinger M, Smith JA, Shi Y, Harper JW, et al. The Brd4 extraterminal domain confers transcription activation independent of pTEFb by recruiting multiple proteins, including NSD3. Mol Cell Biol. 2011;31:2641–52. doi: 10.1128/MCB.01341-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang Z, He N, Zhou Q. Brd4 recruits P-TEFb to chromosomes at late mitosis to promote G1 gene expression and cell cycle progression. Mol Cell Biol. 2008;28:967–76. doi: 10.1128/MCB.01020-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.You J, Li Q, Wu C, Kim J, Ottinger M, Howley PM. Regulation of aurora B expression by the bromodomain protein Brd4. Mol Cell Biol. 2009;29:5094–103. doi: 10.1128/MCB.00299-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nishiyama A, Dey A, Tamura T, Ko M, Ozato K. Activation of JNK triggers release of Brd4 from mitotic chromosomes and mediates protection from drug-induced mitotic stress. PLoS One. 2012;7:e34719. doi: 10.1371/journal.pone.0034719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ai N, Hu X, Ding F, Yu B, Wang H, Lu X, et al. Signal-induced Brd4 release from chromatin is essential for its role transition from chromatin targeting to transcriptional regulation. Nucleic Acids Res. 2011;39:9592–604. doi: 10.1093/nar/gkr698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bartkowiak B, Mackellar AL, Greenleaf AL. Updating the CTD Story: From Tail to Epic. Genet Res Int. 2011;2011:623718. doi: 10.4061/2011/623718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Egloff S, Dienstbier M, Murphy S. Updating the RNA polymerase CTD code: adding gene-specific layers. Trends Genet. 2012;28:333–41. doi: 10.1016/j.tig.2012.03.007. [DOI] [PubMed] [Google Scholar]
- 26.Gegonne A, Weissman JD, Lu H, Zhou M, Dasgupta A, Ribble R, et al. TFIID component TAF7 functionally interacts with both TFIIH and P-TEFb. Proc Natl Acad Sci U S A. 2008;105:5367–72. doi: 10.1073/pnas.0801637105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Price DH. P-TEFb, a cyclin-dependent kinase controlling elongation by RNA polymerase II. Mol Cell Biol. 2000;20:2629–34. doi: 10.1128/MCB.20.8.2629-2634.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Czudnochowski N, Bosken CA, Geyer M. Serine-7 but not serine-5 phosphorylation primes RNA polymerase II CTD for P-TEFb recognition. Nature Communications 2012; DOI:10.1038/ncomms1846. [DOI] [PubMed] [Google Scholar]
- 29.Devaiah BN, Singer DS. Cross-talk among RNA polymerase II kinases modulates C-terminal domain phosphorylation. J Biol Chem. 2012 doi: 10.1074/jbc.M112.412015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang R, Li Q, Helfer CM, Jiao J, You J. Bromodomain protein Brd4 associated with acetylated chromatin is important for maintenance of higher-order chromatin structure. J Biol Chem. 2012;287:10738–52. doi: 10.1074/jbc.M111.323493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Akoulitchev S, Chuikov S, Reinberg D. TFIIH is negatively regulated by cdk8-containing mediator complexes. Nature. 2000;407:102–6. doi: 10.1038/35024111. [DOI] [PubMed] [Google Scholar]
- 32.Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, et al. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–73. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]