Abstract
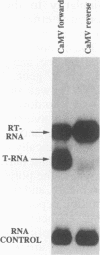
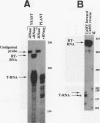
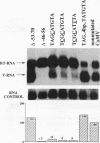
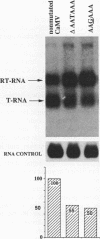
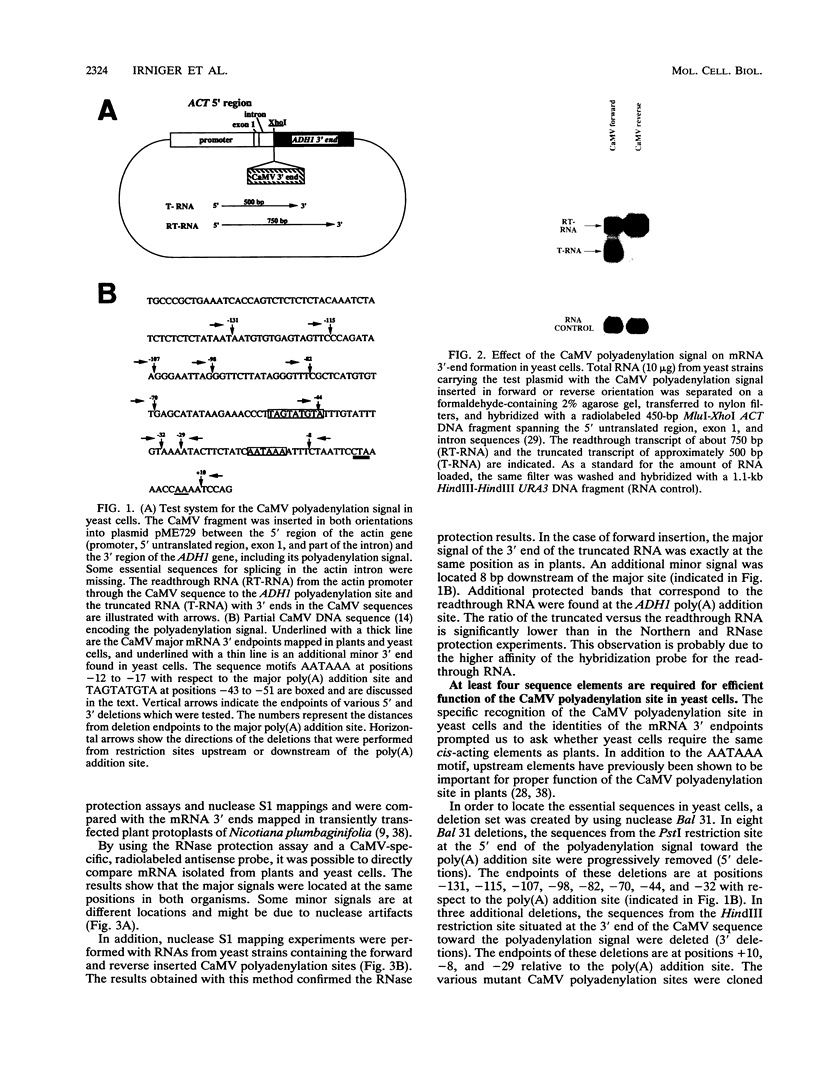
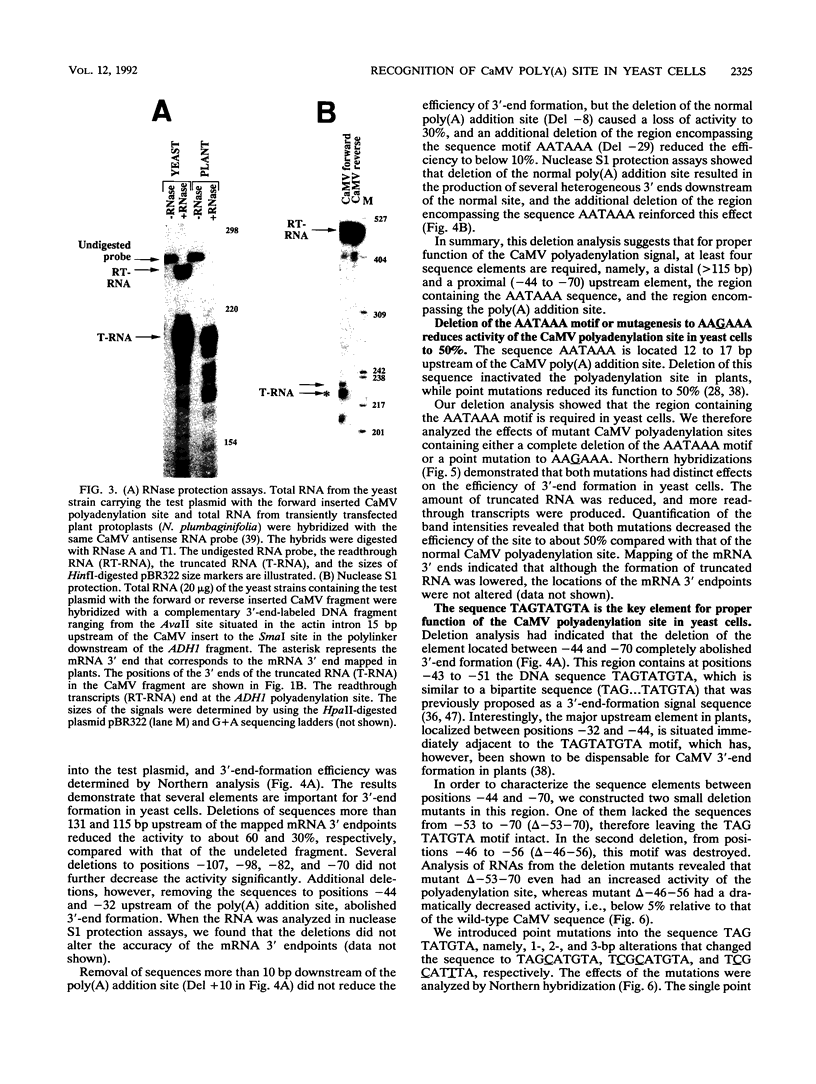
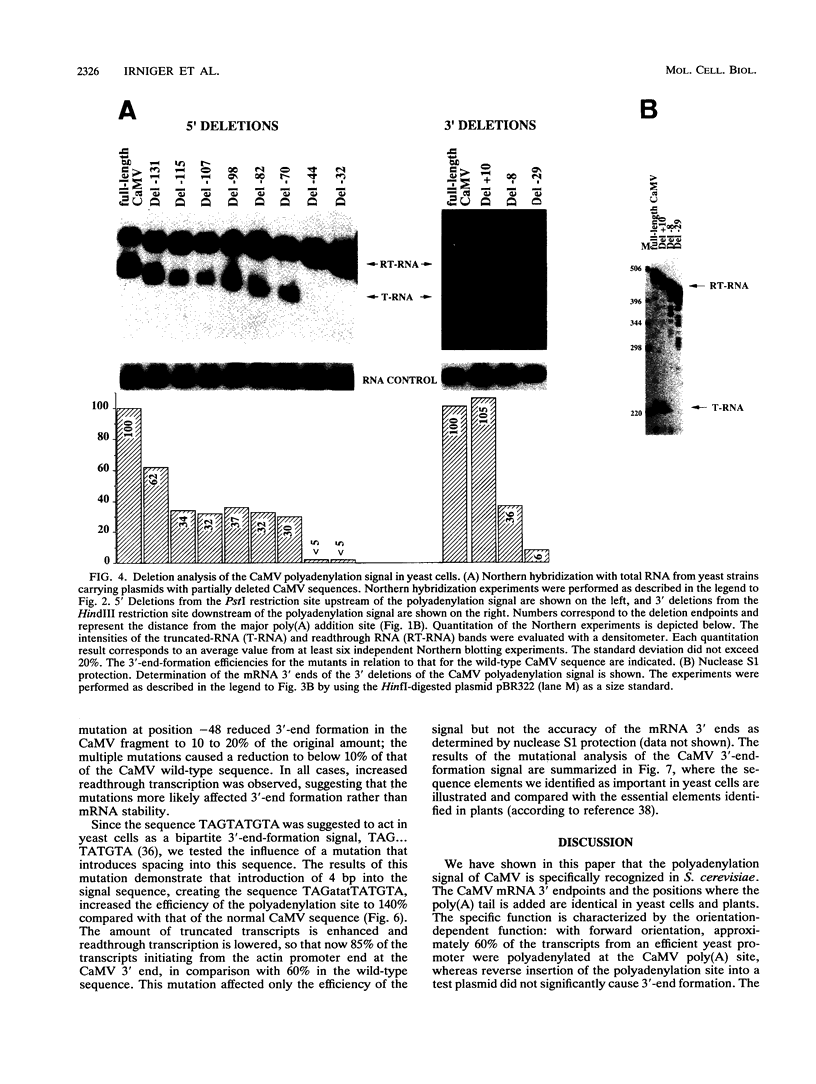
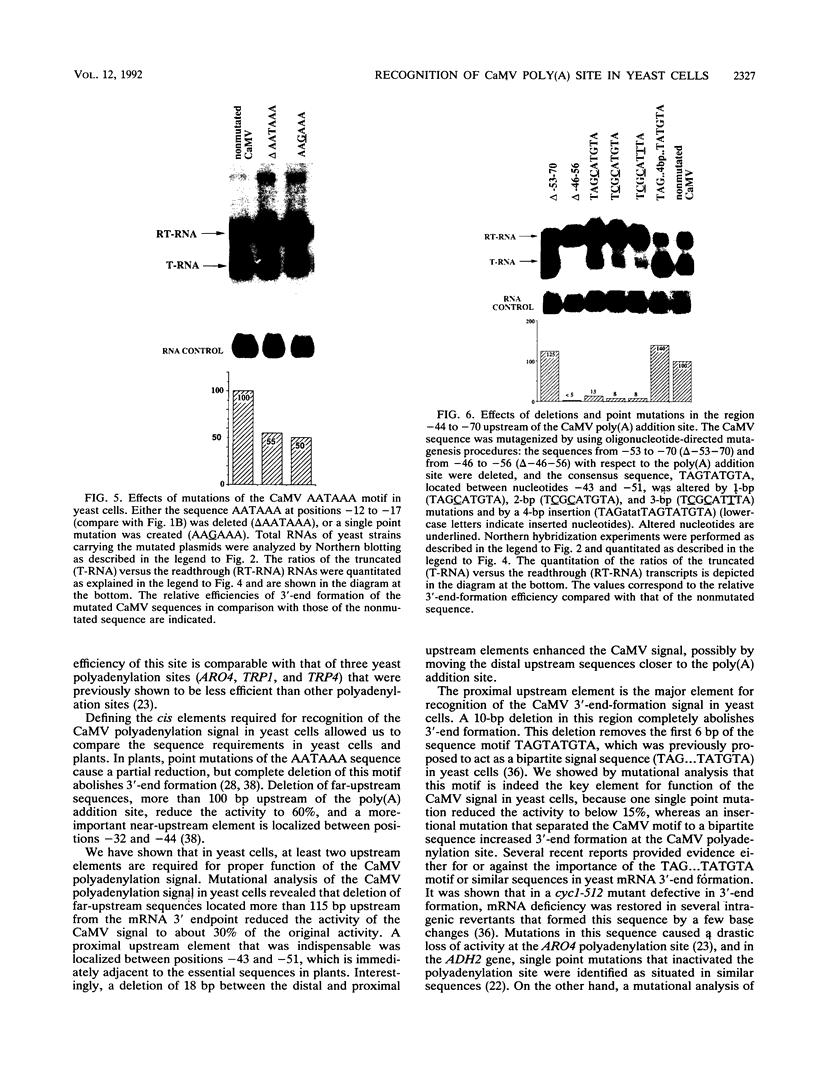
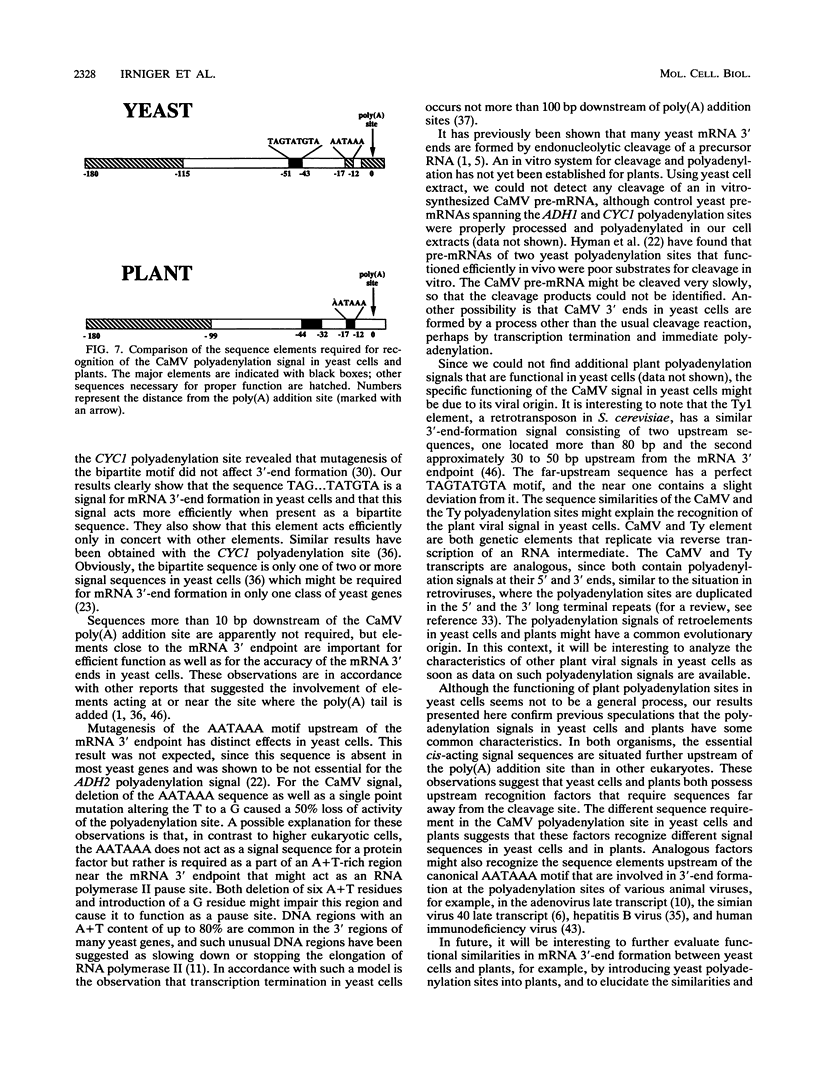
We show that the polyadenylation site derived from the plant cauliflower mosaic virus (CaMV) is specifically functional in the yeast Saccharomyces cerevisiae. The mRNA 3' endpoints were mapped at the same position in yeast cells as in plants, and the CaMV polyadenylation site was recognized in an orientation-dependent manner. Mutational analysis of the CaMV 3'-end-formation signal revealed that multiple elements are essential for proper activity in yeast cells, including two upstream elements that are situated more than 100 and 43 to 51 nucleotides upstream of the poly(A) addition site and the sequences at or near the poly(A) addition site. A comparison of the sequence elements that are essential for proper function of the CaMV signal in yeast cells and plants showed that both organisms require a distal and a proximal upstream element but that these sequence elements are not identical in yeast cells and plants. The key element for functioning of the CaMV signal in yeast cells is the sequence TAGTATGTA, which is similar to a sequence previously proposed to act in yeast cells as a bipartite signal, namely, TAG ... TATGTA. Deletion of this sequence in the CaMV polyadenylation signal abolished 3'-end formation in yeast cells, and a single point mutation in this motif reduced the activity of the CaMV signal to below 15%. These results indicate that the bipartite sequence element acts as a signal for 3'-end formation in yeast cells but only together with other cis-acting elements.