Abstract
MiRNAs have been shown to be involved in regulation of multiple cellular processes including apoptosis. Since a single miRNA can affect the expression of several genes, the utilization of miRNAs for apoptosis engineering in mammalian cells can be more efficient than the conventional approach of manipulating a single gene. Mmu-miR-466h-5p was previously shown to have a pro-apoptotic role in CHO cells by reducing the expression of several anti-apoptotic genes and its transient inhibition delayed both the activation of Caspase-3/7 and the loss of cell viability. The present study evaluates the effect of stable inhibition of mmu-miR-466h-5p in CHO cells on their ability to resist apoptosis onset and their production properties. The expression of mmu-miR-446h-5p in the engineered anti-miR-466h CHO cell line was significantly lower than in the negative control and the parental CHO cells. These engineered cells reached higher maximum viable cell density and extended viability compared with negative control and parental CHO cells in batch cell cultures which resulted in the 53.8 % and 41.6% increase of integral viable cells. The extended viability of anti-miR-466h CHO cells was the result of delayed Caspase-3/7 activation by more than 35 hours, and the increased levels of its anti-apoptotic gene targets (smo, stat5a, dad1, birc6, and bcl2l2) to between 2.1- and 12.5-fold compared with the negative control CHO in apoptotic conditions. The expression of secreted alkaline phosphatase (SEAP) increased 43% and the cell-specific productivity increased 11% in the stable pools of anti-miR-466h CHO compared with the stable pools of negative control CHO cells. The above results demonstrate the potential of this novel approach to create more productive cell lines through stable manipulation of specific miRNA expression.
Introduction
Chinese hamster ovary (CHO) cells are the most commonly used mammalian culture system for production of biologicals due to their ability to make recombinant proteins with human-like post-translational modifications (Baik et al. 2012; Barron et al. 2011b; Kramer et al. 2010; Muller et al. 2008). However, the low tolerance to different stress conditions such as nutrients and growth factors depletion, shear and oxidative stresses, metabolites accumulation, pH, osmolality and hypoxia during production in bioreactors can trigger activation of the apoptosis cascade reducing viable cell concentration and, consequently, the yield and the quality of the produced proteins (Lim et al. 2010; Majors et al. 2007; Muller et al. 2008). As a result, the prevention of apoptosis is a common approach in CHO cells engineering.
Strategies to delay or prevent the onset of apoptosis in mammalian cells cultures are either based on the manipulation of the extracellular environment such as media supplementation, or on genetic engineering methods to change the intracellular environment through over-expressing anti-apoptotic or inhibiting pro-apoptotic proteins (Chiang and Sisk 2005; Lim et al. 2006; Majors et al. 2007; Wong et al. 2006; Zanghi et al. 1999). The major improvements in mammalian cell cultures leading to record viable cell densities and high volumetric productivities were achieved by optimizing media composition and feeding strategies (Hacker et al. 2009; Kim et al. 2012). Genetic and metabolic engineering approaches are being implemented to better understand the cells performance, to improve stress and apoptosis resistance and product quality in mammalian cell cultures (Ishaque et al. 2007; Kim et al. 2012; Lim et al. 2010; Majors et al. 2008b; Nolan and Lee 2011; Suk Ahn and Antnoniewicz 2012; Zhu 2012). Several anti-apoptotic proteins including Bcl-xL, Bcl-2 and Mcl-1 are known to protect the cells from apoptosis onset by maintaining the integrity of mitochondrial membrane. Exogenous over-expression of these proteins protects mammalian cells from apoptosis induction and increases cell density and viability (Fassnacht et al. 1999; Majors et al. 2008b; Majors et al. 2009; Mastrangelo et al. 2000). The over-expression of some other anti-apoptotic proteins such as E1B-19K, Aven, or XIAP enhanced the apoptosis resistance in mammalian cell cultures (Figueroa et al. 2007; Mercille and Massie 1999; Sauerwald et al. 2002). Over-expression of the anti-apoptotic protein, hsp70, increased yield and quality of therapeutic protein production (Ishaque et al. 2007).
RNA interference (RNAi) impacts cellular gene networks by inhibiting mRNA translation (Malphettes and Fussenegger 2006). RNAi inhibition of some pro-apoptotic gene such as Bax and Bak, the apoptosis linked gene Alg-2, and the transcriptional factor Requiem in CHO cells, delayed apoptosis onset resulting in increased cell viability and improved protein production (Lim et al. 2006; Wong et al. 2006). Also, the simultaneous inhibition of Caspase-3 and Caspase-7 enhanced cell viability and thrombopoietin production in CHO cells after sodium butyrate treatment (Sung et al. 2007).
The discovery of microRNAs (miRNAs) offers additional opportunity to improve the performance of mammalian cells for industrial applications. It is estimated that nearly half of all proteins may be affected by miRNAs which are more abundant than transcription factors (Barron et al. 2011b). MiRNAs have been shown to be global regulators of gene expression, affecting almost all central cellular processes and functions including apoptosis regulation. Each miRNA can affect several gene targets by RNAi mechanism and influence the interlinked pathways in parallel or multiple points of the same pathway in the cell. The potential of miRNAs utilization for mammalian cell engineering was shown by analyzing the miRNAs expression profiles under few different physiological conditions (Druz et al. 2011; Gammell 2007; Muller et al. 2008). One of the advantages of using miRNAs rather than regulatory proteins such as transcriptional factors or kinases is that they do not burden the translational machinery and, therefore, reduce the metabolic load in the host cells. As a result, cellular metabolic resources will be better allocated to the production of recombinant protein leading to the increase of cell specific productivity.
Several miRNAs have been shown to be involved in apoptosis regulation (Chan et al. 2005; Cimmino et al. 2005; Druz et al. 2011; Shimizu et al. 2010; Xu et al. 2007). In our previous study, we showed the up-regulation of a large mouse miR 297-669 cluster during apoptotic conditions induced by nutrients depletion in CHO cells. One member of this cluster, mmu-miR-466h-5p, was shown to reduce the expression of five anti-apopototic genes from different apoptosis-initiating pathways (bcl2l2, dad1, birc6, stat5a, and smo) indicating its involvement in apoptosis regulation. Transient antisense knockdown of this miRNA delayed apoptosis activation in nutrient-depleted conditions shown by decreased Caspase-3/7 activation and increased cell viability (Druz et al. 2011).
So far, all the studies associated with miRNA effects on cell behavior were done by transient over-expression or inhibition of specific miRNAs (Barron et al. 2011a; Druz et al. 2011). Although this approach provides quick information on the possible effects of the specific miRNAs, it is limited to small-scale studies and cannot be implemented for industrial applications. The purpose of the presented work was to test the effects of stable inhibition of mmu-miR-466h-5p on the cell growth and apoptosis inactivation, and to evaluate this method as a possible way to delay the activation of the apoptotic cascade and enhance protein production.
