SUMMARY
Generating a balanced network of inhibitory and excitatory neurons during development requires precise transcriptional control. In the dorsal spinal cord, Ptf1a, a basic helix-loop-helix (bHLH) transcription activator, maintains this delicate balance by inducing homeodomain (HD) transcription factors such as Pax2 to specify the inhibitory lineage, while suppressing HD factors such as Tlx1/3 that specify the excitatory lineage. We uncover the mechanism by which Ptf1a represses excitatory cell fate in the inhibitory lineage. We identify Prdm13 as a direct target of Ptf1a and reveal that Prdm13 actively represses excitatory cell fate by binding to regulatory sequences near the Tlx1 and Tlx3 genes to silence their expression. Prdm13 acts through multiple mechanisms including interactions with the bHLH factor Ascl1 to repress Ascl1 activation of Tlx3. Thus, Prdm13 is a key component of a highly coordinated transcriptional network that determines the balance of inhibitory versus excitatory neurons in the dorsal spinal cord.
INTRODUCTION
The central nervous system (CNS) is composed of a balanced number of excitatory and inhibitory neurons that are interconnected to form neuronal networks essential for information processing. In the dorsal spinal cord, which provides the first level of central somatosensory processing, this excitatory/inhibitory balance is required to integrate sensory information including pain, touch and proprioception coming into the spinal cord from dorsal root ganglia, before it is relayed to supraspinal brain regions or locally for reflex responses (Liu and Ma, 2011; Ross, 2011). Disruption in this balance can lead to sensory disorders such as hyperalgesia and allodynia (Fitzgerald, 2005; Tavares and Lima, 2007). Given the importance of the dorsal horn neurons in processing all somatosensory modalities, understanding how these neurons are specified during embryogenesis to generate the correct composition of neurons is critical. Here we identify a key component of the transcriptional machinery that controls how neurons in the somatosensory circuit are generated, and how regulated specification of these neurons leads to a correct excitatory/inhibitory balance during development.
Excitatory and inhibitory neurons in the spinal cord arise from progenitor populations within the ventricular zone of the dorsal neural tube (Gross et al., 2002; Muller et al., 2002). These progenitors are competent to give rise to neurons in either class depending on the basic helix-loop-helix (bHLH) transcription factors that are expressed (Helms and Johnson, 2003; Zhuang and Sockanathan, 2006). For example, ectopic expression of the bHLH factors Ascl1 (previously Mash1) or Atoh1 (previously Math1) result in an increase of excitatory neurons at the expense of neighboring neuronal subtypes (Gowan et al., 2001; Helms et al., 2005). On the other hand, the bHLH factor Ptf1a is required for the generation of local circuit inhibitory neurons, and in its absence excess local circuit excitatory neurons form (Glasgow et al., 2005).
These functions for the bHLH factors in neuronal subtype specification highlight a fundamental question in generating neuronal diversity. A transcription factor with the ability to re-direct the fate of a cell requires two activities; it must activate lineage specific gene expression, and it must repress expression of genes in the alternate lineage. The current understanding of the transcriptional network that controls the excitatory/inhibitory balance in the dorsal horn of the spinal cord has Ptf1a and Ascl1 acting upstream of homeodomain (HD) factors such as Pax2 and Lbx1 to specify GABAergic (inhibitory) lineages, whereas Ascl1 alone acts upstream of Tlx1 and Tlx3 to specify glutamatergic (excitatory) lineages (Batista and Lewis, 2008; Brohl et al., 2008; Cheng et al., 2004; Cheng et al., 2005; Glasgow et al., 2005; Helms et al., 2005; Mizuguchi et al., 2006; Wildner et al., 2006). Since Ascl1 and Ptf1a are transcriptional activators (Beres et al., 2006; Nakada et al., 2004), the simple model has Ascl1 directly activating glutamatergic lineage genes, and Ptf1a directly activating GABAergic lineage genes. However, because both Ascl1 and Ptf1a are expressed in progenitors to the GABAergic lineages, how the glutamatergic genetic program in this lineage is suppressed remains unclear (Beres et al., 2006; Hori et al., 2008; Krapp et al., 1996; Krapp et al., 1998; Masui et al., 2008). Here we identify a PRDM class transcription repressor, Prdm13, as the missing component through which Ptf1a can suppress genes in the glutamatergic lineage in the dorsal spinal cord.
The PRDM family of transcription factors is defined by a PR domain and a variable number of zinc finger domains (for review see Fog et al., 2012; Fumasoni et al., 2007; Hohenauer and Moore, 2012). The PR domain has 20–30% identity to the SET domain that is found in a class of histone methyltransferases (HMTs), which function to silence transcription. Some PRDM members exhibit intrinsic HMT activity, such as Prdm2, 8, and 9, wheras most PRDM members lack this enzymatic activity. However, PRDM factors can recruit other histone-modifying enzymes to mediate their transcriptional activity. Some PRDM factors have been shown to be essential for regulating cell fate decisions. For example, Prdm16 specifies brown fat fate while suppressing white fat or muscle (Kajimura et al., 2010; Seale et al., 2008). In the nervous system, Prdm1 specifies photoreceptor over bipolar neuron fate (Brzezinski et al., 2010; Katoh et al., 2010). Recently, it was reported that Prdm8 regulates Cadherin-11 to ensure correct targeting of neurons in the cortex (Ross et al., 2012), and Prdm14 regulates Isl1 to ensure correct axon growth of motoneurons in zebrafish (Liu et al., 2012).
Despite the emergence of Prdm family members as important regulators of neural development, to date very little is known about their roles in cell fate specification and differentiation in the CNS. In this study, we identify Prdm13 as a critical component of the transcription network that regulates neuronal diversity and the excitatory/inhibitory balance in the dorsal spinal cord. In particular, we show that Prdm13 phenocopies that of Ptf1a in the CNS, where Prdm13 is both necessary and sufficient to promote Pax2+/GABAergic over Tlx1/3+/glutamatergic neuronal fate. Moreover, we present evidence that Prdm13 directly represses the glutamateric neuron specifier genes Tlx1 and Tlx3, thereby indirectly resulting in a derepression of the Pax2+/GABAergic lineage (Cheng et al., 2004; Cheng et al., 2005). Finally, we reveal a mechanism suggesting Prdm13 physically interacts with Ascl1 to silence Ascl1’s transcriptional activity on the Tlx3 gene. Together, our findings put Prdm13 at the center of the bHLH and HD transcription factor cascade that governs cell fate decisions, and its function is central to the generation of a balanced circuitry of excitatory and inhibitory neurons required for somatosensory information processing.
RESULTS
Ptf1a activity is dominant over Ascl1 in specification of dorsal spinal cord neurons
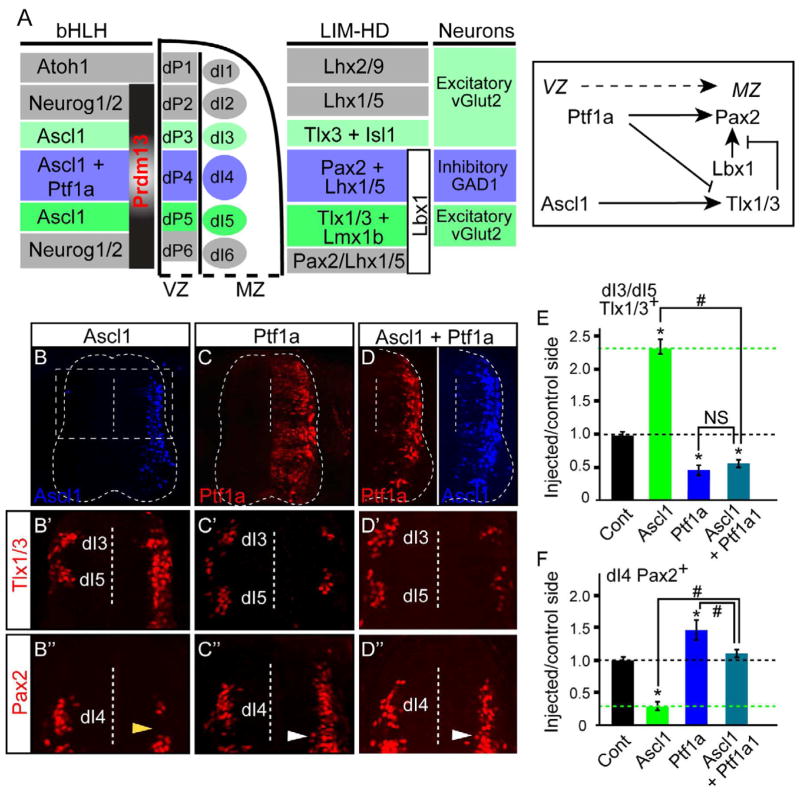
During development of the dorsal spinal cord, progenitor populations are defined by expression of bHLH transcription factors including Ascl1 and Ptf1a (see diagram Fig. 1A) (Glasgow et al., 2005; Helms et al., 2005). At embryonic day 10.5 (E10.5), Ascl1 is present in progenitors to interneurons dI3, dI4, and dI5, whereas Ptf1a is co-expressed with Ascl1 in progenitors to dI4. Functionally, overexpression of Ascl1 in ovo in chick neural tube induces expression of the homeodomain (HD) factors Tlx1/3, Isl1, and Lmx1b (dI3 and dI5) at the expense of Pax2 (dI4) (Fig. 1C′,C″,E,F) (Helms et al., 2005; Mizuguchi et al., 2006; Wildner et al., 2006). In contrast, overexpression of Ptf1a represses these factors, and instead induces Pax2 and Lhx1/5 (Fig. 1B′,B″,E,F) (Glasgow et al., 2005). In addition to the regulation of these HD factors by Ascl1 and Ptf1a, cross-talk between the HD factors also occurs since Tlx1/3 repress the ability of Lbx1 to induce Pax2 (Cheng et al., 2005) (see diagram Fig. 1A). Indeed, ectopic expression of Lbx1 in the chick neural tube induces Pax2, but only in regions outside of Tlx1/3 expression (Fig. S1B″). The output from this regulatory network is the specification of either an excitatory (vGlut2+) or inhibitory (Gad1+) neuronal phenotype (Cheng et al., 2004; Cheng et al., 2005; Glasgow et al., 2005; Gross et al., 2002; Muller et al., 2002).
Figure 1. Opposing activities of Ascl1 and Ptf1a on neuronal subtype specification.
