Abstract
DNAzymes have shown great promise as a general platform for detecting metal ions, as many metal-specific DNAzymes can be obtained using in vitro selection. While DNAzyme-based metal sensors have found many applications in the extracellular environment, no intracellular application of DNAzyme sensors has yet been reported. Here we demonstrate a novel type of metal ion sensor for intracellular metal ion detection. The probe consists of a 13 nm gold nanoparticle (AuNP) core functionalized with a shell consisting of a uranyl-specific 39E DNAzyme whose enzyme strand contains a thiol at the 3′ end for conjugation to the AuNP, and whose substrate strand is modified with a Cy3 fluorophore at the 5′ end and a molecular quencher at the 3′ end. In the absence of uranyl, the fluorescence of the Cy3 is quenched by both AuNP and the molecular quencher. In the presence of uranyl, the DNAzyme cleaves the fluorophore-labeled substrate strand, resulting in release of the shorter product strand containing the Cy3 and increased fluorescence. We demonstrate that this DNAzyme-AuNP probe can readily enter cells and can serve as a metal ion sensor within a cellular environment, making it the first demonstration of DNAzymes as intracellular metal ion sensors. Such a method can be generally applied to the detection of other metal ions using other DNAzymes selected through in vitro selection.
Metal ions are essential for numerous biological processes and their regulation is crucial for maintaining normal functions. However, the beneficial features of many metal ions are often counterbalanced by their toxic effects when the metal ions are in excess, or by the presence of other toxic metal ions in the environment. To gain a better fundamental understanding of how metal ions are regulated and where the potential molecular targets are for toxic metal ions, tools that can monitor localization and concentration of metal ions in living cells are required.1 Toward this goal, tremendous effort has been applied to develop intracellular metal ion sensors. Among them, both small molecular sensors and genetically encoded protein sensors have enjoyed the most success in intracellular metal ion sensing.2 A large number of sensors have been successfully used to detect metal ions that have important biological functions, such as calcium, zinc, copper and iron.3 At the same time, there is also emerging development in intracellular sensors for toxic metal ions, such as mercury, cadmium and lead.4 Despite the advances made over the previous years, it remains a significant challenge to rationally design sensors for metal ions of interest with both high sensitivity and selectivity.
To meet this challenge and design sensors for a much broader range of metal ions, we and others have taken advantage of an emerging field of metalloenzymes called deoxyribozymes (DNAzymes), i.e., DNA molecules with enzymatic activities. Unlike small molecule or protein-based sensors, DNAzymes with high specificity for a specific metal ion of interest can be obtained from a combinatorial process, starting from a large DNA library containing up to 1015 different sequences.5, cause of such high metal ion selectivity, these DNAzymes have been converted into sensors for many metal ions, such as Pb2+, UO22+, Hg2+ and Cu2+, based on either fluorescence, colorimetry, or electrochemistry.6 The development of these sensors has significantly expanded the range of metal ions that can be detected. The biggest advantages of this type of sensor are that it does not require advanced knowledge in order to construct a metal-binding site, and the binding affinity and selectivity toward metal ions can be fine-tuned by introducing different levels of stringency during the selection process. Moreover, it is relatively simple to synthesize DNA and many different modifications and functional groups can be easily introduced into the DNA during synthesis. Furthermore, DNA is naturally water soluble and biocompatible. All of these properties make DNAzyme sensors an attractive candidate for intracellular sensing of metal ions. However, even though DNAzymes have first been demonstrated as metal ion sensors over 10 years ago6a and many sensors have been reported since then6f, 7, all of these sensors are limited to detecting metal ions in extracellular environments.
In this study, we present the design, synthesis, and application of a DNAzyme-gold nanoparticle probe for metal ions in living cells. As an initial demonstration, we chose the 39E DNAzyme, which has exceptional selectivity (more than 1 million-fold over other competing metal ions) and sensitivity (45 pM detection limit) for the uranyl ion (UO22+).8a Uranium has been used in nuclear power and nuclear weapons. However, there is also growing concern about adverse health effects associated with uranium exposure.9 Uranium is known as a highly toxic carcinogen.9a,c High doses of uranium can cause kidney damage,9c and may lead to urinary system disease and lung cancer.9b Chronic low-dose exposure to uranium has been shown to exert negative impacts on many different stages of animal development.9d Uranium can also cross the blood brain barrier and accumulate in regions of the brain, resulting in alterations in behavior.9e Uranyl is the water-soluble form of uranium, and due to its bioavailability, is the form that poses the greatest risk to human health. However, despite its high toxicity, no intracellular sensor for uranyl has been reported.
Based on our previously reported in vitro selection of the uranyl-specific 39E DNAzyme, we and others have transformed these DNAzymes into uranyl sensors with many different methods for signal transduction.8, 10 However, all work to date involves detection outside of cells, and the sensors as designed are not suitable for detection within live cells, in part due to difficulty in delivering the DNAzyme into cells.
