Abstract
Cytochrome oxidase (COX) is the enzyme that constitutes the last step of the mitochondrial electron transport chain for the production of ATP. Measurement of COX activity can be achieved by histochemistry, thus providing information about the metabolic status of the brain. Brain regions with high metabolism will present high COX activity in histochemistry assays and vice versa. Using histochemistry versus biochemistry to assess COX activity presents the advantage of providing a map of the differences in metabolism in discrete brain regions. Moreover, COX histochemistry allows quantifying the activity of a particular brain region, by converting units of optical density into units of activity. In the present work we have devised a methodology that allows not only quantifying differences in COX activity between groups, but also quantifying the amount of COX present in brain tissue sections, by directly relating optical density (OD) measurements to cytochrome C oxidase concentration, something that traditionally is achieved by the use of western blot. For this purpose we created a set of standards of known concentration of COX that were affixed to a nitrocellulose membrane, and this membrane was incubated together with the tissue sections in which COX activity was assessed. A standard curve was created using a gradient of different concentrations of purified bovine heart cytochrome oxidase (from 2 micrograms to 0.1 micrograms in intervals of 0.25 micrograms). This standard curve allowed us to detect changes in optical density as low as 5%, and relate these OD differences with known concentrations of cytochrome C oxidase.
Keywords: Optical density, histochemistry, human brain, substantia nigra, ATP, electron transport chain, image analysis
1. INTRODUCTION
Metabolism has the ultimate goal of producing the energy necessary to maintain all the body functions, and this energy is stored in the form of ATP. The brain is one of the most metabolically demanding organs, requiring high levels of ATP production to maintain its functions, and different brain regions utilize varying amounts of ATP depending on their metabolic status (Hevner and Wong-Riley, 1991). Within the same brain region these differences in metabolic activity can be found at the cellular and subcellular levels (Hevner and Wong-Riley, 1989; 1991; Wong-Riley, 2012). ATP is synthesized in mitochondria by oxidative phosphorylation, a process that takes place at the inner mitochondrial membrane with participation of the electron transport chain. The electron transport chain is composed of a series of enzymes or complexes, of which cytochrome C oxidase (COX or Complex IV) is the terminal enzyme. Cytochrome C oxidase catalyzes the oxidation of its substrate cytochrome C, and the transfer of electrons for the reduction of molecular oxygen. This ultimately produces the electrochemical proton gradient that culminates in the production of ATP, by pumping protons from the matrix to the cytosolic side of the inner mitochondrial membrane (see as a review Wong-Riley, 2012).
Measurement of the metabolic activity of a particular brain region can be reliably achieved by measuring COX activity by histochemistry. Metabolically active brain regions (e.g. with high firing rates) show increased COX histochemistry ratios compared with other regions in which neuronal activity is lower (Hevner and Wong-Riley, 1989; 1991). The advantage of COX histochemistry is that it can provide an accurate mapping of the differences in metabolism in a certain brain region, something that cannot be achieved by the biochemical measurement of COX (see Hevner et al., 1993). The histochemical assay for the detection of COX activity in tissue has been used for decades, however very variable methods have been applied for the conversion of optical density measurements to enzyme amount or to enzymatic activity. When standards have been used, the most commonly reported technique to create these consisted of the preparation of homogenates from tissue that was known to contain a high concentration of COX (i.e. heart tissue; Hess and Pope, 1953), or homogenates from tissue of the same brain area to be studied (Gonzalez-Lima and Garrosa, 1991; Gonzalez-Lima and Jones, 1994). These homogenates are shaped in blocks and sliced using various methodologies. Then, these standards are used to directly compare tissue samples, or their OD values are compared to spectrophotometric measurements of COX activity for the same standards, so they can be transformed into “units of activity” (Gonzalez-Lima and Cada, 1994; Gonzalez-Lima and Jones, 1994). However, none of these methods is able to provide exact measurements of COX expression in the tissue sample, since the concentration of COX in the standards is unknown.
In the present study we show a technique that allows obtaining optical density (OD) measurements of COX histochemistry, and directly convert those to measurements of COX expression in tissue, to then compare COX activity among samples. Our technique has enough resolution to accurately detect changes in COX as low as 5% between samples, in the lower end of the scale. In addition, we show how to analyze the optical data measurements and convert directly to micrograms of COX present in the tissue using NIH Image J software.
2. MATERIALS AND METHODS
2.1 Brain tissue samples
We chose the human substantia nigra/ventral tegmental area (SN/VTA) to test this technique due to our long-term interest in studying SN/VTA pathology in neuropsychiatric and neurological disorders (Perez-Costas et al., 2010, 2012). However, this technique would be equally useful to assess COX expression and activity in other brain areas.
Postmortem human SN/VTA tissue samples (University of Alabama at Birmingham, IRB protocol # N110505002) were obtained from the Alabama Brain Collection (ABC, University of Alabama at Birmingham). These samples were from a 69 year old Caucasian male without any neuropsychiatric or neurological disorder (i.e. control case), with a brain pH of 6.10, and a postmortem interval of 15 hours. The brainstem containing the SN/VTA was dissected immediately upon reception of the brain and frozen in dry ice and stored in a −80°C freezer. The frozen block containing the entire rostro-caudal extent of the SN/VTA was sectioned coronally in five parallel series on a cryostat obtaining 14 micron-thick sections. Four of the series were collected on charged slides, while the fifth series was collected in a vial for protein determination. All the series were preserved at −80°C until use. The first series was stained with thionin (Nissl stain) to assess proper morphological preservation. Using this series for landmark determination, several slides containing the SN/VTA from a parallel series were randomly chosen for COX histochemistry.