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abe A., Hiraoka Y., Fukasawa T. Signal sequence for generation of mRNA 3' end in the Saccharomyces cerevisiae GAL7 gene. EMBO J. 1990 Nov;9(11):3691–3697. doi: 10.1002/j.1460-2075.1990.tb07581.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ammerer G. Expression of genes in yeast using the ADCI promoter. Methods Enzymol. 1983;101:192–201. doi: 10.1016/0076-6879(83)01014-9. [DOI] [PubMed] [Google Scholar]
- Braus G., Furter R., Prantl F., Niederberger P., Hütter R. Arrangement of genes TRP1 and TRP3 of Saccharomyces cerevisiae strains. Arch Microbiol. 1985 Sep;142(4):383–388. doi: 10.1007/BF00491908. [DOI] [PubMed] [Google Scholar]
- Butler J. S., Platt T. RNA processing generates the mature 3' end of yeast CYC1 messenger RNA in vitro. Science. 1988 Dec 2;242(4883):1270–1274. doi: 10.1126/science.2848317. [DOI] [PubMed] [Google Scholar]
- Butler J. S., Sadhale P. P., Platt T. RNA processing in vitro produces mature 3' ends of a variety of Saccharomyces cerevisiae mRNAs. Mol Cell Biol. 1990 Jun;10(6):2599–2605. doi: 10.1128/mcb.10.6.2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carswell S., Alwine J. C. Efficiency of utilization of the simian virus 40 late polyadenylation site: effects of upstream sequences. Mol Cell Biol. 1989 Oct;9(10):4248–4258. doi: 10.1128/mcb.9.10.4248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christofori G., Keller W. 3' cleavage and polyadenylation of mRNA precursors in vitro requires a poly(A) polymerase, a cleavage factor, and a snRNP. Cell. 1988 Sep 9;54(6):875–889. doi: 10.1016/s0092-8674(88)91263-9. [DOI] [PubMed] [Google Scholar]