Materials and methods
Cells Growth and Viability Studies
Chinese Hamster Ovary cells adapted to growth in suspension (CHO-S) were purchased from Life Technologies, Gaithersburg, MD (Cat. No. 11619-012) and grown in commercial CD-CHO serum-free media (Cat. No. 10743-029) supplemented with 8mM of L-glutamine (Cat. No. 25030-164) at 37°C and 5% CO2 in humidified incubator. For the growth kinetic studies, cells were grown in triplicates in 40 ml of media in 125ml vented shake flasks on a rotary shaker at 130 rpm. The initial cell concentration was 2·105 cells/ml and the cultures were sampled daily. Cells were counted using Cedex (Innovatis AG) and viability was assessed using Trypan Blue exclusion method. The integral of viable cells (IVC) was calculated using the sum of trapezoids approximation (Figueroa et al. 2007).
Creation of stable CHO clones
Stable knockouts of mmu-miR-466h-5p were done by using short hairpin RNAs (shRNAs) constructs (Paddison et al. 2002; Sui et al. 2002) designed to target mature sequence of mmu-miR-466h-5p, (target sequence UGUGUGCAUGUGCUUGUGUGU) or pre-miR-466h precursor (target sequence AAUAUAUAUAUCAUACGCACG), and inserted to pSilencer™ 4.1-CMV neo expression vector (Life Technologies, Cat.No. AM5779). Both shRNAs were designed and ligated into the pSilencer™ 4.1-CMV neo vector according to manufacturer's protocol. The full sequences of top and bottom strands for both shRNA constructs are shown in Table 1. The designed pSilencer™ 4.1-CMV-anti miR-466h-5p, pSilencer™ 4.1-CMV-anti pre-miR-466h vectors and the provided pSilencer™ 4.1-CMV-negative control vector (expressing shRNA with limited homology to any known human, mouse or rat sequences) were transfected into CHO-S cells using FreeStyle™ MAX transfection reagent (Life Technologies, Cat. No. 16447100). Prior to transfection of CHO-S cells with pSilencer™ 4.1-CMV vectors, the transfection conditions were optimized with eGFP-expressing vector (pReceiver-M03, GeneCopoeia, Rockville, MD) using the ExpressPlus assay in Guava Easycyte 5HT (Millipore, Billerica, MA). Stable pools expressing anti miR-466h-5p, anti pre-miR-466h and negative control shRNAs were selected with 1mg/ml of G418 sulfate (Mediatech, Manassas, VA, Cat.No. 30-234-CR). Single clones were obtained from the respective stable pools by growing the cells in ClonaCell™-CHO CD Medium (Stem Cell Technologies, Vancouver, BC, Cat.No. 03815). ClonaCell™-CHO CD Medium is semi-solid that contains methylcellulose allowing the suspension CHO cells to grow as separate colonies in a 3-dimensional matrix. Stable CHO-S pools were diluted to 10,000 cells/ml and mixed with equal volume of G418-containing media (1mg/ml final concentration), and 1ml of the resulting mix was combined with 9 ml of ClonaCell™-CHO CD Medium. After mixing and 15min incubation, the cell suspension was aspirated with 12 ml syringe fitted with a 16-gauge blunt end needle (Stem Cell Technologies, Cat.No. 28110), dispensed to 100 mm tissue dishes and incubated for 12 days in 37°C, 5% CO2 well-humidified incubator. Single colonies were then collected and expanded in regular CD-CHO media.
Table 1.
Sequences of shRNAs constructs used to inhibit mmu-miR-466h-5p
| Name of the construct | Strand | Sequence |
|---|---|---|
| anti miR-466h-5p | top | 5′-GATCCTGTGCATGTGCTTGTGTGTTTCAAGAGAACACACAAGCACATGCACACAA-3′ |
| anti miR-466h-5p | bottom | 5′-AGCTTTGTGTGCATGTGCTTGTGTGTTCTCTTGAAACACACAAGCACATGCACAG-3′ |
| anti pre-miR-466h | top | 5′-GATCCTATATATATCATACGCACGTTCAAGAGACGTGCGTATGATATATATATTA-3′ |
| anti pre-miR-466h | bottom | 5′-AGCTTAATATATATATCATACGCACGTCTCTTGAACGTGCGTATGATATATATAG-3′ |
| negative control | top | 5′-GATCCACTACCGTTGTTATAGGTGTTCAAGAGACACCTATAACAACGGTAGTA-3′ |
| negative control | bottom | 5′-AGCTTACTACCGTTGTTATAGGTGTCTCTTGAACACCTATAACAACGGTAGTG-3′ |
RNA isolation and qRT-PCR analysis
Total RNA was isolated from the samples using mirVana™ miRNA isolation kit, Life Technologies (Cat.No. AM1561). qRT-PCR analysis of the smo, stat5a, dad1, birc6, and bcl2l2 genes was performed in Prism 7900H Sequence Detector (Life Technologies) with 40 amplification cycles according to manufacturer's protocols using the TaqMan® mRNA assays from Life Technologies (Assay IDs: Mm01162710_m1, Mm00839861_m1, Mm01319221_m1, Mm00464380_m1, Mm00432054_m1) and normalized to 18S levels (Life Technologies, Assay ID: Hs99999901_s1) in the respective sample. The preamplification (10 cycles) was done using Applied Systems TaqMan® PreAmp Kit (Part No. 4384267) in the PCR Thermal cycler (Applied Biosystems) after reverse transcription and before qRT-PCR reads (Druz et al. 2011). The mmu-miR-466h-5p quantification was done with TaqMan® microRNA assays (Life Technologies, Assay ID: AM002516), normalized to snoRNA202 levels (Life Technologies, Assay ID:4427975) and analyzed as previously described (Druz et al. 2011).