To overcome this limitation, we chose gold nanoparticles (AuNPs) for cellular delivery of the DNAzyme, as the AuNPDNA conjugate has many desirable properties, including stability in serum, ability to enter cells without use of transfection agents, much larger DNA loading efficiency than conventional transfection methods, and increased resistance to enzymatic degradation.11(c)
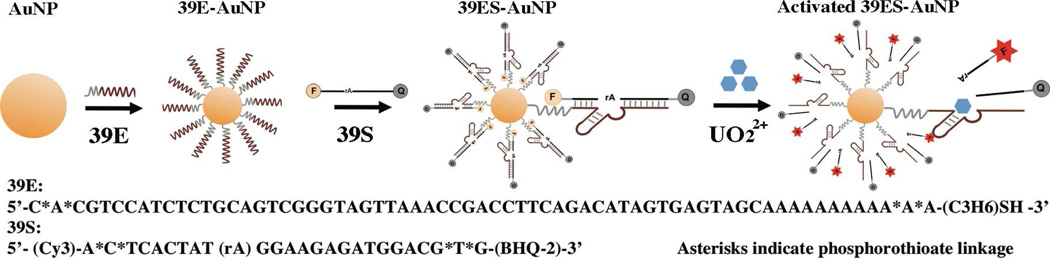
As shown in Scheme 1, the DNAzyme-AuNP cellular sensor (39ES-AuNP) consists of a 13 nm AuNP, 39E enzyme strand, and its 39S substrate strand. The 3′ end of the enzyme strand is functionalized with a thiol group (SH) for immobilization onto the AuNP. In addition, a poly-A spacer is added between the thiol moiety and the enzyme strand to avoid loss of activity due to steric interference of the AuNP. The substrate strand is labeled with a fluorophore (Cy3) at the 5′ end and a quencher (BHQ-2) at the 3′ end (Figure S1). Upon immobilization of enzyme strands onto the AuNP, the substrate strands were hybridized with enzyme strands by heating and annealing. When hybridized, the fluorescence signal from Cy3 should be quenched by the AuNP, as AuNP is known to quench fluorophores.12 However, the poly-A spacer between the enzyme strand and AuNP surface was found to weaken the quenching effect of AuNP, since quenching of a dye’s fluorescence is strongly dependent on the spatial separation of the dye from the nanoparticle surface.12(b) To ensure complete quenching, we also added a quencher (Black Hole Quencher-2, BHQ-2) at the 3′ end, resulting in increased S/N ratio (Figure S5). In the presence of uranyl, the substrate strand is cleaved, resulting in a shorter DNA strand with corresponding lower melting temperature (21°C) than the original full-length substrate strand (60°C). The shorter DNA strand containing Cy3 fluorophore is released. The Cy3 is separated from both the AuNP and BHQ-2 quencher, and the fluorescent signal is enhanced.
Scheme 1.
Design of a fluorescent DNAzyme immobilized onto gold nanoparticles as selective probe of uranyl inside live cells.
