Abstract
DNA lesions pose a physical obstacle to DNA-dependent cellular transactions such as replication and transcription. A great deal is known regarding RNA polymerase II (RNAP II) transcription stalling in the presence of lesions induced by UV, but recent studies have uncovered previously uncharacterized behavior of the RNAP II machinery in the presence of double strand breaks (DSBs). These new data, although contradictory, contribute to our understanding of a vital cellular mechanism that defends against the production of aberrant transcripts and protects cell viability.
Keywords: DNA repair, DNAPK, DSBs, chromatin, transcription
Introduction
DNA is constantly assaulted by exogenous agents, such as mutagenic chemicals and radiation, and by endogenous factors, such as reactive oxygen species, which lead to damage.1 Faulty repair of DNA damage results in mutations, chromosomal rearrangements and aberrant expression, all of which contribute to cancer and aging.1 DNA damage is deleterious because it interferes with the progression of both DNA and RNA polymerases, and thus compromises the fidelity of replication and transcription.1 To minimize the impact of these threats, cells have evolved various DNA repair mechanisms specific to the different types of DNA lesions. For example, UV-induced photoproducts and bulky adducts are recognized and eliminated by nucleotide excision repair (NER),2 oxidative damage by base excision repair (BER)3 and DSBs, which are among the most deleterious DNA lesions, are eliminated by the double strand break repair (DSBR) machinery.4 Each repair pathway exerts its function through the concerted action of multi-subunit complexes composed of units specific to each pathway.2-4
There are several possible outcomes when RNAP II encounters a DNA lesion: the polymerase may be arrested in front of the lesion, it may transcribe over the lesion, or it may be removed from the lesion either by reverse translocation or by release from the DNA.5 RNAP II stalling at bulky lesions acts as a first step for lesion recognition by key factors of transcription coupled repair (TCR).5-7 The TCR repair factor Cockayne Syndrome group B protein (CSB), with DNA-dependent ATPase activity, is required for the assembly of the TCR machinery around the damage-stalled polymerase.5-7 The half-life of an arrested polymerase at cyclobutane pyrimidine dimmers (CPDs) is around 20 h in vitro and its footprint extends 10 nucleotides ahead of the CPD and 25 nucleotides behind.5,8 In vitro experiments have shown that RNAP II backtracks away from the lesion to permit binding by the repair factors.5,9 Alternatively, and as a last resort after prolonged transcription arrest, RNAP II is dislodged from the DNA by ubiquitination and subsequent degradation.5 Recent data have revealed a two-step process in which NEDD4 (Rsp5 in yeast) first monoubiquitylates RNAP II to serve as a docking site for the elongin-cullin complex (Elc1-Cul3), which triggers polyubiquitination of RNAP II at K48 as a signal for degradation.10-12 Certain DNA lesions do not arrest RNAP II but are instead bypassed, which leads to mutated transcripts. Bacterial polymerases can efficiently transcribe over a variety of strand breaks, including one to five base pair long gaps.13 Moreover, non-bulky DNA lesions, such as 8-oxoguanine (8OG), uracil and O6-methylguanine, are known to be efficiently bypassed by RNA polymerases both in vitro and in vivo.13 In these instances, incorporation of incorrect bases opposite the damage leads to mutant transcripts that could direct the synthesis of mutant proteins, a process termed transcriptional mutagenesis. Although this mechanism was described only for particular DNA lesions, recent exciting findings demonstrate that translesion transcription occurs in the presence of thymidine dimers14 and DSBs.15
RNAP II Transcription and DSBs
The most deleterious type of DNA damage is the double-strand break (DSB), because due to the lack of a complementary template to guide repair, it may lead to disruption of genetic information and chromosomal translocations.4 Despite the significant efforts invested in studying the different pathways that mediate DSB response and in identifying the specific repair factors involved, only recently have studies probed how the RNAP II transcription machinery is affected by DSBs. We aim here to discuss these findings, describe their converging points, and give possible explanations for their differences.
