Abstract
Given their medical importance, most attention has been paid towards the venom composition of scorpions of the Buthidae family. Nevertheless, research has shown that the venom of scorpions of other families is also a remarkable source of unique peptidyl toxins. The κ-KTX family of voltage-gated potassium channel (VGPC) scorpion toxins is hereof an example. From the telson of the scorpion Heterometrus laoticus (Scorpionidae), a peptide, Hl15-1-1, with unique primary sequence was purified through HPLC and sequenced by Edman degradation. Based on the amino acid sequence, the peptide could be cloned and the cDNA sequence revealed. Hl15-1-1 was chemically synthesized and functionally characterized on VGPCs of the Shaker-related, Shaw-related and Shal-related subfamilies. Furthermore, the toxin was also tested on small- and intermediate conductance Ca2+-activated K+ channels. From the channels studied, Kv1.1 and Kv1.6 were found to be the most sensitive (Kv1.1 EC50 = 9.9 ± 1.6 μM). The toxin did not alter the activation of the channels. Competition experiments with TEA showed that the toxin is a pore blocker. Mutational studies showed that the residues E353 and Y379 in the pore of Kv1.1 act as major interaction points for binding of the toxin. Given the amino acid sequence, the predicted secondary structure and the biological activity on VGPCs, Hl15-1-1 should be included in the κ-KTX family. Based on a phylogenetic study we rearranged this family of VGPC toxins into five subfamilies and suggest that HI15-1-1 is the first member of the new KTX5 subfamily.
Complex multicellular organisms necessitate rapid and accurate transmission of information among cells and tissues as well as tight coordination of distant functions. Electrical signals control a wide range of physiological processes in both vertebrates and invertebrates. All of these processes are mediated in part by members of the voltage-gated ion channel superfamily (1).Voltage-gated ion channels form the third largest group of signaling molecules in the human genome and more than half the members of this superfamily are K+ channels (2). Within the group of K+ channels, 40 genes encode for voltage-gated potassium channels (VGPC) which can be again divided in 12 subfamilies. In excitable cells, VGPCs are mainly responsible for the repolarization after action potential firing. Furthermore, in both excitable and non-excitable cells, these transmembrane proteins play an important role in Ca2+ signaling, volume regulation, secretion, proliferation and migration (3).
Their crucial involvement in many physiological processes, make VGPCs a major target for peptidyl toxins from venomous animals such as snakes, scorpions, spiders, cone snails and sea anemones. Activation or inhibition of VGPCs in excitable cells will consequently lead either to a reduction or enhancement of excitability and eventually lead to death of the prey or aggressor. However, because of their importance in the overall functioning of every organism, VGPCs constituteimportant potential drug targets. They may have therapeutic utility in disorders ranging from cancer and autoimmune diseases to metabolic, neurological and cardiovascular disorders (3,4). In this way, the venom compounds designed by nature to kill, can for instance re-tune the action potential firing in excitable cells or inhibit the proliferation and suppress cellular activation of lymphocytes and cancer cells. As such these peptidyl toxins form a highly valuable group of potential lead compounds for therapeutic drug candidates.
Scorpionism is a real public health problem in many countries of Africa, America and Asia. Nevertheless, fewer than 25 species out of the more than 1500 described species are lethal to humans. All these deadly species belong to the family Buthidae. All other extant scorpions can be divided in 18 families (5–8). They are all venomous and possess a homologous venom apparatus. This apparatus consists of two symmetrical glands held together in the vesicle and the hypodermic aculeus used to inject the venom (7,9). The venom glands produce a large diversity of toxins from which some have already shown their therapeutic value (3,10).
Given the medical importance of the Buthid scorpions, most research has been focusing on the composition of their venoms. As a consequence, most of the roughly 730 known scorpion peptidyl toxins have been isolated from the venom of Buthid scorpions (11,12). In contrast, the non-Buthid families have often been neglected and are still often overlooked. Nevertheless, both proteomic and transcriptomic studies revealed the existence of peptides with unique primary sequences and biological activity in their venoms (13–18). Therefore, exploring the venom composition of these `forgotten' scorpion families may open a valuable pharmacological treasure chest.
The scorpion Heterometrus laoticus belongs to the family of the Scorpionidae and is commonly found in Cambodia, Thailand, Laos and Vietnam (7). Envenomations by this scorpion are rare and victims suffer at most from severe pain with local inflammation, edema, swelling and redness of the stung area (19). The venom composition remains rarely studied until now. The only peptidyl venom compound isolated so far is Heteroscorpine-1 (P0C2F4) with high homology to members of the β-KTx3 subfamily (20).
Here we report the biochemical isolation, molecular cloning and electrophysiological characterization of a unique peptidyl toxin, Hl15-1-1, from the venom of the scorpion H. laoticus. To the best of our knowledge the toxin bears no similarity with any currently known scorpion toxin. However, based on the presence of two disulphide bridges, the predicted secondary structure and the activity towards VGPCs, Hl15-1-1 should be classified within the scorpion κ-KTx family. In recent years, a couple of new members have been added to this family, however without any clear classification (14,17,21,22). Based on the construction of a phylogenetic tree, we propose the rearrangement of the κ-KTx family into five subfamilies, one of which is formed by Hl15-1-1.
EXPERIMENTAL PROCEDURES
Venom source
Scorpions were obtained from a local pet shop and identified as Heterometrus laoticus (Couzijn, 1981), a species belonging to the family of the Scorpionidae. Upon arrival in the lab, the scorpions were immediately killed after which the telsons (venom gland segments) were cut off, snap frozen in liquid nitrogen and stored at −80°C until use.
Purification of Hl15-1-1
One telson was ground in 4 ml 1 % trifluoroacetic acid (TFA) in dH2O, transferred to an Eppendorf tube and centrifuged for 5 min at 13000 g. The supernatant was placed in a new Eppendorf tube and lipids were removed using ethylacetate/hexane (50/50) extraction. The remaining solvent was dried in a micro concentrator. The sample was dissolved in dH2O, and put on a Solid Phase Extraction Oasis® HLB column (Waters). The column was activated with 100% MeOH and rinsed with dH2O. Then, the sample was put on the column and the column was rinsed with 5 % MeOH. Subsequently, the column was flushed with 100 % CH3CN. The eluate was collected and 1.5 ml 0.1 % TFA in dH2O was added. After CH3CN was evaporated, the sample was filtered (Millex-HV, 0.22 μm, Millipore) and subjected to HPLC.
HPLC analyses were conducted on a Gilson liquid chromatograph using a Waters Symmetry® C18 column (4.6 mm i.d. × 250 mm length, 5 μm particle size). As mobile phases, HPLC H2O with 0.1 % TFA (A) and 60 % CH3CN with 0.1 % TFA (B) were used. The applied gradient elution time profile was as follows (in % B): 0 – 10 min: 2 %; 10 – 70 min: 2 % – 100 %, at a flow rate of 1 ml min−1. Fraction 15 was further purified on a Waters XTerra® phenyl column (2.1 mm i.d. × 150 mm length, 5 μm particle size). The same solvents as described above were used. The applied gradient elution time profile was as follows (in % B): 0 – 15 min: 0 % – 2 %; 15 – 45 min: 2 % – 10 %, at a flow rate of 0.3 ml min−1.
Mass spectrometry analysis and amino acid sequencing
The molecular mass of the fraction eluting between 35.5 and 37.5 min was analyzed by MALDI-TOF MS on a Reflex IV (Bruker Daltonics GmbH). One μl of the fraction was mixed with one μl of a saturated solution of α-cyano-4-hydroxycinnamic acid in acetone on a steel target and air-dried. A quadratic calibration (< 10 ppm) was performed using a standard peptide mixture (Bruker Daltonics GmbH). Positive ion spectra were recorded in the reflectron mode from m/z 500 to 3500.
For amino acid sequence determination, 10 μl of the fraction eluting between 35.5 and 37.5 min was spotted on a precycled Biobrene Plus TM-coated micro TFA filter. The sequencing was performed by Edman degradation on an automated Procise Precise?? protein sequencer (Applied Biosystems). The last amino acid was deduced from the mass difference between the calculated mass and the experimentally obtained mass.
Synthetic Hl15-1-1 was purchased from GenScript Corporation with a purity of 79.5 %.
Preparation of total RNA
One telson was ground in liquid nitrogen and total RNA was prepared using the TRIzol® Reagent following manufacturer's instructions (Invitrogen) (23).