The synthesis of 13 nm AuNP, hybridization of the 39E enzyme and substrate strands, and conjugation of the resulting 39E DNAzyme to the AuNP were carried out using protocols reported previously (Figure S2).10, 13 Quantification of the DNAzyme on the AuNP surface by UV absorption and fluorescence showed that there were about 70 copies of 39E on each AuNP, and the same number of 39S strands hybridized to the enzyme strands. Such a dense loading of DNAzymes and efficient hybridization between the DNAzyme and their substrate strand allows for efficient cellular uptake of many DNAzymes per AuNP and thus maximum dynamic range.
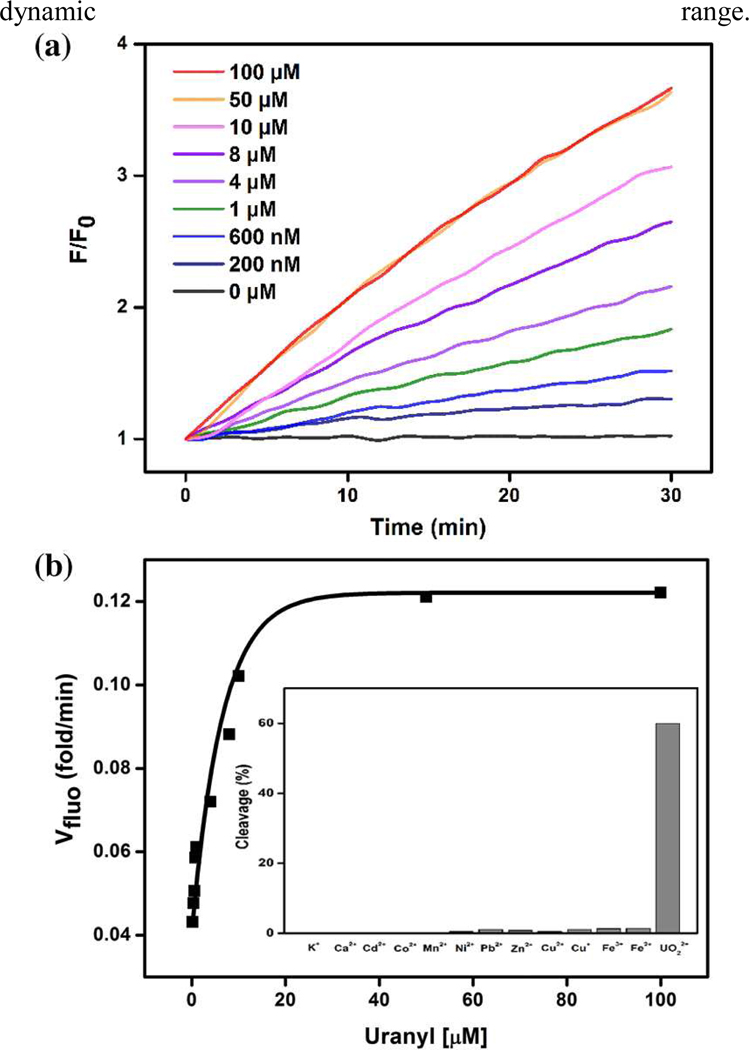
The performance of the 39ES-AuNP cellular sensor was first evaluated in a buffer (20 mM MOPS, 100 mM NaCl, 2 mM MgCl2, pH 7.0) (Figure 1(a)). Uranyl citrate and uranyl bicarbonate are two major uranyl species at physiological conditions.9 To test the response of 39ES-AuNP on both uranyl species, the increase of fluorescent signal with increasing concentrations of both species were measured and the responses are similar for both species up to 10 µM (Figure 1, S3). The rate for fluorescence increase reached plateaus at higher concentrations of uranyl citrate, while the rate for fluorescence decreased if the concentration of uranyl bicarbonate was more than 10 µM. We attribute the different response to different solubility of the two uranyl species in buffer. Because 39ES-AuNP has a wider dynamic range in the presence of uranyl citrate than uranyl bicarbonate, we chose uranyl citrate in our later studies. At pH 5.0, the sensor shows faster responses to the same concentrations of uranyl (Figure S4). Improved sensitivity for uranyl at pH 5.0 also makes the sensor suitable for working in acidic organelles. The sensor maintains excellent selectivity for uranyl over other various biologically relevant metal ions at physiologically relevant concentrations (Figure 1 (b inset), Figure S6).
Figure 1.
(a) Turn-on response of 39ES-AuNP to different concentrations of uranyl citrate over time. All activity tests were carried out in buffer containing 20 mM MOPS, 100 mM NaCl, 2 mM MgCl2, pH 7.0. (b) Rate of turn-on fluorescence increase at different concentrations of uranyl. (b inset) Selectivity of 39ES-AuNP for UO22+ over other metal ions. (100 mM for K+; 2 mM for Ca2+ and Zn2+; 20 µM for all others; reaction with Cu+ and Fe2+ was carried out in an oxygen-free environment to prevent oxidation of metal ions).