(A) Diagram showing the pattern and interacting network of bHLH and LIM-HD transcription factors in the dorsal spinal neural tube. (B–F) Stage HH12-13 chick embryos were electroporated with expression vectors for Ptf1a and/or Ascl1 and harvested at HH24-25. Transverse sections are shown with the electroporated side on the right. Ascl1 (B,D blue) and Ptf1a (C,D red) detects the ectopically expressed proteins. Dashed box in (B) is the region of the neural tube shown in (B′–D″). Dashed vertical line indicates the ventricle. (B′–F) Dorsal neural tube sections show the dI3 and dI5 neuronal populations marked by Tlx1/3 antibody (B′–D′), or dI4 marked by Pax2 (B″–D″). (E,F) the ratio of Tlx1/3+ or Pax2+ neurons on the electroporated side versus the control side showing that overexpression of Ascl1 causes a dramatic increase in Tlx1/3+ and decrease in Pax2+ neurons, while Ptf1a, or Ascl1 plus Ptf1a, causes the opposite. Note that ectopic Pax2+ cells (white arrowheads) are found in the dI5 domain not normally expressing Pax2 (yellow arrowhead). Error bars are reported as SEM. p-values<0.001 between sample and control (*) or between samples (#). NS, not significant. Supplementary Fig. S1 shows Lbx1 also induces Pax2 (associated with panel A).
Since progenitors to dI4 express both Ascl1 and Ptf1a, some mechanism must exist to repress Ascl1 specification activities when Ptf1a is present. Ptf1a appears sufficient to repress Ascl1 activity since ectopic expression of Ptf1a represses Tlx1 and Tlx3 (Fig. 1C′). To test this further, we drove ectopic expression of Ascl1 and Ptf1a simultaneously in the chick neural tube. In this paradigm, Ptf1a was dominant over Ascl1, inhibiting both ectopic and endogenous Tlx1/3 expression while continuing to activate Pax2 (Fig. D′,D″,E,F). Given that Ptf1a acts as a transcriptional activator (Beres et al., 2006), we hypothesized that Ptf1a induces a transcriptional repressor as one of its downstream targets, and this repressor functions to mediate the suppression of glutamatergic lineage genes.
Prdm13 is a direct downstream target of Ptf1a
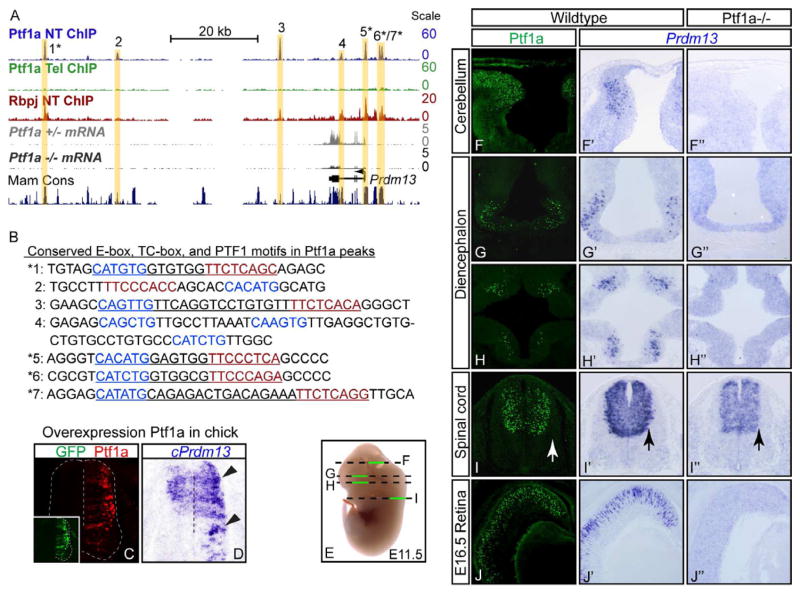
In order to better understand the transcriptional network that controls the specification of inhibitory and excitatory neuronal subtypes in the dorsal spinal cord, we identified direct downstream targets of Ptf1a. Targets were defined by intersecting gene lists identified via Ptf1a ChIP-Seq with genes identified as being Ptf1a-dependent using expression profiling of Ptf1a mutant neural tubes compared to heterozygote or wildtype tissues. For ChIP-Seq experiments localizing Ptf1a to chromatin in vivo, chromatin from mouse E12.5 neural tube was immunoprecipitated with antibodies to Ptf1a. Tissue from the telencephalon of the same embryos was used as a negative neural control as no Ptf1a is present in this tissue. High-throughput sequencing of these samples, followed by peak detection revealed Ptf1a localized to thousands of sites within the genome (DMM, MDB, and JEJ, unpublished). For identifying genes whose expression is Ptf1a-dependent, we performed RNA-Seq from Ptf1a lineage cells sorted from neural tubes of Ptf1a mutants and heterozygotes at E11.5. These analyses revealed Prdm13, a member of the PRDM class of proteins, whose expression decreased 4.5-fold in the Ptf1a mutant, as a particularly interesting target (Fig. 2A). Given the importance of the PRDM family of factors to cell fate decisions in other tissues (for review see Fog et al., 2012; Hohenauer and Moore, 2012) and the lack of functional data for Prdm13, we chose to study this factor to determine its function in neuronal subtype specification in the dorsal neural tube.
Figure 2. Prdm13 is a direct downstream target of Ptf1a.
(A) The mouse genomic region surrounding Prdm13 showing ChIP-Seq for Ptf1a from E12.5 neural tube (blue) and telencephalon (green) or Rbpj from neural tube (red), and RNA-Seq from FACS isolated Ptf1a lineage cells from E11.5 Ptf1a+/− (gray) or Ptf1a−/ − embryos (black). Seven Ptf1a peaks are highlighted with yellow bars. The location of the Prdm13 transcript is shown, as is a histogram of mammalian conserved regions (UCSC Genome Browser). (B) Sequence under the apex of each Ptf1a ChIP-Seq peak. DNA regions 1–7 showing the E-box (blue), TC-box (red), and the PTF1 motifs (underlined). * peaks that are conserved from mouse to zebrafish and contain the consensus PTF1 motif. (C–D) Overexpression of mouse Ptf1a (C, red) in chick neural tube induces cPrdm13 mRNA (D, arrowheads). Inset: GFP indicates the electroporation efficiency. (E) Diagram of section planes for images shown in (F–I). (F–J″) Cross sections of mouse E11.5 ore E16.5 show Ptf1a protein (F–J), and Prdm13 mRNA in wildtype (F′–J′) and Ptf1a−/ − (F″–J″). Note in cerebellum, diencephalon, and retina Prdm13 is lost in the Ptf1a mutant, but in the spinal cord, only the lateral domain that reflects the Ptf1a pattern is lost (arrows). Supplementary Fig. S2 includes expression data for Ptf1a and Prdm13 in the neural tube at additional embryonic stages and genomic coordinates for Ptf1a peaks 1–7 from panel B.
ChIP-Seq for Ptf1a identified 7 binding regions surrounding the Prdm13 gene (Fig. 2A). Ptf1a is a component of a trimeric transcription complex (PTF1) that also includes an E-protein and Rbpj and binds a DNA motif containing an E-box and a TC-box with constrained spacing requirements (Beres et al., 2006; Henke et al., 2009; Hori et al., 2008; Masui et al., 2007). Of the 7 regions surrounding the Prdm13 gene bound by Ptf1a, 5 contain consensus PTF1 binding motifs (Fig. 2B, underlined sequence). To determine if these Ptf1a-bound regions are occupied by the PTF1 complex, we performed ChIP-Seq on neural tube tissue with antibodies to Rbpj (DMM, MDB, and JEJ, unpublished). Examination of the Prdm13 locus confirmed that Rbpj co-localizes with Ptf1a in those regions that contain a consensus PTF1 motif (Fig. 2A). All five of the PTF1 sites are conserved across mammalian species, and 4 of them are conserved to zebrafish (Fig. 2A,B asterisks).
To verify that Prdm13 is a direct target of Ptf1a, we examined the Prdm13 expression pattern relative to Ptf1a, and in response to increased or decreased levels of Ptf1a. In the chick, Prdm13 is expressed in the ventricular zone in the dorsal neural tube (Fig. 2D, control side), and ectopic expression of Ptf1a via in ovo electroporation resulted in a dramatic increase in endogenous cPrdm13 mRNA levels (Fig. 2C,D). In mouse embryos, the pattern of Prdm13 expression is strikingly neural specific and is almost exclusively restricted to Ptf1a domains (Fig. 2E–J′). At E10.5–E12.5, Ptf1a is present in the dorsal neural tube from spinal regions (Fig. 2I,I′, Fig. S2) to the developing cerebellum (Fig. 2F,F′), and in a subset of cells in the diencephalon (Fig. 2G–H′). Prdm13 is present in all these domains, including the retina at E16.5 (Fig. 2J,J′), but is not detected in the pancreas, the other major site of Ptf1a expression. Notably in the caudal neural tube, although Prdm13 is restricted to the dorsal region, it appears to be present more broadly in the ventricular zone than Ptf1a. Prdm13 overlaps progenitor regions from dI2-dI6 (Fig. 2I′, Fig. S2) and is expressed from E9 to E14.5 (Fig. S2). However, even in the neural tube, Prdm13 appears enriched in the Ptf1a domains. In Ptf1a null embryos, Prdm13 is abolished in rostral domains (Fig. 2F″–H″), indicating that expression of Prdm13 in these regions is dependent on Ptf1a. Within the caudal neural tube, Prdm13 mRNA is reduced only at the lateral border of the ventricular zone where Ptf1a is expressed (Fig. 2I–I″, arrows; Fig. S2), suggesting that additional, Ptf1a-independent mechanisms also regulate Prdm13 in this region.
In summary, the neural restricted patterns of Prdm13 and Ptf1a are strikingly similar with a major component of Prdm13 expression being Ptf1a-dependent. Together with the ChIP-Seq results, we conclude that Prdm13 is a neural specific, direct downstream target of Ptf1a.