3' RACE
The 3'RACE (Rapid Amplification of cDNA Ends) was performed using the 3'RACE system for rapid amplification of cDNA ends (Invitrogen) following the manufacturer's instructions. Briefly, total RNA (4 μg) was reverse transcribed into first-strand cDNA using Superscript™ II reverse transcriptase and a custom oligo(dT) primer. Second strand synthesis was performed using a degenerate primer R1 (5'-TGCAAAAAGGAGTGTTCTGG-3') and DNA polymerase I Klenow fragment (Promega). Primer R1 was designed against the N-terminal AA region CKKECSG. cDNA's were further amplified with primer R1, the oligo(dT) primer and Taq DNA polymerase (Fermentas) using the following PCR conditions: 3 min 95 °C, 30× (30 sec 95 °C, 30 sec 62 °C, 120 sec 72 °C) and 5 min 72 °C. The obtained PCR products were purified with the GenElute™ PCR Clean-up Kit (Sigma-Aldrich). 10 μl of the purified PCR was used as template for a second PCR with the same primers and under the same conditions as the initial PCR. PCR products were subsequently subjected to agarose gel electrophoresis and visualized by ethidium bromide staining. cDNA of the appropriate molecular weight size was extracted from the gel using the Qiaex® II Gel extraction kit (Qiagen, Germantown, MD, USA) and ligated into the pGEM-T easy Vector (Promega). XL10-Gold® Ultracompetent Cells (Stratagene) were used for plasmid propagation. Inserts were sequenced by dye terminator-based sequencing (DYEnamic ET Terminator Cycle Sequencing Kit (Amersham Biosciences)) on an automated MegaBACE sequencer (Amersham Biosciences) using the following sequencing primer for the pGEM-T easy vector, 5'-AACGACGGCCAGTGAATTGT-3'.
5' RACE
Based upon the partial cDNA sequence obtained from the 3'RACE, two reverse gene-specific primers (GSP1: 5'-CGTGGCCGTGCTCTCTGTTG-3'; GSP2: 5'-CATTTCTGCATGCATTTCTTCG-3') were designed. The 5'RACE was performed using the 5'RACE system for rapid amplification of cDNA Ends (Invitrogen). Briefly, 2 μg of total RNA were used for synthesis of first strand cDNA using Superscript™ II reverse transcriptase and GSP1. Subsequently, a homopolymeric tail was added to the 3' end of the cDNA using terminal deoxynucleotidyl transferase and dCTP. PCR amplification was accomplished using Taq DNA polymerase, primer GSP2 and oligo(dG)primer under the following “hot start” PCR conditions: 3 min 95 °C, 30 × (30 sec 95 °C, 30 sec 68 °C, 2 min 72 °C) and 5 min 72 °C. The obtained PCR products were analyzed by agarose gel electrophoresis and visualized by ethidium bromide staining. DNA bands of the appropriate molecular weight were purified with the GenElute™ PCR Clean-up Kit and ligated into the pGEM-Teasy Vector for sequencing (vide supra).
Bioinformatics
The amino acid (AA) and DNA sequence of Hl15-1-1 were analyzed by BLAST (http://www.expasy.ch/tools/blast and http://blast.ncbi.nlm.nih.gov). Signalpeptide and propeptide predictions were made respectively with SignalP 3.0 and ProP 1.0 (24,25). The secondary structure and disulphide bridging were predicted with SCRATCH Protein Predictor (26).
A multiple sequence alignment of Hl15-1-1 with existing κ-KTX-subfamily members was done with ClustalW2 (http://www.ebi.ac.uk/Tools/msa/clustalw2). Sequences used were that of Hefutoxin 1 (P82850) and 2 (P82851) (Heterometrus fulvipes) (27); Hefutoxin 3 (P83655) (H. spinifer) (28); OmTx1 (P0C1Z3), OmTx2 (P0C1Z3), OmTx3 (P0C1Z4) and OmTx4 (P0C1Z3) (Opisthacanthus madagascariensis) (29); OcyC8 (P86110) and OcyC9 (FM998751) (Opisthacanthus cayaporum) (17,21,22); HSP009C, HSP040C.1, HSP040C.2, HSP040C.3, HSP040C.4, HSP040C.5, HSP053C.1 and HSP053C.2 (Heterometrus petersii) (14). Based on the alignment, a Bayesian estimation of the phylogeny of these sequences was carried out with mrBayes v. 3.1.2 using 5000000 generations, sampling and print frequency of both 100 and 4 chains for the Markov Chain Monte Carlo analysis (30). The analysis resulted in 50000 trees. Upon examining stationarity, 35000 tree samples were used for the determination of the consensus tree and posterior probabilities. The rooted tree diagrams were visualized using FigTree v.1.3.1 (http://tree.bio.ed.ac.uk/software/figtree).
Two-electrode voltage-clamp experiments on Kv1.3 channels expressed in Xenopus laevis oocytes
hKv1.3 channels were prepared as previously described (31). Stage-V and -VI Xenopus laevis oocytes were harvested by partial ovariectomy under anaesthesia (3-aminobenzoic acid ethyl ester methanesulfonate salt, 0.5 g/l, Sigma). Anaesthetized animals were kept on ice during dissection. The oocytes were defolliculated by treatment with 2 mg/ml collagenase (Sigma) in Ca2+-free ND96 solution (in mM: NaCl 96, KCl 2, MgCl2 1, HEPES 5 adjusted to pH 7.5). Between 1 and 24 h after defolliculation, oocytes were injected with 50 nl at a concentration of 1 ng/μl cRNA. The oocytes were then incubated in ND96 solution (supplemented with 50 mg/l gentamycin sulphate and 1.8 mM CaCl2) at 16 °C for 1–5 days.
Two-electrode voltage-clamp recordings were performed at room temperature using a GeneClamp 500 amplifier (Molecular Devices) controlled by a pClamp data acquisition system (Molecular Devices). Whole-cell currents from oocytes were recorded 1–5 days after injection. Voltage and current electrodes were filled with 3 M KCl. Resistances of both electrodes were kept as low as possible (0.2 – 1 MΩ). Currents were obtained by depolarizing the channels during 500 ms from a holding potential of −90 mV to 0 mV followed by a deactivation step for 500 ms towards −50 mV. The currents records were sampled at 0.5 ms intervals and filtered using a 4-pole Bessel filter at 1 kHz. The leak currents were subtracted using a -P/4 protocol.
Patch clamp experiments on Kv1.x channels expressed in mammalian cells
All VGPCs used were cloned in a pEGFP-N1 expression vector. Mutants were created using the QuickChange Site-Directed Mutagenesis kit (Stratagene) and mutant primers. Double strand sequencing confirmed the presence of the desired modification and the absence of unwanted mutations. Plasmid DNA was amplified in XL2 blue script cells (Stratagene) and isolated using the GenElute HP plasmid maxiprep kit (Sigma-Aldrich).
Ltk− cells (mouse fibroblasts, ATCC CLL.1.3) were cultured in DMEM with 10 % horse serum and 1 % penicillin/streptomycin. Cells were transiently transfected with cDNA for wildtype or mutant constructs using PEI (Sigma-Aldrich).
Patch clamp recordings were done ± 20 hours after transfection at room temperature with an Axopatch-200B amplifier and digitized with a Digidata-1200A (Axon instruments). Command voltages and data storage were controlled with pClamp8 software. Patch pipettes were pulled from 1.2 mm quick-fill borosilicate capillaries (World Precision Instruments) with a P-2000 puller (Sutter Instrument Co.) and heat polished. The bath solution contained (in mM): NaCl 130, KCl 4, CaCl2 1.8, MgCl2 1; HEPES 10 and Glucose 10; and was adjusted to pH 7.35 with NaOH. The pipette solution contained (in mM) KCl 110, K4BAPTA 5, K2ATP 5, MgCl2 1 and HEPES 10; and was adjusted to pH 7.2 with KOH. Junction potentials were zeroed with the filled pipette in the bath solution. Experiments were excluded from the analysis if the voltage error estimate based on the size of the current exceeded 5 mV after series resistance compensation. Toxin concentrations were applied using a fast perfusion system (ALA Scientific Instruments). Tetraethylammonium (TEA) was obtained from Sigma-Aldrich.
Patch clamp experiments on KCa3.1, KCa2.3, Kv3.1, Kv3.2 and Kv4.2 expressed in mammalian cells
HEK-293 cells stably expressing hKCa3.1 were obtained from Khaled Houamed, University of Chicago, IL. The cloning of hKCa2.3 (19 CAG repeats) has been previously described (32); this clone was later stably expressed in COS-7 cells at Aurora Biosciences Corp., San Diego, CA. LTK cells stably expressing rKv4.2 cells were a gift from Michael Tamkun, University of Colorado, Boulder. L929 cells stably expressing mKv3.1 were previously described (33). Rat Kv3.2 in pcDNA3 was obtained from Protinac GmbH (Hamburg, Germany) and transiently transfected into COS-7 cells together with eGFP-C1 with Fugene-6 (Roche) according to the manufacturer's protocol.