- Conway L., Wickens M. A sequence downstream of A-A-U-A-A-A is required for formation of simian virus 40 late mRNA 3' termini in frog oocytes. Proc Natl Acad Sci U S A. 1985 Jun;82(12):3949–3953. doi: 10.1073/pnas.82.12.3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covey S. N., Lomonossoff G. P., Hull R. Characterisation of cauliflower mosaic virus DNA sequences which encode major polyadenylated transcripts. Nucleic Acids Res. 1981 Dec 21;9(24):6735–6747. doi: 10.1093/nar/9.24.6735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeZazzo J. D., Imperiale M. J. Sequences upstream of AAUAAA influence poly(A) site selection in a complex transcription unit. Mol Cell Biol. 1989 Nov;9(11):4951–4961. doi: 10.1128/mcb.9.11.4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enriquez-Harris P., Levitt N., Briggs D., Proudfoot N. J. A pause site for RNA polymerase II is associated with termination of transcription. EMBO J. 1991 Jul;10(7):1833–1842. doi: 10.1002/j.1460-2075.1991.tb07709.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. "A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity". Addendum. Anal Biochem. 1984 Feb;137(1):266–267. doi: 10.1016/0003-2697(84)90381-6. [DOI] [PubMed] [Google Scholar]
- Fitzgerald M., Shenk T. The sequence 5'-AAUAAA-3'forms parts of the recognition site for polyadenylation of late SV40 mRNAs. Cell. 1981 Apr;24(1):251–260. doi: 10.1016/0092-8674(81)90521-3. [DOI] [PubMed] [Google Scholar]
- Franck A., Guilley H., Jonard G., Richards K., Hirth L. Nucleotide sequence of cauliflower mosaic virus DNA. Cell. 1980 Aug;21(1):285–294. doi: 10.1016/0092-8674(80)90136-1. [DOI] [PubMed] [Google Scholar]
- Giebel L. B., Spritz R. A. Site-directed mutagenesis using a double-stranded DNA fragment as a PCR primer. Nucleic Acids Res. 1990 Aug 25;18(16):4947–4947. doi: 10.1093/nar/18.16.4947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil A., Proudfoot N. J. A sequence downstream of AAUAAA is required for rabbit beta-globin mRNA 3'-end formation. 1984 Nov 29-Dec 5Nature. 312(5993):473–474. doi: 10.1038/312473a0. [DOI] [PubMed] [Google Scholar]
