INTRODUCTION
Chronic obstructive pulmonary disease (COPD), a growing health concern, is the fourth leading cause of death in the world (1, 2). COPD is primarily due to irreversible airflow obstruction caused by small airways disease and emphysema. Small airways disease causes structural changes within the lung with loss of airways, airway wall thickening and luminal narrowing. Emphysema leads to destruction of alveolar walls with decreasing lung elastic recoil, loss of blood vessels, airways and loss of extracellular matrix attachment to airway walls. While chronic smokers constitute the highest COPD susceptible population in the United States, worldwide people that are exposed to indoor air pollution using biomass for heating and cooking as well as other environmental lung irritants form another major group of COPD patients (1, 2). Current diagnosis for COPD assessment is done by spirometry or pulmonary function testing (PFT), which is based on global lung volumes. The Global Initiative for Chronic Obstructive Lung Disease (GOLD) defines four severity stages (GOLD1-4) of COPD based on PFT measurements which lump all of the individual phenotypes associated with COPD into a single measure of gas flow at the mouth because PFT parameters ignore regional heterogeneity of the disease and the underlying disease components and disease etiology. Critical to the development of novel, targeted treatments for COPD is the identification of individual phenotypes which have historically been lumped together under the fairly non-descript title, “COPD.”
Computed tomography (CT), has emerged as a tool for quantitatively characterizing parenchymal destruction and small airways involvement. CT allows regional assessment of the disease component and the CT derived measurements have been shown to correlate well with the pathology of the disease (3–9). CT is commonly used to measure the extent of emphysema in the lungs and can be more sensitive than spirometry (10) in quantitating disease progression. Emphysema is quantified using CT densitometry techniques, which for example can calculate the percentage of voxels falling below a given Hounsfield Unit (HU) threshold in the inspiration image. Expiratory CT has also been shown to be useful in calculating the extent of air trapping using CT densitometry techniques (4, 5, 11–13). However, density measurements are influenced by CT reconstruction algorithm or other technical parameters (14, 15) and are dependent on the single threshold value. CT image texture also plays an important role in characterizing lung tissue and its pathologies. Uppaluri et al. proposed the 2D adaptive multiple feature method (AMFM), which captures textural patterns on the CT image. This method has shown good sensitivity in characterizing lung tissue (7, 16, 17) Later, an extension of this method to 3D by Xu et al. further showed good sensitivity in discriminating smoker and nonsmoker subjects (8). Sorensen et al. proposed a multi-scale Gaussian filter bank approach to define the texture on the CT images and has shown better discrimination of COPD and normal subjects with good correlations to the lung function measurements (18). Although density and texture based features serve to map lung destruction and remodeling, these measures do not provide insights into the mechanism of disease onset or disease progression. Mishima et al. (19) have suggested that once emphysema has been initiated with the appearance of small, regionalized tissue destruction, disease progression occurs, in part, because of mechanical factors serving to cause small holes to converge rather than new, isolated small holes emerging. It is important that new imaging-based metrics provide maps of parenchymal mechanics to allow for an improved understanding of subject-specific alterations in lung mechanics and regional parenchymal stresses. This is especially important as new methods emerge, such as endobronchial valves (20–23), to extract or isolate lung regions from the ventilation process. There is a poor understanding in regards to how remaining parenchyma is affected by such interventions.
Mechanical analysis on a regional level can be done from CT images by image registration of a pair of scans at different inflation levels. Different regional ventilation measurements from the registration of inspiration and expiratory CT has been shown useful to determine pulmonary function in COPD subjects (6). Previously, methods have been developed to estimate regional lung tissue expansion and contraction using image registration and biomechanical analysis, and have shown these measures compare well with the other indices of lung function (24). In this study it has been our hypothesis that these regional lung tissue estimates from the image registration will provide valuable information on lung function changes in COPD subjects. We propose a biomechanical feature set comprising three registration based metrics of lung function to describe COPD presence and severity. We also hypothesize the combination of density, texture and biomechanical features can be used to evaluate the severity of COPD more accurately than the individual usage of the feature sets, thus leading to more descriptive measures of the disease. We have used a machine learning framework to evaluate the performance of the obtained biomechanical feature set and compared it to the density and texture based feature sets. Correlations with the PFT parameters and health status metrics were also reported.
MATERIALS AND METHODS
COPDGENE DATA
A database of 162 subjects with varying distribution of COPD severity and nonsmoker subjects without COPD were used in this study. All the subjects were approved by the institutional review boards and provided written consent for participation in the study. The distribution of the subjects was: 27 nonsmokers, 30 GOLD 0, 29 GOLD1, 29 GOLD2, 28 GOLD3, and 19 GOLD4. These subjects were selected from the XXXXXX Cohort of the COPDGene study. The COPDGene Study is a multi-institutional research focused on COPD and other smoking related pulmonary diseases (25). All the patients in this study completed spirometry, health related quality of life questionnaires and CT scanning of the lungs at full inspiration and expiration.
DATA COLLECTION AND PULMONARY FUNCTION TESTING
The COPDGene protocol included the collection of demographic information, smoking history, using self-administered questionnaires. Health related quality of life is estimated using St. George’s respiratory questionnaire (SGRQ), which comprises four categories (symptoms, activity, impact and total score) with scores in each category ranging from 0 to 100. The higher the score the more severe is the disease. Spirometry was performed following the American Thoracic Society guidelines. The metrics reported include forced expiratory volume in 1 second (FEV1), which is the volume of gas forcibly exhaled during the first second of the expiration maneuver, and forced vital capacity (FVC), which is the total volume of gas forcibly exhaled after a full inspiration. The demographic information and PFT measures are shown in table 1.
Table 1.
Demographic information and PFT measures of the subjects in this study. The numbers reported are mean values with standard deviation in parentheses
| Parameters | Non-COPD | COPD |
|---|---|---|
| Age | 67.4 (6.79) | 67.6 (5.87) |
| Gender (M/F) | 34/23 | 57/39 |
| Height (cm) | 168.5 (8.66) | 168.2 (9.02) |
| Weight (kg) | 81 (11.80) | 79.9 (21.30) |
| BMI | 28.5 (4.08) | 28.01 (6.26) |
| Pack years | 16 (8.30) | 39.05 (12.21) |
| FEV1% predicted | 0.9 (0.13) | 0.55 (0.27) |
| FEV1/FVC | 0.7 (0.05) | 0.46 (0.15) |
| GOLD STAGE (N/0/1/2/3/4) | 27/30/0/0/0/0 | 0/0/29/29/28/19 |
IMAGE ACQUISITION
CT scans were acquired at the University of XXXXXX with either a 64 or 128 multidetector CT scanner (Somatom Sensation 64 or Somatom Definition FLASH; Siemens Medical Solutions, Erlangen, Germany) following the COPDGene protocol. Images were acquired with the patient in the supine position during a single breath hold at full inspiration (total lung capacity (TLC)) and a single breath hold at normal expiration (functional residual capacity (FRC)). The scans followed an imaging protocol with a peak tube voltage of 120 kV; tube current time product of 200 mAs for TLC scans and 50 mAs for FRC scans; gantry rotation time of 0.5s; and pitch a pitch of 1. The images were reconstructed at B31f kernel with a slice thickness of 0.75 mm and a reconstruction interval of 0.5 mm respectively, study protocol specifications for Siemens CT scanners (25).