All experiments were conducted with an EPC-10 amplifier (HEKA, Lambrecht/Pfalz, Germany) in the whole-cell configuration of the patch-clamp technique with a holding potential of −80 mV. For measurements of KCa2 and KCa3.1 currents we used an internal pipette solution containing (in mM): 145 K+ aspartate, 2 MgCl2, 10 HEPES, 10 K2EGTA and 5.96 (250 nM free Ca2+) or 8.55 CaCl2 (1 μM free Ca2+), pH 7.2, 290–310 mOsm. Free Ca2+ concentrations were calculated with MaxChelator assuming a temperature of 25°C, a pH of 7.2 and an ionic strength of 160 mM. To reduce currents from native chloride channels in COS-7 and HEK-293 cells, Na+ aspartate Ringer was used as an external solution (in mM): 160 Na+ aspartate, 4.5 KCl, 2 CaCl2, 1 MgCl2, 5 HEPES, pH 7.4, 290–310 mOsm. KCa2 and KCa3.1 currents were elicited by 200-ms voltage ramps from −120 mV to 40 mV applied every 10 sec and the fold-increase of slope conductance at −80 mV by toxin taken as a measure of channel activation. Kv3.1, Kv3.2 and Kv4.2 currents were recorded in normal Ringer solution with Ca2+-free pipette solution containing (in mM): 145 KF, 10 HEPES, 10 EGTA, 2 MgCl2, pH 7.2, 300 mOsm. Currents were elicited by 200-ms depolarizing pulses to 40 mV applied every 10 sec.
Data analysis
The ratio Itoxin/Icontrol was used as a measure for the decrease in current induced by the toxin. For Kv1.1, a dose response curve was constructed by plotting the fraction of current remaining at +40 mV (y) as a function of toxin concentration. The curve was fitted with the Hill equation:
where C is the toxin concentration, p the Hill coefficient and EC50 the concentration needed for 50 % inhibition of the current. The voltage dependence of activation was obtained by plotting the normalized tail currents as a function of the prepulse potential. The activation curves were fitted with a Boltzmann relationship of the form:
where V1/2 is the voltage for half maximal activation and s the slope factor. The activation kinetics were obtained by monoexponential fits to the raw current traces. The retrieved τ-values were plotted as a function of the applied voltage.
Results are expressed as the mean ± SEM with n being the number of cells analyzed.
RESULTS
Purification, primary sequence determination and secondary structure prediction
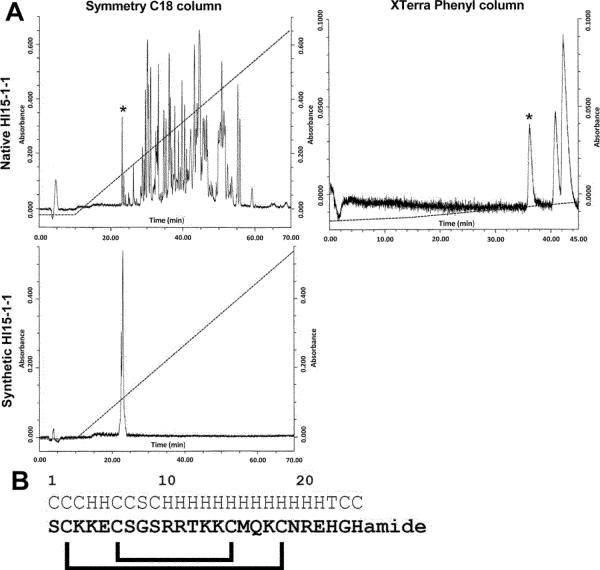
The extract of one telson from H. laoticus was separated by HPLC on a C18 column (Fig. 1A). The obtained fractions were all tested electrophysiologicaly by means of the two-electrode voltage-clamp technique on the VGPC Kv1.3 expressed heterologously in X. laevis oocytes. Fraction 15 showed the highest activity. However, a mass spectrometry analysis using MALDI-TOF MS revealed that the fraction was not pure. Consequently, fraction 15 was further purified by HPLC on a XTerra phenyl column (Fig. 1A). This separation resulted in three fractions each of them blocking Kv1.3. MALDI-TOF MS of the first fraction, Hl15-1-1, showed a doubly-charged ion at m/z 1458.67 (monoisotopic mass) corresponding to a peptide with a molecular mass of 2915.34 Da. Using Edman degradation based sequencing, the first 24 N-terminal amino acids were determined as SCKKECSGSRRTKKCMQKCNREHG. The identity of the remaining amino acid could be further deduced from the mass difference between the calculated mass and the experimental determined mass. In this way the last C-terminal residue could be identified as an amidated histidine (Fig. 1B). The peptide was found cationic (pI = 10.24; 8/25 basic residues; 2/25 acidic residues).
Fig. 1.
Purification and AA sequence determination of Hl15-1-1. A) The filtered extract of the telson of Heterometrus laoticus was first separated on a Waters Symmetry C18 column. Fraction 15 (*) was then further separated on a Waters XTerra Phenyl column. Three pure fractions were obtained containing, in order of retention time, Hl15-1-1 (*), Hl15-1-2 and Hl15-1-3. Synthetic Hl15-1-1 was chromatographically analyzed under the same conditions as the venom and found to elute at the same retention time as the native toxin. B) Amino acid sequence of Hl15-1-1 obtained by Edman degradation. The last amino acid was deduced by the mass difference between the calculated mass and the experimental mass. Above the sequence the predicted secondary structure is shown (C = coil, H = helix, S = strand and T = turn). Under the sequence the predicted disulfide bridge pattern is drawn.
By secondary structure predictions, it could be revealed that amino acids 10–22 form an α-helix as do residues 4 and 5 (Fig. 1B). Furthermore, residues 2–7 (CKKECS) are also found in κ-hefutoxin3 (residues 18–23) (Fig. 3A) in which they align part of the second α-helix (27,28). Hence, it is most likely that Hl15-1-1 consists of two parallel helices encompassing residues 2–7 (helix I) and 10–22 (helix II). Finally, the peptide contains 4 cysteine residues. Predictions showed that the most plausible disulfide bridging is between C2–C19 and C6–C15 (Fig. 1D). Therefore, Hl15-1-1 adopts the cysteine-stabilized helix-loop-helix (CS α/α) fold as found in members of the scorpion VGPC blocker family κ-KTx.
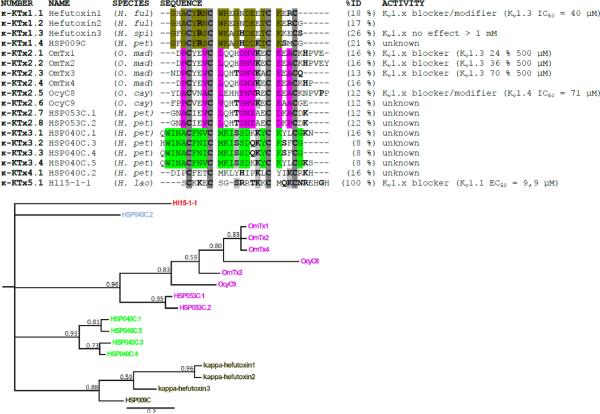
Fig. 3.
Multiple sequence alignment of Hl15-1-1 with the other existing κ-KTx family members. %ID is the identity score of the peptide compared to Hl15-1-1. The alignment is based on the cysteine residues (bold and dark gray background). The bold residues in a light gray background are amino acids identical to Hl15-1-1 and other κ-KTx toxins. Bold residues represent those amino acids having a side chain with a similar chemical nature. For each suggested κ-KTx subfamily, those residues having the same chemical properties are colored. The multiple sequence alignment has been used for constructing a tree topology by Bayesian inference. The consensus tree shown supports the suggested division of the κ-KTx family. The posterior probability indicating the clade credibility is shown at each node. The scale bar gives the expected changes per amino acid site. Species names are H. ful (Heterometrus fulvipus), H. spi (Heterometrus spinnifer), O. mad (Opisthacanthus madagascariensis), O. cay (Opisthacanthus cayaporum), H. pet (Heterometrus petersii) and H. lao (Heterometrus laoticus).