- Gilmartin G. M., McDevitt M. A., Nevins J. R. Multiple factors are required for specific RNA cleavage at a poly(A) addition site. Genes Dev. 1988 May;2(5):578–587. doi: 10.1101/gad.2.5.578. [DOI] [PubMed] [Google Scholar]
- Goodall G. J., Filipowicz W. The AU-rich sequences present in the introns of plant nuclear pre-mRNAs are required for splicing. Cell. 1989 Aug 11;58(3):473–483. doi: 10.1016/0092-8674(89)90428-5. [DOI] [PubMed] [Google Scholar]
- Hill J. E., Myers A. M., Koerner T. J., Tzagoloff A. Yeast/E. coli shuttle vectors with multiple unique restriction sites. Yeast. 1986 Sep;2(3):163–167. doi: 10.1002/yea.320020304. [DOI] [PubMed] [Google Scholar]
- Humphrey T., Christofori G., Lucijanic V., Keller W. Cleavage and polyadenylation of messenger RNA precursors in vitro occurs within large and specific 3' processing complexes. EMBO J. 1987 Dec 20;6(13):4159–4168. doi: 10.1002/j.1460-2075.1987.tb02762.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman L. E., Seiler S. H., Whoriskey J., Moore C. L. Point mutations upstream of the yeast ADH2 poly(A) site significantly reduce the efficiency of 3'-end formation. Mol Cell Biol. 1991 Apr;11(4):2004–2012. doi: 10.1128/mcb.11.4.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irniger S., Egli C. M., Braus G. H. Different classes of polyadenylation sites in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1991 Jun;11(6):3060–3069. doi: 10.1128/mcb.11.6.3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi C. P. Putative polyadenylation signals in nuclear genes of higher plants: a compilation and analysis. Nucleic Acids Res. 1987 Dec 10;15(23):9627–9640. doi: 10.1093/nar/15.23.9627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLauchlan J., Gaffney D., Whitton J. L., Clements J. B. The consensus sequence YGTGTTYY located downstream from the AATAAA signal is required for efficient formation of mRNA 3' termini. Nucleic Acids Res. 1985 Feb 25;13(4):1347–1368. doi: 10.1093/nar/13.4.