Abstract
Glioblastoma (GBM), a malignant brain cancer, is characterized by abnormal activation of receptor tyrosine kinase (RTK) signaling pathways and poor prognosis. Extracellular proteoglycans, including heparan sulfate and chondroitin sulfate, play critical roles in the regulation of cell signaling and migration via interactions with extracellular ligands, growth factor receptors, extracellular matrix components, and intracellular enzymes and structural proteins. In cancer, proteoglycans help drive multiple oncogenic pathways in tumor cells and promote critical tumor-microenvironment interactions. In this review, we summarize the evidence for proteoglycan function in gliomagenesis and we examine the expression of proteoglycans and their modifying enzymes in human GBM using data from The Cancer Genome Atlas (TCGA). Furthermore, we demonstrate an association between specific proteoglycan alterations and changes in RTKs. Based on these data we propose a model in which proteoglycans and their modifying enzymes promote RTK signaling and progression in GBM, and we suggest cancer associated proteoglycans are promising biomarkers for disease and therapeutic targets.
Keywords: Proteoglycans, HSPG, CSPG, SULFs, GBM
INTRODUCTION
Glioblastoma (GBM), the most common primary malignant brain tumor of adults, is characterized by abnormal activation of receptor tyrosine kinase (RTK) signaling pathways and diffuse invasion of tumor cells into the adjacent brain. While advances in current therapeutic strategies have improved outcome for GBM patients, the current median survival for GBM remains less than 2 years [1, 2]. A major impediment to treatment has been the diversity of oncogenic alterations present across tumors as well as within an individual tumor [3–5].
Proteoglycans, including heparan sulfate and chondroitin sulfate proteoglycans (HSPG and CSPG, respectively), regulate the activity of many signaling pathways as well as cell-microenvironment interactions. Due to these diverse functions, proteoglycans and their modifying enzymes have been implicated in tumorigenesis in a number of cancers (reviewed in [6–9]). In brain cancer, data from our laboratory and others suggest that proteoglycans regulate multiple oncogenic pathways in tumor cells and promote critical tumor-microenvironment interactions [10–17]. Thus, proteoglycans and their modifying enzymes are potentially important therapeutic targets and biomarkers of GBM.
A number of fundamental studies important in our understanding of proteoglycan function have focused on processes outside of the central nervous system (CNS), thus we begin the current review with an overview of these studies. We then summarize some of the known functions of proteoglycans in the brain and in cancer, and we examine the expression of proteoglycans and proteoglycan-related enzymes in human GBM. As GBM can be stratified into biologically and clinically relevant subgroups [18–20], we investigate proteoglycan and proteoglycan-related gene expression across tumor subtypes. We further show a correlation between RTK amplification and specific alterations in proteoglycans. We propose a model in which proteoglycans and their modifying enzymes promote RTK signaling and progression in GBM. An improved understanding of proteoglycan function in brain tumors may reveal druggable therapeutic targets for this deadly disease.
Proteoglycans in development and disease
Proteoglycans consist of a core protein and covalently attached glycosaminoglycan (GAG) chains, including heparan sulfate (HS), chondroitin sulfate (CS), and keratan/dermatan sulfate (K/DS). Proteoglycans can be inserted in the plasma membrane, glycophosphatidylinositol (GPI) anchored to the membrane, or secreted (Figure 1). In the brain, the most abundant components of the extracellular environment include HSPGs, CSPGs, and hyaluronan/hyaluronic acid (HA) [21, 22]. While HA is not a proteoglycan, it is a GAG that is directly released extracellularly by hyaluronan synthase (HAS) enzymes located in the plasma membrane. Moreover, HA on neural stem cells and their differentiated progeny binds to a number of proteoglycans, including neurocan (NCAN), aggrecan (ACAN), and versican (VCAN) [23].
Figure 1. Schematic depicting proteoglycan cellular localization and extracellular modification.
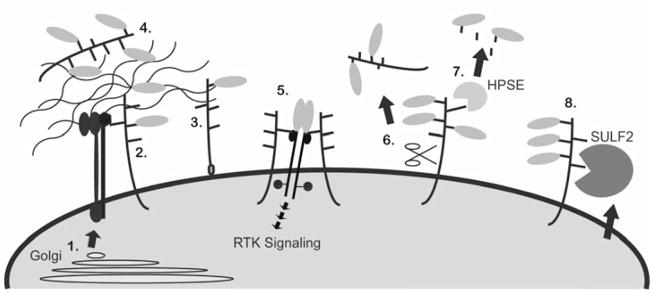
Proteoglycans are post-translationally modified in the Golgi (1) and transported to the plasma membrane where they can remain tethered to the plasma membrane, via a transmembrane domain (2) or a GPI-link (3), or be secreted (4). Extracellular proteoglycans can sequester ligands, such as growth factors and morphogens (green ovals), and bind matrix proteins (4). Transmembrane and GPI-linked proteoglycans can facilitate cell adhesion by interacting with the ECM and with integrins (red and blue structure), or they can act as co-receptors to stabilize ligand-receptor complexes and promote RTK signaling (5). Once in the extracellular environment proteoglycans can be further modified enzymatically. Sheddases can cleave the core protein to generate a soluble fragment (6), HPSE cleaves HS chains to release biologically active GAG chains (7), and the SULFs remove 6O-sulfates from HS (8).