The Role of Chromatin Structure in DSB-Induced Transcription Silencing
The DNA damage response (DDR) following a DSB is initiated by the recruitment and extensive spreading of key players of the repair process around the lesions to form discrete foci.4 DDR involves an extensive set of histone modifications, including ubiquitination, acetylation, methylation, and phosphorylation to facilitate access to the chromatin fiber and to propagate cell cycle checkpoint signals.4 One well-characterized histone modification is the phosphorylation of H2AX at serine 139 (γ-H2AX). This modification extends along the chromatin surrounding a DSB and is mediated by ataxia telangiectasia mutated kinase (ATM), DNA-dependent protein kinase catalytic subunit (DNAPKcs), and Ataxia Telangiectasia and Rad3-related (ATR).4 A set of the histone marks visualized at DSBs are associated with open and decondensed chromatin, a hallmark of actively transcribed genes, whereas another set is associated with transcriptionally silent regions of the genome and heterochromatin.4 Moreover, heterochromatic landmarks, such as HP1 and polycomb proteins, are recruited to the region surrounding DSBs.16 How transcription is affected by DSBs is of foremost importance given that chromatin structure has a major impact on transcription.
Shanbahag et al. investigated the role of ubiquitination of histone H2A that is linked to chromatin compaction and transcription repression in transcriptional silencing upon DSB induction.17 They used an elegant system that allows the simultaneous visualization of transcription and DNA damage response at the single-cell level. Using this system, they generated DSBs distal to the promoter (4–13 kb) of a reporter gene and assessed its transcriptional outcome in relation to the DDR. They found that DSBs led to transcriptional repression of the reporter that was dependent on ATM kinase activity.17 The ATM kinase drives the spreading of chromatin alterations after DSB induction.4 In addition to H2AX phosphorylation, ATM is required for the recruitment of E3 ubiquitin ligases RNF8 and RNF168, which ubiquitinate histones H2A and H2AX at the damaged chromatin.18,19 Since it is known that ubiquitinated H2A covers condensed areas of the genome, such as the inactivated X chromosome20 and promoters of repressed genes,21 the authors postulate that ubiquitination of H2A originating from a DSB and spreading in the surrounding chromatin is responsible for the transcriptional silencing in response to breaks. Although it is not known precisely how far from the lesion this histone mark is detected, enlargement of ubiquitin-enriched chromatin domains results in a reduction of de novo mRNA synthesis,22 suggesting a correlation between ubiquitination and transcriptional repression. Moreover, Shanbahag et al. show that downregulation of RNF8 and RNF168 restores the transcription of the reporter in the presence of DSBs and that depletion of the uH2A deubiquitylating enzyme USP16 prevents the reversal of silencing upon ATM inhibition. At the mechanistic level, the ATM-dependent ubiquitination of H2A leads to the removal of the elongating (S2 phosphorylated) RNAP II from the transcribed region, although the overall RNAP II levels are not affected.17 This observation suggests that transcription is blocked at the elongation rather than the initiation step.
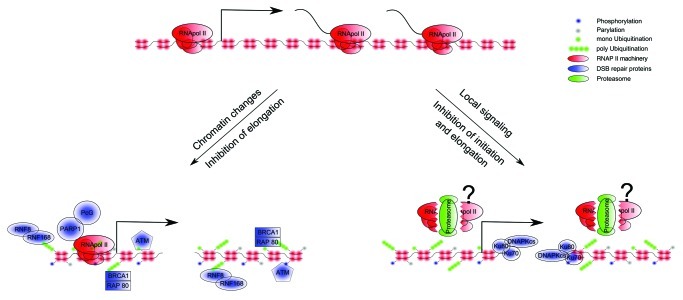
Similar observations were made when RNAP I transcription was monitored in the presence of genotoxic stress.23 DNA damage at the nucleolus resulted in transient inhibition of RNAP I transcription that was not a direct consequence of the damaged template, but rather was mediated by the ATM kinase activity and repair factors that are responsible for the initial recruitment and spreading of the DDR signal in the chromatin surrounding the breaks. In this system, the kinase activity inhibited both the elongation and the re-initiation of RNAP I in response to DSBs.23 The above studies suggest that the DDR spreading around DSBs poses a barrier in the processivity of RNAP I and II by creating a local repressive chromatin structure (Fig. 1).