Prdm13 phenocopies Ptf1a function in the dorsal neural tube
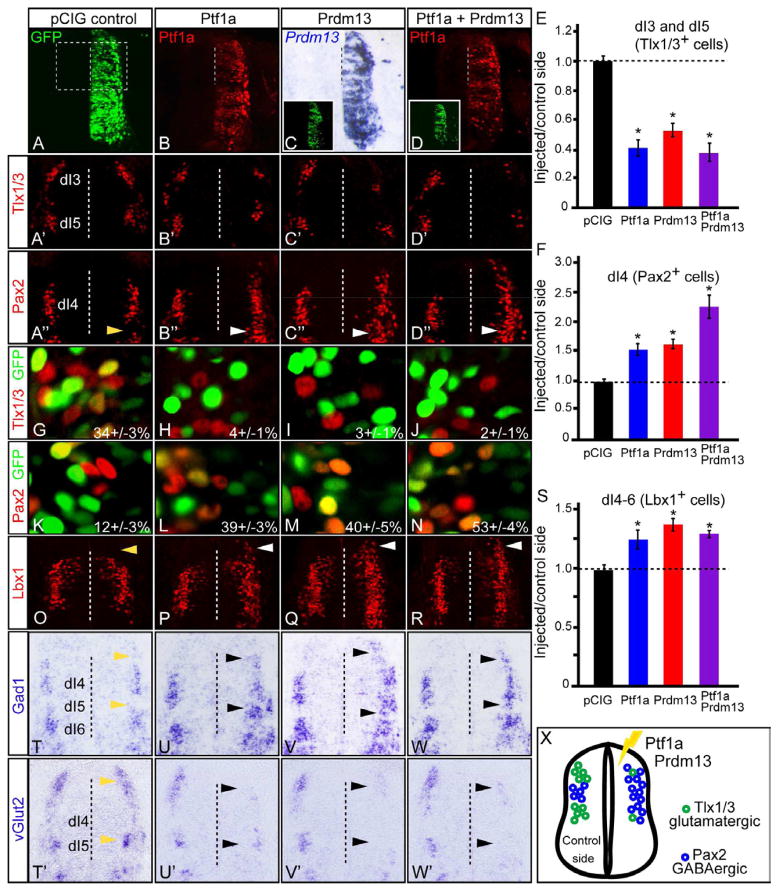
To begin to uncover the function of Prdm13 during neural development, we misexpressed mouse Ptf1a and Prdm13 in chick neural tubes at stage HH12-13 via in ovo electroporation. The exogenous expression was confirmed by specific immunofluorescence for Ptf1a and mRNA in situ hybridization for Prdm13 (Fig. 3B–D), while GFP indicated electroporation efficiency across the dorsoventral axis (Fig. 3A). Transverse sections of thoracic neural tube were collected at HH24-25 and examined by immunofluorescence for alterations in neuronal sub-type specification relative to the non-electroporated side. Ectopic expression of either Ptf1a or Prdm13 dramatically decreased the number of Tlx1/3+ cells (2.5 and 2.0-fold, respectively) while increasing the number of dI4 Pax2+ cells (1.5-fold in each case) (Fig. 3B′–C″,E–F). Electroporation of empty vector, in contrast, did not alter the number of the neurons generated in either population (Fig. 3A′–A″, E–F). Co-electroporation of Prdm13 and Ptf1a together more strongly induced Pax2 expression than either alone (Fig. 3D″–F). Similar trends were seen with additional markers for dorsal interneuron populations including Isl1/2, Lmx1b, and Lhx1/5 (Fig. S3).
Figure 3. Ptf1a and Prdm13 induce inhibitory and suppress excitatory neuronal markers.
Stage HH12-13 chick embryos were electroporated with empty vector (control), Ptf1a, Prdm13, or both. Transverse sections through stage HH24-25 are shown with the electroporated side on the right. (A–D) GFP (green) shows electroporation efficiency along the dorsoventral axis. Ptf1a protein (red) and Prdm13 mRNA show ectopic expression. Dashed box in (A) is the region of the neural tube shown in (A′–D″, O–R). (A′–F) Dorsal neural tube sections from (A–D) indicate the dI3 and dI5 neuronal populations marked by Tlx1/3, or the dI4 neurons marked by Pax2. (E,F) the ratio of Tlx1/3+ or Pax2+ neurons on the electroporated side versus the control side showing that overexpression of Ptf1a and Prdm13 cause a dramatic decrease in Tlx1/3+ and increase in Pax2+ neurons relative to control. Note that ectopic Pax2+ cells (white arrowheads) are found in the dI5 domain not normally expressing Pax2 (yellow arrowhead). (G–N) Electroporated cells, visualized by GFP, co-localized with Tlx1/3 (G–J) or Pax2 (K–N). The percentage of GFP cells that co-express Tlx1/3 or Pax2 in the dorsal mantle zone is shown. Ectopic expression of Ptf1a and/or Prdm13 reduces the percentage of Tlx1/3;GFP double positive cells from 34% to 2–4%, and increases Pax2;GFP double positive cells from 12% to 39–53%. (O–S) Dorsal neural tube sections from (A–D) showing the dI4-6 neuronal populations marked by Lbx1. Quantification (S) showing that Ptf1a and Prdm13 overexpression cause an increase in Lbx1+ neurons relative to control. The white arrowheads indicate ectopic Lbx1 expression detected in the dI3 region not normally expressing Lbx1 (compare to control (O) yellow arrowhead). (T–W′) In situ hybridization with Gad1 or vGlut2 probes showing that Ptf1a and Prdm13 overexpression increase Gad1 and decrease vGlut2 (black arrowheads) when compared to either the empty vector control (yellow arrowheads) or the control side in each neural tube. (X) Diagram summarizing the overexpression phenotypes. Error bars are reported as SEM. * p-values<0.001. Supplementary Fig. S3 includes analysis of the Prdm13 phenotype with additional markers of the dorsal interneuron populations including Isl1/2, Lmx1b and Lhx1/5.
Because other members of the PRDM family are primarily repressors, we hypothesized that Prdm13 might induce Pax2 by indirect means, such as by relieving repression by another factor. For example, derepression of Lbx1, which induces Pax2 in the absence of Tlx1/3 (Cheng et al., 2005), might account for the increase in Pax2 levels seen with Prdm13 misexpression. Indeed, Lbx1 was also induced by ectopic expression of Ptf1a and Prdm13, particularly in regions dorsal to its native domain (Fig. 3O–S, arrows). Since Ptf1a and Prdm13 also inhibit Tlx3 in the dI3 domain, the induction of Pax2 may be indirect through the unopposed activity of ectopic Lbx1.
In the dorsal neural tube, Tlx1/3 cells give rise to vGlut2+ excitatory neurons, and Pax2 cells give rise to Gad1+ inhibitory neurons. To determine whether Ptf1a and Prdm13 are sufficient to modulate the neurotransmitter phenotypes as well, we performed mRNA in situ hybridization for Gad1 and vGlut2 on the electroporated neural tubes. An increase in Gad1 and decrease in vGlut2 confirms that the ectopic Pax2 expressing neurons from Ptf1a and Prdm13 overexpression adopt a GABAergic identity (Fig. 3T–W′, arrows indicating dI3 and dI5 regions with ectopic Gad1 or reduced vGlut2). Thus, Prdm13 serves a similar function as Ptf1a by promoting the Pax2+ GABAergic lineage and suppressing the Tlx1/3+ glutamatergic fate.
A closer look at the pattern of Pax2+ neurons in the Ptf1a and Prdm13 electroporated neural tubes revealed that the normal dI5 gap between dI4 and dI6 was filled with Pax2+ cells (Fig. 3 compare A″ with B″–C″, arrows). This suggested that ectopic expression of Ptf1a and Prdm13 alter the fate of progenitor cells, an interpretation consistent with previous conclusions from analysis of the Ptf1a knock out mouse (Glasgow et al., 2005). Indeed, closer examination of the dI3-dI5 domain revealed that Prdm13 electroporated cells are strongly biased to a Pax2+:Tlx1/3− fate (Fig. 3G–N). In the empty vector control, 34% of the dorsal GFP+ cells were Tlx1/3+. When Ptf1a, Prdm13, or both were ectopically expressed, GFP+:Tlx1/3+ cells were rarely seen and the percentage dropped to 2–4% (Fig. 3H–J). In contrast, the number of dorsal GFP+:Pax2+ cells increased from 12% in the control to 39–40% with ectopic Ptf1a or Prdm13 (Fig. 3L–M). The percentage increased to 53% GFP+;Pax2+ cells when Ptf1a and Prdm13 were introduced simultaneously, consistent with the collaborative effect noted above (Fig. 3N). We confirmed that the loss of Tlx1/3+ cells and enrichment of Pax2+ cells is not an artificial result of selective cell death, as assayed by immunostaining for Caspase 3 (data not shown). These findings indicate both Ptf1a and its target Prdm13 can alter the fate of progenitor cells and bias them to the Pax2+/inhibitory neuronal lineage.
Knockdown of Ptf1a or Prdm13 leads to a reduction of Pax2+ cells
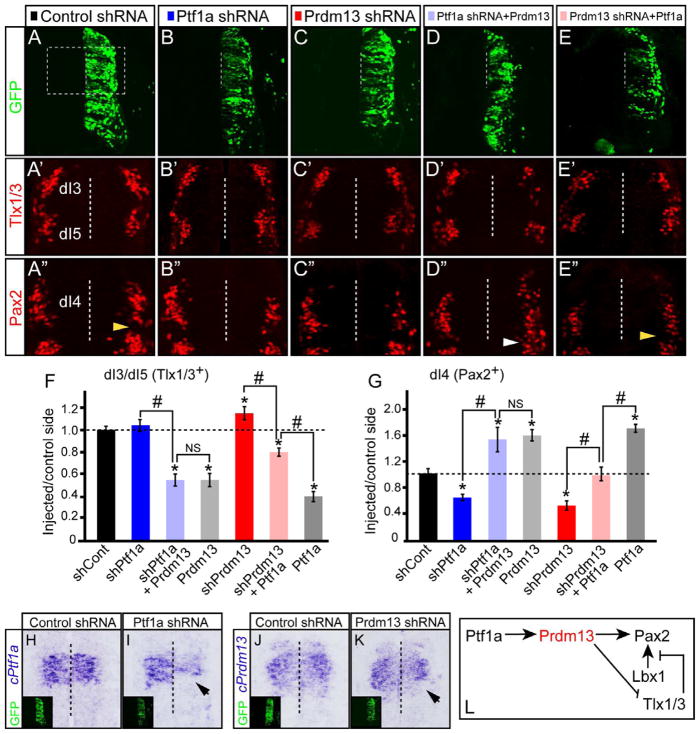
To gain further insight into the role of Prdm13 in neuronal specification, we generated specific short hairpin RNAs (shRNA) to knockdown endogenous Ptf1a or Prdm13 in the chick neural tube. Constructs containing shRNAs to Ptf1a, Prdm13, or DsRed (control) were electroporated as above (Fig. 4A–C), and mRNA in situ hybridization was performed to verify knockdown (Fig. 4H–K, black arrows). Reduction of Ptf1a and Prdm13 levels led to a marked decrease in the number of Pax2+ cells (1.7 and 2.0-fold for Ptf1a and Prdm13, respectively) (Fig. 4B″–C″, G). There was a modest increase in the number of Tlx1/3+ cells with Prdm13 knockdown; however, in the Ptf1a knockdown there was no significant difference (Fig. 4B′–C′,F). Electroporation of control shRNA did not alter the expression of Tlx1/3 nor Pax2 (Fig. 4A′–A″, F,G). There was no increase in cell death in electroporated hemicords as detected by Caspase3 expression (data not shown). Misexpression of mouse Ptf1a or Prdm13 with their respective shRNA constructs rescued the knockdown phenotypes, demonstrating specificity of the shRNA (Fig. S4). These results demonstrate that Prdm13, like Pft1a, is required to generate the correct number of Pax2+ neurons in the dorsal spinal cord.