2.2 Preparation of standards
Cytochrome C oxidase from bovine heart (Sigma-Aldrich, St. Louis, MO, catalog #C5499) was used to create the standards. The concentration of COX in the vial was 5 mg/ml, and this purified COX had 20 units of activity per mg of enzyme.
To prepare the standards it was necessary to determine the highest reasonable concentration of COX to be expected in brain tissue. This was necessary to create an array of standards spanning the high and low ends of COX expression in the brain. We used some assumptions to do this calculation: Since COX is a very abundant protein in brain tissue we assumed that it could represent at least 1% of the total protein content in the brain. Then, we used series #5 to calculate total protein concentration in this brain area, using a spectrophotometer (Bio-Rad SmartSpec Plus, Bio-Rad, Hercules, CA) and the DC protein assay (Bio-Rad, catalog #500-0113 and #500-0114). Since we knew the number of sections collected per series, we were able to estimate the total protein concentration in a single 14 micron-thick section of the SN/VTA, dividing the total protein concentration of series #5 (13,970μg) by the number of sections contained in a series (n=127). Our calculations yielded a total protein concentration of 110μg per section. Using our assumption that COX could represent as much as 1% of the total protein content in the tissue, we estimated that each SN/VTA section could contain 1.1 μg of COX. Since we wanted to guarantee that none of the COX present in the brain could be out of range of our standards, we set the highest concentration of our standards as 2 μg of pure COX. From this highest concentration we created an array of 16 standards (8 of them ranging from 2μg to 0.25μg in decreasing steps of 0.25μg, and 8 additional standards ranging from 0.1μg to 0.03μg, in decreasing steps of 0.01μg).
Once the concentrations of pure COX to be used as standards were determined, the next step was to design a methodology that would allow us to reliably obtain optical density measurements of known concentrations of pure COX. To achieve this objective, we used a dot-blot apparatus (Bio-Dot SF microfiltration apparatus, Bio-Rad, catalog #170-6542) to affix the desired concentrations of pure COX to a nitrocellulose membrane. Table 1 presents the concentrations of COX that were used to create the standards in the nitrocellulose membranes.
TABLE 1.
Final concentrations of pure COX in standards and detailed
| WELL # | Pure COX final concentration (μg) |
COX stock concentration (μg/Ml) |
Volume from COX stock (μl) |
Volume of TBS (μl) |
|---|---|---|---|---|
| 1 | 2.00 | 0.01 | 200 | 0 |
| 2 | 1.75 | 0.01 | 175 | 25 |
| 3 | 1.50 | 0.01 | 150 | 50 |
| 4 | 1.25 | 0.01 | 125 | 75 |
| 5 | 1.00 | 0.01 | 100 | 100 |
| 6 | 0.75 | 0.01 | 75 | 125 |
| 7 | 0.50 | 0.01 | 50 | 150 |
| 8 | 0.25 | 0.01 | 25 | 175 |
| 9 | 0.10 | 0.01 | 10 | 190 |
| 10 | 0.09 | 0.0005 | 180 | 20 |
| 11 | 0.08 | 0.0005 | 160 | 40 |
| 12 | 0.07 | 0.0005 | 140 | 60 |
| 13 | 0.06 | 0.0005 | 120 | 80 |
| 14 | 0.05 | 0.0005 | 100 | 100 |
| 15 | 0.04 | 0.0005 | 80 | 120 |
| 16 | 0.03 | 0.0005 | 60 | 140 |
chart for their preparation
To create the standards, two stock solutions of bovine heart COX were prepared in Tris-Buffered saline (TBS, Bio-Rad, catalog #170-6435). A 1:500 dilution stock (0.01μg/μl of COX) was prepared to create the 2.0 to 0.10μg standards. A second dilution (1:10,000; 0.0005μg/μl of COX) was prepared to create the 0.09 to 0.03μg standards. From these stock solutions the appropriate volumes were taken to create the standards by further diluting the stocks in TBS (Table 1).
Following the instructions on the manufacturer’s manual, the dot-blot apparatus was assembled using the optional slot-blot insert. This allowed applying the samples in rectangular wells instead of the traditional round-shaped wells. Within the apparatus, filter paper (Bio-Dot SF, Bio-Rad, catalog #162-0161) and the nitrocellulose membrane (Bio-Rad, catalog #162-0117) were sandwiched between the top and the bottom parts of the dot-blot apparatus. After proper assemblage, the system was connected to an in-house vacuum system using a vacuum trap. The nitrocellulose membrane was rinsed 2 times using 100μl of TBS per well. For each rinse vacuum was applied to force the TBS through the membrane. Then, 200μl per well of freshly prepared dilutions of the pure COX in TBS (table 1) were applied (from most concentrated to most diluted) to two rows of the dot-blot apparatus. The unused wells were filled with 200μl of TBS. To affix the COX complex to the membrane, vacuum was applied, and immediately after this, 2 consecutive rinses with TBS were applied using the same vacuum method. These steps allowed the immobilization of COX complex into the nitrocellulose membrane and thus, obtaining an easy to handle and durable support for the standards. Care was taken to uniformly load the wells of the dot-blot apparatus to avoid within-well side-to-side gradients of COX. After this process was completed, the membrane was placed in TBS to be immediately incubated for COX histochemistry together with the sections to be tested for COX activity.