Figure 1. Two models for transcriptional silencing in the presence of DSBs. In the first model, repressive chromatin structure induced by DSBs leads to inhibition of RNAP II elongation. In the second model, a phosphorylation event at the vicinity of the break mediated by DNAPKcs leads to the release of the RNAP II complex from the gene and promoter and thus inhibition of transcription elongation and reinitiation.
High resolution mapping of H2AX phosphorylation in chromatin that surrounds a DSB in cis revealed that the signal is not incessant, as holes were observed at RNAP II-enriched regions at which they were actively transcribing.24 Given the negative correlation between reduced γ-H2AX signal and transcriptional activity, one can speculate that there is a mechanism in place to protect transcribing genes from γ-H2AX spreading. Legube and colleagues recently reported that cohesin binding antagonizes spreading of γ-H2AX signal to the actively transcribing genes if they are not damaged.25 More specifically, they showed that downregulation of SCC1, a component of the cohesin complex, leads to an increase in the γ-H2AX signal at cohesin-bound genes and a decrease in their transcriptional activity. Nevertheless, it was shown that cohesin together with the mediator complex physically and functionally connects the enhancers and core promoters of active genes in a cell type-specific manner to guide tissue-specific gene expression.26 Although, one cannot exclude the possibility that the affected gene expression is a direct consequence of cohesin depletion, the fact that the authors identified an additional effect on transcription in damaged cells25 suggests a role of cohesin as an insulator of DDR. It is important to note that the binding of cohesin to active genes reported by Legube and colleagues is constitutive and not triggered by the DSB, while the de novo cohesin recruitment is restricted to only 2 kb around the DSB.25
PARylation, another type of post-translational modification that is driven by the Poly ADP-ribose polymerase 1 (PARP1), was also shown to play a role in removing nascent RNA and elongating RNAP II from sites of DNA damage following laser microirradiation.27 Chou et al. used an antibody that recognizes the 7-methylguanosine (m7G) cap structure of nascent RNA to visualize transcription. They showed that DNA damage inhibits RNAP II transcription, as exemplified by the loss of nascent transcripts from the damaged area. This loss was enhanced upon inhibition or depletion of poly(ADP ribose) glycohydrolase (PARG), an enzyme that counteracts PARP activity by disassembling Poly(ADP-ribose).
This observation is in line with findings by Miller et al. and Beli et al., which showed that sites of microirradiation are depleted from the initiating and elongating form of RNAP II, respectively.28,29 Moreover, the Chou et al. study showed that polycomb group (PcG) proteins, a family of transcriptional repressors, are associated with DSBs in a PARP-dependent manner.27 The authors hypothesized that under their experimental conditions, where a variety of lesions are induced, PARP1 establishes a repressive chromatin structure to block transcription by recruiting members of the polycomb complex (Fig. 1). The recruitment of Polycomb at DSBs and the methylation of H3K27me3 associated with Ezh2, a component of polycomb, were also reported by other groups.27,30-33 However, their role in RNAP II transcriptional arrest was not investigated. In addition to the polycomb complex, which marks facultative heterochromatin and is known to directly promote chromatin compaction, HP1 and Kap1 are also markers of constitutive heterochromatin associated with DSBs.34 They are obvious targets for further research because the functional significance of their recruitment during transcriptional silencing has not been evaluated.
On the other hand, it is well established that chromatin surrounding DSBs undergoes local decondensation, histone eviction, and nucleosome remodeling, which are all hallmarks of active transcription.4 Thus, it is hard to reconcile the reported opposing chromatin alterations that occur at DSBs and construct a unifying model for the role of chromatin structure in DSB-induced transcriptional inhibition.
Taken together, the activation of a DSB at a genomic location that is transcribed by RNAP II with a well-characterized chromatin structure will allow for a better understanding of the involvement of chromatin changes during DSBR in break-induced transcriptional silencing.