Due to their ability to interact with diverse partners, including soluble factors, membrane proteins and the ECM, proteoglycans can regulate processes ranging from ligand-mediated signaling involved in cell proliferation to cell adhesion and cell migration [22, 24, 25] (Figure 1). In addition, the intracellular domain of some transmembrane proteoglycans can interact with the cytoplasmic domain of proteins and help regulate intracellular signaling. For example, the cytoplasmic tail of syndecan-4 (SDC4) directly interacts with alpha-actinin to help regulate cytoskeletal organization [26–28], and the cytoplasmic domain of syndecan-1 (SDC1) interacts with talin to modulate integrin signaling via a SDC1-integrin-IGF1R tri-molecular complex [29].
As extracellular reservoirs for growth factors and as enablers of growth factor-receptor interactions, proteoglycans play a dual role in regulating ligand-mediated signaling. Proteoglycans can bind and sequester soluble ligands, which help to establish and maintain morphogen gradients [30–32]. For example, decreased HSPG synthesis disrupts normal localization of wingless (Wg), Decapentaplegic (Dpp) and hedgehog (Hh) and results in developmental defects in Drosophila [33, 34]. On the other hand, proteoglycans can act as co-receptors for ligand-mediated signaling. Indeed, HSPGs stabilize the fibroblast growth factor (FGF) ligand-receptor complex to promote FGF2 signaling [35–40].
The critical role for proteoglycans, including both the core protein and GAGs, in normal development and growth is well illustrated by data from model organisms and from the study of human disease [41]. In mice, deficiency of perlecan (HSPG2), expressed in the basal lamina of the brain, can result in exencephaly or neuronal ectopias [42–44]. Mutations in human GPC3 (glypican-3) can cause Simpson-Golabi-Behmel syndrome, characterized by overgrowth of multiple tissues and tumor susceptibility [45]. Mice genetically engineered to lack CSPG4/NG2 show multiple phenotypes ranging from delays in the production of mature oligodendrocytes to deficits in brown fat function and adult onset obesity [46, 47]. The data demonstrate proteoglycans play important roles in development and tissue homeostasis.
Proteoglycans and their GAG chains undergo extensive post-translational and post-synthetic enzymatic modifications to generate the necessary structural diversity important for their function. For HSPGs, a major determinant of the specificity and the affinity of ligand interactions is the sulfation pattern of the HS chains, particularly the 6O-sulfate (6OS) of glucosamine [24, 48–51]. Regulation of 6OS levels occurs during biosynthesis and post-synthetically by the extracellular sulfatases (SULFs) [48, 52]. Dhoot et al. [53] first identified the Sulfs and demonstrated their ability to regulate Wnt signaling in myogenic progenitor cells in the quail embryo. Since this time the SULFs have been shown to regulate HSPG-dependent signaling by removing 6OS and liberating protein ligands from HSPG sequestration, including Wnts, FGF2, vascular endothelial growth factor (VEGF), glial cell line-derived growth factor (GDNF), and stromal cell-derived factor 1 (SDF1) [53–62].
The importance of HSPG sulfation in normal development is well illustrated by loss of function studies. Genetic ablation of heparan sulfate 6O-sulfotransferase-1 (HS6ST1) is embryonic lethal and the effected placentas have a profound reduction in microvasculature [63]. Furthermore, mouse embryonic fibroblasts derived from mice with ablation of HS6ST1 and HS6ST2 show reduced FGF-dependent signaling [64]. This is consistent with the co-receptor function for proteoglycans in FGF2 signaling. There are two SULF genes in vertebrates, SULF1 and SULF2, and one or both have been implicated in a number of developmental processes including brain development, esophageal development, dentinogenesis, and bone development [55, 65–67].
Taken-together, alterations in proteoglycan core proteins, biosynthetic enzymes, and the extracellular regulating enzymes, are associated with a number of developmental anomalies and, in some cases, overgrowth or tumor predisposition syndromes.
Proteoglycan functions in the brain
Proteoglycans are abundant in the brain and have known roles in normal development and neurologic disease. Both HSPGs and CSPGs are upregulated in neurogenic brain regions where regulation of growth factor signaling is critical [25, 68, 69]. SDC1 is highly expressed in the neural germinal zone of the developing cortex and can regulate the proliferation and maintenance of neural progenitor cells, at least partially through modulation of Wnt signaling [70]. CSPGs are expressed on neural progenitor cells and their enzymatic removal is associated with decreased proliferation in response to FGF2 and alters cell differentiation in response to epidermal growth factor (EGF) [71].