IMAGE PREPROCESSING AND LUNG SEGMENTATION
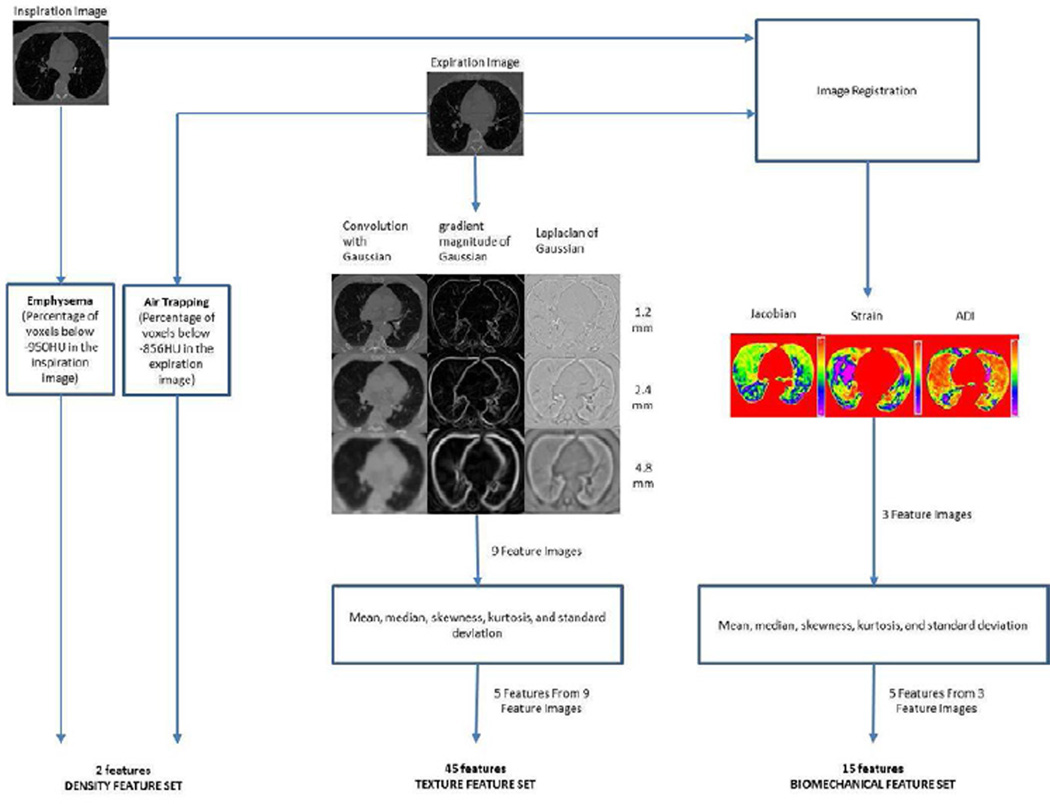
CT inspiratory and expiratory images were downloaded and stored in 16-bit Analyze (Mayo Clinic, Rochester, MN) format. Due to the high computational requirements of image registration, images were down sampled, which is the process of interpolating and resampling the image to produce a smaller and lower resolution of the original. CT images were first segmented to extract lung structures. Segmentation was carried out using a region growing technique where the given image is segmented into regions based on the discontinuities in the gray level and by the selection of initial seed points in the region. The flow of steps from the image preprocessing to machine learning experiments are shown in figure 1.
Figure 1.
Flow chart explaining the steps involved from image acquisition to classification experiments
IMAGE REGISTRATION
The inspiratory and expiratory CT images were registered for each subject. A lung mass preserving registration method was used to capture the large volume changes between these two images (9). This method uses a similarity metric called the sum of squared tissue volume difference (SSTVD), which estimates the local tissue and air fraction by minimizing local tissue mass difference (9, 26). This method has been shown effective in lung image registration protocols (3, 24). Displacement field information corresponding to the tissue deformation patterns in the lung from inspiration to expiration was extracted from the registration process.
FEATURE CALCULATION
In this study, we formed three sets of features from CT images: density based feature set, texture based feature set, and biomechanical feature set. 62 features were calculated from the inspiration and expiration images of a single subject. A summary of the features calculated is shown in figure 1.
DENSITY BASED FEATURE SET
The density based feature set consists of two measures representing the extent of emphysema and air trapping in a subject. These measures were estimated using the threshold technique computing the percent of voxels below a certain threshold in CT images. Emphysema extent is expressed as the percentage of voxels below −950HU in the inspiration image (5, 11, 27, 28). Air trapping extent is expressed as the percentage of voxels below −856HU in the expiration image (4, 29, 30).