Figure 4. Ptf1a requires Prdm13 to specify inhibitory neurons and suppress excitatory neurons.
Stage HH12-13 chick embryos were electroporated with expression or shRNA vectors to modulate levels of Ptf1a and Prdm13. Transverse sections through stage HH24-25 are shown with the electroporated side on the right. (A–E) GFP indicates the electroporation efficiency along the dorsoventral axis. The dashed box is the region shown in (A′–E″). (A′–E′, F) Tlx1/3 shows Prdm13 knockdown results in an increase in dI3/dI5 neurons (C′). Epistasis experiments show Prdm13 does not require Ptf1a for this activity in repressing Tlx1/3 (D′) but Ptf1a activity is abrogated with shPrdm13 (E′). (A″–E″, G) Pax2 shows a significant decrease in the number of dI4 neurons when Ptf1a or Prdm13 are knocked down compared to control shRNA (B″, C″, G). Epistasis experiments show Prdm13 does not require Ptf1a for this activity in inducing Tlx1/3 (D″) but Ptf1a activity is abrogated with shPrdm13 (E″). Note that ectopic Pax2+ cells (white arrowhead) are found in the dI5 domain not normally expressing Pax2 (yellow arrowheads). Error bars are reported as standard error of the mean. p-values<0.001 between sample and control (*), and between samples (#). NS, not significant. (H–K) cPtf1a or cPrdm13 probes indicate the knockdown efficiency (arrows). Inset: GFP indicates the electroporation efficiency. (L) Diagram summarizing the conclusion that Prdm13 is a functional downstream target of Ptf1a that induces Pax2 and suppresses Tlx1/3. Supplementary Fig. S4 includes rescue control experiments for the shRNA.
Ptf1a requires Prdm13 to increase Pax2+ and decrease Tlx1/3+ cells
Results detailed above strongly suggest that Prdm13 functions as a downstream effector of Ptf1a during neuronal sub-type specification. If Prdm13 is required for Ptf1a activity, then knocking down Prdm13 when Ptf1a is ectopically expressed should abrogate Ptf1a activity. Indeed, the increase in Pax2+ cells and the decrease in Tlx1/3+ cells normally seen with ectopic Ptf1a expression was inhibited when Prdm13 was simultaneously knocked down (Fig. 4E′–E″,F,G, compare with gray bar). Conversely, knockdown of Ptf1a did not affect the activity of ectopically expressed Prdm13, consistent with Ptf1a acting upstream of Prdm13 (Fig. 4D′–D″, F–G). These experiments support the requirement for Prdm13 as a downstream effector of Ptf1a.
Prdm13 requires its zinc finger motifs for activity and acts as a repressor
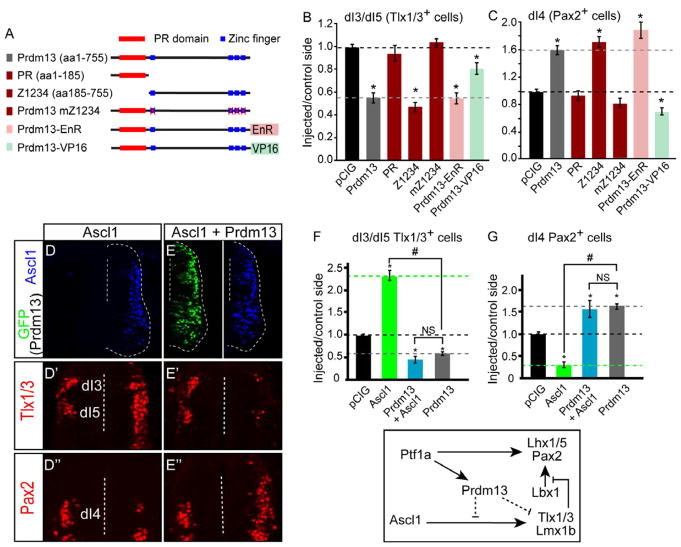
The PRDM family of transcription factors is characterized by a PR domain and a variable number of zinc fingers, but the functional contribution of each of these domains remains unclear for any given member of the family (for review see Fog et al., 2012; Hohenauer and Moore, 2012). To better understand the mechanism behind Prdm13 function, we generated mutants of Prdm13 and tested their activity in chick neural tube (Fig. 5A–C). Although the PR domain has been shown to be critical for the function of other PRDM family members, this domain was dispensable for Prdm13 activity, as the truncated form of Prdm13 containing the four zinc fingers (Z1234) retained full activity (Fig. 5B–C, representative images are shown in Fig. S5I′,I″). Furthermore, the zinc fingers themselves are required for Prdm13 activity since mutations designed to disrupt these domains (Prdm13 mZ1234) also disrupted Prdm13 function (Fig. 5B–C, Fig. S5J′,J″).
Figure 5. Prdm13 Blocks Ascl1 Activity through Repression Requiring Its Zn Fingers.
(A) Diagram of the Prdm13 mutations tested. PR domain (red boxes) and Zn finger domains (blue boxes) are indicated. Engrailed repressor (EnR) or activator VP16 were fused in frame at the C-terminal end of Prdm13.
(B-C) Quantification of experiments testing the Prdm13 mutants indicated. The black dashed line indicates the pCIG control ratio of injected/control side. The gray dashed line indicates the phenotype of the full-length Prdm13. The PR domain is dispensable for Prdm13 activity, but the four Zn fingers are required. The Prdm13-EnR fusion mimics the Prdm13 phenotype, whereas the Prdm13-VP16 does not.
(D-E”) Stage HH12-13 chick embryos were electroporated with Ascl1 (D) or Ascl1 plus Prdm13 (E). Transverse sections through stage HH24-25 are shown with the electroporated side on the right. Ascl1 detects the ectopically expressed protein (blue), and Prdm13 is indicated by GFP. Dashed lines indicate location of the ventricle. Dorsal neural tube sections from (D and E) show Tlx1/3 defining the dI3 and dI5 (D’-E’) and Pax2 defining dI4 neuronal populations (D”-E”). Ectopic Ascl1 causes a dramatic increase in the number of Tlx1/3+ neurons with a decrease in Pax2+ cells, whereas addition of Prdm13 reverses this phenotype.
(F-G) The ability of Prdm13 to block this activity of Ascl1 is quantitatively shown.
All error bars are reported as SEM. p values < 0.001 between sample and control (*), and between samples (#). NS, not significant. Diagram summarizing the conclusion that Prdm13 antagonizes Ascl1 neuronal subtype specification activity. Figure S5 shows Ascl1 expression levels are not altered by overexpression or knockdown of Prdm13. Additionally, representative images for Tlx1/3 and Pax2 for chick electroporation experiments with the Prdm13 truncations are shown.
Members of the PRDM family of factors have been reported to function either as transcriptional repressors or activators depending on the cellular context. To determine whether the neuronal specification activity observed with Prdm13 reflects its function as a transcriptional repressor or an activator, we fused it with engrailed repressor (EnR) (Smith and Jaynes, 1996) or with the VP16 activator (Triezenberg et al., 1988) and tested these chimeric proteins for activity in the chick neural tube. The activity of Prdm13-EnR was indistinguishable from that of wild-type Prdm13 in that the number of Tlx1/3+ neurons was dramatically reduced while Pax2+ neurons increased (Fig. 5B–C, Fig. S5). In contrast, expression of Prdm13-VP16 resulted in a decrease in Pax2+ cells. Prdm13-VP16 also slightly repressed Tlx1/3, although the effect was much less pronounced than that of Prdm13-EnR or Prdm13 (Fig. 5B–C, Fig. S5). Taken together, Prdm13 appears to exert its effects by acting as a transcriptional repressor, an activity that requires its zinc finger domains.
Prdm13 suppresses generation of Tlx1/3+ neurons by antagonizing Ascl1 activity
Given that the bHLH factor Ascl1 specifies opposing programs (dI3 and dI5) to Ptf1a and Prdm13 at this stage (Fig. 1) (Brohl et al., 2008; Helms et al., 2005; Mizuguchi et al., 2006; Nakada et al., 2004), we hypothesized that Prdm13 might function through interfering with Ascl1 activity. We tested this by co-electroporating Ascl1 and Prdm13 into the chick neural tube. Ectopic Ascl1 results in a dramatic increase of Tlx1/3+ neurons and a loss of Pax2+ neurons (Fig. 5D′–D″,F–G). Strikingly, when Ascl1 and Prdm13 are expressed simultaneously, Ascl1 is no longer able to induce Tlx1/3. Instead, we observed a 3.3-fold decrease in Tlx1/3+ neurons and a 1.6-fold increase in Pax2+ cells relative to the control side, similar to effects of Prdm13 misexpression alone (Fig. 5E′–E″,F–G, compare with Fig. 3C′–C″,E–F). Prdm13 does not alter endogenous Ascl1 levels (Fig. S5A′,B′,C); therefore, Prdm13 antagonizes Ascl1 activity without inhibiting expression of Ascl1 itself. Thus, while Ascl1 is expressed in both progenitors to dI4 and dI5 neurons, it is the presence or absence of enriched Prdm13 levels that dictate the decision between these cell fates.