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miozzari G., Niederberger P., Hütter R. Tryptophan biosynthesis in Saccharomyces cerevisiae: control of the flux through the pathway. J Bacteriol. 1978 Apr;134(1):48–59. doi: 10.1128/jb.134.1.48-59.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogen B. D., MacDonald M. H., Graybosch R., Hunt A. G. Upstream sequences other than AAUAAA are required for efficient messenger RNA 3'-end formation in plants. Plant Cell. 1990 Dec;2(12):1261–1272. doi: 10.1105/tpc.2.12.1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng R., Abelson J. Isolation and sequence of the gene for actin in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3912–3916. doi: 10.1073/pnas.77.7.3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne B. I., Guarente L. Mutational analysis of a yeast transcriptional terminator. Proc Natl Acad Sci U S A. 1989 Jun;86(11):4097–4101. doi: 10.1073/pnas.86.11.4097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietrzak M., Shillito R. D., Hohn T., Potrykus I. Expression in plants of two bacterial antibiotic resistance genes after protoplast transformation with a new plant expression vector. Nucleic Acids Res. 1986 Jul 25;14(14):5857–5868. doi: 10.1093/nar/14.14.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt T. Transcription termination and the regulation of gene expression. Annu Rev Biochem. 1986;55:339–372. doi: 10.1146/annurev.bi.55.070186.002011. [DOI] [PubMed] [Google Scholar]
- Proudfoot N. J., Brownlee G. G. 3' non-coding region sequences in eukaryotic messenger RNA. Nature. 1976 Sep 16;263(5574):211–214. doi: 10.1038/263211a0. [DOI] [PubMed] [Google Scholar]
- Proudfoot N. Poly(A) signals. Cell. 1991 Feb 22;64(4):671–674. doi: 10.1016/0092-8674(91)90495-k. [DOI] [PubMed] [Google Scholar]