TEXTURE BASED FEATURE SET
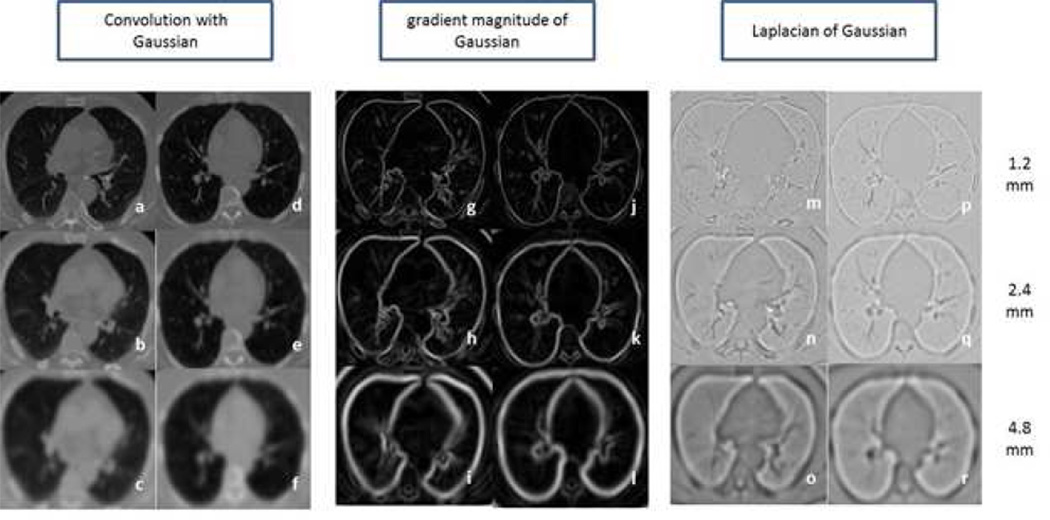
In order to capture the textural patterns, a set of 45 features was computed using three local image descriptors at three different scales. The local descriptors are based on the Gaussian function and its derivatives. The three local descriptors are: 1) convolution with Gaussian, which smooth the image and reduces the noise; 2) gradient magnitude of the Gaussian, which emphasizes edges and region boundaries; and 3) Laplacian of Gaussian, which computes second derivative of the image to highlight the regions of rapid intensity changes. These three filters were calculated at three different scales: 1.2 mm, 2.4 mm and 4.8 mm (18). Three filters at three scales were applied to the expiration images in the dataset giving rise to nine filtered versions of the image. For each of the filtered images, the mean, median, skewness, kurtosis, and standard deviation was computed for voxels in the lung region, thus producing a total of 45 features as a texture-based feature set. Examples of the nine filtered images of a GOLD1 and GOLD4 subjects’ expiration image are shown in figure 2.
Figure 2.
Axial slices of a GOLD1 (Left column in each filter section) and GOLD4 (Right column in each filter section) COPD subject. a, d) Convolution with Gaussian at 1.2mm standard deviation b, e) Convolution with Gaussian at 2.4mm standard deviation c, f) Convolution with Gaussian at 4.8mm standard deviation g, j) Gradient magnitude of Gaussian at 1.2mm standard deviation h, k) Gradient magnitude of Gaussian at 2.4mm standard deviation i, l) Gradient magnitude of Gaussian at 4.8mm standard deviation m, p) Laplacian of Gaussian at 1.2mm standard deviation n, q) Laplacian of at 2.4mm standard deviation o, r) Laplacian of Gaussian at 4.8mm standard deviation
LUNG BIOMECHANICAL FEATURE SET
This feature set is comprised of features calculated from the image registration of inspiration scan to the expiration scan. Mechanical analysis on a regional level is done by finding out the local tissue deformation pattern from the correspondence of each voxel between inspiration and expiration image. Three measures were calculated in this feature set: Jacobian, strain information and anisotropic deformation index (ADI). The Jacobian or Jacobian determinant measures the local volume change under deformation from the inspiration to the expiration. The Jacobian determinant is a measurement to estimate the point wise volume expansion and contraction during the deformation (9, 24). If the Jacobian value is one at a given voxel, then there is no deformation happened. If the value is greater than 1, it represents local expansion. If the value is less than 1, it represents local contraction.
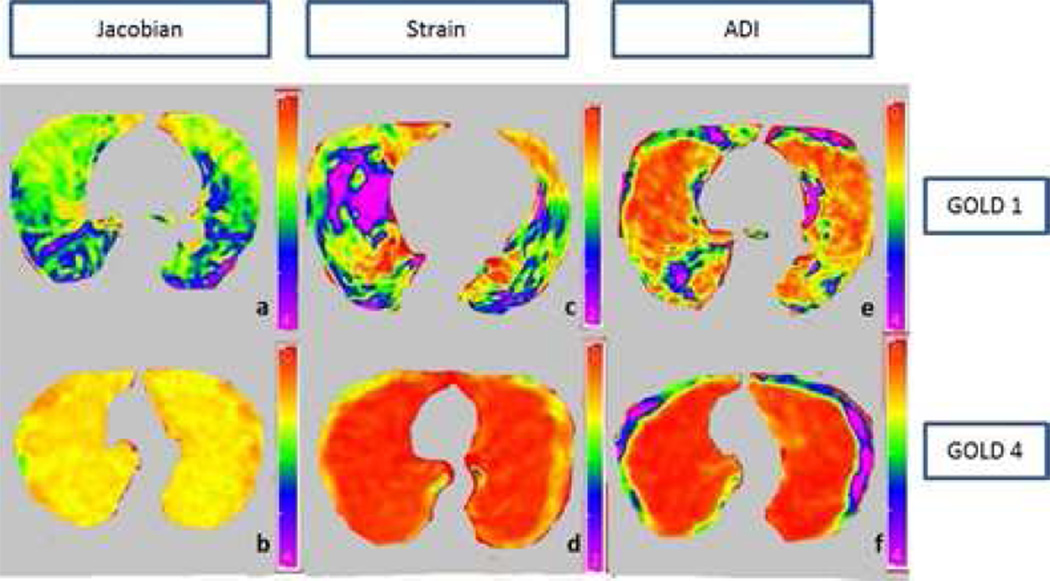
The concept of the strain is used to evaluate how much a given displacement differs locally from the inspiration to the expiration (31). Maximum principle strain is computed using displacement fields from the registration process to represent strain information. Anisotropic deformation index provides the orientation preference of the lung deformation. It calculates the ratio of length in the direction of maximal extension to the length in the direction of minimal extension (3). We have calculated five first order statistical features: mean, median, skewness, kurtosis and standard deviation for each of three feature images. A total of 15 features were computed to form the lung biomechanical feature set. The three biomechanical feature images: Jacobian, strain and ADI of a GOLD1 and GOLD4 subject are shown in figure 3.
Figure 3.