To test the stability of the sensor, 39ES-AuNP was incubated with cell lysate or 80% bovine serum for 3 hours at 37°C. Gel electrophoresis was used to separate the intact 39S strand from the cleaved product based on different lengths of the strands. No obvious cleavage of substrate strand was observed, suggesting the stability of the sensor is sufficient for application in cellular environments (Figure S10).
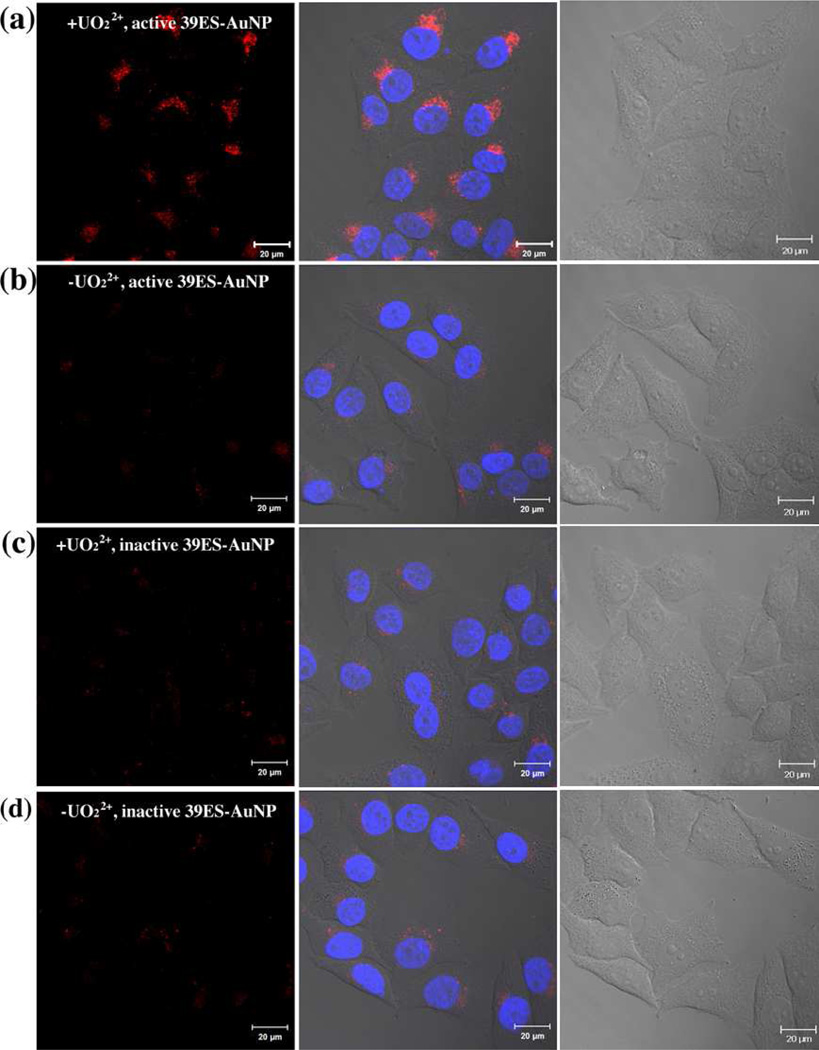
Having demonstrated the effectiveness of the 39ES-AuNP cellular sensor in buffer, we next tested its cellular uptake and fluorescence changes using HeLa cells (human cervical cancer) as a model. HeLa cells were first treated with 750 µM uranyl citrate for 12 hours to allow sufficient uranyl uptake.9 The cell viability tested via MTT assay suggests no obvious toxicity up to 1,000 µM of uranyl citrate or uranyl bicarbonate (Figure S7). Based on ICP-MS, the intracellular concentration of uranyl under these conditions is estimated to reach 100 µM (Supporting Information). HeLa cells that were pretreated with uranyl citrate were incubated with the probes for another 2 hours before images were taken using confocal microscopy. The amount of 39ES-AuNP was estimated to be 1×106/cell based on ICP-MS measurement (Supporting Information). As shown in Figure 2a, HeLa cells incubated with uranyl citrate showed more fluorescence than those without uranyl citrate. To further demonstrate that the fluorescence observed was due to the activity of the DNAzyme, an inactive DNAzyme substrate strand was prepared in which the adenosine ribonucleotide at the cleavage site was replaced with a deoxyribonucleotide. HeLa cells using such an inactive probe showed less fluorescence than those with the active probe (Figure 2c, 2d). Based on z-stack images, the Cy3 fluorescence signal was located at the same focal plane as the nucleus staining, suggesting intracellular localization of 39ES-AuNP probes (Figure S8).
Figure 2.
(a, b) Confocal microscopy images of HeLa cells treated (a) with or (b) without uranyl and incubated with active 39ES-AuNP. Cells incubated with inactive 39ES-AuNP probes and (c) with or (d) without uranyl were also imaged. The red channel is Cy3 fluorescence from activated 39ES-AuNP and the blue channel is Hoechst 33258 for nucleus staining. Differential interference contrast microscopy (DIC) images of cells are shown in the third column. Scale bar = 20 µm.