Apoptotic assay
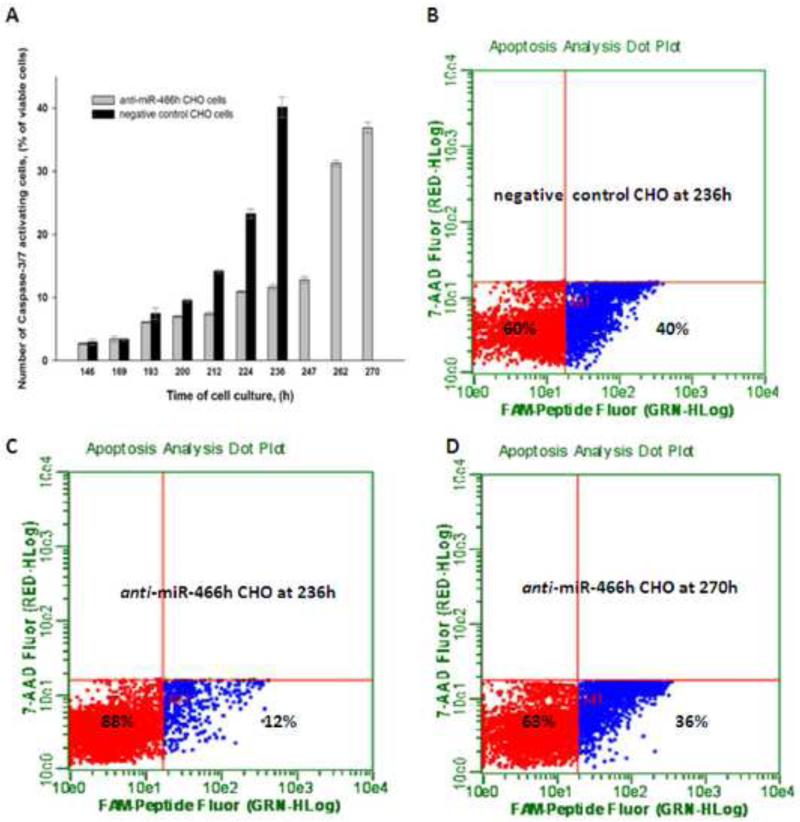
Apoptosis onset was determined by assaying the intracellular activation of Caspase-3/7 using Guava Caspase-3/7 FAM kit (Millipore, Cat.No.4500-0540). The cells were collected at the indicated time points, re-suspended in warm PBS and the concentration was adjusted to 4×105 cells/ml. 100μL of each sample was then incubated with 10μL of Caspase-3/7 carboxyfluorescin (FAM) reagent in U-bottom 96 well plates in 37°C, 5% CO2 humidified incubator with gentle shaking (130rpm) for 1h . The Caspase-3/7 FAM reagent is a non-toxic, cell permeable molecule with FAM-DEVD-FMK sequence which binds to the activated Caspase-3/7 via Asp-Glu-Val-Asp (DEVD) group, and fluoromethyl ketone group (FMK) then covalently links this reagent to Caspase-3/7. The covalently linked FAM reagent is retained in the cell as the non-bound reagent diffuses out of the cell and gets washed away. After 1h of incubation the cells were washed and stained with 7-ADD viability stain. Cells were analyzed using the Caspase assay in Guava Easycyte 5HT (Millipore) where the fluorescence signals and cell gating were previously adjusted with viable, non FAM-labeled cells with more than 99% of the cells in lower left quadrant (red population in Figure 3). Dead cells were gated out, and the apoptosis onset was monitored as the percentage of Caspase-3/7 positive cells in both negative control and anti-466h CHO cells at indicated time points (blue population in Figure 3).
Figure 3.
Activation of Caspase-3/7 in anti-miR-466h and negative control CHO cells. (A) Time course of Caspase-3/7 activation in both cell lines (B), (C) and (D) FACS images of the ratio of viable cells negative for Caspase-3/7 activity (shown in red) and viable cells positive for Caspase-3/7 activity (shown in blue). (B) Negative control CHO at 236h. (C) Anti-miR-466h CHO at 236h. (D) Anti-miR-466h CHO at 270h.
Cells treatment with 4-phenylbutyric acid
Single CHO-S cell clones and non-transfected CHO-S were seeded in 2ml of media at ~2×106 cells/ml in 12-well plates (Corning) and treated with 4-phenylbutyric acid, PBA, (Sigma-Aldrich, St. Louis, MO) (Druz et al. 2012; Saito et al. 2006). PBA working concentration was optimized to 10mM in order to achieve at least the same mmu-miR-466h-5p activation levels as was previously shown in apoptotic conditions (Druz et al. 2011). PBA was dissolved in 70% ethanol at 1M prior to treatment. 20μl of PBA or 70% ethanol was added to the wells for each clone and cells were incubated for 24h. After 24h cells were collected, washed, and RNA was isolated. The expression of mmu-miR-466h-5p in each clone after PBA treatment was related to its expression in the same clone treated with the ethanol.
Generation of SEAP-expressing pools and detection of SEAP
The sequence encoding secreted alkaline phosphatase (SEAP) was excised from the pReceiver-M61-SEAP vector (GeneCopoeia S.O. No.617379) using EcoRI and NotI and inserted directionally into the pcDNA3.1+ zeo vector (Life Technologies, Cat.No. V860-20). The anti-miR-466h, negative control and parental CHO cells were transfected with pcDNA3.1+zeo/SEAP vector using 25kDA linear polyethylenimine (PEI) (Polysciences, Cat.No. 23966-2 ) as previously described (Majors et al. 2008b). Stable pools were selected with 1.1mg/ml of zeocin (Life Technologies, Cat. No. R250-01). SEAP activity in media samples was assessed using the fluorometric alkaline phosphatase assay kit (Abcam, Cambridge, MA Cat.No. ab83371) according to manufacturer's protocol and fluorescence was determined in the SPECTRAmax GEMINI-XS spectrofluorometer. The SEAP activity was calculated by the formula:
Where A is the amount of 4-Methylumbelliferyl phosphate disodium salt substrate (4-MUP) in the samples determined based on the standard curve, V is volume of added sample in milliliters, T is reaction time in minutes. Cell-specific productivity (q) was calculated by the formula:
Results
1. Screening and selection of anti-miR-466h-5p clone
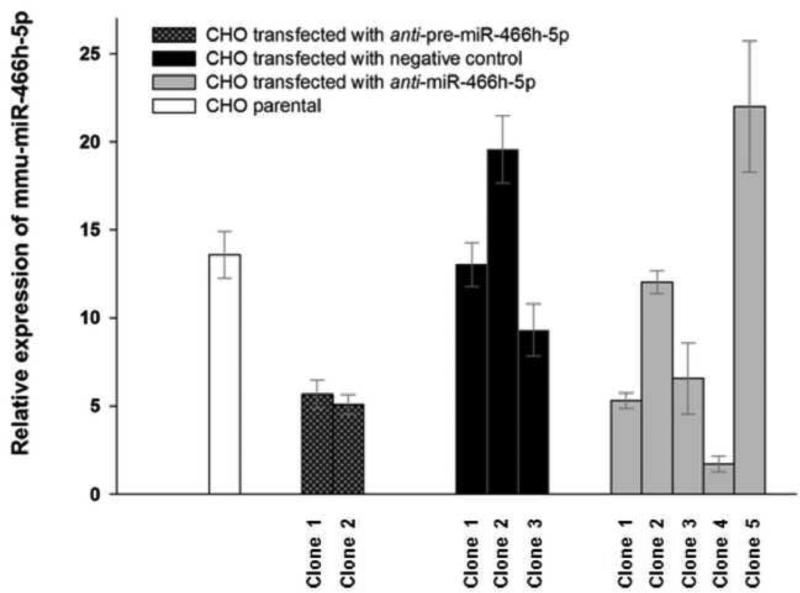
The screening and selection of the CHO cells expressing anti mmu-miR-466h-5p (the oligonucleotide sequence which stably inhibits mmu-miR-466h-5p expression) was based on measuring the extent of mmu-miR-466h-5p activation after treating the cells with 4-phenylbutyric acid (PBA), a compound which prevents histone deacetylation and causes transcriptional activation of mmu-miR-466h-5p expression (Druz et al. 2012). In cells expressing anti mmu-miR-466h-5p, the transcriptional activation by PBA was expected to be lower than in cells which did not express anti mmu-miR-466h-5p. The relative activation of mmu-miR-466h-5p by PBA in the tested single CHO clones is shown in Figure 1. Following PBA treatment, 13.6 fold activation of mmu-miR-466h-5p was observed in the parental (non- transfected) CHO, while lower activation was observed in both clones targeting the mmu-miR-466h precursor sequence (termed anti-pre-miR-466h clones), and the clones 1, 3 and 4 expressing an antisense of mature mmu-miR-466h-5p sequence (termed anti-miR-466h-5p clones). The lowest mmu-miR-466h-5p activation (1.71 fold) was seen in anti-miR-466h-5p clone 4 (from here on anti-miR-466h CHO) which was selected for subsequent analysis. The derivative of CHO cell line stably transfected with negative control, clone 1, (scrambled short hairpin RNA sequence with limited homology to any of the known mouse, human or rat sequences) that showed similar activation of mmu-miR-466h-5p compared to parental CHO after addition of PBA (13 fold activation) was selected for comparative analysis.