Axial slices of a mild COPD (GOLD1) and severe COPD (GOLD4) subject. a) Jacobian map of GOLD1 subject b) Jacobian map of GOLD4 subject c) Strain map of GOLD1 subject d) Strain map of GOLD4 subject e) Anisotropic deformation index map of GOLD1 subject f) Anisotropic deformation of GOLD4 subject
FEATURE SELECTION AND CLASSIFICATION
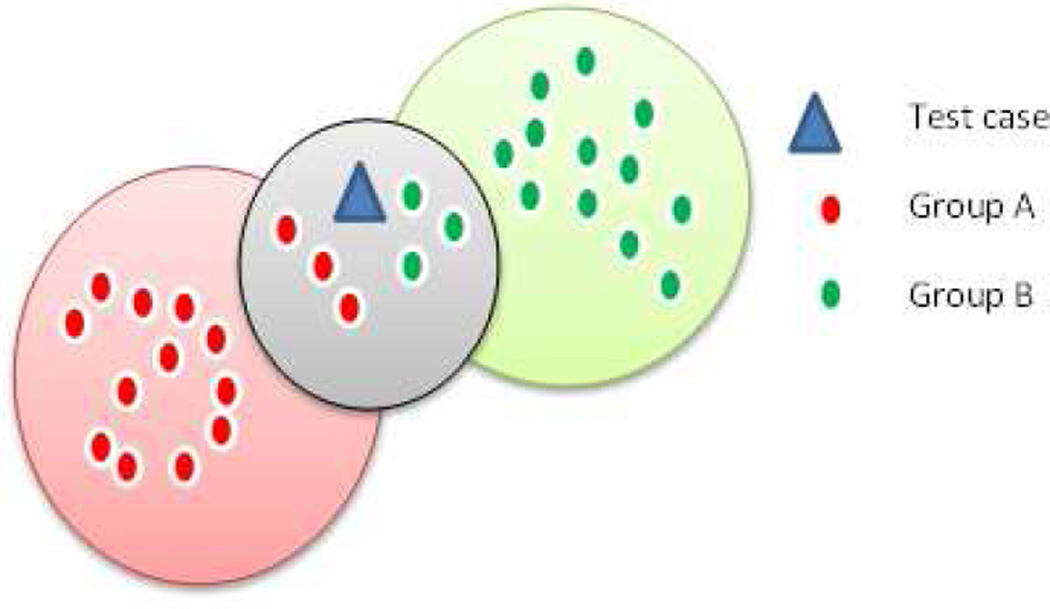
Feature selection was carried out on 62 features using a linear forward feature selection technique based on correlation based feature selection evaluator (32). This technique is a modified version of the sequential forward selection technique (32, 33). The subset of features which are high correlated with the class while having low intercorrelation considered as an optimal subset.. In the linear forward selection, the user will be able to limit the number of features that are considered in each step which in turn reduces the run time and the number of evaluations. With the selection of optimal set of features from each set, we have performed two classification experiments using the k nearest neighbor learning algorithm (KNN) (33). This algorithm is a non-parametric approach based directly on distances computed between the test and training data points. For any given test data point, KNN searches its nearest neighbors formed by the training sets. The classifier returns the selected number of neighbors, k, which are closest in the distance. The decision is made based on the majority vote of its neighbors, with the test point assigned to the group most common among its nearest neighbors, shown in figure 4. For instance, in figure 4, there are 15 data points in group A (red), 15 in group B (green) and one test data point (blue). Nearest neighbor algorithm computes the Euclidean distance to each data point in group A and group B from the test data point. In this example, the k value is chosen as 7. It selects 7 nearest neighbors closest to it based on the distance calculation. Since there are 4 data points from Group B out of 7 nearest neighbors, the given test data point is labeled as group B by the classifier
Figure 4.
K nearest neighbor algorithm
STATISTICAL ANALYSIS AND CLASSIFICATION EXPERIMENTS
Statistical experiments were performed using Microsoft Excel (Microsoft, Redmond, WA) and MATLAB software (MATLAB 7.12, The MathWorks Inc., Natick, MA, 2011). P values < 0.05 were considered statistically significant. Pearson’s correlation coefficients were calculated to investigate the correlation between the PFT parameters and the optimal CT based features. Spearman correlation coefficient was calculated to investigate the correlation between SGRQ scores and CT based features since the score is scaled based variable.
First experiment is to classify a given subject into either COPD or non COPD class using the three feature sets. Nonsmoker and GOLD0 subjects were considered as non COPD group in this experiment. Feature selection is carried out on 90 subjects (30 non COPD/ 60 COPD) out of 162 subjects used in the experiment. Second experiment is to classify a given subject into their corresponding severity stage. Feature selection is carried out on 75 subjects (15/severity stage) out of 135 subjects ranging from GOLD0 to GOLD4 stage.
For both training and test subjects, the optimal features from the feature selection process were used in further classification experiments. Feature selection and classification experiments were implemented using WEKA machine learning tool (34, 35) which provides a framework with a wide range of visualization tools and machine learning algorithms for data analysis. For training purposes, dataset was divided into training and testing sets and repeated ten times, with each split randomly selected each time. For each split, area under receiver operator characteristic curve (AUC) is reported. The best k (number of neighbors) value in the kNN algorithm is selected by cross validation. Predictions of test data are ranked by the probability of the class label and AUC for each class is separately calculated using one versus all approach. By considering the number of instances of a particular class label as the weight, average AUC of the ten splits is computed. Multiple regression analysis was performed using EXCEL software (Microsoft, Redmond, WA) to find the correlation between optimal features from each feature set and PFT measurements. Adjusted R squared correlation coefficient is reported for each combination of optimal features which takes the sample size and number of predictor variables into account
RESULTS
CORRELATION RESULTS
Pearson’s linear correlation coefficient, r, and corresponding P values were calculated between the PFT measurements and the optimal set of features from each feature set which were selected in the feature selection process. Spearman’s correlation coefficient was reported to find the correlation between SGRQ total score and CT based features. The results are shown in table 2 and all the correlations have shown statistical significance of P < 0.001. Air trapping measure in the density based feature set showed strong negative correlation with FEV1/FVC (r = −0.84) and also showed higher correlations than the emphysema extent measure. Texture based features have also shown strong correlations with FEV1/FVC measure. A strong positive correlation is seen between the mean Jacobian and PFT parameters (FEV1%, r = 0.80; FEV1/FVC, r = 0.76). Similarly, of all the features, mean Jacobian (r = −0.63) and median Jacobian (r = −0.63) features showed strong negative correlations with the SGRQ total scores. Median feature of Laplacian of Gaussian at 4.8mm (rho = −0.61) in the texture based feature set showed good negative correlations with the SGRQ total score whereas emphysema (rho = 0.46) and air trapping (rho = 0.56) scores were poorly correlated.
Table 2.