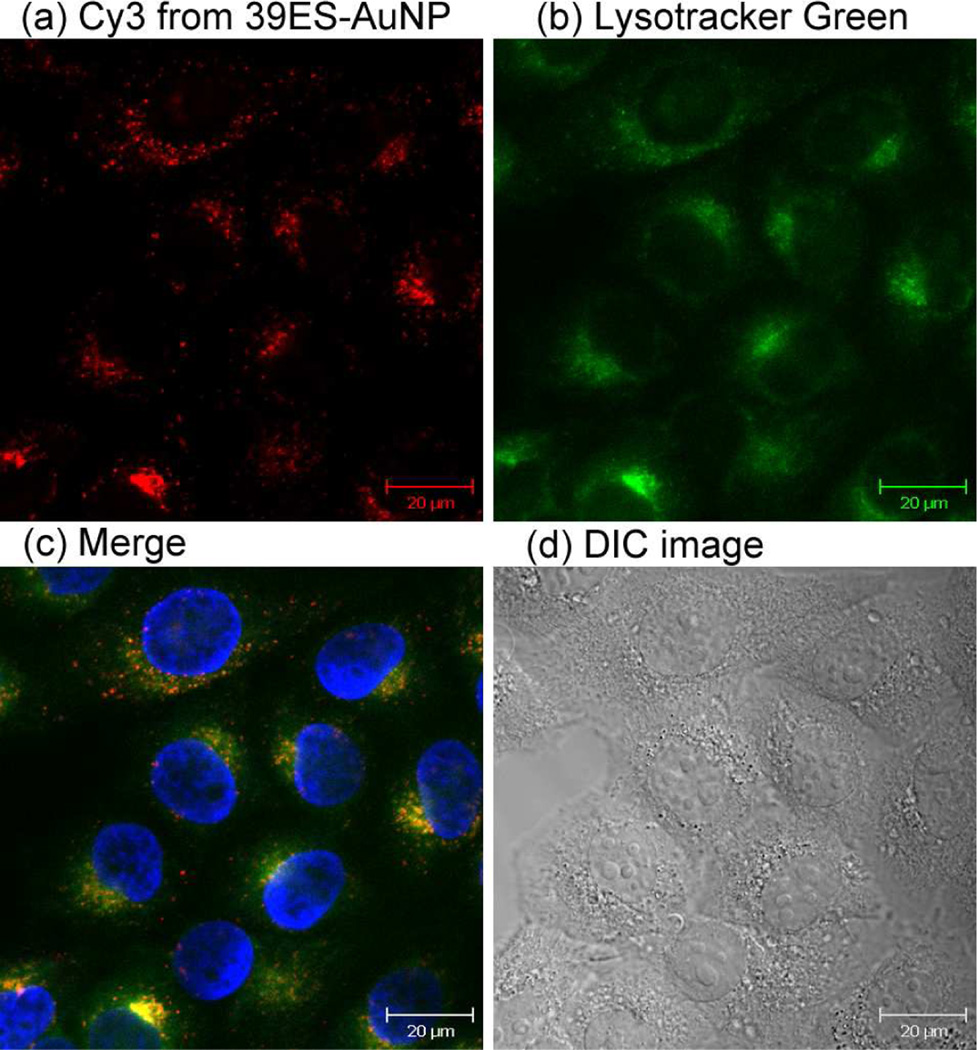
To study the distribution of 39ES-AuNP inside cells, Lysotracker was used to specifically stain the lysosomes of cells. Fluorescence from 39ES-AuNP and Lysotracker showed good colocalization, and this conclusion is further supported by the calculated Pearson's correlation coefficient of 0.61 using an ImageJ plugin14 (see Supporting Information), which indicates that the 39ES-AuNP probe is mainly transported to the lysosomes (Figure 3). This result is consistent with DNA-AuNPs being known to enter HeLa cells via receptor-mediated endocytosis (Figure S9).15 Such colocalization may have implications on the mechanism of uranyl detoxification inside cells. Lysosomes are known to play ubiquitous sequestration and detoxification roles for heavy metals, such as copper, zinc, cadmium and mercury.16 Although the mechanism for uranyl detoxification inside mammalian cells is not well understood, accumulation of uranyl inside lysosomes as detected by our sensor suggests that the cells may use similar strategies to sequester uranyl inside lysosomes as a way for detoxification. Further study is required to confirm this suggestion.
Figure 3.
Localization of 39ES-AuNP inside cells. (a) Cy3 fluorescence from a 39ES-AuNP probe. (b) Fluorescence from Lysotracker Green staining. (c) Overlay of Cy3 and Lysotracker Green fluorescence. Nucleus staining is shown in blue. (d) DIC images of cells.
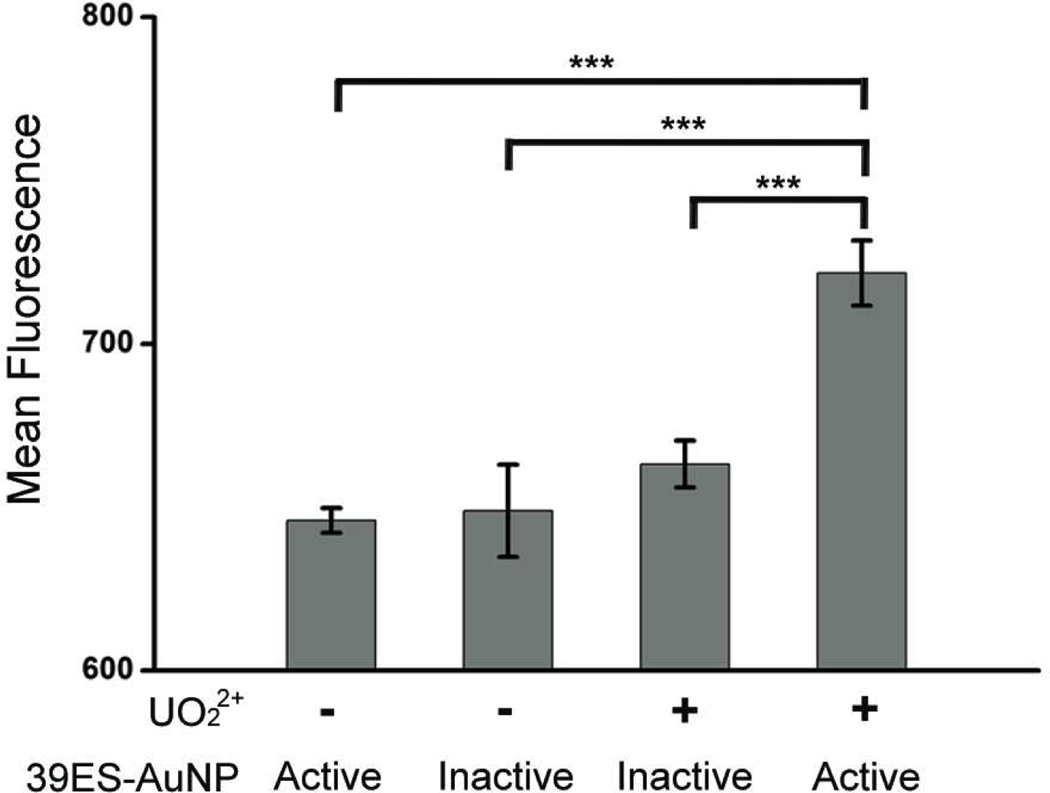
Finally, to ensure that the observations made via confocal microscopy apply to the whole cell population, we examined the fluorescence coming from a population of cells and quantified intracellular fluorescence in cells with or without uranyl using flow cytometry. The results shown in Figure 3 suggest that HeLa cells pretreated with uranyl citrate had a higher level of fluorescence than untreated cells. Compared with the active 39ES-AuNP probe, the inactive 39ES-AuNP probe showed less fluorescence in both uranyl-treated and untreated cells. The difference between the positive group and all three control groups is statistically significant (p<0.001). These results demonstrate retention of signaling ability of the probes within live cells.