Furthermore, Hl15-1-1 was made synthetically and the obtained synthetic peptide eluted at the same retention time as the mature peptide (Fig 1C).
cDNA sequence
Using 3'RACE and 5'RACE strategies, the cDNA sequence of Hl15-1-1 could be obtained (Fig. 2). The clone consisted of an ORF, a 5' UTR and 3' UTR (Fig. 2). The polyadenylation signal AATAA was found 40 nucleotides downstream of the stop codon. The ORF encoded a peptide consisting of 72 amino acids. The deduced amino acid sequence starts with a N-terminal 23-residue signal peptide, followed by a 22-residue propeptide and ends with a C-terminal 27-residue mature peptide (Hl15-1-1) (Fig. 2). The propeptide was found to be rich in negatively charged amino acids (2D+6E/22 AA) and ended in the typical processing signal KR (Fig. 2). The deduced mature peptide was found similar as the biochemical purified one. However it contained C-terminally two extra residues GR, which are removed during posttranslational processing resulting in an amidated histidine (H).
Fig. 2.

cDNA sequence of Hl15-1-1. The nucleotides (nt) in capital letters are those of the ORF, the lower case nt are the 5' and 3' UTR. The polyadenylation signal is shown in italic and underlined. The deduced amino acid (AA) sequence consist of three domains: a signal peptide (italic), a propeptide (underlined) and the mature peptide (bold).
Sequence comparison and phylogenetic analysis of the κ-KTx family
Both the amino acid sequence and cDNA sequence were used to carry out a BLAST search. While for the cDNA sequence no hits were found, the amino acid sequence returned one hit. Hl15-1-1 shows 77% similarity with OcyKTx3 (P86117) isolated from the venom of Opisthacanthus cayaporum (21). However, from this toxin only the N-terminal 13 amino acids (SCKRECSGSKRQK) are known.
Since no similarity was found with any other known toxin and taking into account the predicted secondary structure and activity towards VGPCs (vide infra), an alignment of Hl15-1-1 with all known scorpion toxins belonging to the κ-KTx family of VGPC scorpion toxins was conducted (Fig. 3). The highest identity received was 26 %. Consequently, based on the criteria published by Tytgat et al. for the α-KTx family of VGPC scorpion toxins, Hl15-1-1 is the member of a new subfamily within the κ-KTx family (34). Recently, some new members have been added to the κ-KTx family, however without a clear classification. Therefore, a phylogenetic analysis of this family was performed (Fig. 3). The tree topology was reconstructed by Bayesian inference, on the basis of the above mentioned alignment of all κ-KTx family members. The obtained consensus tree and posterior probabilities support the clear division of the family into five subfamilies (Fig. 3). The first subfamily κ-KTx1 consist of 4 members being the hefutoxins 1, 2 and 3 and HSP009C. κ-KTx2 is formed by the 8 toxins OmTx 1–4, OcyC8 and 9 and HSP053C.1 and 2. Furthermore, the 4 peptides HSP040C.1 and 3–5 make up κ-KTx3. Finally, HSP040C.2 and Hl15-1-1 form each respectively the subfamilies κ-KTx4 and κ-KTx5.
Electrophysiological characterization of Hl15-1-1
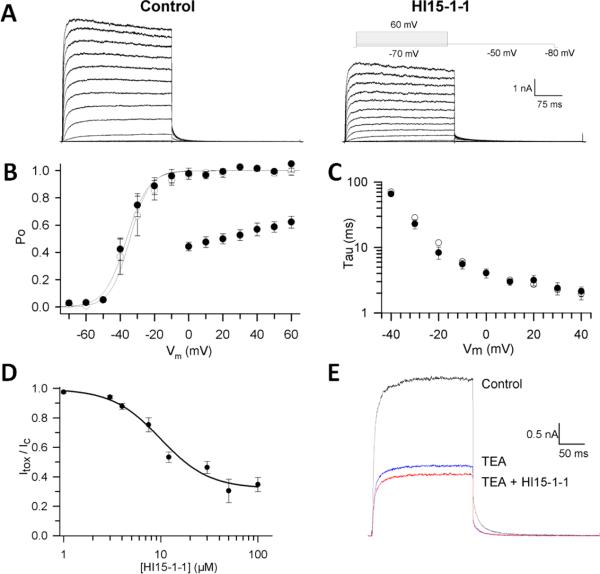
Because Hl15-1-1 was found to block Kv1.3 channels in X. laevis oocytes and because of the known activity of κ-KTx family members as VGPC toxins, the peptide was tested on a wider range of K+ channels expressed in mammalian cell lines. First, the activity of Hl15-1-1 was studied on other members of the Shaker-related Kv1.x subfamily being Kv1.1 – Kv1.6 and Kv1.8. At a concentration of 30 μM, Hl15-1-1 inhibited the tested channels as follow: Kv1.1 53 ± ? % (n =); Kv1.2 1 ± ? % (n =); Kv1.3 20 ± ? % (n =); Kv1.4 13 ± ? %(n =); Kv1.5 1± ? % (n =); Kv1.6 60 ± ? % (n =) and Kv1.8 6 ± ? % (n =) (Fig. 4). Kv1.1 and Kv1.6 were found to be the most sensitive. For Kv1.1, a dose response curve was constructed using the Hill equation (Fig. 5D). The EC50 was 9.9 ± 1.6 μM with a Hill-coefficient of 1.75 ± 0.53 (n = 3–7). Towards higher toxin concentrations, the dose-response curve shows saturation at ± 65% inhibition of the K+ current, indicating that the toxin binds the channel with a low efficacy. Furthermore, at higher membrane potentials, the reduction of the K+ current by the toxin was decreased from 56 ± 3% at 0 mV till 38 ± 4% at +60 mV (n = 4), indicating a clear voltage-dependent mode of action (Fig. 5B). Since some members of the κ-KTx family besides inhibiting the K+ currents also modify the gating kinetics, the effect of Hl15-1-1 on the activation was evaluated (Fig. 5A) (22,27). As shown in Fig. 5B and C, 9.9 μM Hl15-1-1 did not shift the voltage-dependence of activation, nor did it shift the time constants of activation.
Fig. 4.
Bar diagram showing the percentage block of 30 µM Hl15-1-1 on the different K+ channels tested (mean ± SEM). From each Shaker-related VGPC, a current trace in control (black) and in the presence of 30 μM Hl15-1-1 (red) is shown. Current recordings were elicited by a depolarizing step (0 mV for Kv1.3; +10 mV for Kv1.1 and Kv1.6; +30 mV for Kv1.5; +40 mV for Kv1.8 and + 60mV for Kv1.2) from a holding potential of 80 mV followed by a repolarization to −50 mV. Scale bars are given as inset.
Fig. 5.
Electrophysiological characterization of Hl15-1-1 on Kv1.1. A) Kv1.1 current recordings elicited by 300-ms steps to various potentials (−50 mV to +60 mV) in control conditions (left) and in presence of 9.9 μM Hl15-1-1 (right). B) Voltage dependence of activation. Activation curves were obtained by plotting the normalized tail currents as a function of the prepulse potential. The solid lines represent the average Boltzmann fits in control conditions (open symbols) and in presence of 9.9 μM Hl15-1-1 (closed symbols). Additionally, the voltage dependence of block is shown by plotting the normalized currents at the end of the prepulse in the presence of 9.9 μM Hl15-1-1 as a function of the prepulse voltage (scatter plot). C) Activation time constants derived from mono-exponential fits to the raw current traces were plotted as a function of applied voltage in control conditions (open symbols) and in presence of 9.9 μM Hl15-1-1 (closed symbols). D) Concentration dependence of inhibition by Hl15-1-1 obtained from the normalized current suppression at +40 mV as a function of the Hl15-1-1 concentration, fitted with the Hill equation (solid line). E) Competition between TEAext and Hl15-1-1. Shown are the currents for 250-ms steps to +10 mV for control, steady-state block by 0.4 mM TEAext and the subsequent effect of 9.9 μM Hl15-1-1 together with 0.4 mM TEAext.