Figure 1.
Relative expression of mmu-miR-466h-5p in single CHO cell clones after PBA treatment. Single colonies of parental CHO and CHO transfected with negative control, anti-pre-miR-466h and anti-miR-466h-5p shRNAs were treated with PBA to compare activation of mmu-miR-466h-5p TaqMan qRT-PCR assays were used for analysis with snoRNA202 used as control for 2-ΔΔCt analysis. The levels of mmu-miR-466h-5p in each clone after PBA treatment were related to its levels in the same clone treated with solvent.
2. Growth and viability comparison of anti-miR-466h, negative control and non-transfected CHO cells
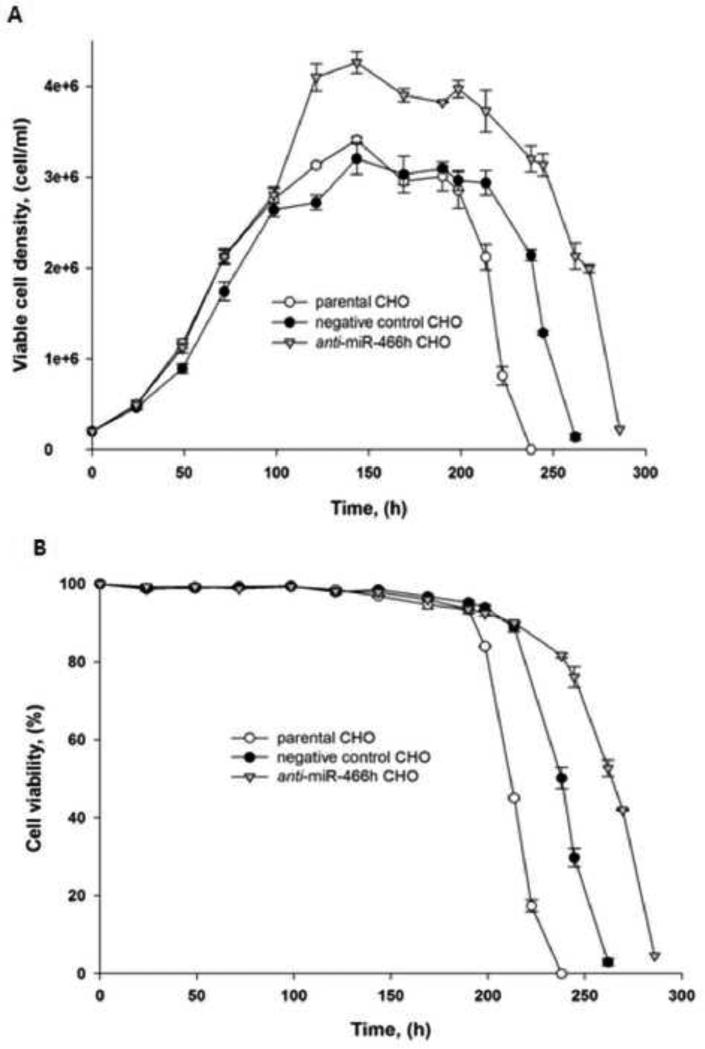
Growth and viability of anti-miR-466h, negative control, and non-transfected CHO cells are shown in Figure 2. All three cell lines grew at the same rate and reached their maximum cell density 144 h after seeding. The CHO cells expressing anti-miR-466h reached the maximum viable cell density of 4.26×106 cells/ml compared with 3.41×106 and 3.20×106 cells/ml for nontransfected and negative control transfected CHO cells respectively (Figure 2A). In addition, the anti-miR-466h CHO cells also remained viable for longer period of time compared with negative control and non-transfected CHO cells (Figure 2B). The higher number of viable cells and the extended viability of anti-miR-466h CHO cells resulted in the 53.8 % and 41.6% increase of integral viable cells (IVC) compared with non-transfected and negative control transfected CHO cells respectively.
Figure 2.
Comparison of growth and viability of anti-miR-466h, negative control and parental CHO cells in batch cell culture. (A) Growth. (B) Viability.
3. Activation of the apoptotic cascade in negative control and anti-miR-466h CHO cells
To find out if the delayed viability loss and increased integral of viable cells in anti-miR-466h CHO cells was the result of enhanced resistance to apoptosis, the Caspase-3/7 activation was monitored as a function of time in anti-miR-466h and negative control CHO cells. The time-dependent activation of Caspase-3/7 in these two cell lines is shown in Figure 3. The activation of Caspase-3/7 for both cell lines correlated with the viability loss shown in Figure 2A. The sharp increase of Caspase-3/7 activity in negative control CHO cells between 212 and 236 hours indicated the onset of apoptosis in these cells (Figure 3A and B). 40.2% of viable negative control CHO cells were undergoing apoptosis at 236 hours compared to only 11.2% of anti-miR-466h CHO cells (Figure 3B and C). Significant increase of Caspase-3/7 activity in anti-miR-466h CHO cells was observed between 247 and 270 hours , approximately 35 hours later than in the negative control CHO cells (Figure 3A and D).