RNAP II Transcriptional Silencing in Response to a Single DSB: Local to Global Effect
Induction of a single DSB at the yeast MAT locus revealed that transcription is inhibited only at the gene harboring the DNA break site; no change in gene expression was observed for genes within 50 kb of the DSB, where H2AX is phosphorylated. On the other hand, as the DNA within this region becomes single strand due to 5′ to 3′ resection, the mRNA expression level of the neighboring genes decreases progressively as a function of distance from the break site.35
Similarly, studies in mammalian cells of transcription and RNAP II binding after DSB induction showed that gene expression is accurately maintained within γ-H2AX domains and only transcription of genes that contained the break is arrested.15,24 To search for a mechanism mediating the break-induced transcriptional arrest, our group used meganuclease I-PpoI, which has a unique recognition sequence at the coding regions of several RNAP II-transcribed genes in human cells.15 We showed that DNA-dependent protein kinase (DNAPK), but not ATM kinase, is required to inhibit transcription during DSB repair.15 Moreover, we reported that the DSB infliction leads to the eviction of RNAP II from the coding region as well as the promoter and that upon inhibition of the kinase activity of DNAPKcs, RNAP II bypasses the break and continues transcription elongation.15 Although both ATM and DNAPKcs are phosphatidylinositol 3-kinase-related kinases (PIKKs) that bind at sites of DNA damage and share several common substrates, they each associate with DSBs in a different manner: DNAPKcs is recruited near the broken ends, whereas ATM spreads from the DSB to a very large region of the surrounding chromatin.4 Moreover, we showed that inhibition of DNAPKcs does not significantly affect the γ-H2AX spreading around the breaks.15 Therefore, our results suggested that an event near the break, rather than global chromatin structure changes upon DDR signaling, is involved in blocking RNAP II transcription in response to DSBs (Fig. 1). To support this hypothesis, depletion of Ku70 or Ku80 of the Ku heterodimer complex, which recruits DNAPKcs to the breaks, reversed transcriptional arrest in response to DSBs.15 Given the proposed action of Ku80 in transcription and its interaction with the elongating RNAP II,36 it is tempting to speculate that the Ku heterodimer is in a complex with RNAP II and travels with it. When a lesion occurs, it is immediately recognized by the Ku heterodimer and DNAPKcs is recruited to promote transcriptional arrest and DNA repair.
The E3 ubiquitin ligase BRCA1 is another example of a DSBR protein that stably associates with RNAP II.37 BRCA1 binds to elongating RNAP II via BRCA1 associated RING protein 1 (BARD1).38 Earlier studies showed that BRCA1 ubiquitinates RPB1, the largest RNAP II subunit, upon UV exposure and facilitates rapid proteasome-dependent degradation.39 More recent studies have challenged these results and proposed that two different ubiquitin ligases, NEDD4 and Cul3, mediate RNAP II ubiquitination.11 Our data implicate the proteasome in the eviction of RNAP II from the damaged template.15 It will be very exciting to investigate whether the mechanism is similar to UV-induced transcriptional arrest and RNAP II degradation.
Defining the Specificity of DSB-Induced Transcriptional Silencing
All the studies reviewed above described fascinating, albeit dissimilar, mechanisms for transcriptional silencing in response to DSBs. This discrepancy may be due to the different number of breaks induced and the topology of the breaks in the different studies. While ATM signaling may block transcription elongation when the breaks are located near the promoter regions, DNAPKcs appears to be essential for arresting transcription when the RNAP II machinery comes across a single DSB. While ATM signaling may block transcription elongation when the breaks are located near the promoter regions, DNAPKcs appears to be essential for arresting transcription when the RNAP II machinery comes across a single DSB. Another possible explanation for the discrepancy is the different transcriptional status and chromatin structures of the genes that existed prior to break induction in the different studies. Shanbahag et al. induced the breaks proximal to an inducible gene that is highly transcribed upon doxycycline addition, whereas we used I-PpoI target genes that are constitutively transcribed at moderate levels. Furthermore, laser microirradiation is applied to a region of the nucleus that presumably contains both silent and active genes transcribed at different rates. In addition, the type of lesions induced in the different studies may explain the dependency on different factors for transcriptional silencing. For example, the I-PpoI endonuclease cleaves DNA with 3′ overhangs, while the FokI nuclease utilized by the Greenberg laboratory (Shanbahag et al.) leaves 5′ overhangs. Laser microirradiation also generates a variety of DSBs with different complexity along with other types of lesions, including oxidative damage and single-strand breaks.