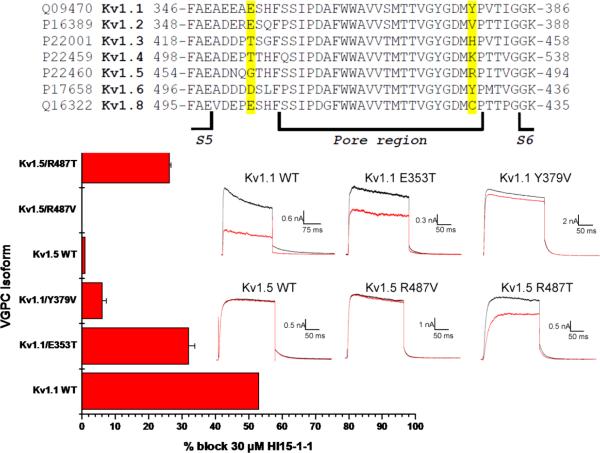
In order to test whether Hl15-1-1 is an external pore blocker, a competition experiment with tetraethylammonium (TEA) was performed. First, 0.4 mM (IC50) TEA was applied to Kv1.1 and resulted in a reduction of the K+ current with 55 ± 2 % (n = 6) (Fig. 5E). Next, a mixture of 0.4 mM TEA and 9.9 μM Hl15-1-1 was added. The observed additional block of only 15.5 ± 2.3 % (n = 6) indicates a clear competition of both ligands (Fig. 5E). For that reason, it can be concluded that Hl15-1-1, just as TEA, inhibits Kv1.1 by binding to the pore and occluding it. Knowing the domain on Kv1.1 for interaction with Hl15-1-1, an alignment of the pore region of all tested Kv1.x channels was made (Fig. 6). The most possible dramatic changes in amino acids between the sensitive (Kv1.1 and Kv1.6) and insensitive (Kv1.2, Kv1.3, Kv1.4, Kv1.5 and Kv1.8) VGPCs are a) from a negatively charged residue E346/D503 (Kv1.1/Kv1.6) into a neutral residue G501 (Kv1.5) and b) from a polar residue T425/T505 (Kv1.3/Kv1.4) or aromatic residue Y379/Y429 (Kv1.1/Kv1.6) into a nonpolar residue V381/C428 (Kv1.2/Kv1.8) or positively charged residue H451/K531/R487 (Kv1.3/Kv1.4/Kv1.5) (Fig. 6). In order to test the importance of these amino acids for interaction with Hl15-1-1, two mutant Kv1.1 channels Kv1.1 E353T and Kv1.1 Y379V were constructed. Compared to wildtype Kv1.1 channels, 30 μM Hl15-1-1 induced only 32 ± 1.9 % (n = 3) inhibition of the K+-efflux through Kv1.1 E353T and 6.1 ± 1.3 % (n = 3) through Kv1.1 Y379V. To further study the importance of residue Y379, the corresponding R487 in Kv1.5 was mutated into a valine keeping the channel insensitive to 30 μM Hl15-1-1 (Fig. 6). However, when the same amino acid R487 was mutated into a polar threonine, the K+ current was inhibited by 26.3 ± 0.5 % (n = 3) using 30 μM toxin (Fig. 6).
Fig. 6.
Multiple sequence alignment of the pore region of the tested Shaker-related VGPCs. The amino acids considered most important for the selectivity of Hl15-1-1 towards Kv1.1 and Kv1.6 are highlighted in yellow. Those residues have been mutated resulting in the channels Kv1.1 E353T, Kv1.1 Y379V, Kv1.5 R487V and Kv1.5 R487T. The bar diagram shows the inhibition caused by 30 μM Hl15-1-1 on the wildtype (WT) and mutated channels (mean ± SEM). Current recordings of Kv1.1 and Kv1.5 wildtype and mutant channels were elicited by a depolarizing step to +10 mV from a holding potential of −80 mV followed by a repolarization to −50 mV. Raw current traces are shown in control (black) and after application of 30 μM Hl15-1-1 (red). Scale bars are given as inset.
Hl15-1-1 was also tested on members of the Shaw-related Kv3.x and Shal-related Kv4.x subfamilies. However, 30 μM of the toxin did not affect the channels (Fig. 4). Additionally, the activity of Hl15-1-1 was evaluated on the “small conductance” Ca2+-activated K+-channel KCa2.3 and the “intermediate-conductance” Ca2+-activated K+-channel KCa3.1. Once more, 30 μM toxin did not alter the K+ current through these channels (Fig. 4).
DISCUSSION
The sting of the Asian scorpion Heterometrus laoticus (Scorpionidae) causes rarely envenomations and therefore the venom composition remains poorly known (19). So far only a member of the VGPC scorpion toxin β-KTx3 subfamily has been isolated (20). Separation of an extract of a telson from H. laoticus led to the identification of three peptidyl toxins with VGPC inhibiting properties (Fig. 1A). Through a combination of amino acid sequencing by Edman degradation and molecular cloning techniques, the sequences Hl15-1-1, Hl15-1-2 and Hl15-1-3 could be revealed. Hl15-1-1 showed the most unique sequence (Fig. 1B) and was consequently chosen for further functional characterization. Hl15-1-2 and Hl15-1-3 both belong to the well known VGPC scorpion toxin α-KTx6 subfamily (35). Hl15-1-2 was found to have an amino acid sequence similar to HsTx1 (P59867) from Heterometrus spinifer (36). However, instead of Y21 it bears a histidine at that position due to a point mutation on the DNA level. Hl15-1-3 (ISCTGSKQCYDPCKRKTGCPNAKCMNKSCK CYGC) showed79 % similarity with Maurotoxin (P80719) from Scorpio maurus (37). Interestingly, during the molecular cloning of Hl15-1-3, also Spinoxin (P84094) was found. Spinoxin was for the first time isolated from the venom of the scorpion Heterometrus spinnifer and was recently found through a transcriptomic analysis of the venom gland of Heterometrus petersii (14). Although all above mentioned scorpions belong to the same genus and share the same habitat/prey, this is to the best of our knowledge the first time the same peptidyl toxin has been isolated from three different scorpion species. Therefore further studies on this particular toxin, may give important information on scorpion toxin evolution and ecology.
Although, the BLAST analysis of the cDNA sequence of Hl15-1-1 did not reveal any hits, the translation of the ORF into a precursor peptide containing the three domains signal peptide, propeptide and mature peptide (Fig. 2) is quite interesting. Such arrangement is common in transcripts encoding peptidyl ion channel toxins from cone snails and spiders, but is rare among the scorpion toxin transcripts (38). So far, a propeptide between the signal peptide and the mature toxin has only been found in members of the β-KTx family (39), the κ-KTx family (14,17), the calcines (40) and in the DDH-family (18). All other scorpion ion channel toxin precursors bear only a signal peptide immediately followed by the mature peptide. Similar to most other propeptides found in animal ion channel toxin transcripts, the propeptide of Hl15-1-1 is rich in acidic amino acids. Many secretory peptides are synthesized by the cell as inactive precursors containing a propeptide. This ancient mechanism enables cells to regulate the level of specific bioactive peptides. Several functions have been found for these prodomains such as mediating correct folding as intramolecular chaperones, mediating transport/localization, regulation of activity and quality control of folding (25). What the exact function of the prodomains in ion channel toxin precursors is and why they only occur in certain classes of these toxins remains to be elucidated.
Blast results of the amino acid sequence and of Hl15-1-1 showed that the toxin has a new, unique sequence that has not been seen before during scorpion venom analysis. Based on the amino acid sequence, however, one single hit was found. This hit is the partially sequenced peptide OcyKTx3 isolated from the venom of Opisthacanthus cayaporum (21). The sequenced N-terminal 13 amino acids (SCKRECSGSKRQK) show 77% similarity with those of Hl15-1-1 and the not identical amino acids (not bold) have a side chain with similar chemical nature (K/R and Q/T). As such it would not be surprising that upon full sequencing of the amino acid sequence an almost identical peptide as Hl15-1-1 will be found. Schwartz et al. however found that OcyKTx3 bears 87% identity to the 6 – 13 region (CSGSCRQK) of OsK2 (P83244) (21). OsK2 was isolated from the venom of the Buthid scorpion Ortochirus scrobiculosus and classified as VGPC scorpion toxin α-KTx 13.2 (41). Alignment of Hl15-1-1 with OsK2 reveals 40 % similarity (data not shown). However, OsK2 contains six cysteines and most probably adapts just as Tc1 (P83243) (α-KTx 13.1 - Tityus cambridgei) the cystine-stabilized α/β (CSα/β) scaffold (41,42). Since Hl15-1-1 only contains 4 cysteines and most likely adapts the cystine-stabilized helix-loop-helix (CS α/α) fold, it should definitely not be classified within this α-KTx subfamily.