The proteoglycan CSPG4/NG2, encoded by the cspg4 gene, is an example of a molecule with pleiotropic functions in the postnatal mammalian brain. CSPG4/NG2 is an integral membrane proteoglycan and has a large extracellular and small cytoplasmic domain. CSPG4/NG2 is found on the surface of several immature progenitor cells, including oligodendrocyte progenitor cells (OPC) and pericytes (for review see [72]). On OPCs, the largest population of dividing cells in the adult brain, CSPG4/NG2 promotes cell proliferation and cell migration [46, 73, 74]. Early studies suggested that CSPG4/NG2 cooperates with platelet-derived growth factor receptor-α (PDGFRα), which mediates OPC proliferation in response to its ligand PDGF [75]. Subsequent studies in mice lacking the cspg4 gene have demonstrated a role for CSPG4/NG2 in promoting proliferation of PDGFRα-positive OPC at postnatal stages [46]. Indeed, PDGFAA and FGF-2 show high affinity binding to the CSPG4/NG2 core protein [73] suggesting the proteoglycan may act as a reservoir and co-receptor for these growth factors, thereby modulating RTK signaling. In addition to influencing ligand bioavailability, direct interactions between CSPG4/NG2 and the RTK itself are thought to promote mitogenic signaling, as observed for FGFR1 and FGFR3 in pericytes and smooth muscle cells [76].
Recently, Sugiarto et al. demonstrated that CSPG2/NG2 regulates EGF-dependent proliferation and self-renewal of OPCs [12]. Moreover, CSPG4/NG2 itself exhibits polarized localization in OPCs prior to mitosis and is unequally inherited to the self-renewing progeny of OPCs but not the progeny destined to differentiate. The study provides unprecedented evidence that OPC divide asymmetrically to regulate self-renewal and differentiation. CSPG4/NG2 is also required to set-up OPC polarity in part by achieving asymmetric segregation of active epidermal growth factor receptor (EGFR). These data not only show that CSPG4/NG2 is a marker of polarity and self-renewing cell fate, but that it may actively participate in regulating such fundamental processes as asymmetric progenitor divisions.
CSPG4/NG2 further interacts with cytoplasmic and ECM components such as collagen V and VI [77] and forms signaling complexes with α3β1 integrin and galectin-3, possibly acting as a co-receptor [74, 78]. Other partner molecules include matrix metalloproteinase-16 (MT3MMP), plasminogen and tissue-type plasminogen activator (reviewed in [79]). Moreover, several binding partners of the C-terminal, PDZ containing cytoplasmic domain include the multiple PDZ domain protein (MUPP1), the glutamate receptor interacting protein (GRIP) and Syntenin-1 [80–82]. The interaction with GRIP suggests a function for CSPG4/NG2 in synapse formation, while its interaction with syntenin-1 provides connection to the cell cytoskeleton. In addition, differential phosphorylation of the CSPG4/NG2 cytoplasmic domain, and subsequent changes in cellular localization, are determinants of β1-integrin signaling in glioma [83, 84].
During brain development, the sulfation of HS on neural progenitor cells changes, and this may be important in regulating their differentiation [38]. Indeed, undersulfation of HS on embryonic stem cells restricts their differentiation potential and blocks maturation along the neural lineage [85]. In the ventral spinal cord, the Sulfs establish a morphogen gradient for Shh, which is required for neural progenitors to switch from a neuronal to an oligodendroglial fate [58, 86, 87]. Knockdown of CSPG biosynthetic enzymes has shown that sulfation of CS chains is a critical determinant of cell migration from the ventricular zone into the cortical plate [88].
In addition to their role in development, proteoglycans also play important roles in response to nervous system injury and neurologic disease. In the CNS, CSPGs are upregulated in response to injury and demyelination, and they can limit axonal regeneration and remyelination [89–91]. Interestingly, despite their inhibitory action on neural repair, exogenous application of CSPG fragments has been shown to reduce excitotoxicity and repress neural cell death induced by glutamate analogs [92]. Furthermore, in disease models, CSPG4/NG2 deficiency leads to reduced myelin repair due to decreased proliferation of normally CSPG4/NG2+ cells, such as oligodendrocyte progenitor cells, pericytes and macrophages/microglia [93]. The 6O-sulfation levels of HS are also important in regulating signaling in neural injury. In a recent study, increased HS sulfation was shown to promote the formation of the glial scar suggesting that manipulation of HS sulfation may be therapeutically useful following CNS injury [94].
Proteoglycan alterations are clearly a normal response to CNS injury and modulation of this response may be important to promote repair or ameliorate disease.
Proteoglycans in cancer growth, angiogenesis, and inflammation
Consistent with their diverse roles in normal growth and development, proteoglycans have been implicated in influencing several aspects of tumor biology, including cell proliferation, tumor cell adhesion and migration, inflammation, and angiogenesis; reviewed in [6, 8, 95, 96].
Changes in proteoglycan core proteins are observed in many cancers and are often associated with changes in cell signaling and invasion. For example, glypican-1 (GPC1) expression is elevated in human pancreatic cancer and attenuation or ablation of GPC1 expression in pancreatic tumor cells can confer decreased response to FGF2 and heparin-binding EGF-like growth factor (HB-EGF), decreased downstream MAPK signaling, decreased tumor cell proliferation, decreased production of angiogenic factors, and decreased tumor growth and metastasis in vivo [97–99]..
Alterations in proteoglycan biosynthetic and modifying enzymes are also associated with carcinogenesis. Heparanase (HPSE) specifically liberates biologically active GAG chains from HSPGs, and its activity is associated with increased tumorigenesis, angiogenesis and invasiveness in diverse cancers including, multiple myeloma, breast, and pancreatic cancer (reviewed in [100]). In multiple myeloma, a number of oncogenic functions for HPSE have been identified including direct effects on tumor cells and indirect effects on tumor-associated endothelial cells [101–103]. For example, HPSE can drive hepatocyte growth factor (HGF) signaling via its ability to increase the production of HGF and increase the production and shedding of SDC1, which forms a SDC1-HGF complex able to activate the c-Met receptor [101, 104].