Relationship between CT derived features and clinical diagnostic measures of COPD. Pearson linear correlation coefficient is represented with r and Spearman correlation coefficient is represented with ρ. All the correlations have shown statistical significance of P < 0.0001
| Features | γ FEV1% predicted |
γ FEV1/FVC | ρ SGRQ total score |
|---|---|---|---|
| Mean Jacobian | 0.80 | 0.76 | −0.63 |
| Median Jacobian | 0.76 | 0.75 | −0.63 |
| Skewness Jacobian | −0.42 | −0.52 | 0.35 |
| Standard deviation of Jacobian | 0.74 | 0.64 | −0.59 |
| Standard deviation of Strain | 0.70 | 0.59 | −0.53 |
| Median convolution with Gaussian at 1.2mm | 0.67 | 0.76 | −0.52 |
| Kurtosis convolution with Gaussian at 2.4mm | −0.59 | −0.60 | −0.39 |
| Mean convolution with Gaussian at 2.4mm | 0.68 | 0.77 | −0.51 |
| median convolution with Gaussian at 2.4mm | 0.69 | 0.78 | −0.51 |
| Skewness convolution with Gaussian at 4.8mm | −0.57 | 0.60 | 0.41 |
| Skewness gradient magnitude of Gaussian at 1.2mm | −0.55 | −0.61 | 0.47 |
| Skewness gradient magnitude of Gaussian at 4.8mm | −0.61 | −0.66 | 0.55 |
| Median Laplacian of Gaussian at 4.8mm | 0.70 | 0.72 | −0.61 |
| Kurtosis Laplacian of Gaussian at 4.8mm | 0.24 | 0.26 | −0.2 |
| Emphysema (percentage of voxels below −950HU) | −0.65 | −0.73 | 0.46 |
| Air trapping (percentage of voxels below −856HU) | −0.78 | −0.84 | 0.56 |
CLASSIFICATION EXPERIMENT 1 (COPD vs. Non COPD Classification)
As an initial experiment, the performance of biomechanical and the combination feature set in detecting the presence and absence of COPD was evaluated. The dataset is divided into two classes: COPD and non COPD. Classifier is asked to classify a given subject into either of these classes using the four feature sets. AUC values were reported for each feature set from the ROC analysis with p- values for difference in AUC with ALL feature set according to the test proposed by Hanley JA and McNeil BJ (36). The results of this experiment are shown in table 3. Biomechanical feature set (AUC = 0.85), density (AUC = 0.83) and texture (AUC = 0.89) based feature sets. Texture (R2= 0.72) based features showed higher correlations with FEV1/FVC measures whereas the biomechanical feature set (R2 = 0.71) showed higher correlations with FEV1% measure. All these correlations have shown a statistical significance of P < 0.001.
Table 3.
Classification results of COPD/non COPD classification. AUC values reported from the ROC analysis with p- values for difference in AUC with ALL feature set according to the test proposed by Hanley JA and McNeil BJ (36) shown in parenthesis. Correlation results from multiple regression analysis between optimal features of density, texture, lung biomechanical features with PFT parameters and SGRQ scores
| Feature Sets | AUC | FEV1%predicted (R2) | FEV1/FVC (R2) | SGRQ Total score (R2) |
|---|---|---|---|---|
| Density | 0.83 (p< 10−4) | 0.57 | 0.68 | 0.30 |
| Texture | 0.89 (p< 10−4) | 0.58 | 0.72 | 0.37 |
| Biomechanical | 0.85 (p< 10−4) | 0.71 | 0.68 | 0.41 |
| ALL | 0.87 (−) | 0.73 | 0.77 | 0.47 |
CLASSIFICATION EXPERIMENT 2 (COPD Severity Classification)
In the second experiment, COPD severity classification was performed using the four feature sets. The subjects used in this experiment ranges from GOLD0 to GOLD4 stages of COPD. Classifier is asked to classify a given subject into their corresponding GOLD stage. Similar to the previous classification, AUC value significance were reported between the feature sets. The results of this experiment are shown in table 4. Biomechanical features were more effective in recognizing COPD severity than the density and texture feature sets by achieving an AUC of 0.81 and also correlated well with the FEV1% predicted measure (r = 0.71), which is a COPD severity measure from PFT diagnosis. The combination feature set (ALL) achieved an AUC of 0.80 by showing higher correlations with all the diagnostic measurements. All these correlations have shown a statistical significance of P < 0.001.
Table 4.
COPD severity classification results. AUC values reported from the ROC analysis with p- values for difference in AUC with ALL feature set according to the comparison test proposed by Hanley JA and McNeil BJ (36) shown in parenthesis. Correlation results from multiple regression analysis between optimal features of density, texture, lung biomechanical features with PFT parameters and SGRQ scores
| Feature Sets | AUC | FEV1%predicted (R2) | FEV1/FVC (R2) | SGRQ Total score (R2) |
|---|---|---|---|---|
| Density | 0.76 (p< 10−4) | 0.57 | 0.69 | 0.26 |
| Texture | 0.73 (p< 10−4) | 0.57 | 0.70 | 0.30 |
| Biomechanical | 0.81 (p< 10−3) | 0.71 | 0.63 | 0.35 |
| ALL | 0.80 (−) | 0.72 | 0.73 | 0.35 |
DISCUSSION
The two classification experiments conducted in this study show that the estimates of regional lung tissue expansion and contraction can serve as a useful parameter to describe COPD in pulmonary CT scans. Such COPD quantification, based on regional lung mechanics, is a step forward in understanding the disease impact on the lung function. The relationship between the CT derived biomechanical features and clinical diagnostic measures (PFT measurements, SGRQ scores) were estimated by correlation, as shown in table 2. The jacobian features, which capture local volume changes, have shown strong correlations with FEV1% predicted (r = 0.80), FEV1/FVC measures (r = 0.76). It has also shown strong negative correlations (ρ=-0.63) with the SGRQ scores, which depicts the disease impact on patient’s quality of life. Overall, the biomechanical measures have shown strong correlations with the severity diagnostic measure, FEV1% predicted. This suggests a strong relation between estimates of lung mechanics and severity level of COPD. On the other hand, density and texture based features have shown higher degree of correlation towards diagnostic measure, FEV1/FVC.
Density based features have been previously shown to be effective in COPD diagnosis (4, 5, 11, 12, 37,38). Earlier studies using CT textural patterns have also been shown to be successful in judging the presence of COPD (7, 8, 18, 39–41). The proposed lung biomechanical features were tested against these existing CT derived features. Moreover, in our study, all these three feature sets were combined to form a single feature set (ALL) and the combined performance was thereby evaluated.
As a first classification experiment, a two-class problem was defined by dividing the dataset into two groups; COPD (GOLD1-4) and non COPD (nonsmokers/GOLD0). The biomechanical features showed similar performance with the existing density and texture features in discriminating subjects with and without COPD, with an AUC of 0.85, as shown in table 3. Also, a good correlation with the PFT measurements and SGRQ scores was shown. However, it should be noted that texture based features performed better than the biomechanical features. Since, the subject range in this classification is from nonsmokers to GOLD4 (very severe); there is a possibility of minimal lung functional changes happening at the initial stages. As a consequence, this may lead to a higher number of misclassifications between nonsmokers, GOLD0 and GOLD1 groups, resulting in overall reduction of the classifier performance with biomechanical features. In this study, density and texture based features been shown to be effective in recognizing COPD presence or absence by achieving an AUC of 0.83 and 0.89. The optimal features from these two feature sets showed strong correlations (R2=0.68, 0.72) with the diagnostic measure, FEV1/FVC.