Based on the presence of 4 cysteines in Hl15-1-1 and the secondary structure prediction in addition to the similarity of residues 2-–7 (CKKECS) with residues 18–23 (Fig 3A) in Hefutoxin3, it was assumed that Hl15-1-1 adapts the CS α/α scaffold. Together with the activity on VGPCs, this suggests the classification of Hl15-1-1 into the VGPC scorpion toxin κ-KTx family. The highest identity obtained during alignment of Hl15-1-1 with all known members was 26% (Fig. 3). The only residues found identical with some other κ-KTx toxins were S9, K13 and K18 whereas K4, R10, T12, Q17, N20, R21 and G24 have a side chain with a similar chemical nature as other residues found at the same position in other κ-KTx toxins (Fig. 3). Remarkable, is the somewhat conserved basic residue at position 13 (Hl15-1-1 numbering). The function of this residue is not completely clear, however in some κ-KTx toxins it has been suggested to form part of the functional dyad together with an aromatic residue (if present) in the N-terminal (22). K4 in Hl15-1-1 corresponds to R6 in Hefutoxin1 (Fig. 3). This residue has been shown to be important for interaction with Shaker-related VGPCs (43). It might therefore be possibe that K4 in Hl15-1-1 plays a similar role. However, this should be further investigated through mutagenesis.
Because of the low similarity between Hl15-1-1 and the other κ-KTx family members, and based on the criteria of classification proposed by Tytgat et al. (34), Hl15-1-1 should be classified into a new κ-KTx subfamily. So far only two subfamilies have been recognized in the κ-KTx family. These are κ-KTx-subfamily 1 and 2 respectively containing hefutoxin1-3 and OmTx1-4 and OcyC8 and 9 (22). From the venom gland of the scorpion Heterometrus petersii, new κ-KTx toxins have been cloned without being properly classified (14). Only the homology between HSP009C and the hefutoxins and HSP053C with the OmTxs has been noted. Furthermore, HSP040C was interestingly labeled as an intermediate between the two κ-KTx subfamilies. Therefore, a phylogenetic study was performed to evaluate the relationship between all existing κ-KTx-toxins. As shown in Fig. 3, the consensus tree obtained by Bayesian inference clearly suggests a division of the κ-KTx family into 5 subfamilies. The first subfamily κ-KTx1 is formed by the hefutoxins 1-3 and HSP009C. It has a consensus signature GX(Np)C(Ar)R(Po)CW(Ba)XGXD(Ac)XTCKXX C in which Np, Ar, Po, Ba and Ac respectively stand for non-polar residue, aromatic residue, polar residue, basic residue and acidic residue. X represents any amino acid. Hefutoxin1 (and 2) was found to block Kv1.2 and Kv1.3 channels and additionally slow the activation kinetics of Kv1.3 (27). In silica mutation studies and docking/MD simulations showed that Y5, R6, W9, R10 and K19 in hefutoxin1 and Y400, D402, D386, G380 (respectively Y447, D449, D433, G427 in Fig. 6 Kv1.3 numbering) in each subunit of the Shaker-related VGPCs and H404 (H451) in Kv1.3, as well as the residue E378 (E425) in Kv1.1 and Kv1.2 are the key elements for the toxin-channel interactions (43). Y5 and K19 form the functional dyad commonly found, but not required for scorpion VGPC blockers (44). The functional dyad is usually formed by a lysine and an aromatic residue separated from each other by 6–7 Å. Hefutoxin3 was found inactive on Shaker-related channels because of an additional lysine at position 20 (28). The activity of HSP009C still has to be verified; besides a lysine at position 19, it also bears a lysine at position 16 which is aligned with the lysine forming the functional dyad in OmTx1 (29). It would be interesting to see which lysine participates in forming the functional dyad. Additionally, the role of a hydrophilic residue (S) flanking K20 needs also to be further investigated. The κ-KTx2 subfamily consists of the OmTx1-4, OcyC8-9 and HSP053.1–2 (Fig. 3). A consensus sequence XX(Np)CXX(Np)C(Np)XXX(Np)N(Np)XXC(Ac)X(Np)CX can be recognized. OmTx1-4 were found to interact with Kv1.1, Kv1.2 and Kv1.3 (29). Most likely Y4 and K15 in OmTx1, 2 and 4 form the functional dyad. Interestingly, Chagot et al. claim that the dyad is not present in OmTx3 because of the lack of the aromatic residue on the first helix (29). However, in the amino acid sequence submitted to the UniProtKB database (http://www.uniprot.org/uniprot/P0C1Z4), there is a tyrosine present at position 5 similar as to the other OmToxins. The confusion is provoked by the appearance of two sequences for OmTx3 in the manuscript. In accordance with the publications by the group of L. Possani and E. Schwartz (17,22), the sequence submitted to the database was used for the analysis presented here. Recently, OcyC8 was functionally characterized and found to block Kv1.1 and Kv1.4 channels (22). Additionally, it also altered the activation kinetics similar to hefutoxin1. Interestingly, the toxin only affected the human channels expressed in mammalian cells and not the rat homologues expressed in X. laevis oocytes. This toxin does not contain the functional dyad. Furthermore through docking simulations with Kv1.2, N24 and K23 were found to interact with D348 (D363 in Fig. 6) of Kv1.2. This residue is highly conserved among all members of the Shaker-related K+ channel subfamily. OcyC9, HSP05053.1 and 2 still need to be functionally characterized. Interestingly, just as OcyC9 they are lacking the functional dyad. The third κ-KTx subfamily consists of HSP040C.1, 3, 4 and 5 and has as consensus XWINACFN(Np)CMKISSDXKYCKXXCG (Fig. 3). All four peptides have the functional dyad, however it might be formed as in hefutoxin1 (F7/K21) or as in OmTx1 (F7/K18). Furthermore, both HSP040C.1 and 5 contain an additional lysine flanking K18. This might cause insensitivity towards VGPCs as observed with hefutoxin3. Finally, HSP040C.2 and Hl15-1-1 have both quite unique sequences showing very low similarity with the other κ-KTx family members. As a result, they each respectively represent the κ-KTx subfamilies 4 and 5 (Fig. 3). As a κ-KTx subfamily 3 member, HSP040C.2 does also contain the functional dyad (F5/K16) similar as the one observed in OmTx1. Although κ-KTx toxins are classified according to their activity on VGPCs it has to be stressed that their IC50s range from 10 – 70 μM and concentrations up to 500 μM are sometimes needed to achieve an effect (Fig. 3). Therefore it is very unlikely that VGPCs are the primary target of these peptides. Testing κ-KTx for other activities than blocking VGPCs is a major point for further research. In this manuscript Hl15-1-1 has been tested on Ca2+ activated potassium channels, likewise Srinivasan et al. tested hefutoxin1 on two-pore background K+ channels, pacemaker channels and VGSCs and Camargos et al. evaluated the activity of OcyC8 on VGSCs and tested its bradikinin potentiating and antibacterial activity (22,27). However, the toxins did not show any effect.
The sharing of one scaffold between (till now) solely two scorpion families (Scorpionidae and Liochelidae) may support their evolutionary relationship and taxonomical grouping within the superfamily Scorpionoidea (7,8,45). Therefore, it would be highly interesting to investigate the occurrence of similar peptidyl toxins in the other scorpion families belonging to this superfamily.
The CS α/α fold is so far scarcely found among all known scorpion toxins. However, VGPCs toxins adapting the same scaffold have been isolated from the cone snails Conus floridanus floridensis and C. villepinii (46). Furthermore, the cytotoxic viscotoxins isolated from the European mistletoe (Viscum album) directly interact with cell membranes and also fold in a similar way (47). Baring in mind the explanation for the common evolutionary origin of the inhibitor cystine knot peptides, it would not be surprising that the CS α/α toxins found in animals have a common ancestor and that divergent evolution took place (40). Since related functions may act as indicators of common ancestry, this supports the divergence evolution in animals, but indicates that the structural similarity observed between animal and plant CS α/α toxins might be the result of convergent evolution. However, this hypothesis needs to be further proven by analyzing the precursor organization and gene structures which are not in all cases elucidated yet.
It should be noted that besides the κ-KTx family, other scorpion venom peptides containing 4 cysteine residues have been isolated (17,18,48,49). These have not been included in the present phylogenetic analysis because first of all there is no information on their pharmacological activity. They are known to act against insects, but the molecular target has not been found yet. For sure, they do not affect voltage gated sodium, potassium and calcium channels (18) (unpublished data of U1-LITX-Lw1a on VGICs expressed in Xenopus laevis oocytes, obtained in the lab of Toxicology (KULeuven)). Secondly, they contain a propeptide that, however, atypically does not end in one of the well known processing signals recognized by the evolutionary conserved dibasic- and monobasic-specific Ca2+-dependent serine proteases (so called subtilisin/kenin-like proprotein convertases) (50). Finally, the tertiary structure of U1-LITX-Lw1a (2KXJ-A) has been recently solved (18). It adapts a novel scorpion toxin scaffold being the disulfide-directed β-hairpin (DDH). This finding totally supports the exclusion of these intriguing scorpion peptides from the κ-KTx family. We suggest calling this family the “DDH-family” till the molecular target has been found and they can be properly classified.