As mentioned above, the levels of 6O-sulfation are critical for HSPG function, and altered SULF levels have been associated with a large number of human cancers including brain, breast, non-small cell lung carcinoma (NSCLC), multiple myeloma, gastric, pancreatic cancer and hepatocellular carcinoma [8, 9, 56, 105–110]. In brain cancer, SULF2 has been directly implicated in driving tumorigenesis in murine and human malignant glioma [11, 111]. SULF2 protein is highly expressed in primary human GBM, and SULF2 levels are inversely related to HSPG 6O-sulfation in a murine model for GBM. Furthermore, ablation of SULF2 decreases tumor growth, prolongs host survival, and decreases the activity of PDGFRα and as well as related downstream signaling pathways [11].
Importantly, proteoglycans also regulate ligand-mediated signaling in non-neoplastic components of the tumor. In multiple myeloma, SDC1 shed from tumor cells that express high levels of HPSE promotes VEGF signaling in endothelial cells [102]. Conditional knockout of N-acetylglucosamine N-deacetylase/N-sulfotransferase 1 (Ndst1) in endothelial cells alters their HSPG composition, decreases FGF2 and VEGF signaling in tumors, and reduces tumor angiogenesis in vivo [112]. Interestingly, vessel formation during normal development was not affected in these mice, suggesting that proteoglycan involvement in angiogenesis is different in disease, providing an opportunity to specifically target tumor vascularity. In ovarian cancer, increased HSPG 6O-sulfation, due to increased HS6OST levels in tumor-associated endothelial cells, promotes VEGF and FGF signaling [113]. CSPG4/NG2 knockout mice also have reduced vascularization of syngeneic tumors due to decreased integrin-dependent pericyte recruitment and altered ECM collagen deposition [114]. In addition, proteoglycans have important roles regulating the immune response to cancer [115]. Indeed, tumor-derived VCAN has been shown to activate macrophages and increase metastatic tumor growth in a model for lung carcinoma [116]. Furthermore, overexpression of HPSE results in chronic inflammation and is involved in immune cell recruitment in colitis-associated colon cancer [117]. Thus, tumor proteoglycans can promote tumor progression via direct effects on tumor cells and effects on endothelial cells and immune cells that contribute to tumor-associated angiogenesis and inflammation, respectively.
Brain cancer
Diffuse glioma, which include astrocytoma, oligodendroglioma, and oligoastrocytoma, are characterized by invasive growth of tumor cells into adjacent non-neoplastic brain, making complete surgical excision impossible. GBM, an astrocytoma, is the most malignant and the most common type of diffuse glioma. Common to virtually all GBM is dysregulation of RTK signaling pathways. Despite the importance of RTK signaling in GBM, therapies targeting RTKs have had little clinical success. One explanation may be that tumors are driven by the summation of multiple signaling inputs [3–5]. In addition, GBM is a molecularly heterogeneous disease and tumors can be stratified into molecularly defined subtypes with differences in signaling pathway activation [18–20, 118–121]. Abnormal RTK activity can be driven by alterations in receptor expression or by altered ligand availability. As described above, proteoglycans regulate the extracellular availability of oncogenic factors; receptor localization and activity; immune cell recruitment; and tumor cell migration, all of which contribute to tumorigenesis and are likely to be involved in GBM.
Collectively the data from developmental biology and from other cancers suggest proteoglycans have the potential to regulate multiple determinants of tumorigenesis in GBM. We therefore examined common proteoglycan alterations in GBM and whether they are associated with abnormalities of specific signaling pathways.
Altered proteoglycan expression in human GBM
A comprehensive analysis of the expression pattern of proteoglycans and their modifying enzymes in human GBM is lacking. Using data from The Cancer Genome Atlas (http://cancergenome.nih.gov/ [122]) we investigated the expression of proteoglycan and proteoglycan-related genes across 424 human tumors. Multiple proteoglycan core proteins and related enzymes were differentially expressed in GBM tumors relative to normal brain (Figure 2A and B).
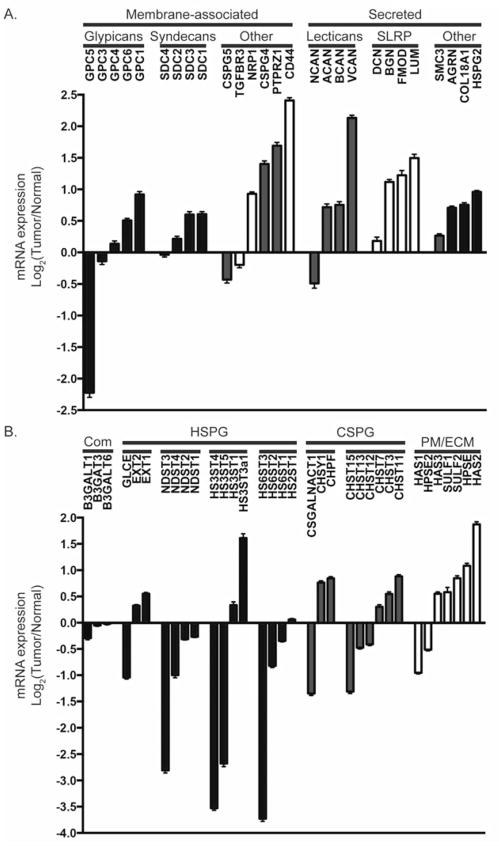
Figure 2. Proteoglycan and proteoglycan-modifying enzyme gene expression is altered in GBM.