Prdm13 complexes with Ascl1 to directly repress Tlx3 expression
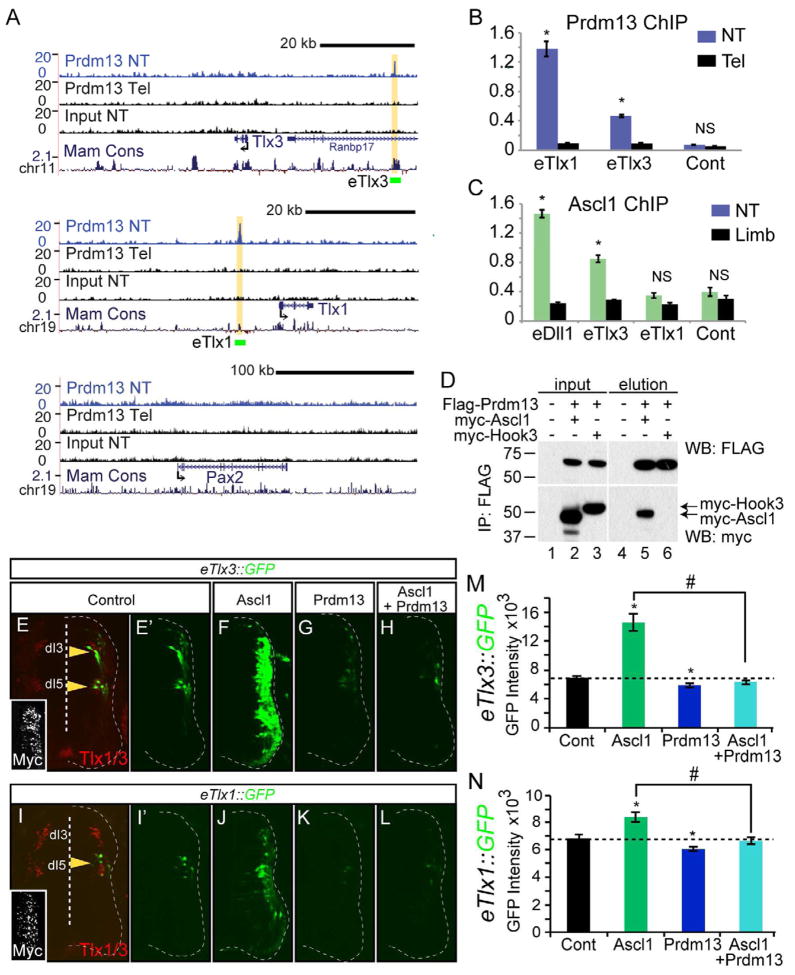
We used ChIP-Seq to localize Prdm13 to chromatin in vivo as a means to uncover how Prdm13 suppresses expression of Tlx1 and Tlx3. We performed ChIP-Seq with antibodies to Prdm13 using chromatin from E11.5 neural tubes. We found Prdm13 enriched at regions upstream of both the Tlx1 and Tlx3 genes (Fig. 6A). No enrichment was detected with these antibodies in chromatin isolated from E11.5 mouse telencephalon tissue where Prdm13 is absent (Fig. 6A). Furthermore, Prdm13 was not found within a 300 kb region surrounding the Pax2 gene consistent with Prdm13 induction of this gene being indirect. Enrichment of the Tlx1 and Tlx3 regions was confirmed in ChIP-qPCR experiments. Again, there was enrichment of the Tlx1 and Tlx3 sites in neural tube that was not seen with telencephalon tissue or with a control genomic region (Fig. 6B). These data suggest that Prdm13 inhibits glutamatergic identity by binding near the glutamatergic specifier genes Tlx1 and Tlx3, and directly repressing their transcription. This in turn may facilitate Lbx1-mediated induction of Pax2 and GABAergic neuronal specification (see Fig. 7 for model).
Figure 6. Prdm13 directly suppresses Tlx1 and Tlx3 through multiple mechanisms.
(A) UCSC Genome Browser showing mouse genomic regions surrounding Tlx3, Tlx1, and Pax2 including Prdm13 ChIP-Seq from neural tube (NT, blue), and two negative controls including Prdm13 ChIP-Seq from telencephalon (Tel) and input from NT, all from E11.5 mouse embryos. Prdm13 occupies chromatin near the Tlx1 and Tlx3 genes at sequence conserved across mammals (yellow highlight). Green bars labeled eTlx1 and eTlx3 are the regions used in the GFP reporter constructs in (E–N). (B) ChIP-qPCR validating Prdm13 ChIP-Seq at the eTlx1 and eTlx3 loci in (A). Tel tissue is negative for Prdm13 and serves as a control for specificity of the antibody. (C) ChIP-qPCR for Ascl1 at the eTlx1 and eTlx3 loci in (A). Limb tissue is negative for Ascl1 and serves as a control for specificity of the antibody. eDll1 is a known target of Ascl1. (D) Co-IP experiments of epitope tagged versions of the proteins transfected into HEK 293 cells indicate specific interaction between Ascl1 and Prdm13 (lane 5), but not the negative control, myc-Hook3 (lane 6). (E–N) Chick neural tube electroporated with either eTlx3::GFP or eTlx1::GFP reporter and expression vectors for Ascl1, Prdm13, or both as indicated. Only the electroporated side is shown and the neural tube is outlined with a dashed line. (E) The eTlx3::GFP signals are restricted and co-localize with dI3/dI5 markers, Tlx1/3 (yellow arrowheads). (E′–H) Ascl1 dramatically induces eTlx3::GFP and this is blocked by Prdm13. (I) The eTlx1::GFP signals are restricted to dI5 neurons (yellow arrowhead). (I′–L) Induction by Ascl1 is more subtle than that seen with eTlx3::GFP but Prdm13 is still repressive. Inset: myc immunofluorescence indicates the electroporation efficiency. (M and N) shows the quantification of these data by measuring pixel intensity. Error bars are reported as SEM. * and # p-value<0.001 relative to control or between samples indicated.
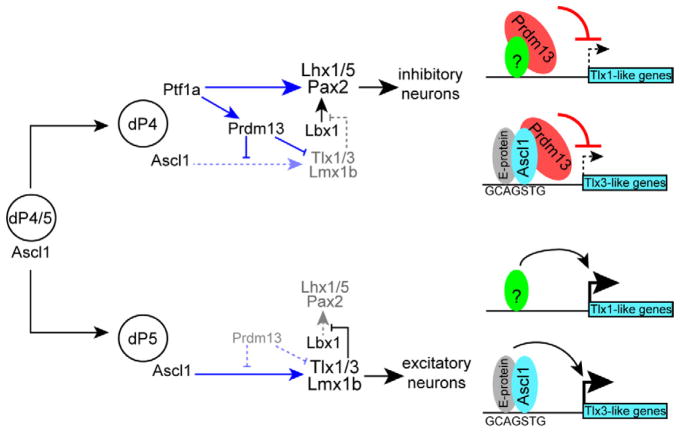
Figure 7. Transcriptional network controlling the balance of inhibitory and excitatory neurons in the dorsal spinal cord.
Progenitors in the ventricular zone express Ascl1. In the absence of Ptf1a, these cells become excitatory neurons (dP5 lineage) through Ascl1 induction of the HD factors Tlx1/3 and Lmx1b. Tlx1/3 repress the Lbx1-dependent induction of Pax2 and the inhibitory neuronal program. Ascl1 regulates Tlx3 directly whereas Tlx1 may be indirect. Ptf1a is upregulated in a subset of the Ascl1 progenitor cells directing their fate to inhibitory neurons (dP4 lineage) through induction the HD factors Pax2 and Lhx1/5. Additionally, Ptf1a directly increases levels of Prdm13 that in turn suppresses Tlx1 and Tlx3, ensuring a suppression of the glutamatergic program in the inhibitory neurons. Prdm13 works through at least two mechanisms; one being Ascl1-dependent possibly involving a novel transcriptional repressor complex for Tlx3-like genes, and the other Ascl1-independent requiring some other activator (green circle) for Tlx1-like genes.
To explore how Prdm13 antagonizes Ascl1, we asked whether Ascl1 was also found at the Prdm13 bound Tlx1 and Tlx3 sites. ChIP with Ascl1 antibodies from E11.5 neural tubes showed significant enrichment at the Tlx3 site but not the Tlx1 (Fig. 6C). This enrichment was similar to that seen with a known target of Ascl1, Dll1 (Castro et al., 2006). Enrichment at the Tlx3 site was not detected from limb tissue where Ascl1 is not expressed or from a control genomic region. This finding suggests a model whereby Ascl1 binds and activates expression of Tlx3, but when Prdm13 levels increase downstream of Ptf1a, a novel transcription repressor complex that includes Ascl1 and Prdm13 is formed. Supporting this model, specific binding between Ascl1 and Prdm13 is detected in co-IP experiments when epitope-tagged versions of the proteins are expressed in HEK 293 cells (Fig. 6D).
Two distinct mechanisms, one Ascl1-dependent and one Ascl1-independent, are suggested for Prdm13 suppression of the glutamatergic gene expression program in cells fated to become GABAergic neurons (see Fig. 7). To verify that Prdm13 and Ascl1-bound regions function as enhancers, we tested reporter constructs derived from the Tlx1 and Tlx3 genomic loci. eTlx1::GFP and eTlx3::GFP reporters were electroporated into the chick neural tube at HH12-13 and assayed 48 hours later. Strikingly, eTlx3::GFP was largely restricted to the dI3 and dI5 domains and eTlx1::GFP to the dI5 domain reflecting the expression of their respective gene loci (Fig. 6E,I). Furthermore, co-electroporation of the reporter constructs with Ascl1 resulted in a dramatic induction of eTlx3::GFP and only a slight induction of eTlx1::GFP, consistent with Ascl1 localizing to Tlx3 but not Tlx1 (Fig. 6F,J,M,N). This dramatic activation of eTlx3::GFP by Ascl1 was abrogated when Prdm13 was also expressed (Fig. 6H,L). Misexpression of Prdm13 alone with the reporter constructs similarly inhibited their activity in dI3 and dI5 domains (Fig. 6G,K). Thus, in addition to identifying enhancers for Tlx1 and Tlx3 that are directly repressed by Prdm13, we suggest a mechanism whereby Prdm13 may be recruited to the Tlx3 enhancer by Ascl1 to directly repress Tlx3.