4. Activation of mmu-miR-466h-5p and inhibition of its target genes in negative control and anti-miR-466h CHO cells during apoptosis
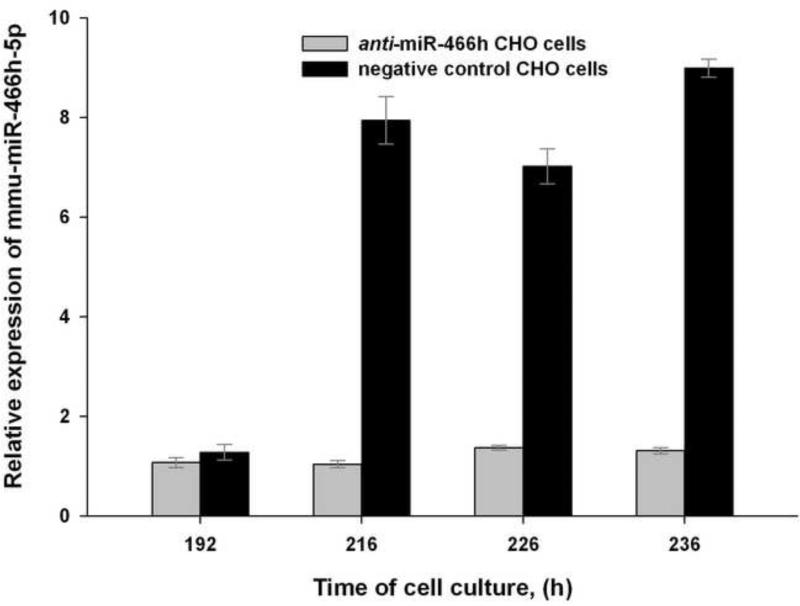
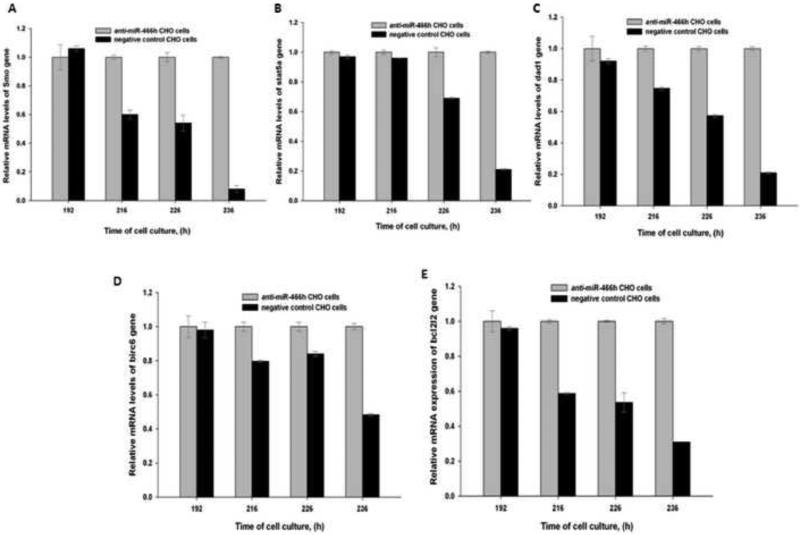
To investigate if the activation of mmu-miR-466h-5p expression correlated with the onset of apoptosis, the levels of this miRNA and its target genes were measured before and during apoptosis in the anti-miR-466h-5p and negative control CHO cells. Mmu-miR-466h-5p expression in negative control CHO cells was not increased before apoptosis onset at 192 hours of cell culture, but it was 7.6, 5.1 and 6.9 fold greater than in anti-miR-466h CHO cells at 216, 226, and 236 hours respectively (Figure 4). The expression of the previously reported putative gene targets of mmu-miR-466h-5p, smo, stat5a, dad1, birc6, and bcl2l2 (Druz et al. 2011) in anti-miR-466h and negative control CHO cells is shown in Figure 5. The mRNA levels of smo, stat5a, dad1, birc6, and bcl2l2 genes were not significantly changed at 192 hours in the negative control CHO cells compared to anti-miR-466h CHO cells , but were significantly reduced at 216, 226, and 236 hours of cell culture. The mRNA levels of these genes were reduced between 2.1- and 12.5-fold in the negative control CHO compared with their levels in the anti-miR-466h CHO at 236 hours of the beginning of the cells culture. It is, therefore, likely that the inhibition of the activated mmu-miR-466h-5p in anti-miR-466h CHO cells shown in Figure 4 leads to increased levels of the anti-apoptotic genes: smo, stat5a, dad1, birc6, and bcl2l2 as seen in Figure 5 which delays the activation of Caspase-3/7 and onset of apoptotic cascade shown in Figure 3.
Figure 4.
Relative levels of mmu-miR-466h-5p in anti-miR-466h and negative control CHO cells at selected time points. The levels of mmu-miR-466h-5p before apoptosis onset (192 h) and after apoptosis onset (216, 226 and 236h) were related to their respective levels after 24 hours. TaqMan microRNA qRT-PCR analysis was used to assess mmu-miR-466h-5p levels with snoRNA 202 as control for 2-ΔΔCt analysis.
Figure 5.
Relative expression of the mmu-miR-466h-5p target genes in anti-miR-466h and negative control CHO cells at selected time points. The relative changes in genes expression are compared at 192, 216, 226 and 236h. (A) smo, (B) stat5a, (C) dad1, (D) birc6, (E) bcl2l2. TaqMan qRT-PCR assays were used for analysis with 18S used as control for 2-ΔΔCt analysis.
5. Enhancement of SEAP production in anti-miR-466h CHO cells
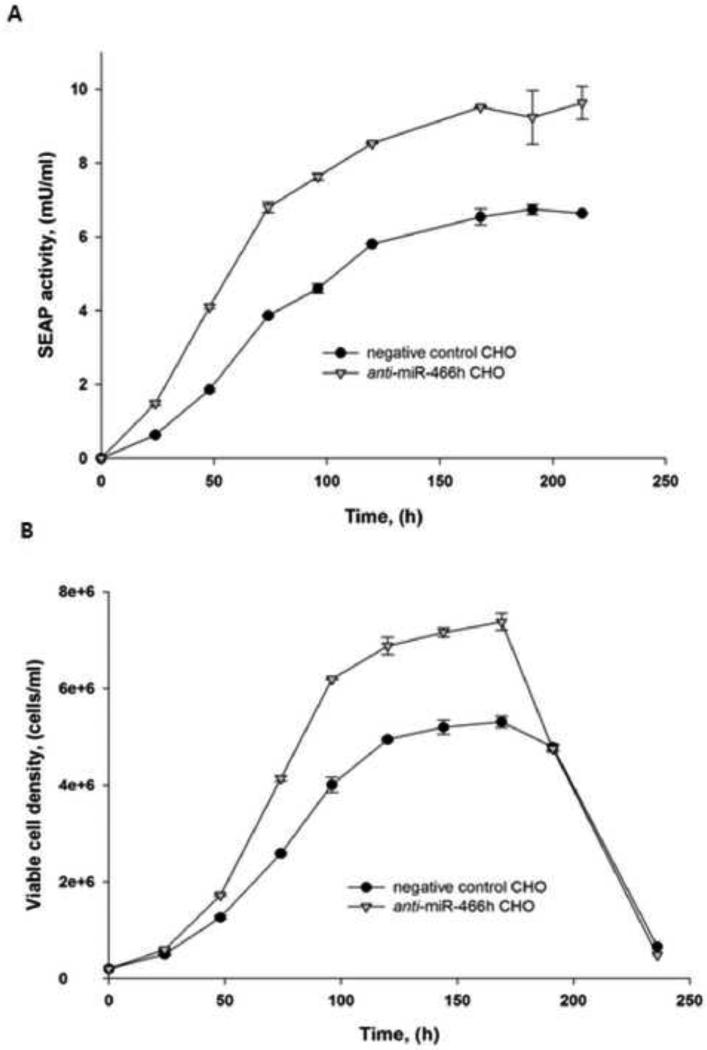
To evaluate if the enhanced apoptosis resistance in the anti-miR-466h CHO cells can increase protein production; time-course accumulation of secreted alkaline phosphatase (SEAP) was measured in stable pools of SEAP-expressing anti-miR-466h and in the negative control CHO cells. SEAP accumulation and cells growth profiles in the media are shown in Figure 6. An increase of SEAP activity in the anti-miR-466h CHO culture media compared with the negative control CHO culture media was observed after 24h of growth. SEAP activity reached 9.63mU/ml in anti-miR-466h CHO culture compared with 6.75 mU/ml in negative control CHO culture which is 42.7 increase (Figure 6A). SEAP activity in the culture of the stable pools of parental CHO cells was comparable with SEAP activity in negative control CHO culture (data not shown). Based on the cell growth profile (Figure 6 B) the cell-specific productivity of the anti-miR-466h CHO was 11.1% higher than the cell-specific productivity of the negative control CHO.