The activity of Hl15-1-1 has been evaluated on VGPCs of the Shaker-, Shaw- and Shal- related subfamilies as well as on “small conductance” and “intermediate-conductance” Ca2+-activated K+-channels. From the channels tested, Kv1.1 and Kv1.6 were the most sensitive. The obtained EC50 on Kv1.1 (~ 10 μM) makes it the most potent κ-KTx toxin so far. The finding of a Hill-coefficient close to two is interesting. A similar Hill coefficient was obtained for OcyC8 on Kv1.1 and Kv1.4 (22). This could explain the observation that one molecule OcyC8 binds to one subunit of the K+ channel leaving the channel pore unbarred. A second molecule OcyC8 binds than to another channel subunit and blocks the pore by toxin-toxin interactions. This mode of interaction between toxins and channel may be the same for Hl15-1-1 and as suggested by Camargos et al. explain the high concentrations needed for activity. However, the authors also suggest that OcyC8 interacts with the outer region of the channel, close to the lipid environment. In this way, the absence of block in X. laevis oocytes by differences in cell membrane composition can be explained. However, Hl15-1-1 blocks both VGPCs in mammalian cells as in oocytes (data not shown). Whether Hl15-1-1 truly follows this mode of interaction, will need to be addressed through tertiary structure studies of the toxin. To the best of our knowledge, the Hill coefficient for hefutoxin1 is not mentioned, however, the docking simulations done do not seem to support the binding of two toxin molecules to the channel (27,43). In contrast to hefutoxin1 and OcyC8, Hl15-1-1 did not alter the activation kinetics of Kv1.1 (Fig. 5). Furthermore, it is the first κ-KTx toxin showing a voltage-dependence of block. This can be explained to the much higher content of basic residues in Hl15-1-1 (pI 10.24 vs 4.92 of OcyC8) causing repulsions at higher membrane voltages. The cationic nature of Hl15-1-1 may as well explain the observed maximal obtained inhibition of 65 %. TEA competition experiments showed that Hl15-1-1 is a pore blocker, so this behavior cannot be explained through “turret block” as observed with the ErgToxins on HERG channels (51). Most probably electrostatic repulsion between toxin and channel causes the low efficacy. Through mutagenesis experiments, E353 and Y379 in the Kv1.1 pore were demonstrated as important interaction points for toxin binding (Fig. 6). Especially, mutations in Y379 have a drastic effect and the requirement of an amino acid with a side chain aromatic/polar in chemical nature has been demonstrated. Binding of scorpion toxins to VGPCs involves a combination of hydrophobic, electrostatic and hydrogen bonding interactions (51). Since Hl15-1-1 mainly consists of positively charged and polar residues, the importance of E/D353 (electrostatic) and Y379 (hydrogen binding) is in fact not surprising and explains the toxin's preference for Kv1.1 and Kv1.6. It has been shown that R6 in hefutoxin1 interacts with E353 in Kv1.1 and Kv1.2 (43). Hl15-1-1 has at the same position a lysine. Therefore, it would be possible that this lysine interacts with E353 and as suggested for hefutoxin1, is important for recognition and correct positioning of the toxin on Kv1.1. The selectivity towards Kv1.1 and Kv1.6 may be explained in addition to the presence of V381, by the presence of an arginine flanking E355 in Kv1.2 repulsing the positively charged toxin. Finally, K23 and N24 in OcyC8 interact with D363 in Kv1.2. This residue is highly conserved in the pore region of all Shaker-related VGPCs studied and as such does not explain the selectivity of OcyC8 towards Kv1.1 and Kv1.4, but nevertheless stabilizes the toxin on the channels (22). Hl15-1-1 has also an aspargine at position 20 flanked by a basic residue at position 21. Hence, it is possible that these residues interact as well with D361 in Kv1.1 and Kv1.6 and stabilize the toxin interaction. Although, mutational studies have been done in the pore region of Kv1.1, mutagenesis of the toxin should be performed in the future to confirm the above stated hypotheses.
In conclusion, the study presented here proves the relevance of focusing on venom analysis of non-Buthid scorpions. A peptidyl toxin, Hl15-1-1, with unique sequence has been isolated from the venom of Heterometrus laoticus. The functionality of the peptide has been extensively studied on VGPCs demonstrating its pore blocking mechanism through interaction with E353 and Y379 in Kv1.1 and Kv1.6. Furthermore, the VGPC scorpion toxin κ-KTx family has been revised and a new classification proposed with Hl15-1-1 representing subfamily 5. Finally, although a relative high EC50 value, Hl15-1-1 can be further used as molecular tool for studying the physiology of VGPCs. It might as well serve as an example scaffold for designing novel, potential drug candidates.
Acknowledgments
We are thankful to Maria L. Garcia, Merck Research Laboratories, USA for sharing Kv1.3. We are also grateful to L. Prendini, American Museum of Natural History, NY, USA for his help in the identification of the scorpions. We would like to thank Sarah Debaveye and Luc Vanden Bosch for technical assistance. TV and EC are respectively research assistant and postdoctoral fellow of the Fund for Scientific Research (FWO)-Flanders.
REFERENCES
- 1.Yu FH, Yarov-Yarovoy V, Gutman GA, Catterall WA. Pharmacol Rev. 2005;57:387–395. doi: 10.1124/pr.57.4.13. [DOI] [PubMed] [Google Scholar]
- 2.Harmar AJ, Hills RA, Rosser EM, Jones M, Buneman OP, Dunbar DR, Greenhill SD, Hale VA, Sharman JL, Bonner TI, Catterall WA, Davenport AP, Delagrange P, Dollery CT, Foord SM, Gutman GA, Laudet V, Neubig RR, Ohlstein EH, Olsen RW, Peters J, Pin JP, Ruffolo RR, Searls DB, Wright MW, Spedding M. Nucleic Acids Res. 2009;37:D680–D685. doi: 10.1093/nar/gkn728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wulff H, Castle NA, Pardo LA. Nat Rev Drug Discov. 2009;8:982–1001. doi: 10.1038/nrd2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wickenden AD. Pharmacol Therapeut. 2002;94:157–182. doi: 10.1016/s0163-7258(02)00201-2. [DOI] [PubMed] [Google Scholar]
- 5.Brownell P, Polis G. Scorpion Biology and Research. Oxford University Press Inc; New York: 2001. [Google Scholar]
- 6.Fet V, Sissom WD, Lowe G, Braumwalder ME. Catalog of the Scorpions of the World (1758 – 1998. The New York Entomological society; New York: 2000. [Google Scholar]
- 7.Stockmann R, Ythier E. Scorpions of the World. N.A.P. Editions Verrièresle-Buisson; France: 2010. [Google Scholar]
- 8.Prendini L. Cladistics. 2005;21:446–494. doi: 10.1111/j.1096-0031.2005.00073.x. [DOI] [PubMed] [Google Scholar]
- 9.Polis GA. The Biology of Scorpions. Stanford University Press; Stanford, California: 1990. [Google Scholar]
- 10.Mouhat S, Andreotti N, Jouirou B, Sabatier JM. Curr Pharm Design. 2008;14:2503–2518. doi: 10.2174/138161208785777441. [DOI] [PubMed] [Google Scholar]