DISCUSSION
A balance of excitatory and inhibitory neuronal input is crucial for normal nervous system function. Since dorsal horn neurons in the spinal cord form the first level of central processing of somatosensory input from the periphery, this balance is required for normal perception of pain, touch, and sense of body position. Generation of neuronal diversity in the spinal cord involves the specification of GABAergic and glutamatergic neurons from neighboring and interspersed progenitors in the dorsal neural tube ventricular zone just as they are transitioning to post-mitotic neurons. The network of transcription factors required for this process includes members of the bHLH and HD families. In particular, Ascl1+ proliferating progenitors in the dorsal neural tube are competent to become either glutamatergic during early neurogenesis (around E10.5) or both GABAergic and glutamatergic after E12.5. In the absence of Ptf1a, Ascl1 induces expression of Tlx1/3 and Lmx1b. These HD factors in turn induce the glutamatergic program while suppressing the ability of Lbx1 to induce Pax2 and the GABAergic program (Cheng et al., 2005; Helms et al., 2005) (Fig. 7 for model). In contrast, a subset of these Ascl1+ progenitors begins to express Ptf1a, which activates expression of the HD factors Pax2 and Lhx1/5 and the resulting GABAergic program (Glasgow et al., 2005; Wildner et al., 2006) (Fig. 7). However, for correct specification of these neurons, the glutamatergic program must be repressed. Here we identify the zinc finger transcription factor Prdm13 that provides this critical function through repression of Ascl1 activity and direct repression of Tlx1 and Tlx3 expression (Fig. 7).
Mechanisms of Prdm13 action in neuronal specification
The PRDM family of factors is interesting in the variety of mechanisms they reportedly use to regulate transcription (for review Fog et al., 2012; Hohenauer and Moore, 2012). They contain two major recognizable motifs, the PR domain and zinc fingers. The PR domain is similar to the SET domain found in histone lysine methyltransferases (HMT), and thus, some members of the PRDM family have been shown to have this enzymatic activity. However, the sequence homology between PR domains and the SET domain reveals that the PR domain of Prdm13 is substantially divergent. We show that the PR domain is dispensable for Prdm13 activity in our assays since a truncated Prdm13 (amino acids 185-755) that lacks the PR domain but contains all four zinc fingers is sufficient to induce Pax2+ cells and repress Tlx1/3 at levels similar to that seen with the full-length protein. Furthermore, the zinc fingers are required for Prdm13 function since mutations designed to disrupt each zinc finger abolished Prdm13 activity (Fig. 5).
Zinc finger domains in other PRDM factors have been shown to be involved in a variety of activities including DNA binding, protein-protein interaction, intrinsic HMT activity, and recruitment of other histone-modifying proteins. Prdm1, 3, 5, 9, and 14 can directly bind DNA (Fog et al., 2012). In contrast, Prdm8, which shows the highest similarity to Prdm13, does not directly bind DNA but rather binds chromatin in a complex with Bhlhb5 (Ross et al., 2012). Results from Prdm13 ChIP indicate it localizes with chromatin, but like Prdm8, it may do so indirectly. This is supported by three findings in our data. First, for efficient ChIP, a long-arm cross-linker compound treatment, ethylene glycol-bis (succinimidyl succinate) (Zeng et al., 2006) was required. Second, no zinc finger type binding motif was identified from the ChIP-Seq results in regions bound by Prdm13 (data not shown). And third, we provide evidence for Prdm13 interacting with Ascl1 to repress Ascl1 target genes (Fig. 6). However, these results do not exclude the possibility that Prdm13 also binds DNA directly. Indeed, ChIP for Prdm13 in Ascl1 mutant neural tubes still showed Prdm13 bound to the Tlx3 enhancer (data not shown). Thus, Prdm13 is functioning through mechanisms distinct from those seen with Prdm8, and may repress its targets by combinations of direct and indirect binding to DNA.
We demonstrate here that Prdm13 is a key factor in ensuring glutamatergic lineage genes are silenced during generation of GABAergic neurons. Prdm13 suppresses the glutamatergic lineage and induces the GABAergic lineage through transcriptional repression of the glutamatergic specifier factors Tlx1 and Tlx3 (Fig. 7). This is supported by 1) ectopically expressed Prdm13 reduces the number of Tlx1/3+ cells in the dorsal neural tube, 2) Prdm13 functions as a transcriptional repressor, 3) Prdm13 occupies genomic regions near both the Tlx1 and Tlx3 genes in sequence conserved across mammalian species, and 4) Prdm13 bound Tlx1 and Tlx3 regions have enhancer activity that was blocked by ectopic Prdm13. Since Prdm13 was not localized to the Pax2, Lhx1/5, or Lbx1 genes (Fig. 6 and data not shown), and Prdm13 is acting as a transcriptional repressor, the increase in expression of these GABAergic lineage genes upon overexpression of Prdm13 is likely indirect. Indeed, induction of Pax2 can be explained by Prdm13’s direct suppression of Tlx1 and Tlx3, known to block Lbx1 induction of Pax2 as mentioned above (Cheng et al., 2005) (see Model Fig. 7).
Additional functions for Prdm13
Ptf1a not only functions in specifying the excitatory/inhibitory balance in the dorsal spinal cord, but it also controls the balance of GABAergic interneurons with that of the glutamatergic granule cells in the cerebellum (Hoshino et al., 2005; Pascual et al., 2007). Similarly, Ptf1a is required in the retina for generation of GABAergic and glycinergic amacrine and horizontal neurons and suppression of glutamatergic retinal ganglia neurons (Dullin et al., 2007; Fujitani et al., 2006; Jusuf and Harris, 2009; Nakhai et al., 2007). By mRNA in situ hybridization, Prdm13 reflects Ptf1a in cerebellum and retina, and in Ptf1a mutants, Prdm13 is dramatically attenuated in these domains. This suggests Prdm13 may suppress non-Ptf1a lineage gene expression in these regions as it does in the dorsal neural tube.
Non-Ptf1a dependent activities are also suggested by Prdm13 expression detected as early as E9 in the dorsal neural tube ventricular zone (Fig. S1). This ventricular zone expression spans the region where dI2 to dI6 arise, and it is not affected in the Ptf1a mutant. A hint that Prdm13 may also function in neuronal proliferation and differentiation was seen in the Prdm13 overexpression experiments in chick. Normally, Ascl1 drives neuronal differentiation when overexpressed, however, when Prdm13 was co-expressed with Ascl1, it disrupted this phenotype (Fig. 5, compare the distribution of Ascl1+ cells in D and E). Thus, Prdm13 may function to modulate Ascl1 activity (and other bHLH factors) in their role in neuronal differentiation as well as their role in controlling cell fate decisions.
Switching Ascl1 from transcriptional activator to repressor
Ascl1 has been shown to be essential in the development of multiple neural and neuroendocrine lineages, particularly during early stages of differentiation from progenitors (Bertrand et al., 2002). Most recently it was identified as an essential component of a transcription factor cocktail that can directly reprogram or transdifferentiate fibroblasts to neurons (Yang et al., 2011). At a mechanistic level, Ascl1 heterodimerizes with E-proteins (such as Heb and E2a), binds DNA, and activates transcription (Johnson et al., 1992). Interactions with the global co-activator p300, a histone acetyltransferase, have been reported (Yamamoto et al., 2001). Our current findings suggest Ascl1 may make a repressor complex with Prdm13, switching Ascl1 from an activator to a repressor with respect to the glutamatergic specifier gene Tlx3. Supporting this model (Fig. 7) we show 1) Ascl1 and Prdm13 localize to the same genomic location upstream of the Tlx3 gene, 2) Ascl1 and Prdm13 co-immunoprecipitate, and 3) Ascl1 induces expression of the Tlx1 and Tlx3 enhancer reporters, activities repressed by Prdm13. An alternative model would have Prdm13 blocking Ascl1 from binding the Tlx3 enhancer. Further experiments are required to definitively distinguish between these models. Nevertheless, Prdm13 is instrumental to silencing the gene program for the glutamatergic fate via Ascl1 as cells progress towards the GABAergic neuronal lineage.
In summary, PRDM factors utilize diverse mechanisms to modulate transcription, and broadly function in progenitor populations to control lineage decisions. This study places Prdm13, a zinc finger transcription factor, in a critical role connecting the neuronal specification functions of bHLH and HD factors, and highlights the importance of silencing transcription of gene programs in the opposing lineage.
EXPERIMENTAL PROCEDURES
Mouse strains and tissue preparation
Mutant mouse strains have been previously described. Ptf1aCre (p48Cre), where the Ptf1a coding region is replaced by that coding for Cre recombinase was used as the Ptf1a null (Kawaguchi et al., 2002). 12.4Ptf1a::mCherry transgenic mice, where a 12.4 kb regulatory sequence from 3′ of the Ptf1a gene drives expression of mCherry (Meredith et al., 2009), was used for fluorescence activated cell sorting (FACS) of Ptf1a lineage cells from E11.5 wildtype or Ptf1a null neural tubes. PCR genotyping was performed as previously described (Glasgow et al., 2005; Meredith et al., 2009). All procedures on animals follow NIH Guidelines and were approved by the UT Southwestern Institutional Animal Care and Use Committee.
Chromatin Immunoprecipitation (ChIP-Seq)
Tissue source for ChIP was mouse E11.5 or 12.5 neural tubes. Ptf1a, Ascl1 and Prdm13 negative tissue for control included telencephalon and/or limbs from the same embryos. ChIP was performed as previously described (Holmstrom et al., 2011; Masui et al., 2007), with details in the Supplemental Experimental Procedures (SEP). Notably, the Prdm13 ChIP was treated with formaldehyde along with the long-arm cross-linker, ethylene glycol-bis (succinimidyl succinate), to increase ChIP efficiency (Zeng et al., 2006). Antibodies used for Ptf1a and Ascl1 ChIP are as published (Castro et al., 2006)(Beres et al., 2006). Lab generated antisera were used for Rbpj (TX857, rabbit anti-Rbpj) and Prdm13 (TX970, rabbit anti-Prdm13), details provided in the SEP. NEBNext ChIP-Seq Sample Prep Master Mix Set 1 was used for library generation. Detailed methods and primers used for ChIP qPCR are listed in SEP. ChIP efficiency (CE) was calculated relative to input as CE = (2Ct input − Ct ChIP) × DF × 100%, where DF is the dilution factor between input and ChIP sample.