- Russnak R., Ganem D. Sequences 5' to the polyadenylation signal mediate differential poly(A) site use in hepatitis B viruses. Genes Dev. 1990 May;4(5):764–776. doi: 10.1101/gad.4.5.764. [DOI] [PubMed] [Google Scholar]
- Russo P., Li W. Z., Hampsey D. M., Zaret K. S., Sherman F. Distinct cis-acting signals enhance 3' endpoint formation of CYC1 mRNA in the yeast Saccharomyces cerevisiae. EMBO J. 1991 Mar;10(3):563–571. doi: 10.1002/j.1460-2075.1991.tb07983.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo P., Sherman F. Transcription terminates near the poly(A) site in the CYC1 gene of the yeast Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1989 Nov;86(21):8348–8352. doi: 10.1073/pnas.86.21.8348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanfaçon H., Hohn T. Proximity to the promoter inhibits recognition of cauliflower mosaic virus polyadenylation signal. Nature. 1990 Jul 5;346(6279):81–84. doi: 10.1038/346081a0. [DOI] [PubMed] [Google Scholar]
- Skolnik-David H., Moore C. L., Sharp P. A. Electrophoretic separation of polyadenylation-specific complexes. Genes Dev. 1987 Sep;1(7):672–682. doi: 10.1101/gad.1.7.672. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Takagaki Y., Manley J. L., MacDonald C. C., Wilusz J., Shenk T. A multisubunit factor, CstF, is required for polyadenylation of mammalian pre-mRNAs. Genes Dev. 1990 Dec;4(12A):2112–2120. doi: 10.1101/gad.4.12a.2112. [DOI] [PubMed] [Google Scholar]
- Valsamakis A., Zeichner S., Carswell S., Alwine J. C. The human immunodeficiency virus type 1 polyadenylylation signal: a 3' long terminal repeat element upstream of the AAUAAA necessary for efficient polyadenylylation. Proc Natl Acad Sci U S A. 1991 Mar 15;88(6):2108–2112. doi: 10.1073/pnas.88.6.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickens M. How the messenger got its tail: addition of poly(A) in the nucleus. Trends Biochem Sci. 1990 Jul;15(7):277–281. doi: 10.1016/0968-0004(90)90054-f. [DOI] [PubMed] [Google Scholar]