In conclusion, we have developed the first DNAzyme-based probe of metal ions in living cells by conjugating a fluorescent DNAzyme onto AuNPs. Since a number of DNAzymes specific for different metal ions have been obtained and additional DNAzymes for other metal ions can be selected using in vitro selection, the method demonstrated here provides us with a simple and general platform to convert any of these DNAzymes into intracellular probes for a wide range of metal ions. Continued development of these DNAzyme-AuNP probes will allow for a better understanding of the localization and distribution of metal ions in biological systems.
Supplementary Material
Figure 4.
Flow cytometric quantification of cell associated fluorescence. Cells with or without uranyl treatment, and containing either active or inactive 39ES-AuNP were measured. For each condition, averaged mean fluorescence intensity is plotted, with error bars indicating standard deviation. Asterisks indicate statistically significant differences (p<.001).
Acknowledgments
Funding Sources
This work was supported by the U.S. National Institute of Health (Grant ES016865) and by the Office of Science (BER), the U.S. Department of Energy (DE-FG02-08ER64568). P.W. was supported by NSF Grant 0965918 IGERT: Training the Next Generation of Researchers in Cellular and Molecular Mechanics and BioNanotechnology. K.H. is supported by the NIH Molecular Biophysics Training Grant (Grant T32GM008276), and by the Lester E. and Kathleen A. Coleman Fellowship at the University of Illinois at Urbana-Champaign.
Footnotes
ASSOCIATED CONTENT
Supporting Information.
Detailed synthesis and characterization data as well as supplementary results. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.(a) Finney L, Mandava S, Ursos L, Zhang W, Rodi D, Vogt S, Legnini D, Maser J, Ikpatt F, Olopade OI, Glesne D. Proc. Natl. Acad. Sci. U.S.A. 2007;104:2247. doi: 10.1073/pnas.0607238104. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Dodani SC, Domaille DW, Nam CI, Miller EW, Finney LA, Vogt S, Chang CJ. Proc. Natl. Acad. Sci. U.S.A. 2011;108:5980. doi: 10.1073/pnas.1009932108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.(a) Zhang J, Campbell RE, Ting AY, Tsien RY. Nat. Rev. Mol. Cell. Biol. 2002;3:906. doi: 10.1038/nrm976. [DOI] [PubMed] [Google Scholar]; (b) Domaille DW, Que EL, Chang CJ. Nat. Chem. Biol. 2008;4:168. doi: 10.1038/nchembio.69. [DOI] [PubMed] [Google Scholar]; (c) Que EL, Domaille DW, Chang CJ. Chem. Rev. 2008;108:4328. doi: 10.1021/cr078203u. [DOI] [PubMed] [Google Scholar]; (d) Marbella L, Serli-Mitasev B, Basu P. Angew. Chem. Int. Ed. 2009;48:3996. doi: 10.1002/anie.200806297. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) McRae R, Bagchi P, Sumalekshmy S, Fahrni CJ. Chem. Rev. 2009;109:4780. doi: 10.1021/cr900223a. [DOI] [PMC free article] [PubMed] [Google Scholar]; (f) Pluth MD, Tomat E, Lippard SJ. Annu. Rev. Biochem. 2011;80:333. doi: 10.1146/annurev-biochem-061009-091643. [DOI] [PMC free article] [PubMed] [Google Scholar]; (g) Palmer AE, Qin Y, Park JG, McCombs JE. Trends Biotechnol. 2011;29:144. doi: 10.1016/j.tibtech.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]; (h) Chan J, Dodani SC, Chang CJ. Nat. Chem. 2012;4:973. doi: 10.1038/nchem.1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.(a) Kerr R, Lev-Ram V, Baird G, Vincent P, Tsien RY, Schafer WR. Neuron. 2000;26:583. doi: 10.1016/s0896-6273(00)81196-4. [DOI] [PubMed] [Google Scholar]; (b) Zeng L, Miller EW, Pralle A, Isacoff EY, Chang CJ. J. Am. Chem. Soc. 2006;128:10. doi: 10.1021/ja055064u. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Liu J, Lu Y. Chem. Commun. 2007:4872. doi: 10.1039/b712421j. [DOI] [PubMed] [Google Scholar]; (d) Qian F, Zhang CL, Zhang YM, He WJ, Gao X, Hu P, Guo ZJ. J. Am. Chem. Soc. 2009;131:1460. doi: 10.1021/ja806489y. [DOI] [PubMed] [Google Scholar]; (e) Tomat E, Lippard SJ. Curr. Opin. Chem. Biol. 2010;14:225. doi: 10.1016/j.cbpa.2009.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]; (f) Wegner SV, Arslan H, Sunbul M, Yin J, He C. J. Am. Chem. Soc. 2010;132:2567. doi: 10.1021/ja9097324. [DOI] [PubMed] [Google Scholar]; (g) Qin Y, Dittmer PJ, Park JG, Jansen KB, Palmer AE. Proc. Natl. Acad. Sci. U.S.A. 2011;108:7351. doi: 10.1073/pnas.1015686108. [DOI] [PMC free article] [PubMed] [Google Scholar]; (h) Hirayama T, Van de Bittner GC, Gray LW, Lutsenko S, Chang CJ. Proc. Natl. Acad. Sci. U.S.A. 2012;109:2228. doi: 10.1073/pnas.1113729109. [DOI] [PMC free article] [PubMed] [Google Scholar]; (i) Liu J, Lu Y. J. Am. Chem. Soc. 2007;129:9838. doi: 10.1021/ja0717358. [DOI] [PubMed] [Google Scholar]; (j) Sen S, Sarkar S, Chattopadhyay B, Moirangthem A, Basu A, Dhara K, Chattopadhyay P. Analyst. 2012;137:3335. doi: 10.1039/c2an35258c. [DOI] [PubMed] [Google Scholar]; (k) Wei YB, Aydin Z, Zhang Y, Liu ZW, Guo ML. Chembiochem. 2012;13:1569. doi: 10.1002/cbic.201200202. [DOI] [PubMed] [Google Scholar]
- 4.(a) Deo S, Godwin HA. J. Am. Chem. Soc. 2000;122:174. [Google Scholar]; (b) He QW, Miller EW, Wong AP, Chang CJ. J. Am. Chem. Soc. 2006;128:9316. doi: 10.1021/ja063029x. [DOI] [PubMed] [Google Scholar]; (c) Cheng TY, Xu YF, Zhang SY, Zhu WP, Qian XH, Duan LP. J. Am. Chem. Soc. 2008;130:16160. doi: 10.1021/ja806928n. [DOI] [PubMed] [Google Scholar]; (d) Nolan EM, Lippard SJ. Chem. Rev. 2008;108:3443. doi: 10.1021/cr068000q. [DOI] [PubMed] [Google Scholar]; (e) Dodani SC, He QW, Chang C. J. Am. Chem. Soc. 2009;131:18020. doi: 10.1021/ja906500m. [DOI] [PMC free article] [PubMed] [Google Scholar]; (f) Au-Yeung HY, New EJ, Chang CJ. Chem. Commun. 2012;48:5268. doi: 10.1039/c2cc31681a. [DOI] [PubMed] [Google Scholar]
- 5.Breaker RR, Joyce GF. Chem. Biol. 1994;1:223. doi: 10.1016/1074-5521(94)90014-0. [DOI] [PubMed] [Google Scholar]
- 6.(a) Li J, Lu Y. J. Am. Chem. Soc. 2000;122:10466. [Google Scholar]; (b) Liu ZJ, Mei SHJ, Brennan JD, Li YF. J. Am. Chem. Soc. 2003;125:7539. doi: 10.1021/ja035208+. [DOI] [PubMed] [Google Scholar]; (c) Xiao Y, Rowe AA, Plaxco KW. J. Am. Chem. Soc. 2007;129:262. doi: 10.1021/ja067278x. [DOI] [PubMed] [Google Scholar]; (d) Wang ZD, Lee JH, Lu Y. Chem. Commun. 2008:6005. doi: 10.1039/b812755g. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Hollenstein M, Hipolito C, Lam C, Dietrich D, Perrin DM. Angew. Chem. Int. Ed. 2008;47:4346. doi: 10.1002/anie.200800960. [DOI] [PubMed] [Google Scholar]; (f) Liu JW, Cao ZH, Lu Y. Chem. Rev. 2009;109:1948. doi: 10.1021/cr030183i. [DOI] [PMC free article] [PubMed] [Google Scholar]; (g) Li T, Dong SJ, Wang E. Anal. Chem. 2009;81:2144. doi: 10.1021/ac900188y. [DOI] [PubMed] [Google Scholar]; (h) Yin BC, Ye BC, Tan WH, Wang H, Xie CC. J. Am. Chem. Soc. 2009;131:14624. doi: 10.1021/ja9062426. [DOI] [PMC free article] [PubMed] [Google Scholar]; (i) Lan T, Furuya K, Lu Y. Chem. Commun. 2010;46:3896. doi: 10.1039/b926910j. [DOI] [PMC free article] [PubMed] [Google Scholar]; (j) Torabi SF, Lu Y. Faraday Discuss. 