The mean expression of a number of proteoglycan core protein genes (A) and proteoglycan-modifying enzymes (B) are altered in GBM relative to non-neoplastic controls. Bars represent the mean log2(Tumor/Normal) ratio +/− SEM gene expression from n=424 primary GBM. TCGA Data Portal [122]; http://cancergenome.nih.gov. Proteoglycan genes include HSPGs (blue); CSPGs (red); and part-time proteoglycans or those commonly modified by KS and DS (white). Enzymes in (B) include those common to both CSPG and HSPG biosynthesis (Com), HSPG biosynthetic enzymes (blue), and CSPG biosynthetic enzymes (red) including those involved in chain elongation and sulfation. The plasma membrane associated and the extracellular enzymes (PM/ECM) include HPSE, extracellular SULFs and HAS family members. For gene names and more information on the proteoglycan core proteins and enzymes, please refer to GeneCards at http://www.genecards.org/. For reviews on proteoglycan synthesis see [176]: HSPG [24]; CSPG [25]; SULF [8]; HPSE [100]; HAS [141]
These included many genes previously implicated in promoting tumor cell invasion or tumor development, including GPC1, SDC1, HSPG2, CSPG4, PTPRZ1, CD44, and VCAN [12, 16, 99, 123–128]. As previously mentioned, GPC1, a GPI-linked HSPG, can regulate signaling of a number of heparin-binding ligands, including FGF and VEGF [99, 129, 130]. In brain, breast, and pancreatic cancer, increased expression of GPC1 on tumor cells or on tumor-associated endothelial cells is associated with alterations in RTK signaling and promotes tumorigenesis [16, 97, 99, 130, 131].
Of the membrane-associated proteoglycans CSPG4/NG2, PTPRZ1 and CD44 were the most upregulated, and all three have been implicated in gliomagenesis [124, 132, 133]. CSPG4/NG2 has both cell autonomous and non-cell autonomous functions in GBM and its expression has been associated with shorter survival [13, 114, 132]. PTPRZ1, expressed in the normal brain, can be modified by CS/DS chains and exists as 3 isoforms, full-length PTPRZ1-A, the truncated PTPRZ1-B and a secreted isoform comprising the ectodomain, known as phosphacan. In embryonic oligodendrocyte progenitors PTPRZ1 acts to maintain self-renewal and suppress differentiation and this could have important implications in cancer [134]. High CD44 expression is also common in GBM and it is one of a panel of genes used to define a subset of human GBM with particularly poor survival [20, 118].
Of the secreted proteoglycans VCAN was the most upregulated. A member of the lectican family, VCAN contains an amino-terminal hyaluronan-binding domain, a C-terminal globular lectin domain that interacts with GAGs and other proteins, and a domain that can be modified with CS. VCAN is often upregulated in response to brain injury, and in GBM, VCAN can promote TGF-β2 induced tumor cell migration [89, 127]. Consistent with the pleiotropic role for proteoglycans in cell signaling, alterations in VCAN expression are not exclusively associated with tumor promotion. In malignant melanoma a VCAN splice variant interferes with the interaction of EGFR and CD44/ERBB2 and confers decreased tumor growth [135]. Lumican (LUM), a small leucine-rich proteoglycan, was also upregulated in GBM. While alterations in LUM expression in cancer are common, their role may be tissue specific and include anti-tumor effects [95, 136, 137]. While many proteoglycans are upregulated in GBM, some are downregulated such as glypican-5 (GPC5). Interestingly, a single nucleotide polymorphism, associated with downregulation of GPC5, was recently linked with increased susceptibility to lung cancer in non-smokers [138]. Overall both HSPG and CSPG core proteins are largely upregulated in GBM relative to normal brain (Figure 2A).
In contrast, the HS and CS biosynthetic enzymes appear to be differentially regulated (Figure 2B). While CS biosynthetic enzyme expression is predominantly upregulated in GBM, HS biosynthetic enzyme expression, including HS6ST1-3, is predominantly downregulated, with the striking exception of HS3ST3a1. Increased HS3ST3a1 expression has previously been reported in GBM cell lines [16]. As the sulfation status of HSPGs is a critical determinant for ligand interactions, these data suggest increased 3O-sulfation levels and relatively low 6O-sulfation levels may be important in GBM. Moreover, the SULFs, which act to decrease HS 6O-sulfation levels in the extracellular environment, are upregulated in GBM, Figure 2B and [9, 11].
In addition, the extracellular HS modifying enzyme HPSE and the membrane bound biosynthetic enzyme HAS2 are upregulated in GBM. As discussed above, HPSE is upregulated in a number of cancers and its expression is associated with increased oncogenic signaling and promotion of inflammation [117]; reviewed in [100]. In mammary epithelial cells, HAS2 regulates epithelial-mesenchymal transition induced by transforming growth factor-β (TGFβ) and this appears to be independent of extracellular hyaluronan [139]. Increased HAS2 activity and HA production have been associated with malignant transformation and increased RTK activity ([140, 141]; reviewed in [142]).