- Wickens M., Stephenson P. Role of the conserved AAUAAA sequence: four AAUAAA point mutants prevent messenger RNA 3' end formation. Science. 1984 Nov 30;226(4678):1045–1051. doi: 10.1126/science.6208611. [DOI] [PubMed] [Google Scholar]
- Yu K., Elder R. T. Some of the signals for 3'-end formation in transcription of the Saccharomyces cerevisiae Ty-D15 element are immediately downstream of the initiation site. Mol Cell Biol. 1989 Jun;9(6):2431–2444. doi: 10.1128/mcb.9.6.2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaret K. S., Sherman F. DNA sequence required for efficient transcription termination in yeast. Cell. 1982 Mar;28(3):563–573. doi: 10.1016/0092-8674(82)90211-2. [DOI] [PubMed] [Google Scholar]
- Zaret K. S., Sherman F. Mutationally altered 3' ends of yeast CYC1 mRNA affect transcript stability and translational efficiency. J Mol Biol. 1984 Jul 25;177(1):107–135. doi: 10.1016/0022-2836(84)90060-3. [DOI] [PubMed] [Google Scholar]
- Zarkower D., Stephenson P., Sheets M., Wickens M. The AAUAAA sequence is required both for cleavage and for polyadenylation of simian virus 40 pre-mRNA in vitro. Mol Cell Biol. 1986 Jul;6(7):2317–2323. doi: 10.1128/mcb.6.7.2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarkower D., Wickens M. Formation of mRNA 3' termini: stability and dissociation of a complex involving the AAUAAA sequence. EMBO J. 1987 Jan;6(1):177–186. doi: 10.1002/j.1460-2075.1987.tb04736.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zitomer R. S., Hall B. D. Yeast cytochrome c messenger RNA. In vitro translation and specific immunoprecipitation of the CYC1 gene product. J Biol Chem. 1976 Oct 25;251(20):6320–6326. [PubMed] [Google Scholar]