- 11.de la Vega RCR, Schwartz EF, Possani LD. Toxicon. 2010;56:1155–1161. doi: 10.1016/j.toxicon.2009.11.010. [DOI] [PubMed] [Google Scholar]
- 12.He QY, He QZ, Deng XC, Yao L, Meng E, Liu ZH, Liang SP. Nucleic Acids Res. 2008;36:D293–297. doi: 10.1093/nar/gkm832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miyashita M, Otsuki J, Hanai Y, Nakagawa Y, Miyagawa H. Toxicon. 2007;50:428–437. doi: 10.1016/j.toxicon.2007.04.012. [DOI] [PubMed] [Google Scholar]
- 14.Ma YB, Zhao Y, Zhao RM, Zhang WP, He YW, Wu YL, Cao ZJ, Guo L, Li WX. Proteomics. 2010;10:2471–2485. doi: 10.1002/pmic.200900763. [DOI] [PubMed] [Google Scholar]
- 15.Ma YB, Zhao RM, He YW, Li SR, Liu J, Wu YL, Cao ZJ, Li WX. Bmc Genomics. 2009;10 doi: 10.1186/1471-2164-10-290. - [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schwartz EF, Diego-Garcia E, de la Vega RCR, Possani LD. Bmc Genomics. 2007;8 doi: 10.1186/1471-2164-8-119. - [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Silva ECN, Camargos TS, Maranhao AQ, Silva-Pereira I, Silva LP, Possani LD, Schwartz EF. Toxicon. 2009;54:252–261. doi: 10.1016/j.toxicon.2009.04.010. [DOI] [PubMed] [Google Scholar]
- 18.Smith JJ, Hill JM, Little MJ, Nicholson GM, King GF, Alewood PF. Proc Natl Acad Sci U S A. 2011 doi: 10.1073/pnas.1103501108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Uawonggul N, Chaveerach A, Thammasirirak S, Arkaravichien T, Chuachan C, Daduang S. J Ethnopharmacol. 2006;103:201–207. doi: 10.1016/j.jep.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 20.Uawonggul N, Thammasirirak S, Chaveerach A, Arkaravichien T, Bunyatratchata W, Ruangjirachuporn W, Jearranaiprepame P, Nakamura T, Matsuda M, Kobayashi M, Hattori S, Daduang S. Toxicon. 2007;49:19–29. doi: 10.1016/j.toxicon.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 21.Schwartz EF, Camargos TS, Zamudio FZ, Silva LP, Bloch C, Caixeta F, Schwartz CA, Possani LD. Toxicon. 2008;51:1499–1508. doi: 10.1016/j.toxicon.2008.03.029. [DOI] [PubMed] [Google Scholar]
- 22.Camargos TS, Restano-Cassulini R, Possani LD, Peigneur S, Tytgat J, Schwartz CA, Alves EM, de Freitas SM, Schwartz EF. Peptides. 2011 doi: 10.1016/j.peptides.2011.05.017. [DOI] [PubMed] [Google Scholar]
- 23.Chomczynski P, Sacchi N. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 24.Bendtsen JD, Nielsen H, von Heijne G, Brunak S. J Mol Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- 25.Duckert P, Brunak S, Blom N. Protein Eng Des Sel. 2004;17:107–112. doi: 10.1093/protein/gzh013. [DOI] [PubMed] [Google Scholar]
- 26.Cheng J, Randall AZ, Sweredoski MJ, Baldi P. Nucleic Acids Res. 2005;33:W72–76. doi: 10.1093/nar/gki396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Srinivasan KN, Sivaraja V, Huys I, Sasaki T, Cheng B, Kumar TKS, Sato K, Tytgat J, Yu C, San BCC, Ranganathan S, Bowie HJ, Kini RM, Gopalakrishnakone P. J Biol Chem. 2002;277:30040–30047. doi: 10.1074/jbc.M111258200. [DOI] [PubMed] [Google Scholar]
- 28.Nirthanan S, Pil J, Abdel-Mottaleb Y, Sugahara Y, Gopalakrishnakone P, Joseph JS, Sato K, Tytgat J. Biochem Pharmacol. 2005;69:669–678. doi: 10.1016/j.bcp.2004.10.018. [DOI] [PubMed] [Google Scholar]
- 29.Chagot B, Dai L, Pil J, Tytgat J, Nakajima T, Corzo G, Darbon H, Ferrat G. Biochem J. 2005;388:263–271. doi: 10.1042/BJ20041705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huelsenbeck JP, Ronquist F. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 31.Vandendriessche T, Abdel-Mottaleb Y, Maertens C, Cuypers E, Sudau A, Nubbemeyer U, Mebs D, Tytgat J. Toxicon. 2008;51:334–344. doi: 10.1016/j.toxicon.2007.10.011. [DOI] [PubMed] [Google Scholar]
- 32.Wulff H, Miller MJ, Haensel W, Grissmer S, Cahalan MD, Chandy KG. Proc Natl Acad Sci USA. 2000;97:8151–8156. doi: 10.1073/pnas.97.14.8151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grissmer S, Nguyen AN, Aiyar J, Hanson DC, Mather RJ, Gutman GA, Karmilowicz MJ, Auperin DD, Chandy KG. Mol. Pharmacol. 1994;45:1227–1234. [PubMed] [Google Scholar]
- 34.Tytgat J, Chandy KG, Garcia ML, Gutman GA, Martin-Eauclaire MF, van der Walt JJ, Possani LD. Trends Pharmacol Sci. 1999;20:444–447. doi: 10.1016/s0165-6147(99)01398-x. [DOI] [PubMed] [Google Scholar]
- 35.Rodriguez de la Vega RC, Possani LD. Toxicon. 2004;43:865–875. doi: 10.1016/j.toxicon.2004.03.022. [DOI] [PubMed] [Google Scholar]
- 36.Lebrun B, Romi-Lebrun R, Martin-Eauclaire MF, Yasuda A, Ishiguro M, Oyama Y, Pongs O, Nakajima T. Biochem J. 1997;328(Pt 1):321–327. doi: 10.1042/bj3280321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rochat H, Kharrat R, Sabatier JM, Mansuelle P, Crest M, Martin-Eauclaire MF, Sampieri F, Oughideni R, Mabrouk K, Jacquet G, Van Rietschoten J, El Ayeb M. Toxicon. 1998;36:1609–1611. doi: 10.1016/s0041-0101(98)00153-6. [DOI] [PubMed] [Google Scholar]
- 38.Sollod BL, Wilson D, Zhaxybayeva O, Gogarten JP, Drinkwater R, King GF. Peptides. 2005;26:131–139. doi: 10.1016/j.peptides.2004.07.016. [DOI] [PubMed] [Google Scholar]
- 39.Possani LD, Diego-Garcia E, Abdel-Mottaleb Y, Schwartz EF, de la Vega RCR, Tytgat J. Cell Mol Life Sci. 2008;65:187–200. doi: 10.1007/s00018-007-7370-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhu SY, Darbon H, Dyason K, Verdonck F, Tytgat J. Faseb J. 2003;17:1765. doi: 10.1096/fj.02-1044fje. + [DOI] [PubMed] [Google Scholar]
- 41.Dudina EE, Korolkova YV, Bocharova NE, Koshelev SG, Egorov TA, Huys I, Tytgat J, Grishin EV. Biochem Bioph Res Co. 2001;286:841–847. doi: 10.1006/bbrc.2001.5492. [DOI] [PubMed] [Google Scholar]
- 42.Wang IR, Wu SH, Chang HK, Shieh RC, Yu HM, Chen CP. Protein Science. 2002;11:390–400. doi: 10.1110/ps.33402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Naderi-Manesh H, Zarrabi A. Proteins. 2008;71:1441–1449. doi: 10.1002/prot.21833. [DOI] [PubMed] [Google Scholar]
- 44.de la Vega RCR, Possani LD. Toxicon. 2004;43:865–875. doi: 10.1016/j.toxicon.2004.03.022. [DOI] [PubMed] [Google Scholar]
- 45.Prendini L. Cladistics. 2000;16:1–78. doi: 10.1111/j.1096-0031.2000.tb00348.x. [DOI] [PubMed] [Google Scholar]
- 46.Moller C, Rahmankhah S, Lauer-Fields J, Bubis J, Fields GB, Mari F. Biochemistry. 2005;44:15986–15996. doi: 10.1021/bi0511181. [DOI] [PubMed] [Google Scholar]
- 47.Sheldrick GM, Pal A, Debreczeni JE, Sevvana M, Gruene T, Kahle B, Zeeck A. Acta Crystallogr D. 2008;64:985–992. doi: 10.1107/S0907444908022646. [DOI] [PubMed] [Google Scholar]
- 48.Miyashita M, Matsushita N, Sakai A, Nakagawa Y, Miyagawa H. Toxicon. 2007;50:861–867. doi: 10.1016/j.toxicon.2007.06.014. [DOI] [PubMed] [Google Scholar]
- 49.Lazarovici P, Yanai P, Pelhate M, Zlotkin E. J Biol Chem. 1982;257:8397–8404. [PubMed] [Google Scholar]
- 50.Seidah NG, Day R, Marcinkiewicz M, Chretien M. Ann Ny Acad Sci. 1998;839:9–24. doi: 10.1111/j.1749-6632.1998.tb10727.x. [DOI] [PubMed] [Google Scholar]
- 51.Xu CQ, Zhu SY, Chi CW, Tytgat J. Trends in Pharmacological Sciences. 2003;24:446–448. doi: 10.1016/S0165-6147(03)00223-2. [DOI] [PubMed] [Google Scholar]