In the second experiment, a five class problem was then defined to categorize COPD subjects into their corresponding GOLD stage. Biomechanical features are more effective in COPD severity classification (AUC = 0.81) than the density (AUC = 0.76) and texture (AUC = 0.73) feature sets, as shown in table 4. The strong correlations with FEV1% predicted show the sensitivity of biomechanical features to the level of COPD severity. Another interesting observation that can be made from this experiment is the performance of these features at different stages of the disease. In table 5, the AUC values of the feature sets in classifying each severity stage of COPD is shown. It shows that the biomechanical feature set performance is improved at the later stage of the disease than at the initial stages. All the feature sets were less effective in classifying GOLD1 and GOLD2 stage subjects. Especially, in classifying GOLD2 subjects, lung biomechanical features showed better performance than the density and texture based features. This suggests the possibility of major lung functional changes at GOLD2 stage, which were captured by lung biomechanical features. This shows that the lung mechanics provide valuable information at later stages of the disease which was not possible to capture using the density and textural features.
Table 5.
AUC values at each GOLD stage of COPD from COPD severity classification with p- values for difference in AUC with Biomechanical feature set according to the comparison test proposed by Hanley JA and McNeil BJ [36] shown in parenthesis. The standard deviation values are represented in parenthesis.
| Feature Sets | GOLD0 | GOLD1 | GOLD2 | GOLD3 | GOLD4 |
|---|---|---|---|---|---|
| Density | 0.80 (p=0.0521) | 0.67 (p< 10−4) | 0.63 (p<10−4) | 0.75 (p<10−4) | 0.89 (p=0.0781) |
| Texture | 0.79 (p<0.05) | 0.74 (p=0.6524) | 0.58 (p<10−4) | 0.75 (p<10−4) | 0.83 (p<10−4) |
| Biomechanical | 0.82 (−) | 0.75 (−) | 0.76 (−) | 0.89 (−) | 0.92 (−) |
In both the experiments, another feature set (ALL) is evaluated, which is formed by combining all the three feature sets together. While recognizing COPD presence or absence in the first experiment, ALL achieved similar AUC (0.87, table 3) but it has shown better correlations with the PFT measurements and SGRQ total scores. Similarly, in the second experiment (table 4), adding biomechanical features to the density (AUC = 0.76) and textural features (AUC = 0.73), further showed an improvement in the classifier performance (AUC = 0.80). Also, there is a strong correlation with the clinical diagnostic measures of the disease. This shows biomechanical features add useful measures to the density and texture based features for more accurate diagnosis of COPD severity from CT images.
The proposed features performed comparatively well with the previous methods of COPD diagnosis and severity classifications. The adaptive multiple feature method (AMFM), proposed by Uppaluri et al., based on textural patterns of 2D CT images achieved an accuracy of 100% in classifying normal and severe emphysema subjects but with no significant correlation with PFT measures of emphysema (7). The extension of 2D AMFM to 3D AMFM proposed by Xu et al. showed better results in discriminating normal smoker and nonsmoker lung parenchyma (8). Another texture based approach proposed by Sorensen et al. based on gaussian filter versions of CT, achieved an AUC of 0.713 in classifying COPD and Non-COPD subjects (18). In COPD severity classification, registration based ventilation measures proposed by Murphy et al. achieved 67% classification accuracy (6). In COPD diagnosis and severity experiments, the biomechanical feature set achieved an AUC of 0.85 and 0.81, as shown in table 3 and 4, and also correlated well with the PFT measures and SGRQ scores.
However, in addition to the density and texture based features that were used in this study, there are several other CT derived features providing robust quantification of COPD (6–8, 18, 42). The texture based feature set consists of only three basic Gaussian filtered versions of the image at multiple scales. There are several other textural features which have been proven to be effective in COPD quantification (7, 8, 18). The number of features used in this study was less which gives a definite scope in testing the effectiveness of several other features either individually or in combination with the proposed biomechanical features. A complete system consisting of all the CT derived features related to both emphysema and small airway disease may result in more accurate diagnosis of COPD. In this study, all of the CT scans were acquired from one of two scanners, both from the same manufacturer, and acquired using a carefully specified protocol. In a larger study involving more sites and a variety of scanner manufacturers, it will be necessary to test for scanner-to-scanner variations in the image features used for classification.
CONCLUSION
This study demonstrates the effectiveness of the registration based estimates of lung tissue expansion and contraction in COPD diagnosis. Three measures were extracted from the registered scans and the features based on these three measures showed good correlations with the pulmonary function. The classification experiments illustrated that the proposed measurements perform equally well or better than the density and texture feature sets in assessing COPD presence and severity. Also, the inclusion of biomechanical features to the density and texture improved the classifier performance with higher correlation to pulmonary function indices. With further testing on larger databases, the proposed approach may be used for accurate measure of the pulmonary function and disease.
Acknowledgments
Grants
This work was supported by Grants HL079406 and HL064368 from the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Sandeep Bodduluri, Graduate Student, Department of Biomedical Engineering, 1402 Seamans Center for the Engineering Arts and Sciences, The University of Iowa, Iowa City, Iowa, 52242, Phone: (949) 338 – 2356.
John D. Newell, Jr, Professor, Department of Radiology, Biomedical Engineering, C751GH, The University of Iowa Hospitals and Clinics, 200 Hawkins Drive, Iowa City, IA 52242, Phone: (319) 356-1037 Fax: (319) 356-2220.