Figure 6.
SEAP activity and growth of SEAP-expressing anti-miR-466h and negative control CHO stable pools. Anti-miR-466h and negative control CHO cells were stably transfected with SEAP and stable pools were grown in batch conditions (A) SEAP activity in the culture media, (B) Cell growth
Discussion
MiRNAs have been shown to be involved in regulation of multiple cellular processes and have the potential to globally affect gene expression (Cimmino et al. 2005; Garofalo et al. 2008; Lynam-Lennon et al. 2009; Matsubara et al. 2007; Ovcharenko et al. 2007). The strategy to affect the expression of several genes by a single molecule is more efficient for mammalian cell engineering than the conventional approach of manipulating a single gene. Since one of the most common approaches in mammalian cells gene engineering is modulation of the apoptosis cascade, the utilization of miRNAs to affect apoptosis can potentially generate stress-tolerant mammalian cells better suitable for production of biologicals (Barron et al. 2011b; Druz et al. 2011; Muller et al. 2008).
The purpose of this study was to create more stress-tolerant CHO cells by the stable inhibition of mmu-miR-466h-5p which was shown to be involved in apoptosis regulation in CHO cells. This miRNA simultaneously targeted the expression of several anti-apoptotic genes (smo, stat5a, dad1, birc6, and bcl2l2) and delayed Caspase-3/7 activation and viability loss in apoptotic conditions (Druz et al. 2011). This study extended previous findings, based on the transient inhibition of mmu-miR-466h-5p, to the engineering of CHO cell line with stable inhibition of mmu-miR-466h-5p expression (anti-miR-466h CHO cells). Since mmu-miR-466h-5p is epigenetically silenced in the regular growth conditions and because of the complexity of the molecular events leading to its activation, the screening and selection of anti-miR-466h CHO clone was conducted by following the degree of mmu-miR-466h-5p activation in response to 4-phenylbutyric acid (PBA) rather than by glucose deprivation-dependent induction of the oxidative stress (Druz et al. 2012). The activation of mmu-miR-466h-5-p in the selected anti-miR-466h CHO cells was 87.4% lower than in the parental CHO cells.
The comparison of the growth of anti-miR-466h CHO cells with negative control and non-transfected CHO cells in batch cell cultures showed higher maximum viable cell density and extended viability of anti-miR-466h CHO cells which resulted in the significant increase of the integral of viable cells (IVC). Although the maximum viable cell densities obtained in the shake flask experiments presented here are lower than the values achieved in industrial applications, the significant increase in maximum viable cells density in anti-miR-466h CHO culture provided evidence for the practicability of described engineering approach. The extended viability of anti-miR-466h CHO cells in batch cell culture was a consequence of delayed Caspase-3/7 activation by more than 35 hours in anti-miR-466h CHO compared to negative control CHO cells. Parental CHO were less resistant to viability loss than both anti-miR-466h and negative control CHO (Figure 2B) probably due to the increased load on the miRNA processing machinery which could delay the activation of other pro-apoptotic miRNAs up-regulated in apoptotic conditions (Barron et al. 2011b; Druz et al. 2011; Martello et al. 2010). Since the higher IVC and apoptosis resistance are known to correlate with higher titers and better quality of recombinant proteins (Figueroa et al. 2007; Majors et al. 2008b; Majors et al. 2009), the anti-miR-466h CHO cells are likely to enhance the yields and quality of monoclonal antibodies and other biologicals. More research is needed to determine if this miRNA engineering strategy will improve protein production in the industrial-scale applications.
The activation of mmu-miR-466h-5p and the levels of putative targets of this miRNA (Druz et al. 2011) were then compared in anti-miR-466h and negative control CHO cells in apoptotic conditions. The levels of mmu-miR-466h-5p were increased almost 7-fold and the levels of smo, stat5a, dad1, birc6, and bcl2l2 genes were reduced between 2.1- and 12.5-fold in the negative control CHO compared to the anti-miR-466h CHO. Therefore, the extended viabilities in anti-miR-466h-5p CHO cells culture can be explained by the inhibition of mmu-miR-466h-5p activation which prevents the inhibition of its anti-apoptotic gene targets leading to delay of Caspase-3/7 activation.
In the next step, the expression of secreted alkaline phosphatase (SEAP) was compared in stable pools of anti-miR-466h and negative control CHO cells. The total SEAP titer and cell-specific productivity were significantly higher in anti-miR-466h CHO cell pools than in negative control CHO pools. The latter indicates the increased production capacity of anti-miR-466h CHO cell lines. The protein expression in this cell line is likely to be improved further once the single protein-expressing clones are isolated (Figueroa et al. 2007; Majors et al. 2008a; Majors et al. 2008b).
This study is the first report of the stable alteration of miRNA(s) expression in mammalian cells with effects on the apoptosis cascade. It showed that the alteration of the expression of a single miRNA (mmu-miR-466h-5p) can be useful for creating apoptosis resistant CHO cell lines with higher protein production capacity. It may be worthwhile to extend this study to investigate the role of other members of miR 297-669 cluster and their involvement in regulation of apoptosis (Druz et al. 2011). Since all detected miRNAs of this cluster were previously shown to be up-regulated in CHO during apoptosis, the stable knockouts of these miRNAs (or the whole miR 297-669) may further improve the stress resistance of CHO cells. In addition, the stable inhibition of mmu-miR-466h-5p (or other members of miR 297-669 cluster) together with alteration of the expression of other miRNAs with key roles in protein expression or secretion, such as miR-7 (Barron et al. 2011a; Meleady et al. 2012) may further improve protein production in CHO cells. While the current industrial protocols to obtain high cell densities and g/L recombinant protein titers rely on media and process optimization coupled with site-specific integration of recombinant DNA and efficient cell screening (Hacker et al. 2009; Kim et al. 2012), miRNAs engineering approach described here can be complementary to these industrial techniques.
Highlights.