2.3 Preparation of negative controls
These controls serve the purpose of monitoring the specificity of the COX reaction by inactivating the activity of the enzyme prior to the histochemical reaction using sodium azide as the inactivating agent (Seligman et al, 1968). Negative controls were prepared by randomly selecting some of the SN/VTA sections from the same case and pretreating them to inactivate the COX enzyme present in the tissue. The control sections were removed from the −80°C freezer and immediately fan-dried for 25 minutes at room temperature (RT). Sections were immersed in 4% paraformaldehyde in 0.1M phosphate buffer (PB) pH 7.4 for 60 minutes at RT. After five rinses in PB (5 minute each), the slides were incubated in a 10mM sodium azide solution in PB for 17 hours at RT to inactivate COX. After this, sections were rinsed in PB (5 times, 5 minute each) and they were ready to be incubated together with the standards and the sections to be assayed for COX activity.
2.4 COX histochemistry assay
We used a previously published COX histochemistry assay that produces reliable and consistent results in frozen brain samples (Divac et al., 1995; Poeggeler et al., 1998). Prior to start the COX assay, SN/VTA sections were removed from the −80°C freezer and fan-dried as described for the negative controls (see section 2.3). After that, these sections, together with the negative controls and COX standards were placed in the same cuvette for incubation. The incubation medium contained the following components: 22.4 mg cytochrome C (Sigma-Aldrich, catalog #C3131), 115.23 mg diaminobenzidine (DAB, Sigma-Aldrich, catalog #D5637), 4.5 g sucrose (Sigma-Aldrich, catalog #S0389), 12.51 ml of a 1% nickel ammonium sulfate hexahydrate (Aldrich, catalog #A1827) solution, all mixed in 100 ml of 0.1M Hepes buffer (Sigma-Aldrich, catalog #H3375), pH 7.4. This solution was prepared in an aluminum foil-covered beaker under constant stirring adding the cytochrome C, DAB and the sucrose to the Hepes buffer first, and then adding the nickel ammonium sulfate solution dropwise under constant stirring. The sections and the standards were incubated with this solution for 2 hours in an incubator at 37°C in the dark. The COX enzymatic reaction was terminated by immersing the sections and standards in 4% paraformaldehyde in PB for 1 hour at RT. After that, sections and standards were rinsed in PB (4 times, 5 minute each). Sections were then dehydrated in ethanol, cleared in xylene and coverslipped with Eukitt (Electron Microscopy Sciences, catalog #15322). The nitrocellulose membrane containing the standards was rinsed in distilled water and dried at RT between two sheets of filter paper.
2.5 Image acquisition and analysis
Images from SN/VTA sections and standards were acquired using a flatbed scanner at a resolution of 600 dpi and 16 bits grayscale. For optical density analysis all images were saved as uncompressed TIFF files, and then imported into NIH Image J software version 1.46r (http://rsbweb.nih.gov/ij/). In Image J, prior to measuring optical density (OD), background was subtracted for all images first (process>subtract background), using a “rolling ball radius” of 25 pixels for light background. After this, images were inverted to negative (edit>invert) in order to obtain OD values.
A standard curve was created by measuring the OD values for the standards in Image J and then inputting the known values of COX protein concentration in micrograms (table 1) for each standard. To obtain the OD values for each standard, a rectangular selection tool was used to define the “region of interest” (ROI). Once all the OD values for the standards were obtained, Image J calibration tool (image>measure>calibrate) was used to input manually the COX microgram values for each standard versus their OD values. The appropriate type of function (equation) that relates OD versus COX micrograms was selected (i.e. exponential), and the standard curve was plotted, bringing the standard curve graph and the goodness of fit of the curve into view. Once the calibration curve was created, images for analysis were opened in image J, and OD values for ROIs were measured. Rectangular ROIs were used to select several areas within the section, and using the measurement tools (analyze>tools>ROI manager, and then, analyze>measure), OD values were measured and immediately converted by the software to micrograms of COX. Afterwards, micrograms of COX were converted manually to COX activity units, using the information provided by the manufacturer regarding the COX activity units present in the COX used for our standards (i.e. 20 activity units per microgram of COX).
2.6 Statistical analysis
Graphpad Prism 5.04 software (Graphpad Inc., San Diego, CA, USA) was used to perform a regression analysis of optical density values versus COX concentration loaded for the standards. A paired t-test was also performed for the analysis of the interpolated values of COX amount obtained with Image J.