It is fundamental to fully understand processes that may produce aberrant transcripts in essential genes and alter vital cellular functions. Inducing DSBs at different genomic loci using site-specific nucleases (meganucleases, transcription activator-like effector nucleases (TALENS), etc.) and in vivo transcription visualization methods in single cells will facilitate our understanding of how the participants of DSB repair coordinate with the RNAP II machinery to regulate transcription after DNA damage.
Footnotes
Previously published online: www.landesbioscience.com/journals/transcription/article/22879
References
- 1.Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461:1071–8. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Compe E, Egly JM. TFIIH: when transcription met DNA repair. Nat Rev Mol Cell Biol. 2012;13:343–54. doi: 10.1038/nrm3350. [DOI] [PubMed] [Google Scholar]
- 3.Svilar D, Goellner EM, Almeida KH, Sobol RW. Base excision repair and lesion-dependent subpathways for repair of oxidative DNA damage. Antioxid Redox Signal. 2011;14:2491–507. doi: 10.1089/ars.2010.3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Misteli T, Soutoglou E. The emerging role of nuclear architecture in DNA repair and genome maintenance. Nat Rev Mol Cell Biol. 2009;10:243–54. doi: 10.1038/nrm2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hanawalt PC, Spivak G. Transcription-coupled DNA repair: two decades of progress and surprises. Nat Rev Mol Cell Biol. 2008;9:958–70. doi: 10.1038/nrm2549. [DOI] [PubMed] [Google Scholar]
- 6.Fousteri M, Mullenders LH. Transcription-coupled nucleotide excision repair in mammalian cells: molecular mechanisms and biological effects. Cell Res. 2008;18:73–84. doi: 10.1038/cr.2008.6. [DOI] [PubMed] [Google Scholar]
- 7.Fousteri M, Vermeulen W, van Zeeland AA, Mullenders LH. Cockayne syndrome A and B proteins differentially regulate recruitment of chromatin remodeling and repair factors to stalled RNA polymerase II in vivo. Mol Cell. 2006;23:471–82. doi: 10.1016/j.molcel.2006.06.029. [DOI] [PubMed] [Google Scholar]
- 8.Selby CP, Drapkin R, Reinberg D, Sancar A. RNA polymerase II stalled at a thymine dimer: footprint and effect on excision repair. Nucleic Acids Res. 1997;25:787–93. doi: 10.1093/nar/25.4.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Donahue BA, Yin S, Taylor JS, Reines D, Hanawalt PC. Transcript cleavage by RNA polymerase II arrested by a cyclobutane pyrimidine dimer in the DNA template. Proc Natl Acad Sci U S A. 1994;91:8502–6. doi: 10.1073/pnas.91.18.8502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Anindya R, Aygün O, Svejstrup JQ. Damage-induced ubiquitylation of human RNA polymerase II by the ubiquitin ligase Nedd4, but not Cockayne syndrome proteins or BRCA1. Mol Cell. 2007;28:386–97. doi: 10.1016/j.molcel.2007.10.008. [DOI] [PubMed] [Google Scholar]
- 11.Harreman M, Taschner M, Sigurdsson S, Anindya R, Reid J, Somesh B, et al. Distinct ubiquitin ligases act sequentially for RNA polymerase II polyubiquitylation. Proc Natl Acad Sci U S A. 2009;106:20705–10. doi: 10.1073/pnas.0907052106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Malik S, Bagla S, Chaurasia P, Duan Z, Bhaumik SR. Elongating RNA polymerase II is disassembled through specific degradation of its largest but not other subunits in response to DNA damage in vivo. J Biol Chem. 2008;283:6897–905. doi: 10.1074/jbc.M707649200. [DOI] [PubMed] [Google Scholar]