GBM subtype-specific alterations in proteoglycan expression
GBM is a heterogeneous disease associated with multiple oncogenic alterations. The two most commonly altered RTKs in GBM are EGFR and PDGFRA [18, 143, 144]. A number of strategies have been proposed to subgroup GBM [19, 20, 118–121], and a common strategy, proposed by Verhaak et al., [18] is based on gene expression analysis. As the oncogenic pathways thought to drive disease appear to differ across tumor subtypes, we hypothesized that proteoglycan alterations associated with these oncogenic pathways may be different between the tumor subtypes. Indeed, previous studies have demonstrated high expression of SULF2, which can promote PDGFRα signaling, in the proneural GBM subtype characterized by alterations in this signaling pathway [11].
The gene expression of proteoglycan and proteoglycan-modifying enzymes in 170 previously subtyped [18] GBM was examined. Consistent with the diverse role for proteoglycans in gliomagenesis, expression of multiple genes was altered in a subtype-specific manner. Representative examples are illustrated in Figure 3, where the normalized expression score for each gene is compared across tumor subtypes.
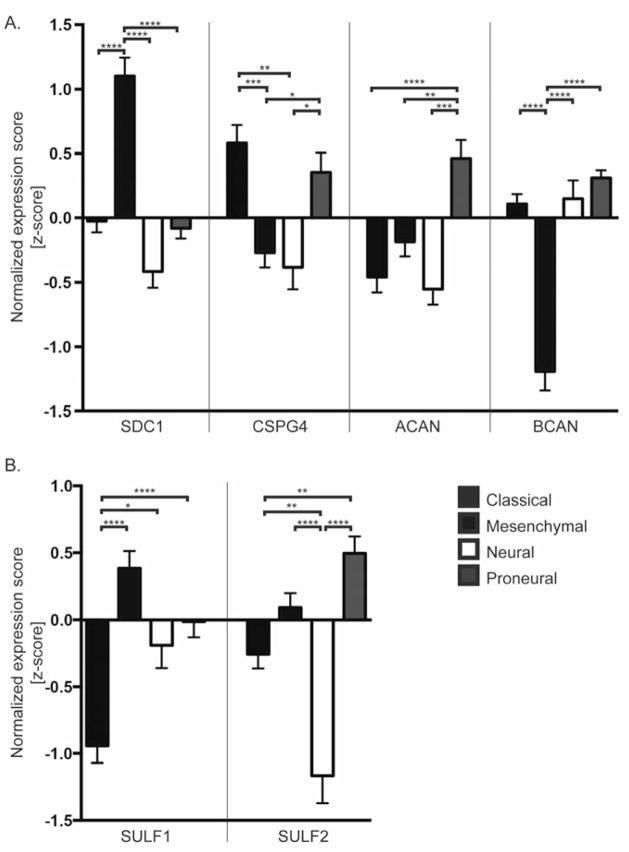
Figure 3. GBM subtype-specific expression of proteoglycans.
Comparison of normalized expression scores (z-scores) for proteoglycan core proteins (A) and the extracellular sulfatases (B) across GBM subtypes. Bars denote mean z-score +/− SEM for tumors in the specified subtype as determined in Verhaak et al. [18] with Classical (black), n=38; Mesenchymal (blue), n=56; Neural (white) n=23; and Proneural (red) n=53; TCGA Data Portal [122]. A negative z-score denotes the expression value was below the GBM population mean. Data were analyzed using 1-way ANOVA; p<0.0001. Tukey’s multiple comparisons test revealed significant differences in gene expression between subtypes; * p< 0.05, ** p<0.01, *** p<0.001, **** P<0.0001.
For many proteoglycans and proteoglycan-modifying enzymes we identified selective and significant upregulation in specific GBM subtypes (Figure 3A). SDC1 expression was specifically increased in the mesenchymal GBM subtype, which is characterized by high expression of genes in the tumor necrosis factor super family pathway, the NF-kB pathway, and increased expression of inflammatory related genes [18, 145]. Interestingly, in a glioma cell line, SDC1 expression has been shown to be dependent on NF-κB activation [146]. Together these data suggest cell surface SDC1 expression levels may reflect NF-κB activation, and it can be speculated that SDC1 function may be particularly relevant in GBM with NF-κB activation. In multiple myeloma, SDC1 can promote tumor growth and tumor angiogenesis, and high levels of shed SDC1 in the blood is a poor prognostic indicator [128]. SDC1 interactions with integrins can promote tumor invasion, as has been shown in breast cancer [147], and disruption of this interaction decreases tumor growth and angiogenesis [148]. SDC1 can also modulate the inflammatory response via binding to chemokines, cytokines, and integrins, and in models of allergic disease shed SDC1 can attenuate inflammation [149–151]. Together, this suggests SDC1 could modulate multiple tumorigenic pathways in GBM and this may be of particular relevance in mesenchymal GBM.