3. RESULTS AND DISCUSSION
In the present work we tested a set of 16 different concentrations of pure COX for the creation of our standards (from 2 μg as the highest value to 0.03 μg as the lowest, see Table 1). Our tests showed that in the low end of the curve, the limit of resolution for reliable measurement of COX optical density values was 0.1 μg of COX, since below this value the measurement of OD levels using Image J software became unreliable for reproducibility. Thus, a set of 9 different COX concentrations ranging from 2 to 0.1 μg was the optimal range to build our standard curve to assess COX expression in brain tissue sections (Figure 1), which was enough to cover the whole range of COX amount present in our brain tissue sections. Traditionally, COX standards were created using brain homogenates sectioned at different thicknesses that were incubated in the same conditions as the brain sections to be analyzed, thus allowing a quantitative analysis of COX activity (Gonzalez-Lima and Cada, 1994; Gonzalez-Lima and Jones, 1994). Since sample preparation can significantly influence enzymatic activity of COX (Hevner et al., 1993) the use of pure COX enzyme that is affixed to an inert matrix such as nitrocellulose (thus requiring a minimal amount of manipulation) can eliminate those issues. However, in our method it is important to avoid gradients in the distribution of COX within the well when creating the standards. The uniformity in the amount of COX in the well allows the use of different voxel sizes for the measurement of COX amount in the standards. In our case, the use of different voxel size within the same sample gave identical results.
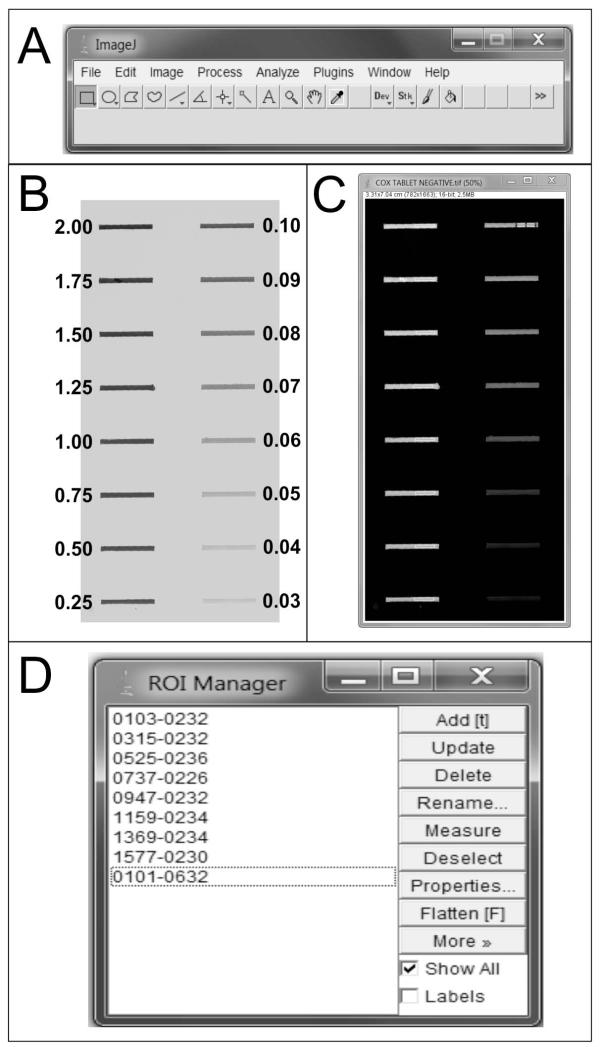
Figure 1. Preparation of standards and Image J.
After opening Image J (A), the scanned image of the nitrocellulose membrane containing the COX standards is imported into the software, and background is subtracted (see section 2.5). B) Scanned image of the nitrocellulose membrane after background subtraction. The numbers indicate the amount of pure COX loaded for each standard, in micrograms. C) Image of the standards after it has been inverted in Image J, ready to be sampled. Note that bands of 0.09 to 0.03 micrograms of COX are also visible, but they are not reliably detected by the software. D) The ROI manager records the location of each measurement within the image, thus allowing having all the ROIs visible in the image.
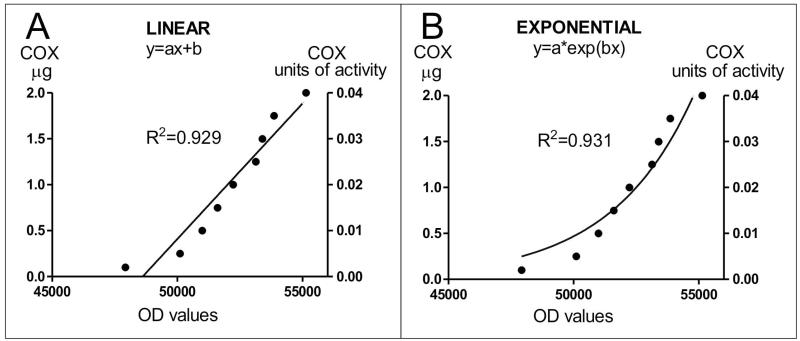
Our COX histochemical experiments show that the concentration of our standards and their corresponding OD values fit well a linear relation (R2 value=0.929 Figure 2A). Since the amount of histochemical reaction for COX is mainly dependent on the amount of enzyme (Hevner and Wong-Riley, 1989), a linear relation is usually predicted for the histochemical reaction of standards, as it occurs for the spectrophotometric measurement of COX activity (Hevner et al., 1993). However, with our experimental conditions, and for the range of COX enzyme contained in our standards, the mathematical analysis of our results reveals that the predicted relation between the amount of COX in the standards and their OD values also fits an exponential curve as revealed by the R2 value (see figures 2B and 3). For the range of our standards, an analysis of the interpolated values of COX amount obtained from the two curves using Image J revealed no significant differences (paired t-test assuming equal variances; n=27 values per curve; P=0.3516, df=26). For the two curves, the interpolated data were normally distributed, as revealed by the Kolmogorov-Smirnov normality test (KS=0.15, P>0.10). Finally, a point by point assessment of the interpolated values revealed a high correlation between the two curves (r=0.9662, P<0.0001). Differences in turnover of individual enzymes may account for the exponential COX reaction observed for our standards in our experimental conditions. Total COX activity is equal to the product of COX amount and COX turnover (Hevner and Wong-Riley, 1989). Although a minor role for enzyme turnover has been postulated (Hevner and Wong-Riley, 1989), this cannot be discarded when we consider a reaction taking place in a biological material (see the differences between ROI #5 and ROI #8 in figure 4).