Eric A. Hoffman, Professor, Department of Radiology, Biomedical Engineering, C748GH, The University of Iowa Hospitals and Clinics, 200 Hawkins Drive, Iowa City, IA 52242, Phone: (319) 353-6199 Fax: (319) 356-2220
Joseph M. Reinhardt, Professor, Department of Biomedical Engineering, 1402A Seamans Center for the Engineering Arts and Sciences, The University of Iowa, Iowa City, Iowa, 52242, Phone: (319) 335-5634 Fax: (319) 335-5631
REFERENCES
- 1.From the Global Strategy for the Diagnosis, Management and Prevention of COPD. Global Initiative for Chronic Obstructive Lung Disease (GOLD) 2011 [Google Scholar]
- 2.Rabe KF, Hurd S, Anzueto A, Barnes PJ, Buist SA, Calverley P, et al. Global strategy for the diagnosis, management, and prevention of chronic obstructive pulmonary disease: GOLD executive summary. American journal of respiratory and critical care medicine. 2007;176(6):532–555. doi: 10.1164/rccm.200703-456SO. Epub 2007/05/18. [DOI] [PubMed] [Google Scholar]
- 3.Amelon R, Cao K, Ding K, Christensen GE, Reinhardt JM, Raghavan ML. Three-dimensional characterization of regional lung deformation. Journal of biomechanics. 2011;44(13):2489–2495. doi: 10.1016/j.jbiomech.2011.06.009. Epub 2011/08/02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Busacker A, Newell JD, Jr, Keefe T, Hoffman EA, Granroth JC, Castro M, et al. A multivariate analysis of risk factors for the air-trapping asthmatic phenotype as measured by quantitative CT analysis. Chest. 2009;135(1):48–56. doi: 10.1378/chest.08-0049. Epub 2008/08/12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Muller NL, Staples CA, Miller RR, Abboud RT. "Density mask". An objective method to quantitate emphysema using computed tomography. Chest. 1988;94(4):782–787. doi: 10.1378/chest.94.4.782. Epub 1988/10/01. [DOI] [PubMed] [Google Scholar]
- 6.Murphy K, Pluim JP, van Rikxoort EM, de Jong PA, de Hoop B, Gietema HA, et al. Toward automatic regional analysis of pulmonary function using inspiration and expiration thoracic CT. Medical physics. 2012;39(3):1650–1662. doi: 10.1118/1.3687891. Epub 2012/03/03. [DOI] [PubMed] [Google Scholar]
- 7.Uppaluri R, Mitsa T, Sonka M, Hoffman EA, McLennan G. Quantification of pulmonary emphysema from lung computed tomography images. American journal of respiratory and critical care medicine. 1997;156(1):248–254. doi: 10.1164/ajrccm.156.1.9606093. Epub 1997/07/01. [DOI] [PubMed] [Google Scholar]
- 8.Xu Y, Sonka M, McLennan G, Guo J, Hoffman EA. MDCT-based 3-D texture classification of emphysema and early smoking related lung pathologies. IEEE transactions on medical imaging. 2006;25(4):464–475. doi: 10.1109/TMI.2006.870889. Epub 2006/04/13. [DOI] [PubMed] [Google Scholar]
- 9.Yin Y, Hoffman EA, Lin CL. Mass preserving nonrigid registration of CT lung images using cubic Bspline. Medical physics. 2009;36(9):4213–4222. doi: 10.1118/1.3193526. Epub 2009/10/09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Spaggiari E, Zompatori M, Verduri A, Chetta A, Bna C, Ormitti F, et al. Early smoking-induced lung lesions in asymptomatic subjects. Correlations between high resolution dynamic CT and pulmonary function testing. La Radiologia medica. 2005;109(1–2):27–39. Epub 2005/02/25. [PubMed] [Google Scholar]
- 11.Gevenois PA, de Maertelaer V, De Vuyst P, Zanen J, Yernault JC. Comparison of computed density and macroscopic morphometry in pulmonary emphysema. American journal of respiratory and critical care medicine. 1995;152(2):653–657. doi: 10.1164/ajrccm.152.2.7633722. Epub 1995/08/01. [DOI] [PubMed] [Google Scholar]
- 12.Gould GA, MacNee W, McLean A, Warren PM, Redpath A, Best JJ, et al. CT measurements of lung density in life can quantitate distal airspace enlargement--an essential defining feature of human emphysema. The American review of respiratory disease. 1988;137(2):380–392. doi: 10.1164/ajrccm/137.2.380. Epub 1988/02/01. [DOI] [PubMed] [Google Scholar]
- 13.Gould GA, Redpath AT, Ryan M, Warren PM, Best JJ, Flenley DC, et al. Lung CT density correlates with measurements of airflow limitation and the diffusing capacity. The European respiratory journal : official journal of the European Society for Clinical Respiratory Physiology. 1991;4(2):141–146. Epub 1991/02/01. [PubMed] [Google Scholar]
- 14.Boedeker KL, McNitt-Gray MF, Rogers SR, Truong DA, Brown MS, Gjertson DW, et al. Emphysema: effect of reconstruction algorithm on CT imaging measures. Radiology. 2004;232(1):295–301. doi: 10.1148/radiol.2321030383. Epub 2004/06/29. [DOI] [PubMed] [Google Scholar]
- 15.Sieren JP, Newell JD, Judy PF, Lynch DA, Chan KS, Guo J, et al. Reference standard and statistical model for intersite and temporal comparisons of CT attenuation in a multicenter quantitative lung study. Medical physics. 2012;39(9):5757–5767. doi: 10.1118/1.4747342. Epub 2012/09/11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Uppaluri R, Hoffman EA, Sonka M, Hartley PG, Hunninghake GW, McLennan G. Computer recognition of regional lung disease patterns. American journal of respiratory and critical care medicine. 1999;160(2):648–654. doi: 10.1164/ajrccm.160.2.9804094. Epub 1999/08/03. [DOI] [PubMed] [Google Scholar]
- 17.Uppaluri R, Hoffman EA, Sonka M, Hunninghake GW, McLennan G. Interstitial lung disease: A quantitative study using the adaptive multiple feature method. American journal of respiratory and critical care medicine. 1999;159(2):519–525. doi: 10.1164/ajrccm.159.2.9707145. Epub 1999/02/02. [DOI] [PubMed] [Google Scholar]
- 18.Sorensen L, Nielsen M, Lo P, Ashraf H, Pedersen JH, de Bruijne M. Texture-based analysis of COPD: a data-driven approach. IEEE transactions on medical imaging. 2012;31(1):70–78. doi: 10.1109/TMI.2011.2164931. Epub 2011/08/24. [DOI] [PubMed] [Google Scholar]