Plasmid description
FLAG-tagged Prdm13 and variations used for expression in the chick neural tube were inserted into the pCIG vector, which drives expression through a combined CMV early enhancer/chicken β-actin promoter and contains an IRES-NLS-GFP (Megason and McMahon, 2002). Prdm13 mZ1234 contains point mutations in each zinc finger (Z1;C185A, H207A, Z2;C622A, H638A, Z3;C650A, H666A, Z4;C679A, H695A). Ptf1a and Ascl1 expression constructs used pMiWIII as previously described (Hori et al., 2008; Nakada et al., 2004). The Lbx1 expression vector was as described (Muller et al., 2002). The engrailed repressor (EnR) (Smith and Jaynes, 1996) or the VP16 activator (Triezenberg et al., 1988) were fused at the C-terminal end of Prdm13 in pMiWIII. The eTlx1::GFP and eTlx3::GFP reporter constructs include mm9 coordinates (eTlx1, chr19:45217747-45218583) and (eTlx3, chr11:33134077-33134502), respectively, cloned into a GFP reporter cassette with a β-globin basal promoter (Lai et al., 2011). All constructs used were sequence verified.
The pSilencer 1.0-U6 vector (Ambion) was used in knock down experiments where several sets of 21-mer oligonucleotides were selected from chick Ptf1a or Prdm13 mRNA sequences (listed in the SEP). Additionally, shRNA targeting DSred was used as control (Rao et al., 2004). The shRNA constructs were co-electroporated with pCIG so the GFP could be used to assess electroporation efficiency.
In ovo chick electroporation
Fertilized white Leghorn eggs were obtained from the Texas A&M Poultry Department (College Station, TX) and incubated at 37°C for 48 hours until stage HH12-13 (Hamburger and Hamilton, 1992). Supercoiled plasmid DNA (1–2.5 μg/μl each) were injected into the lumen of the closed neural tube, and embryos were electroporated as previously described (Timmer et al., 2001). After 48 hours incubation at 37°C, stage HH24-25 embryos were harvested and processed for immunofluorescence or in situ hybridization.
Immunofluorescence and In Situ hybridization
Immunofluorescence was performed as previously described (Glasgow et al., 2005; Hori et al., 2008). A list of primary antibodies used is listed in SEP. Fluorescence imaging was carried out on a Zeiss LSM 510 confocal microscope. For each experiment, multiple sections from at least six different embryos were analyzed and used for quantification. Cell counting was blind to the condition and was performed manually using ImageJ. The quantitative results are presented as a ratio of the number of marker positive cells on the electroporated side divided by the number on the non-electroporated side. Only sections with confirmed high efficiency expression across the dorsal ventral axis were used in the analysis. Mean GFP pixel intensity for eTlx1/3::GFP expression was measured by ImageJ. Significant differences between control and experimental samples were calculated using a two-tailed two-sample equal variance (homoscedastic) Student’s t test (* indicates P < 0.001) in Microsoft Excel. SEM is shown.
In situ hybridization was performed as previously described (Lai et al., 2011). Chick Gad1 and vGlut2 (Cheng et al., 2004), and chick Ptf1a (gift T. Reh) probes were used. Plasmids for generating chick and mouse Prdm13 probes were cloned by PCR into pBluescript with the primers listed in SEP.
Co-immunoprecipitation
HEK293T cells were transfected with Flag-tagged Prdm13, myc-tagged Ascl1 or Hook3 (control) expression constructs using FuGene 6 (Roche Life Sciences), and collected 48 hours later. Anti-Flag antibodies conjugated to protein A/G beads (Santa Cruz) were added to cleared lysate. Co-immunoprecipitation was assessed by western blot using anti-myc (A-14, Santa Cruz, 1:2000).
Supplementary Material
Acknowledgments
We acknowledge the many hours of helpful discussions with members of the Johnson laboratory particularly Dr. Tou Yia Vue. We are grateful for the generosity of colleagues for antibody reagents including Drs. T. Muller, C. Birchmeier, E. Turner, T. Jessell, and R. MacDonald. This work was supported by NIH R21 NS067553, R01 HD037932, and R01 NS032817 to JEJ; NIH F31 NS061440 to DMM; and NIH F31 NS705592 to MDB.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Batista MF, Lewis KE. Pax2/8 act redundantly to specify glycinergic and GABAergic fates of multiple spinal interneurons. Dev Biol. 2008;323:88–97. doi: 10.1016/j.ydbio.2008.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beres TM, Masui T, Swift GH, Shi L, Henke RM, MacDonald RJ. PTF1 is an organ-specific and Notch-independent basic helix-loop-helix complex containing the mammalian Suppressor of Hairless (RBP-J) or its paralogue, RBP-L. Mol Cell Biol. 2006;26:117–130. doi: 10.1128/MCB.26.1.117-130.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand N, Castro DS, Guillemot F. Proneural genes and the specification of neural cell types. Nat Rev Neurosci. 2002;3:517–530. doi: 10.1038/nrn874. [DOI] [PubMed] [Google Scholar]
- Brohl D, Strehle M, Wende H, Hori K, Bormuth I, Nave KA, Muller T, Birchmeier C. A transcriptional network coordinately determines transmitter and peptidergic fate in the dorsal spinal cord. Dev Biol. 2008;322:381–393. doi: 10.1016/j.ydbio.2008.08.002. [DOI] [PubMed] [Google Scholar]
- Brzezinski JAt, Lamba DA, Reh TA. Blimp1 controls photoreceptor versus bipolar cell fate choice during retinal development. Development. 2010;137:619–629. doi: 10.1242/dev.043968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro DS, Skowronska-Krawczyk D, Armant O, Donaldson IJ, Parras C, Hunt C, Critchley JA, Nguyen L, Gossler A, Gottgens B, et al. Proneural bHLH and Brn proteins coregulate a neurogenic program through cooperative binding to a conserved DNA motif. Dev Cell. 2006;11:831–844. doi: 10.1016/j.devcel.2006.10.006. [DOI] [PubMed] [Google Scholar]
- Cheng L, Arata A, Mizuguchi R, Qian Y, Karunaratne A, Gray PA, Arata S, Shirasawa S, Bouchard M, Luo P, et al. Tlx3 and Tlx1 are post-mitotic selector genes determining glutamatergic over GABAergic cell fates. Nat Neurosci. 2004;7:510–517. doi: 10.1038/nn1221. [DOI] [PubMed] [Google Scholar]
- Cheng L, Samad OA, Xu Y, Mizuguchi R, Luo P, Shirasawa S, Goulding M, Ma Q. Lbx1 and Tlx3 are opposing switches in determining GABAergic versus glutamatergic transmitter phenotypes. Nat Neurosci. 2005;8:1510–1515. doi: 10.1038/nn1569. [DOI] [PubMed] [Google Scholar]
- Dullin JP, Locker M, Robach M, Henningfeld KA, Parain K, Afelik S, Pieler T, Perron M. Ptf1a triggers GABAergic neuronal cell fates in the retina. BMC Dev Biol. 2007;7:110. doi: 10.1186/1471-213X-7-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald M. The development of nociceptive circuits. Nat Rev Neurosci. 2005;6:507–520. doi: 10.1038/nrn1701. [DOI] [PubMed] [Google Scholar]
- Fog CK, Galli GG, Lund AH. PRDM proteins: Important players in differentiation and disease. Bioessays. 2012;34:50–60. doi: 10.1002/bies.201100107. [DOI] [PubMed] [Google Scholar]
- Fujitani Y, Fujitani S, Luo H, Qiu F, Burlison J, Long Q, Kawaguchi Y, Edlund H, MacDonald RJ, Furukawa T, et al. Ptf1a determines horizontal and amacrine cell fates during mouse retinal development. Development. 2006;133:4439–4450. doi: 10.1242/dev.02598. [DOI] [PubMed] [Google Scholar]
- Fumasoni I, Meani N, Rambaldi D, Scafetta G, Alcalay M, Ciccarelli FD. Family expansion and gene rearrangements contributed to the functional specialization of PRDM genes in vertebrates. BMC Evol Biol. 2007;7:187. doi: 10.1186/1471-2148-7-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasgow SM, Henke RM, Macdonald RJ, Wright CV, Johnson JE. Ptf1a determines GABAergic over glutamatergic neuronal cell fate in the spinal cord dorsal horn. Development. 2005;132:5461–5469. doi: 10.1242/dev.02167. [DOI] [PubMed] [Google Scholar]
- Gowan K, Helms AW, Hunsaker TL, Collisson T, Ebert PJ, Odom R, Johnson JE. Crossinhibitory activities of Ngn1 and Math1 allow specification of distinct dorsal interneurons. Neuron. 2001;31:219–232. doi: 10.1016/s0896-6273(01)00367-1. [DOI] [PubMed] [Google Scholar]
- Gross MK, Dottori M, Goulding M. Lbx1 specifies somatosensory association interneurons in the dorsal spinal cord. Neuron. 2002;34:535–549. doi: 10.1016/s0896-6273(02)00690-6. [DOI] [PubMed] [Google Scholar]
- Hamburger V, Hamilton HL. A series of normal stages in the development of the chick embryo. 1951. Dev Dyn. 1992;195:231–272. doi: 10.1002/aja.1001950404. [DOI] [PubMed] [Google Scholar]
- Helms AW, Battiste J, Henke RM, Nakada Y, Simplicio N, Guillemot F, Johnson JE. Sequential roles for Mash1 and Ngn2 in the generation of dorsal spinal cord interneurons. Development. 2005;132:2709–2719. doi: 10.1242/dev.01859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helms AW, Johnson JE. Specification of dorsal spinal cord interneurons. Curr Opin Neurobiol. 2003;13:42–49. doi: 10.1016/s0959-4388(03)00010-2. [DOI] [PubMed] [Google Scholar]
- Henke RM, Savage TK, Meredith DM, Glasgow SM, Hori K, Dumas J, MacDonald RJ, Johnson JE. Neurog2 is a direct downstream target of the Ptf1a-Rbpj transcription complex in dorsal spinal cord. Development. 2009;136:2945–2954. doi: 10.1242/dev.035352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohenauer T, Moore AW. The Prdm family: expanding roles in stem cells and development. Development. 2012;139:2267–2282. doi: 10.1242/dev.070110. [DOI] [PubMed] [Google Scholar]