Figure 2. Relation between micrograms of COX, optical density and units of activity.
In our experimental conditions and for the values of the standards, the analysis of the best fit equation for the relation between micrograms of COX loaded in the standards and OD values (as a surrogate of the reaction product) showed that both a linear (A) and an exponential (B) curve provided a good fit for the data, as shown by the R2 values. For both graphs, the equivalence of COX micrograms to COX activity units is shown in the right Y axis (see section 2.5).
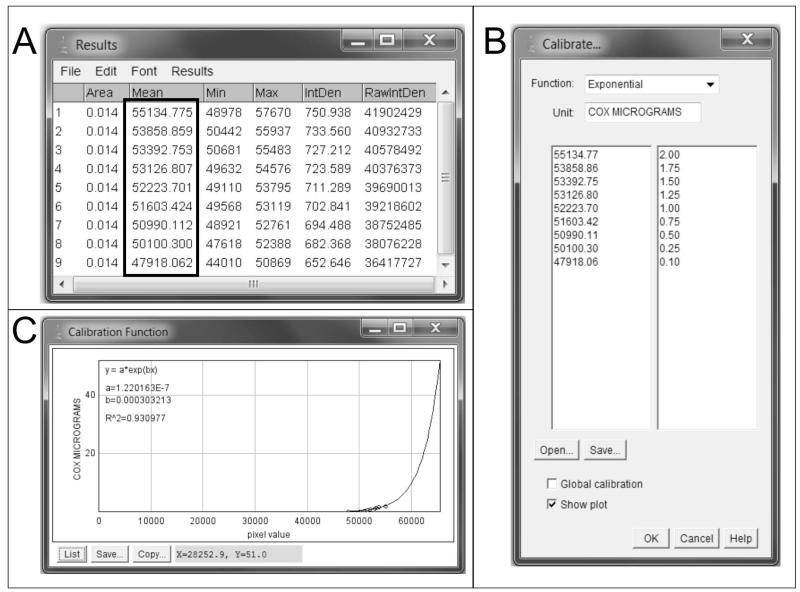
Figure 3. Creation of a standard curve in Image J.
A) OD measurements are obtained for each standard (see box). B) Using the calibration tools of Image J, the appropriate function (equation) is selected (i.e. exponential). The OD values for each standard obtained in A are inputted in the table together with the amount of COX that was loaded for each standard, in micrograms. C) The curve is then plotted and ready to start measuring COX activity in the brain tissue samples.
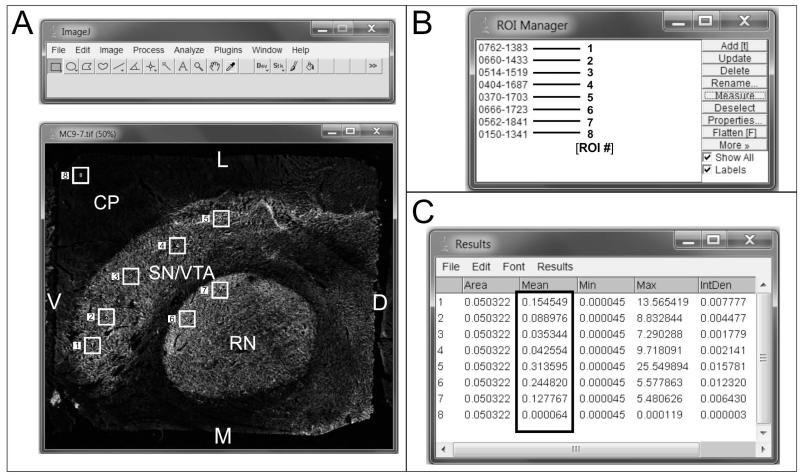
Figure 4. Measurement of COX activity in brain sections.
A) A digitized image of COX histochemistry in a section through the human brainstem containing the SN/VTA and the red nucleus (RN) is opened in Image J. This image has already been processed for background subtraction (see section 2.5) and inverted. A total of eight ROIs were randomly selected, 1-5 in the SN/VTA, 6 and 7 in the RN, and 8 in the cerebellar peduncle (CP), a tissue area that presented little to no COX activity. B) The ROI manager records the location of each ROI selected for measurement. For informative purposes we have noted the number of the ROI next to each location. C) The results of the measurements of OD in the 8 ROIs are converted directly to micrograms of COX (boxed area). Note the big difference in COX amount in micrograms between ROI #5 and ROI #8, calculated by Image J. This can also be inferred qualitatively by the difference in “brightness” between those two areas. Other data in the results include the area of the ROIs, maximum and minimum pixel intensity, and integrated density (IntDen), which is the result of multiplying the area by the mean intensity.