The classical and the proneural GBM subtype are characterized by high-level amplification of EGFR and alterations in the PDGFRα signaling pathway, respectively [18, 120]. CSPG4/NG2 is significantly elevated in both the classical and proneural subtypes relative to the remaining subtypes (Figure 3A), and CSPG4/NG2 has been implicated in promoting both of these RTK signaling pathways [12, 75]. As reviewed above, CSPG4/NG2 in glial progenitor cells co-localizes with PDGFRα and EGFR and it is required for the normal signaling response to ligand [12, 75, 152]; for a review see [15]. CSPG4/NG2 is also significantly upregulated in low grade oligodendroglioma [12], suggesting potential functions early in gliomagenesis. It is intriguing to speculate that sequential upregulation of CSPG4/NG2 and EGFR or PDGFRα may further tumor progression.
Our analysis also revealed interesting GBM subtype differences in expression of the CSPG lecticans, including ACAN and BCAN (Figure 3A), both of which are normally expressed in the brain and bind HA [153, 154]. ACAN was significantly increased in the proneural GBM subtype relative to the other subtypes. BCAN expression was also increased in the proneural subtype and decreased in the mesenchymal GBM subtype relative to the other subtypes. Striking inter-tumoral differences in BCAN expression have been previously reported in GBM, including upregulation of BCAN in proneural GBM and selective downregulation of BCAN in mesenchymal GBM [18, 20]. In fact, downregulation of BCAN is a signature alteration of the mesenchymal subtype used in our analysis [18]. Like other proteoglycans, BCAN can be enzymatically cleaved, and its shed hyaluronan-binding domain can promote glioma cell invasion and EGFR signaling [155–159]. In contrast, VCAN was not differentially expressed between tumor subtypes suggesting a function independent of tumor subtype (data not shown).
The extracellular sulfatases, SULF1 and SULF2, also demonstrate subtype-specific differences in expression (Figure 3B). SULF1 expression is significantly decreased in classical GBM relative to the other subtypes. In contrast, SULF2 expression is significantly elevated in proneural GBM, which is characterized by alterations in PDGFRα signaling. In recent studies, we identified SULF2 as an important determinant of PDGFRα signaling in GBM [11]. SULF1 and SULF2 are both extracellular sulfatases with similar reported substrate specificities; however, they exhibit partially non-overlapping tissue and cellular expression patterns [55, 160]. As both SULFs can also undergo alternative splicing, a combination of differences in cellular localization and isoform heterogeneity may contribute to differences in SULF function in vivo [161].
The GBM subtypes in Figure 3 are defined based on gene expression profiles not specific oncogenic alterations. While these subtypes generally correspond to tumors with different patterns of RTK signaling [19], we wanted to determine whether specific oncogenic events are associated with alterations in proteoglycan or proteoglycan-modifying enzymes. As EGFR and PDGFRA are the most common RTKs altered in GBM we stratified tumors based on their amplification status and compared proteoglycan expression.
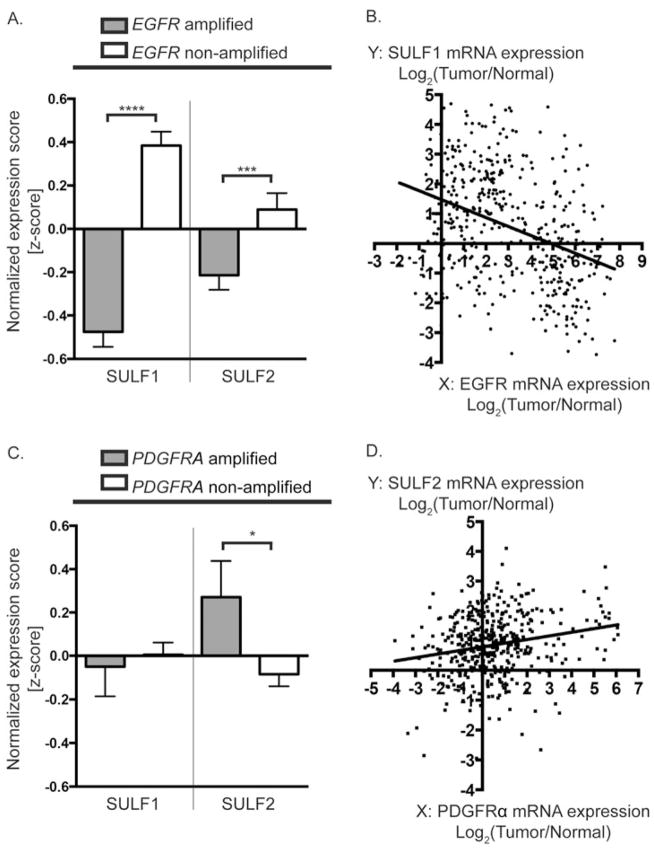
The expression of SULF1 and SULF2 were strikingly different in tumors with amplification of different RTKs (Figure 4). SULF1 expression was significantly decreased in GBM with EGFR amplification relative to GBM without amplification, and this association held true at the mRNA expression level as SULF1 expression was significantly negatively correlated with EGFR expression (r =−0.3554; Figure 4A, B). In contrast, SULF2 expression was significantly elevated in PDGFRA-amplified GBM relative to tumors without amplification (Figure 4C). SULF2 expression was also positively correlated with PDGFRα expression across all GBM tumors (r=0.2115; Figure 4D). These data suggest SULF2 may be particularly relevant in tumors with PDGFRα pathway activation, consistent with our previous studies in human glioma and in a murine model for glioma, in which loss of SULF2 decreases PDGFRα signaling, decreases proliferation, and prolongs mouse survival [11].