- 19.Mishima M, Hirai T, Itoh H, Nakano Y, Sakai H, Muro S, et al. Complexity of terminal airspace geometry assessed by lung computed tomography in normal subjects and patients with chronic obstructive pulmonary disease. Proceedings of the National Academy of Sciences of the United States of America. 1999;96(16):8829–8834. doi: 10.1073/pnas.96.16.8829. Epub 1999/08/04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barua A, Vaughan P, Wotton R, Naidu B. Do endobronchial valves improve outcomes in patients with emphysema? Interactive cardiovascular and thoracic surgery. 2012 doi: 10.1093/icvts/ivs371. Epub 2012/09/05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brown MS, Kim HJ, Abtin FG, Strange C, Galperin-Aizenberg M, Pais R, et al. Emphysema lung lobe volume reduction: effects on the ipsilateral and contralateral lobes. European radiology. 2012;22(7):1547–1555. doi: 10.1007/s00330-012-2393-6. Epub 2012/04/03. [DOI] [PubMed] [Google Scholar]
- 22.Herth FJ, Eberhardt R, Gompelmann D, Ficker JH, Wagner M, Ek L, et al. Radiological and clinical outcomes of using chartis to plan endobronchial valve treatment. The European respiratory journal : official journal of the European Society for Clinical Respiratory Physiology. 2012 doi: 10.1183/09031936.00015312. Epub 2012/05/05. [DOI] [PubMed] [Google Scholar]
- 23.Herth FJ, Noppen M, Valipour A, Leroy S, Vergnon JM, Ficker JH, et al. Efficacy predictors of lung volume reduction with Zephyr valves in a European cohort. The European respiratory journal : official journal of the European Society for Clinical Respiratory Physiology. 2012;39(6):1334–1342. doi: 10.1183/09031936.00161611. Epub 2012/01/28. [DOI] [PubMed] [Google Scholar]
- 24.Reinhardt JM, Ding K, Cao K, Christensen GE, Hoffman EA, Bodas SV. Registration-based estimates of local lung tissue expansion compared to xenon CT measures of specific ventilation. Medical image analysis. 2008;12(6):752–763. doi: 10.1016/j.media.2008.03.007. Epub 2008/05/27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Regan EA, Hokanson JE, Murphy JR, Make B, Lynch DA, Beaty TH, et al. Genetic epidemiology of COPD (COPDGene) study design. Copd. 2010;7(1):32–43. doi: 10.3109/15412550903499522. Epub 2010/03/11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cao K, Christensen GE, Ding K, Reinhardt JM, editors. Second International Workshop on Pulmonary Image Analysis. London, UK: 2009. Intensity-and-Landmark-Driven, Inverse Consistent, B-Spline Registration and Analysis forLung Imagery. [Google Scholar]
- 27.Muller NL, Thurlbeck WM. Thin-section CT, emphysema, air trapping, and airway obstruction. Radiology. 1996;199(3):621–622. doi: 10.1148/radiology.199.3.8637975. Epub 1996/06/01. [DOI] [PubMed] [Google Scholar]
- 28.Newell JD., Jr Quantitative computed tomography of lung parenchyma in chronic obstructive pulmonary disease: an overview. Proceedings of the American Thoracic Society. 2008;5(9):915–918. doi: 10.1513/pats.200804-034QC. Epub 2008/12/06. [DOI] [PubMed] [Google Scholar]
- 29.Foreman MG, DeMeo DL, Hersh CP, Reilly JJ, Silverman EK. Clinical determinants of exacerbations in severe, early-onset COPD. The European respiratory journal : official journal of the European Society for Clinical Respiratory Physiology. 2007;30(6):1124–1130. doi: 10.1183/09031936.00009307. Epub 2007/08/24. [DOI] [PubMed] [Google Scholar]
- 30.Newman KB, Lynch DA, Newman LS, Ellegood D, Newell JD., Jr Quantitative computed tomography detects air trapping due to asthma. Chest. 1994;106(1):105–109. doi: 10.1378/chest.106.1.105. Epub 1994/07/01. [DOI] [PubMed] [Google Scholar]
- 31.Lubliner J. Plasiticity Theory. Mineola, NY: Dover Publication; 2008. [Google Scholar]
- 32.Hall MA. Correlation-based Feature Subset Selection for Machine Learning. Hamilton, New Zealand: University of Waikato; 1998. [Google Scholar]
- 33.Aha DW, KD, Albert MK. Instance-based learning algorithms. Machine Learning. 1991 [Google Scholar]
- 34.Mark Hall EF, Holmes Geoffrey, Pfahringer Bernhard, Reutemann Peter, Witten Ian H. The WEKA Data Mining Software: An Update. SIGKDD Explorations. 2009 [Google Scholar]
- 35.Holmes GDA, Witten IH, editors. WEKA: a machine learning workbench. Proceedings of the 1994 Second Australian and New Zealand Conference; 29 Nov-2 Dec.1994. [Google Scholar]
- 36.Hanley JA, McNeil BJ. A method of comparing the areas under receiver operating characteristic curves derived from the same cases. Radiology. 1983;148(3):839–843. doi: 10.1148/radiology.148.3.6878708. Epub 1983/09/01. [DOI] [PubMed] [Google Scholar]
- 37.Gurney JW, Jones KK, Robbins RA, Gossman GL, Nelson KJ, Daughton D, et al. Regional distribution of emphysema: correlation of high-resolution CT with pulmonary function tests in unselected smokers. Radiology. 1992;183(2):457–463. doi: 10.1148/radiology.183.2.1561350. Epub 1992/05/01. [DOI] [PubMed] [Google Scholar]
- 38.Matsuoka S, Kurihara Y, Yagihashi K, Hoshino M, Watanabe N, Nakajima Y. Quantitative assessment of air trapping in chronic obstructive pulmonary disease using inspiratory and expiratory volumetric MDCT. AJR American journal of roentgenology. 2008;190(3):762–769. doi: 10.2214/AJR.07.2820. Epub 2008/02/22. [DOI] [PubMed] [Google Scholar]
- 39.Lynch DA, Newell JD. Quantitative imaging of COPD. Journal of thoracic imaging. 2009;24(3):189–194. doi: 10.1097/RTI.0b013e3181b31cf0. Epub 2009/08/26. [DOI] [PubMed] [Google Scholar]
- 40.Newell JD, Jr, Hogg JC, Snider GL. Report of a workshop: quantitative computed tomography scanning in longitudinal studies of emphysema. The European respiratory journal : official journal of the European Society for Clinical Respiratory Physiology. 2004;23(5):769–775. doi: 10.1183/09031936.04.00026504. Epub 2004/06/05. [DOI] [PubMed] [Google Scholar]
- 41.Sorensen L, Shaker SB, de Bruijne M. Quantitative analysis of pulmonary emphysema using local binary patterns. IEEE transactions on medical imaging. 2010;29(2):559–569. doi: 10.1109/TMI.2009.2038575. Epub 2010/02/05. [DOI] [PubMed] [Google Scholar]
- 42.Van Rikxoort EMDJP, Mets OM, Van Ginneken B, editors. Automatic classification of pulmonary function in COPD patients using trachea analysis in chest CT scans. SPIE; 2012. [Google Scholar]