D: dorsal; L: lateral; M: medial; V: ventral.
Several agents have been shown to inhibit COX activity. Among these are chemical fixatives (Chalmers and Edgerton, 1989), as well as sodium azide (Seligman et al., 1968). In our experiments, we used a combination of both methods to ensure a complete abolition of COX activity in our controls (see section 2.3). Our negative controls performed this way showed no residual activity of COX in sections of the SN/VTA (not shown).
The use of COX OD measurements together with standards created using known amounts of enzyme allows the quantification of the amount of enzyme present in brain sections (Figure 4). Our methodology allows for the detection of differences as low as 5% in COX concentration, and hence activity, in the lower end of the standard scale. This is important because subtle differences in COX amount in different subregions can thus be quantified, since there is a close relation between COX OD measured by histochemistry in sections, and the amount of enzyme present in those sections (Hevner and Wong-Riley, 1989). As it can be seen in figure 4, different ROIs within the SN/VTA show different amounts of COX in micrograms (compare the value of micrograms of COX for adjacent ROIs # 4 and 5, with ROI#8 located in the cerebellar peduncle), which indicates the existence of differences in the metabolic status of different subnuclei and between different regions (Hevner and Wong-Riley, 1989). The fact that these differences can be measured with our methodology allows sampling in brain subregions to detect differences in COX activity that otherwise would be masked by the use of biochemical assays for its measurement (Hevner et al., 1993). In addition, with our methodology the amount of COX present in the tissue is easily converted to units of activity using the information provided by the manufacturer, and does not require the use of any biochemical assay.
Our methodology employs metal enhancement to increase the contrast between gray and white matter in the histochemical reaction, something that is not achieved using DAB alone (Divac et al., 1995). The use of metal enhancement can negatively affect the COX reaction by making regions with low amount of COX appear stained qualitatively equal to regions with high COX activity, due to the saturation of the system (Gonzalez-Lima and Cada, 1994; Gonzalez-Lima and Jones, 1994). Our quantitative analysis shows that this is not the case for our samples, since different ROIs within the same section present different amounts of COX OD (compare ROI#5 and ROI#4 with ROI#8; Figure 4), which allows quantifying COX amount and activity in different subregions of the SN/VTA.
Quantification of proteins is important, since the expression of a particular protein in the brain can reflect a pathological status in a disease (see e.g. Perez-Costas et al., 2012). Proteins are usually quantified by their immunological detection in western blot techniques. However, western blots can mask the subtle differences in protein expression in a particular neuronal subpopulation in a brain region, and at the same time does not provide the anatomical distribution of that protein in the tissue. The use of a method that can give an accurate measurement of protein quantity and activity and at the same time provides a map of the expression of that protein is desirable, especially if regional differences are expected or observed (Hevner and Wong-Riley, 1993). That said, there is a series of assumptions that have to be taken into account when quantifying OD with histochemistry for COX. This type of analysis works with the assumption that the entire COX present in the tissue is functional, thus the differences observed by histochemistry are based on amount of enzyme. In the case of a structural deficiency of COX that impairs the functioning of the complex, histochemistry will underestimate the amount of COX present in the tissue, and it will only determine the amount of functional COX present based on its ability to produce a reaction product that is measurable by OD.
Another important parameter to take into account when quantifying enzymatic activity is the fact that the enzyme has to be active for the reaction to take place. The method of preservation of the tissue has to ensure that the full activity of the enzyme is kept. Our samples were all fresh-frozen human postmortem brain samples, not subjected to chemical fixation. Chemical fixation has been shown to diminish the enzymatic activity of COX at various degrees and a reduction of up to 10.5% has been reported, with some neurons being more affected than others (Chalmers and Edgerton, 1989). Although some authors have described a small decrease of COX activity after chemical fixation of the sections (Gonzalez-Lima and Jones, 1994), this small reduction in activity can mask subtle differences in COX activity within and between samples, especially if differential effects are produced. Fixation is usually utilized to avoid detachment of sections from slides through the histochemical procedure (Gonzalez-Lima and Jones, 1994). In our case, our samples were all frozen, and no detachment of sections from the slides occurred. In our protocol fixation was only used as the last step of the histochemical reaction to stop the enzymatic activity of COX. Moreover, freezing of the samples has been shown not to have an effect in COX activity (Hevner at al., 1993).
A variety of other methods have been used to assess cell metabolism. These include studying the expression of mRNA levels for specific subunits of the COX enzyme (Vila et al., 1997; Perier et al., 2000; Bacci et al., 2002; Rolland et al., 2007), assessing the activity of the enzyme succinate dehydrogenase that participates both in the electron transport chain and in the tricarboxylic acid cycle (Bertoni-Freddari et al., 2001; Bubber et al., 2011), the measurement of 2-deoxyglucose levels (see e.g. Porrino et al., 1987; Mitchell et al., 1989, 1992), and the analysis of c-fos expression by immunohistochemistry (see e.g. Joh and Weiser, 1993; Schulte et al., 2006; Reyes and Mitrofanis, 2008). Among these methods, the measurement of COX subunit I mRNA levels has been shown to be a very reliable marker that correlates well with cytoplasmic COX activity, although it requires the use of radioactive mRNA probes and dedicated laboratory space. Succinate dehydrogenase assays provide a good measurement of general metabolic status, but do not assess specifically the health of the electron transport chain. On the other hand, 2-deoxyglucose measurements have been used reliably to measure short term changes in metabolism, but COX measurements are more reliable for the assessment of long term metabolic changes (Vila et al., 1997). Finally, c-fos immunohistochemistry has been used to assess neuronal activity. However, the use of c-fos as a metabolic marker presents several problems (Dragunow and Faull, 1989), and is only reliable for the study of acute metabolic changes (Hoffman et al., 1993; Hoffman and Lyo, 2002).