- 13.Saxowsky TT, Doetsch PW. RNA polymerase encounters with DNA damage: transcription-coupled repair or transcriptional mutagenesis? Chem Rev. 2006;106:474–88. doi: 10.1021/cr040466q. [DOI] [PubMed] [Google Scholar]
- 14.Walmacq C, Cheung AC, Kireeva ML, Lubkowska L, Ye C, Gotte D, et al. Mechanism of translesion transcription by RNA polymerase II and its role in cellular resistance to DNA damage. Mol Cell. 2012;46:18–29. doi: 10.1016/j.molcel.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pankotai T, Bonhomme C, Chen D, Soutoglou E. DNAPKcs-dependent arrest of RNA polymerase II transcription in the presence of DNA breaks. Nat Struct Mol Biol. 2012;19:276–82. doi: 10.1038/nsmb.2224. [DOI] [PubMed] [Google Scholar]
- 16.Soria G, Polo SE, Almouzni G. Prime, repair, restore: the active role of chromatin in the DNA damage response. Mol Cell. 2012;46:722–34. doi: 10.1016/j.molcel.2012.06.002. [DOI] [PubMed] [Google Scholar]
- 17.Shanbhag NM, Rafalska-Metcalf IU, Balane-Bolivar C, Janicki SM, Greenberg RA. ATM-dependent chromatin changes silence transcription in cis to DNA double-strand breaks. Cell. 2010;141:970–81. doi: 10.1016/j.cell.2010.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Doil C, Mailand N, Bekker-Jensen S, Menard P, Larsen DH, Pepperkok R, et al. RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins. Cell. 2009;136:435–46. doi: 10.1016/j.cell.2008.12.041. [DOI] [PubMed] [Google Scholar]
- 19.Mailand N, Bekker-Jensen S, Faustrup H, Melander F, Bartek J, Lukas C, et al. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell. 2007;131:887–900. doi: 10.1016/j.cell.2007.09.040. [DOI] [PubMed] [Google Scholar]
- 20.Fang J, Chen T, Chadwick B, Li E, Zhang Y. Ring1b-mediated H2A ubiquitination associates with inactive X chromosomes and is involved in initiation of X inactivation. J Biol Chem. 2004;279:52812–5. doi: 10.1074/jbc.C400493200. [DOI] [PubMed] [Google Scholar]
- 21.Nakagawa T, Kajitani T, Togo S, Masuko N, Ohdan H, Hishikawa Y, et al. Deubiquitylation of histone H2A activates transcriptional initiation via trans-histone cross-talk with H3K4 di- and trimethylation. Genes Dev. 2008;22:37–49. doi: 10.1101/gad.1609708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gudjonsson T, Altmeyer M, Savic V, Toledo L, Dinant C, Grøfte M, et al. TRIP12 and UBR5 suppress spreading of chromatin ubiquitylation at damaged chromosomes. Cell. 2012;150:697–709. doi: 10.1016/j.cell.2012.06.039. [DOI] [PubMed] [Google Scholar]
- 23.Kruhlak M, Crouch EE, Orlov M, Montaño C, Gorski SA, Nussenzweig A, et al. The ATM repair pathway inhibits RNA polymerase I transcription in response to chromosome breaks. Nature. 2007;447:730–4. doi: 10.1038/nature05842. [DOI] [PubMed] [Google Scholar]
- 24.Iacovoni JS, Caron P, Lassadi I, Nicolas E, Massip L, Trouche D, et al. High-resolution profiling of gammaH2AX around DNA double strand breaks in the mammalian genome. EMBO J. 2010;29:1446–57. doi: 10.1038/emboj.2010.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Caron P, Aymard F, Iacovoni JS, Briois S, Canitrot Y, Bugler B, et al. Cohesin protects genes against γH2AX Induced by DNA double-strand breaks. PLoS Genet. 2012;8:e1002460. doi: 10.1371/journal.pgen.1002460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430–5. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chou DM, Adamson B, Dephoure NE, Tan X, Nottke AC, Hurov KE, et al. A chromatin localization screen reveals poly (ADP ribose)-regulated recruitment of the repressive polycomb and NuRD complexes to sites of DNA damage. Proc Natl Acad Sci U S A. 2010;107:18475–80. doi: 10.1073/pnas.1012946107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Beli P, Lukashchuk N, Wagner SA, Weinert BT, Olsen JV, Baskcomb L, et al. Proteomic investigations reveal a role for RNA processing factor THRAP3 in the DNA damage response. Mol Cell. 2012;46:212–25. doi: 10.1016/j.molcel.2012.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller KM, Tjeertes JV, Coates J, Legube G, Polo SE, Britton S, et al. Human HDAC1 and HDAC2 function in the DNA-damage response to promote DNA nonhomologous end-joining. Nat Struct Mol Biol. 2010;17:1144–51. doi: 10.1038/nsmb.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seiler DM, Rouquette J, Schmid VJ, Strickfaden H, Ottmann C, Drexler GA, et al. Double-strand break-induced transcriptional silencing is associated with loss of tri-methylation at H3K4. Chromosome Res. 2011;19:883–99. doi: 10.1007/s10577-011-9244-1. [DOI] [PubMed] [Google Scholar]
- 31.Hong Z, Jiang J, Lan L, Nakajima S, Kanno S, Koseki H, et al. A polycomb group protein, PHF1, is involved in the response to DNA double-strand breaks in human cell. Nucleic Acids Res. 2008;36:2939–47. doi: 10.1093/nar/gkn146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O’Hagan HM, Wang W, Sen S, Destefano Shields C, Lee SS, Zhang YW, et al. Oxidative damage targets complexes containing DNA methyltransferases, SIRT1, and polycomb members to promoter CpG Islands. Cancer Cell. 2011;20:606–19. doi: 10.1016/j.ccr.2011.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sustáčková G, Kozubek S, Stixová L, Legartová S, Matula P, Orlova D, et al. Acetylation-dependent nuclear arrangement and recruitment of BMI1 protein to UV-damaged chromatin. J Cell Physiol. 2012;227:1838–50. doi: 10.1002/jcp.22912. [DOI] [PubMed] [Google Scholar]
- 34.Baldeyron C, Soria G, Roche D, Cook AJ, Almouzni G. HP1alpha recruitment to DNA damage by p150CAF-1 promotes homologous recombination repair. J Cell Biol. 2011;193:81–95. doi: 10.1083/jcb.201101030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim JA, Kruhlak M, Dotiwala F, Nussenzweig A, Haber JE. Heterochromatin is refractory to gamma-H2AX modification in yeast and mammals. J Cell Biol. 2007;178:209–18. doi: 10.1083/jcb.200612031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mo X, Dynan WS. Subnuclear localization of Ku protein: functional association with RNA polymerase II elongation sites. Mol Cell Biol. 2002;22:8088–99. doi: 10.1128/MCB.22.22.8088-8099.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bennett CB, Westmoreland TJ, Verrier CS, Blanchette CA, Sabin TL, Phatnani HP, et al. Yeast screens identify the RNA polymerase II CTD and SPT5 as relevant targets of BRCA1 interaction. PLoS One. 2008;3:e1448. doi: 10.1371/journal.pone.0001448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Starita LM, Horwitz AA, Keogh MC, Ishioka C, Parvin JD, Chiba N. BRCA1/BARD1 ubiquitinate phosphorylated RNA polymerase II. J Biol Chem. 2005;280:24498–505. doi: 10.1074/jbc.M414020200. [DOI] [PubMed] [Google Scholar]
- 39.Kleiman FE, Wu-Baer F, Fonseca D, Kaneko S, Baer R, Manley JL. BRCA1/BARD1 inhibition of mRNA 3′ processing involves targeted degradation of RNA polymerase II. Genes Dev. 2005;19:1227–37. doi: 10.1101/gad.1309505. [DOI] [PMC free article] [PubMed] [Google Scholar]