2011;149:125. doi: 10.1039/c005404f. [DOI] [PMC free article] [PubMed] [Google Scholar]; (k) Wang ZD, Lu Y. J. Mater. Chem. 2009;19:1788–1798. doi: 10.1039/B813939C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Navani NK, Li YF. Curr. Opin. Chem. Biol. 2006;10:272. doi: 10.1016/j.cbpa.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 8.(a) Liu J, Brown AK, Meng X, Cropek DM, Istok JD, Watson DB, Lu Y. Proc. Natl. Acad. Sci. U.S.A. 2007;104:2056. doi: 10.1073/pnas.0607875104. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Liu J, Lu Y. Angew. Chem. Int. Ed. 2007;46:7587. doi: 10.1002/anie.200702006. [DOI] [PubMed] [Google Scholar]; (c) Moshe M, Elbaz J, Willner I. Nano Lett. 2009;9:1196. doi: 10.1021/nl803887y. [DOI] [PubMed] [Google Scholar]; (d) Xiang Y, Wang ZD, Xing H, Wong NY, Lu Y. Anal. Chem. 2010;82:4122. doi: 10.1021/ac100244h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.(a) Lin RH, Wu LJ, Lee CH, Lin-Shiau SY. Mutat. Res. 1993;319:197. doi: 10.1016/0165-1218(93)90079-s. [DOI] [PubMed] [Google Scholar]; (b) Domingo JL. Reprod. Toxicol. 2001;15:603. doi: 10.1016/s0890-6238(01)00181-2. [DOI] [PubMed] [Google Scholar]; (c) Craft ES, Abu-Qare AW, Flaherty MM, Garofolo MC, Rincavage HL, Abou-Donia MB. J. Toxicol. Env. Heal. 2004;7:297. doi: 10.1080/10937400490452714. [DOI] [PubMed] [Google Scholar]; (c) Carriere M, Thiebault C, Milgram S, Avoscan L, Proux O, Gouget B. Chem. Res. Toxicol. 2006;19:1637. doi: 10.1021/tx060206z. [DOI] [PubMed] [Google Scholar]; (d) Kathren RL, Burklin RK. Health Physics. 2008;94:170. doi: 10.1097/01.HP.0000288043.94908.1f. [DOI] [PubMed] [Google Scholar]; (e) Briner W. Int. J. Env. Res. Public Health. 2010;7:303. doi: 10.3390/ijerph7010303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee JH, Wang ZD, Liu JW, Lu Y. J. Am. Chem. Soc. 2008;130:14217. doi: 10.1021/ja803607z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.(a) Rosi NL, Giljohann DA, Thaxton CS, Lytton-Jean AKR, Han MS, Mirkin CA. Science. 2006;312:1027. doi: 10.1126/science.1125559. [DOI] [PubMed] [Google Scholar]; (b) Zheng D, Seferos DS, Giljohann DA, Patel PC, Mirkin CA. Nano Lett. 2009;9:3258. doi: 10.1021/nl901517b. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Seferos DS, Prigodich AE, Giljohann DA, Patel PC, Mirkin CA. Nano Lett. 2009;9:308. doi: 10.1021/nl802958f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.(a) Dulkeith E, Morteani AC, Niedereichholz T, Klar TA, Feldmann J, Levi SA, van Veggel FCJM, Reinhoudt DN, Moller M, Gittins DI. Phys. Rev. Lett. 2002;89 doi: 10.1103/PhysRevLett.89.203002. [DOI] [PubMed] [Google Scholar]; (b) Obliosca JM, Wang P, Tseng F. J. Colloid Interface Sci. 2012;371:34. doi: 10.1016/j.jcis.2011.12.026. [DOI] [PubMed] [Google Scholar]
- 13.Liu J, Lu Y. Nat. Protoc. 2006;1:246. doi: 10.1038/nprot.2006.38. [DOI] [PubMed] [Google Scholar]
- 14.French AP, Mills S, Swarup R, Bennett MJ, Pridmore TP. Nat. Protoc. 2008;3:619. doi: 10.1038/nprot.2008.31. [DOI] [PubMed] [Google Scholar]
- 15.Patel PC, Giljohann DA, Daniel WL, Zheng D, Prigodich AE, Mirkin CA. Bioconjugate Chem. 2010;21:2250. doi: 10.1021/bc1002423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.(a) Ahearn GA, Mandal PK, Mandal A. J. Comp. Physiol. B. 2004;174:439. doi: 10.1007/s00360-004-0438-0. [DOI] [PubMed] [Google Scholar]; (b) Pourahmad J, Ghashang M, Ettehadi HA, Ghalandari R. Environ. Toxicol. 2006;21:349. doi: 10.1002/tox.20196. [DOI] [PubMed] [Google Scholar]; (c) Ahearn G, Sterling K, Mandal P, Roggenbeck B. In: Epithelial Transport Physiol. Gerencser GA, editor. Humana Press; 2010. p. 49. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.