We were able to stably expressed microRNA inhibitor in CHO cells.
We created new engineered CHO cell line “anti miR466h CHO“ that is apoptotic resistant
This cell line demonstrate 43% increase in expression of secreted recombinant protein
Acknowledgments
Funding was provided by the intramural program of the National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health. The authors would like to thank Mrs. D. Livnat for critical editorial assistance.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Baik JY, Gasimli L, Yang B, Datta P, Zhang F, Glass CA, Esko JD, Linhardt RJ, Sharfstein ST. Metabolic engineering of Chinese hamster ovary cells: towards a bioengineered heparin. Metab Eng. 2012;14(2):81–90. doi: 10.1016/j.ymben.2012.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barron N, Kumar N, Sanchez N, Doolan P, Clarke C, Meleady P, O'Sullivan F, Clynes M. Engineering CHO cell growth and recombinant protein productivity by overexpression of miR-7. J Biotechnol. 2011a;151(2):204–11. doi: 10.1016/j.jbiotec.2010.12.005. [DOI] [PubMed] [Google Scholar]
- Barron N, Sanchez N, Kelly P, Clynes M. MicroRNAs: tiny targets for engineering CHO cell phenotypes? Biotechnol Lett. 2011b;33(1):11–21. doi: 10.1007/s10529-010-0415-5. [DOI] [PubMed] [Google Scholar]
- Chan JA, Krichevsky AM, Kosik KS. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005;65(14):6029–33. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- Chiang GG, Sisk WP. Bcl-x(L) mediates increased production of humanized monoclonal antibodies in Chinese hamster ovary cells. Biotechnol Bioeng. 2005;91(7):779–92. doi: 10.1002/bit.20551. [DOI] [PubMed] [Google Scholar]
- Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, Shimizu M, Wojcik SE, Aqeilan RI, Zupo S, Dono M. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci U S A. 2005;102(39):13944–9. doi: 10.1073/pnas.0506654102. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Druz A, Betenbaugh M, Shiloach J. Glucose depletion activates mmu-miR-466h-5p expression through oxidative stress and inhibition of histone deacetylation. Nucleic Acids Res. 2012 doi: 10.1093/nar/gks452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Druz A, Chu C, Majors B, Santuary R, Betenbaugh M, Shiloach J. A novel microRNA mmu-miR-466h affects apoptosis regulation in mammalian cells. Biotechnol Bioeng. 2011;108(7):1651–61. doi: 10.1002/bit.23092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fassnacht D, Rossing S, Singh RP, Al-Rubeai M, Portner R. Influence of bcl-2 on antibody productivity in high cell density perfusion cultures of hybridoma. Cytotechnology. 1999;30(1-3):95–106. doi: 10.1023/A:1008055702079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa B, Jr., Ailor E, Osborne D, Hardwick JM, Reff M, Betenbaugh MJ. Enhanced cell culture performance using inducible anti apoptotic genes E1B-19K and Aven in the production of a monoclonal antibody with Chinese hamster ovary cells. Biotechnol Bioeng. 2007;97(4):877–92. doi: 10.1002/bit.21222. [DOI] [PubMed] [Google Scholar]
- Gammell P. MicroRNAs: recently discovered key regulators of proliferation and apoptosis in animal cells : Identification of miRNAs regulating growth and survival. Cytotechnology. 2007;53(1-3):55–63. doi: 10.1007/s10616-007-9049-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garofalo M, Quintavalle C, Di Leva G, Zanca C, Romano G, Taccioli C, Liu CG, Croce CM, Condorelli G. MicroRNA signatures of TRAIL resistance in human non-small cell lung cancer. Oncogene. 2008;27(27):3845–55. doi: 10.1038/onc.2008.6. [DOI] [PubMed] [Google Scholar]
- Hacker DL, De Jesus M, Wurm FM. 25 years of recombinant proteins from reactor-grown cells - where do we go from here? Biotechnol Adv. 2009;27(6):1023–7. doi: 10.1016/j.biotechadv.2009.05.008. [DOI] [PubMed] [Google Scholar]
- Ishaque A, Thrift J, Murphy JE, Konstantinov K. Over-expression of Hsp70 in BHK-21 cells engineered to produce recombinant factor VIII promotes resistance to apoptosis and enhances secretion. Biotechnol Bioeng. 2007;97(1):144–55. doi: 10.1002/bit.21201. [DOI] [PubMed] [Google Scholar]
- Kim JY, Kim YG, Lee GM. CHO cells in biotechnology for production of recombinant proteins: current state and further potential. Appl Microbiol Biotechnol. 2012;93(3):917–30. doi: 10.1007/s00253-011-3758-5. [DOI] [PubMed] [Google Scholar]
- Kramer O, Klausing S, Noll T. Methods in mammalian cell line engineering: from random mutagenesis to sequence-specific approaches. Appl Microbiol Biotechnol. 2010;88(2):425–36. doi: 10.1007/s00253-010-2798-6. [DOI] [PubMed] [Google Scholar]
- Lim SF, Chuan KH, Liu S, Loh SO, Chung BY, Ong CC, Song Z. RNAi suppression of Bax and Bak enhances viability in fed-batch cultures of CHO cells. Metab Eng. 2006;8(6):509–22. doi: 10.1016/j.ymben.2006.05.005. [DOI] [PubMed] [Google Scholar]
- Lim Y, Wong NS, Lee YY, Ku SC, Wong DC, Yap MG. Engineering mammalian cells in bioprocessing - current achievements and future perspectives. Biotechnol Appl Biochem. 2010;55(4):175–89. doi: 10.1042/BA20090363. [DOI] [PubMed] [Google Scholar]
- Lynam-Lennon N, Maher SG, Reynolds JV. The roles of microRNA in cancer and apoptosis. Biol Rev Camb Philos Soc. 2009;84(1):55–71. doi: 10.1111/j.1469-185X.2008.00061.x. [DOI] [PubMed] [Google Scholar]
- Majors BS, Arden N, Oyler GA, Chiang GG, Pederson NE, Betenbaugh MJ. E2F-1 overexpression increases viable cell density in batch cultures of Chinese hamster ovary cells. J Biotechnol. 2008a;138(3-4):103–6. doi: 10.1016/j.jbiotec.2008.08.003. [DOI] [PubMed] [Google Scholar]
- Majors BS, Betenbaugh MJ, Chiang GG. Links between metabolism and apoptosis in mammalian cells: applications for anti-apoptosis engineering. Metab Eng. 2007;9(4):317–26. doi: 10.1016/j.ymben.2007.05.003. [DOI] [PubMed] [Google Scholar]