4. CONCLUSIONS
Here we have devised a method to accurately measure the amount of COX enzyme present in brain tissue sections, using standards of known COX concentration. The amount of COX can be easily converted into activity units without the need of complex biochemical assays. Our methodology can detect differences as low as 5% in COX amount. This is useful to analyze and compare the relative abundance of COX between different regions within a brain, or between subregions within a brain region.
Highlights.
Our technique allows the quantification of COX amount directly on brain sections
Changes of as low as 5% in COX amount between samples can be detected
Our technique is simple and does not require specialized equipment
ACKNOWLEDGMENTS
The authors wish to thank the Alabama Brain Collection (University of Alabama at Birmingham) for providing the human brain tissue used in this work. This work was supported by NIMH RO1 grant MH066123 to MMF, EPC and RCR.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Bacci JJ, Kerkerian-Le Goff L, Salin P. Effects of intralaminar thalamic nuclei lesion on glutamic acid decarboxylase (GAD65 and GAD67) and cytochrome oxidase subunit I mRNA expression in the basal ganglia of the rat. Eur. J. Neurosci. 2002;15:1918–28. doi: 10.1046/j.1460-9568.2002.02039.x. [DOI] [PubMed] [Google Scholar]
- Bertoni-Freddari C, Fattoretti P, Casoli T, Di Stefano G, Solazzi M, Gracciotti N, Pompei P. Mapping of mitochondrial metabolic competence by cytochrome oxidase and succinic dehydrogenase cytochemistry. J. Histochem. Cytochem. 2001;49:1191–2. doi: 10.1177/002215540104900915. [DOI] [PubMed] [Google Scholar]
- Bubber P, Hartounian V, Gibson GE, Blass JP. Abnormalities in the tricarboxylic acid (TCA) cycle in the brains of schizophrenia patients. Eur. Neuropsychopharmacol. 2011;21:254–60. doi: 10.1016/j.euroneuro.2010.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalmers GR, Edgerton VR. Marked and variable inhibition by chemical fixation of cytochrome oxidase and succinate dehydrogenase in single motoneurons. J. Histochem. Cytochem. 1989;37:899–901. doi: 10.1177/37.6.2542395. [DOI] [PubMed] [Google Scholar]
- Divac I, Mojsilovic-Petrovic J, Lopez-Figueroa MO, Petrovic-Minic B, Moller M. Improved contrast in histochemical detection of cytochrome oxidase: metallic ions protocol. J. Neurosci. Methods. 1995;56:105–13. doi: 10.1016/0165-0270(94)00112-t. [DOI] [PubMed] [Google Scholar]
- Dragunow M, Faull R. The use of c-fos as a metabolic marker in neuronal pathway tracing. J. Neurosci. Methods. 1989;29:261–5. doi: 10.1016/0165-0270(89)90150-7. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Lima F, Cada A. Cytochrome oxidase activity in the auditory system of the mouse: a qualitative and quantitative histochemical study. Neuroscience. 1994;63:559–78. doi: 10.1016/0306-4522(94)90550-9. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Lima F, Jones D. Quantitative mapping of cytochrome oxidase activity in the central auditory system of the gerbil: a study with calibrated activity standards and metal-intensified histochemistry. Brain Res. 1994;660:34–49. doi: 10.1016/0006-8993(94)90836-2. [DOI] [PubMed] [Google Scholar]
- Hess HH, Pope A. Ultramicrospectrophotometric determination of cytochrome oxidase for quantitative histochemistry. J. Biol. Chem. 1953;204:295–306. [PubMed] [Google Scholar]
- Hevner RF, Liu S, Wong-Riley MTT. An optimized method for determining cytochrome oxidase activity in brain tissue homogenates. J. Neurosci. Methods. 1993;50:309–19. doi: 10.1016/0165-0270(93)90038-s. [DOI] [PubMed] [Google Scholar]
- Hevner RF, Wong-Riley MTT. Brain cytochrome oxidase: purification, antibody production, and immunohistochemical/histochemical correlations in the CNS. J. Neurosci. 1989;9:3884–98. doi: 10.1523/JNEUROSCI.09-11-03884.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hevner RF, Wong-Riley MTT. Neuronal expression of nuclear and mitochondrial genes for cytochrome oxidase (CO) subunits analyzed by in situ hybridization: comparison with CO activity and protein. J. Neurosci. 1991;11:1942–58. doi: 10.1523/JNEUROSCI.11-07-01942.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hevner RF, Wong-Riley MTT. An optimized method for determining cytochrome oxidase activity in brain tissue homogenates. J. Neurosci. Methods. 1993;50:309–19. doi: 10.1016/0165-0270(93)90038-s. [DOI] [PubMed] [Google Scholar]