- Holmstrom SR, Deering T, Swift GH, Poelwijk FJ, Mangelsdorf DJ, Kliewer SA, MacDonald RJ. LRH-1 and PTF1-L coregulate an exocrine pancreas-specific transcriptional network for digestive function. Genes Dev. 2011;25:1674–1679. doi: 10.1101/gad.16860911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori K, Cholewa-Waclaw J, Nakada Y, Glasgow SM, Masui T, Henke RM, Wildner H, Martarelli B, Beres TM, Epstein JA, et al. A nonclassical bHLH Rbpj transcription factor complex is required for specification of GABAergic neurons independent of Notch signaling. Genes Dev. 2008;22:166–178. doi: 10.1101/gad.1628008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino M, Nakamura S, Mori K, Kawauchi T, Terao M, Nishimura YV, Fukuda A, Fuse T, Matsuo N, Sone M, et al. Ptf1a, a bHLH transcriptional gene, defines GABAergic neuronal fates in cerebellum. Neuron. 2005;47:201–213. doi: 10.1016/j.neuron.2005.06.007. [DOI] [PubMed] [Google Scholar]
- Johnson JE, Birren SJ, Saito T, Anderson DJ. DNA binding and transcriptional regulatory activity of mammalian achaete-scute homologous (MASH) proteins revealed by interaction with a muscle-specific enhancer. Proc Natl Acad Sci U S A. 1992;89:3596–3600. doi: 10.1073/pnas.89.8.3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jusuf PR, Harris WA. Ptf1a is expressed transiently in all types of amacrine cells in the embryonic zebrafish retina. Neural Dev. 2009;4:34. doi: 10.1186/1749-8104-4-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajimura S, Seale P, Spiegelman BM. Transcriptional control of brown fat development. Cell Metab. 2010;11:257–262. doi: 10.1016/j.cmet.2010.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Omori Y, Onishi A, Sato S, Kondo M, Furukawa T. Blimp1 suppresses Chx10 expression in differentiating retinal photoreceptor precursors to ensure proper photoreceptor development. J Neurosci. 2010;30:6515–6526. doi: 10.1523/JNEUROSCI.0771-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi Y, Cooper B, Gannon M, Ray M, MacDonald RJ, Wright CV. The role of the transcriptional regulator Ptf1a in converting intestinal to pancreatic progenitors. Nat Genet. 2002;32:128–134. doi: 10.1038/ng959. [DOI] [PubMed] [Google Scholar]
- Krapp A, Knofler M, Frutiger S, Hughes GJ, Hagenbuchle O, Wellauer PK. The p48 DNA-binding subunit of transcription factor PTF1 is a new exocrine pancreas-specific basic helix-loop-helix protein. Embo J. 1996;15:4317–4329. [PMC free article] [PubMed] [Google Scholar]
- Krapp A, Knofler M, Ledermann B, Burki K, Berney C, Zoerkler N, Hagenbuchle O, Wellauer PK. The bHLH protein PTF1-p48 is essential for the formation of the exocrine and the correct spatial organization of the endocrine pancreas. Genes Dev. 1998;12:3752–3763. doi: 10.1101/gad.12.23.3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai HC, Klisch TJ, Roberts R, Zoghbi HY, Johnson JE. In vivo neuronal subtype-specific targets of Atoh1 (Math1) in dorsal spinal cord. J Neurosci. 2011;31:10859–10871. doi: 10.1523/JNEUROSCI.0445-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Ma W, Su W, Zhang J. Prdm14 acts upstream of islet2 transcription to regulate axon growth of primary motoneurons in zebrafish. Development. 2012 doi: 10.1242/dev.083055. [DOI] [PubMed] [Google Scholar]
- Liu Y, Ma Q. Generation of somatic sensory neuron diversity and implications on sensory coding. Curr Opin Neurobiol. 2011;21:52–60. doi: 10.1016/j.conb.2010.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masui T, Long Q, Beres TM, Magnuson MA, MacDonald RJ. Early pancreatic development requires the vertebrate Suppressor of Hairless (RBPJ) in the PTF1 bHLH complex. Genes Dev. 2007;21:2629–2643. doi: 10.1101/gad.1575207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masui T, Swift GH, Hale MA, Meredith DM, Johnson JE, Macdonald RJ. Transcriptional autoregulation controls pancreatic Ptf1a expression during development and adulthood. Mol Cell Biol. 2008;28:5458–5468. doi: 10.1128/MCB.00549-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megason SG, McMahon AP. A mitogen gradient of dorsal midline Wnts organizes growth in the CNS. Development. 2002;129:2087–2098. doi: 10.1242/dev.129.9.2087. [DOI] [PubMed] [Google Scholar]
- Meredith DM, Masui T, Swift GH, MacDonald RJ, Johnson JE. Multiple transcriptional mechanisms control Ptf1a levels during neural development including autoregulation by the PTF1-J complex. J Neurosci. 2009;29:11139–11148. doi: 10.1523/JNEUROSCI.2303-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuguchi R, Kriks S, Cordes R, Gossler A, Ma Q, Goulding M. Ascl1 and Gsh1/2 control inhibitory and excitatory cell fate in spinal sensory interneurons. Nat Neurosci. 2006;9:770–778. doi: 10.1038/nn1706. [DOI] [PubMed] [Google Scholar]
- Muller T, Brohmann H, Pierani A, Heppenstall PA, Lewin GR, Jessell TM, Birchmeier C. The homeodomain factor lbx1 distinguishes two major programs of neuronal differentiation in the dorsal spinal cord. Neuron. 2002;34:551–562. doi: 10.1016/s0896-6273(02)00689-x. [DOI] [PubMed] [Google Scholar]
- Nakada Y, Hunsaker TL, Henke RM, Johnson JE. Distinct domains within Mash1 and Math1 are required for function in neuronal differentiation versus neuronal cell-type specification. Development. 2004;131:1319–1330. doi: 10.1242/dev.01008. [DOI] [PubMed] [Google Scholar]
- Nakhai H, Sel S, Favor J, Mendoza-Torres L, Paulsen F, Duncker GI, Schmid RM. Ptf1a is essential for the differentiation of GABAergic and glycinergic amacrine cells and horizontal cells in the mouse retina. Development. 2007;134:1151–1160. doi: 10.1242/dev.02781. [DOI] [PubMed] [Google Scholar]
- Pascual M, Abasolo I, Mingorance-Le Meur A, Martinez A, Del Rio JA, Wright CV, Real FX, Soriano E. Cerebellar GABAergic progenitors adopt an external granule cell-like phenotype in the absence of Ptf1a transcription factor expression. Proc Natl Acad Sci U S A. 2007;104:5193–5198. doi: 10.1073/pnas.0605699104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao M, Baraban JH, Rajaii F, Sockanathan S. In vivo comparative study of RNAi methodologies by in ovo electroporation in the chick embryo. Dev Dyn. 2004;231:592–600. doi: 10.1002/dvdy.20161. [DOI] [PubMed] [Google Scholar]
- Ross SE. Pain and itch: insights into the neural circuits of aversive somatosensation in health and disease. Curr Opin Neurobiol. 2011;21:880–887. doi: 10.1016/j.conb.2011.10.012. [DOI] [PubMed] [Google Scholar]
- Ross SE, McCord AE, Jung C, Atan D, Mok SI, Hemberg M, Kim TK, Salogiannis J, Hu L, Cohen S, et al. Bhlhb5 and prdm8 form a repressor complex involved in neuronal circuit assembly. Neuron. 2012;73:292–303. doi: 10.1016/j.neuron.2011.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seale P, Bjork B, Yang W, Kajimura S, Chin S, Kuang S, Scime A, Devarakonda S, Conroe HM, Erdjument-Bromage H, et al. PRDM16 controls a brown fat/skeletal muscle switch. Nature. 2008;454:961–967. doi: 10.1038/nature07182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ST, Jaynes JB. A conserved region of engrailed, shared among all en-, gsc-, Nk1-, Nk2- and msh-class homeoproteins, mediates active transcriptional repression in vivo. Development. 1996;122:3141–3150. doi: 10.1242/dev.122.10.3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavares I, Lima D. From neuroanatomy to gene therapy: searching for new ways to manipulate the supraspinal endogenous pain modulatory system. J Anat. 2007;211:261–268. doi: 10.1111/j.1469-7580.2007.00759.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmer J, Johnson J, Niswander L. The use of in ovo electroporation for the rapid analysis of neural-specific murine enhancers. Genesis. 2001;29:123–132. doi: 10.1002/gene.1015. [DOI] [PubMed] [Google Scholar]
- Triezenberg SJ, Kingsbury RC, McKnight SL. Functional dissection of VP16, the trans-activator of herpes simplex virus immediate early gene expression. Genes Dev. 1988;2:718–729. doi: 10.1101/gad.2.6.718. [DOI] [PubMed] [Google Scholar]
- Wildner H, Muller T, Cho SH, Brohl D, Cepko CL, Guillemot F, Birchmeier C. dILA neurons in the dorsal spinal cord are the product of terminal and non-terminal asymmetric progenitor cell divisions, and require Mash1 for their development. Development. 2006;133:2105–2113. doi: 10.1242/dev.02345. [DOI] [PubMed] [Google Scholar]
- Yamamoto N, Yamamoto S, Inagaki F, Kawaichi M, Fukamizu A, Kishi N, Matsuno K, Nakamura K, Weinmaster G, Okano H, et al. Role of Deltex-1 as a transcriptional regulator downstream of the Notch receptor. J Biol Chem. 2001;276:45031–45040. doi: 10.1074/jbc.M105245200. [DOI] [PubMed] [Google Scholar]
- Yang N, Ng YH, Pang ZP, Sudhof TC, Wernig M. Induced neuronal cells: how to make and define a neuron. Cell Stem Cell. 2011;9:517–525. doi: 10.1016/j.stem.2011.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng PY, Vakoc CR, Chen ZC, Blobel GA, Berger SL. In vivo dual cross-linking for identification of indirect DNA-associated proteins by chromatin immunoprecipitation. Biotechniques. 2006;41:694, 696, 698. doi: 10.2144/000112297. [DOI] [PubMed] [Google Scholar]
- Zhuang B, Sockanathan S. Dorsal-ventral patterning: a view from the top. Curr Opin Neurobiol. 2006;16:20–24. doi: 10.1016/j.conb.2005.11.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.