- Majors BS, Betenbaugh MJ, Pederson NE, Chiang GG. Enhancement of transient gene expression and culture viability using Chinese hamster ovary cells overexpressing Bcl-x(L). Biotechnol Bioeng. 2008b;101(3):567–78. doi: 10.1002/bit.21917. [DOI] [PubMed] [Google Scholar]
- Majors BS, Betenbaugh MJ, Pederson NE, Chiang GG. Mcl-1 overexpression leads to higher viabilities and increased production of humanized monoclonal antibody in Chinese hamster ovary cells. Biotechnol Prog. 2009;25(4):1161–8. doi: 10.1002/btpr.192. [DOI] [PubMed] [Google Scholar]
- Malphettes L, Fussenegger M. Impact of RNA interference on gene networks. Metab Eng. 2006;8(6):672–83. doi: 10.1016/j.ymben.2006.07.005. [DOI] [PubMed] [Google Scholar]
- Martello G, Rosato A, Ferrari F, Manfrin A, Cordenonsi M, Dupont S, Enzo E, Guzzardo V, Rondina M, Spruce T. A MicroRNA targeting dicer for metastasis control. Cell. 2010;141(7):1195–207. doi: 10.1016/j.cell.2010.05.017. others. [DOI] [PubMed] [Google Scholar]
- Mastrangelo AJ, Hardwick JM, Zou S, Betenbaugh MJ. Part II. Overexpression of bcl-2 family members enhances survival of mammalian cells in response to various culture insults. Biotechnol Bioeng. 2000;67(5):555–64. doi: 10.1002/(sici)1097-0290(20000305)67:5<555::aid-bit6>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- Matsubara H, Takeuchi T, Nishikawa E, Yanagisawa K, Hayashita Y, Ebi H, Yamada H, Suzuki M, Nagino M, Nimura Y. Apoptosis induction by antisense oligonucleotides against miR-17-5p and miR-20a in lung cancers overexpressing miR-17-92. Oncogene. 2007;26(41):6099–105. doi: 10.1038/sj.onc.1210425. others. [DOI] [PubMed] [Google Scholar]
- Meleady P, Gallagher M, Clarke C, Henry M, Sanchez N, Barron N, Clynes M. Impact of miR-7 over-expression on the proteome of Chinese hamster ovary cells. J Biotechnol. 2012;160(3-4):251–62. doi: 10.1016/j.jbiotec.2012.03.002. [DOI] [PubMed] [Google Scholar]
- Mercille S, Massie B. Apoptosis-resistant E1B-19K-expressing NS/0 myeloma cells exhibit increased viability and chimeric antibody productivity under perfusion culture conditions. Biotechnol Bioeng. 1999;63(5):529–43. doi: 10.1002/(sici)1097-0290(19990605)63:5<529::aid-bit3>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- Muller D, Katinger H, Grillari J. MicroRNAs as targets for engineering of CHO cell factories. Trends Biotechnol. 2008;26(7):359–65. doi: 10.1016/j.tibtech.2008.03.010. [DOI] [PubMed] [Google Scholar]
- Nolan RP, Lee K. Dynamic model of CHO cell metabolism. Metab Eng. 2011;13(1):108–24. doi: 10.1016/j.ymben.2010.09.003. [DOI] [PubMed] [Google Scholar]
- Ovcharenko D, Kelnar K, Johnson C, Leng N, Brown D. Genome-scale microRNA and small interfering RNA screens identify small RNA modulators of TRAIL-induced apoptosis pathway. Cancer Res. 2007;67(22):10782–8. doi: 10.1158/0008-5472.CAN-07-1484. [DOI] [PubMed] [Google Scholar]
- Paddison PJ, Caudy AA, Bernstein E, Hannon GJ, Conklin DS. Short hairpin RNAs (shRNAs) induce sequence-specific silencing in mammalian cells. Genes Dev. 2002;16(8):948–58. doi: 10.1101/gad.981002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito Y, Liang G, Egger G, Friedman JM, Chuang JC, Coetzee GA, Jones PA. Specific activation of microRNA-127 with downregulation of the proto-oncogene BCL6 by chromatin-modifying drugs in human cancer cells. Cancer Cell. 2006;9(6):435–43. doi: 10.1016/j.ccr.2006.04.020. [DOI] [PubMed] [Google Scholar]
- Sauerwald TM, Betenbaugh MJ, Oyler GA. Inhibiting apoptosis in mammalian cell culture using the caspase inhibitor XIAP and deletion mutants. Biotechnol Bioeng. 2002;77(6):704–16. doi: 10.1002/bit.10154. [DOI] [PubMed] [Google Scholar]
- Shimizu S, Takehara T, Hikita H, Kodama T, Miyagi T, Hosui A, Tatsumi T, Ishida H, Noda T, Nagano H. The let-7 family of microRNAs inhibits Bcl-xL expression and potentiates sorafenib-induced apoptosis in human hepatocellular carcinoma. J Hepatol. 2010;52(5):698–704. doi: 10.1016/j.jhep.2009.12.024. others. [DOI] [PubMed] [Google Scholar]
- Sui G, Soohoo C, Affar el B, Gay F, Shi Y, Forrester WC. A DNA vector-based RNAi technology to suppress gene expression in mammalian cells. Proc Natl Acad Sci U S A. 2002;99(8):5515–20. doi: 10.1073/pnas.082117599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suk Ahn W, Antnoniewicz MR. Parallel labeling experiments with [1,2-(13)C]glucose and [U-(13)C]glutamine provide new insights into CHO cell metabolism. Metab Eng. 2012 doi: 10.1016/j.ymben.2012.10.001. [DOI] [PubMed] [Google Scholar]
- Sung YH, Lee JS, Park SH, Koo J, Lee GM. Influence of co-down-regulation of caspase-3 and caspase-7 by siRNAs on sodium butyrate-induced apoptotic cell death of Chinese hamster ovary cells producing thrombopoietin. Metab Eng. 2007;9(5 6):452–64. doi: 10.1016/j.ymben.2007.08.001. [DOI] [PubMed] [Google Scholar]
- Wong DC, Wong KT, Nissom PM, Heng CK, Yap MG. Targeting early apoptotic genes in batch and fed-batch CHO cell cultures. Biotechnol Bioeng. 2006;95(3):350–61. doi: 10.1002/bit.20871. [DOI] [PubMed] [Google Scholar]
- Xu C, Lu Y, Pan Z, Chu W, Luo X, Lin H, Xiao J, Shan H, Wang Z, Yang B. The muscle-specific microRNAs miR-1 and miR-133 produce opposing effects on apoptosis by targeting HSP60, HSP70 and caspase-9 in cardiomyocytes. J Cell Sci. 2007;120(Pt 17):3045–52. doi: 10.1242/jcs.010728. [DOI] [PubMed] [Google Scholar]
- Zanghi JA, Fussenegger M, Bailey JE. Serum protects protein-free competent Chinese hamster ovary cells against apoptosis induced by nutrient deprivation in batch culture. Biotechnol Bioeng. 1999;64(1):108–19. doi: 10.1002/(sici)1097-0290(19990705)64:1<108::aid-bit12>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- Zhu J. Mammalian cell protein expression for biopharmaceutical production. Biotechnol Adv. 2012;30(5):1158–70. doi: 10.1016/j.biotechadv.2011.08.022. [DOI] [PubMed] [Google Scholar]