- Hoffman GE, Lyo D. Anatomical markers of activity in neuroendocrine systems: are we all ‘fos-ed out’? J. Neuroendocrinol. 2002;14:259–68. doi: 10.1046/j.1365-2826.2002.00775.x. [DOI] [PubMed] [Google Scholar]
- Hoffman GE, Smith MS, Verbalis JG. c-Fos and related immediate early gene products as markers of activity in neuroendocrine systems. Front. Neuroendocrinol. 1993;14:173–213. doi: 10.1006/frne.1993.1006. [DOI] [PubMed] [Google Scholar]
- Joh TH, Weiser M. Early and late molecular events occurring following neuronal degeneration in the dopamine system. Adv. Neurol. 1993;60:316–20. [PubMed] [Google Scholar]
- Mitchell IJ, Boyce S, Sambrook MA, Crossman AR. A 2-deoxyglucose study of the effects of dopamine agonists on the parkinsonian primate brain. Implications for the neural mechanisms that mediate dopamine agonist-induced dyskinesia. Brain. 1992;115:809–24. doi: 10.1093/brain/115.3.809. [DOI] [PubMed] [Google Scholar]
- Mitchell IJ, Clarke CE, Boyce S, Robertson RG, Peggs D, Sambrook MA, Crossman AR. Neural mechanisms underlying parkinsonian symptoms based upon regional uptake of 2-deoxyglucose in monkeys exposed to 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine. Neuroscience. 1989;32:213–26. doi: 10.1016/0306-4522(89)90120-6. [DOI] [PubMed] [Google Scholar]
- Perez-Costas E, Melendez-Ferro M, Rice MW, Conley RR, Roberts RC. Dopamine pathology in schizophrenia: analysis of total and phosphorylated tyrosine hydroxylase in the substantia nigra. Front. Psychiatry. 3:31. doi: 10.3389/fpsyt.2012.00031. doi:10.3389/fpsyt.2012.00031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Costas E, Melendez-Ferro M, Roberts RC. Basal ganglia pathology in schizophrenia: dopamine connections and anomalies. J. Neurochem. 2010;113:287–302. doi: 10.1111/j.1471-4159.2010.06604.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perier C, Vila M, Feger J, Agid Y, Hirsch EC. Functional activity of zona incerta neurons is altered after nigrostriatal denervation in hemiparkinsonian rats. Exp. Neurol. 2000;162:215–24. doi: 10.1006/exnr.1999.7331. [DOI] [PubMed] [Google Scholar]
- Poeggeler B, Rassoulpour A, Guidetti P, Wu HQ, Schwarcz R. Dopaminergic control of kynurenate levels and N-Methyl-D-Aspartate toxicity in the developing rat striatum. Dev. Neurosci. 1998;20:146–53. doi: 10.1159/000017309. [DOI] [PubMed] [Google Scholar]
- Porrino LJ, Burns RS, Crane AM, Palombo E, Kopin IJ, Sokoloff L. Local cerebral metabolic effects of L-dopa therapy in 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine-induced parkinsonism in monkeys. Proc. Natl. Acad. Sci. USA. 1987;84:5995–9. doi: 10.1073/pnas.84.16.5995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes S, Mitrofanis J. Patterns of FOS expression in the spinal cord and periaqueductal grey matter of 6OHDA-lesioned rats. Int. J. Neurosci. 2008;118:1053–79. doi: 10.1080/00207450701239210. [DOI] [PubMed] [Google Scholar]
- Rolland AS, Herrero MT, Garcia-Martinez V, Ruberg M, Hirsch EC, François C. Metabolic activity of cerebellar and basal ganglia-thalamic neurons is reduced in parkinsonism. Brain. 2007;130:265–75. doi: 10.1093/brain/awl337. [DOI] [PubMed] [Google Scholar]
- Schulte T, Brecht S, Herdegen T, Illert M, Mehdorn HM, Hamel W. Induction of immediate early gene expression by high-frequency stimulation of the subthalamic nucleus in rats. Neuroscience. 2006;138:1377–85. doi: 10.1016/j.neuroscience.2005.12.034. [DOI] [PubMed] [Google Scholar]
- Seligman AM, Karnowsky MJ, Wasserkrug HL, Hanker JS. Nondroplet ultrastructural demonstration of cytochrome oxidase activity with a polymerizing osmiophilic reagent, diaminobenzidine (DAB) J. Cell Biol. 1968;38:1–14. doi: 10.1083/jcb.38.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vila M, Levy R, Herrero MT, Ruberg M, Faucheux B, Obeso JA, Agid Y, Hirsch EC. Consequences of nigrostriatal denervation on the functioning of the basal ganglia in human and nonhuman primates: an in situ hybridization study of cytochrome oxidase subunit I mRNA. J. Neurosci. 1997;17:765–73. doi: 10.1523/JNEUROSCI.17-02-00765.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong-Riley MTT. Bigenomic regulation of cytochrome c oxidase in neurons and the tight coupling between neuronal activity and energy metabolism. In: Kadenbach B, editor. Mitochondrial oxidative phosphorylation, Advances in Experimental Medicine and Biology. Vol. 748. Springer; New York: 2012. pp. 283–304. [DOI] [PMC free article] [PubMed] [Google Scholar]