Figure 4. Alterations in SULF1 and SULF2 expression associated with RTK amplification.
SULF1 expression is significantly decreased in EGFR amplified tumors relative to EGFR non-amplified tumors as reflected by the normalized expression scores (z-scores) (A). Linear regression demonstrates a negative correlation between SULF1 and EGFR gene expression across all GBM (B) (n=424, Linear regression slope = −0.3003; Pearson correlation coefficient r = −0.3554; p<0.0001). In contrast, SULF2 expression is significantly upregulated in GBM with PDGFRA amplification relative to non-amplified tumors, as reflected by the normalized expression scores (z-scores) (C). SULF2 gene expression is significantly positively correlated with PDGFRα expression (D); (n=424, Linear regression slope = 0.1222; Pearson correlation coefficient r = 0.2115; p<0.0001). All data are from TCGA Data Portal [122]. (A, C) Bars denote mean z-score +/− SEM for tumors with RTK amplification defined as Log2(Tumor/Normal) >1; EGFR amplified tumors (grey) n=167; EGFR non-amplified tumors (white) n=205; PDGFRA amplified tumors (grey) n=40; PDGFRA non-amplified tumors (white) n=332. Data was analyzed using unpaired Students t-test if the D’Agostino & Pearson omnibus normality test was passed, or the Mann-Whitney U test, if not. * p< 0.05, ** p<0.01, *** p<0.001, **** P<0.0001.
We performed a similar analysis with the proteoglycan core proteins and two representative examples are illustrated (Figure 5A, B). CSPG4/NG2 expression was significantly elevated in both PDGFRA amplified and EGFR amplified tumors relative to non-amplified tumors. These data are consistent with our subtype analysis (Figure 3A), and suggest joint alterations in CSPG4/NG2 and EGFR or PDGFRA may contribute to gliomagenesis. ACAN expression was significantly decreased in EGFR-amplified tumors but significantly increased in PDGFRA amplified tumors relative to non-amplified tumors suggesting potential tumor-specific differences in ACAN function (Figure 5A, B).
Figure 5. Alterations in proteoglycan core protein expression associated with RTK amplification.
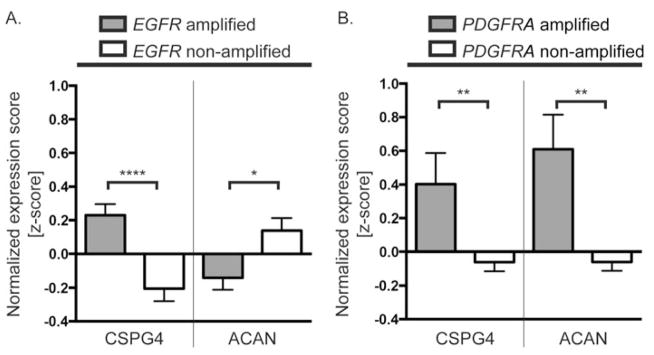
CSPG4 expression is significantly increased in both EGFR and PDGFRA amplified tumors relative to non-amplified tumors, as reflected by the normalized expression scores (z-scores) (A, B, left). In contrast, ACAN expression is significantly decreased in EGFR amplified tumors and increased in PDGFRA amplified tumors relative to non-amplified tumors (A, B, right). Bars denote mean z-score +/− SEM from TCGA Data Portal [122]; amplification defined as Log2(tumor/normal) >1; EGFR amplified tumors (grey) n=167; EGFR non-amplified tumors (white) n=205; PDGFRA amplified tumors (grey) n=40; PDGFRA non-amplified tumors (white) n=332. Data was analyzed using unpaired Students t-test if the D’Agostino & Pearson omnibus normality test was passed, or the Mann-Whitney U test, if not. * p< 0.05, ** p<0.01, *** p<0.001, **** P<0.0001.
By stratifying tumors based on RTK amplification status, it is clear that the SULFs may well have different functions in GBM subgroups, and that this consideration will be important to incorporate into future studies on SULF function in GBM.
Tools to dissect proteoglycan function
Proteoglycans represent an excellent potential therapeutic target in GBM, however, determination of the mechanisms by which they promote disease is critical. This is especially important given the subtype-specific differences in proteoglycan expression and possibly function. Models for GBM that reflect the heterogeneity of the human disease are required for the study of proteoglycan function and for conducting preclinical studies of potential therapeutic agents.
Human GBM xenografts, isolated from primary tumors and sustained in vivo in mice, provide a robust model system to study human disease. When passaged in mice, human xenografts are molecularly and phenotypically stable over time, they retain much of their genomic and transcriptional heterogeneity, and they can be transcriptionally stratified into GBM subtypes similar to human GBM [18, 162]. Indeed, using human GBM xenografts we demonstrate diverse proteoglycan and SULF2 expression profiles (Figure 6A, B). In addition to human xenografts, murine neural stem cell (NSC)-derived models for high-grade astrocytoma provide a genetically tractable, immunocompetent system to study the role of proteoglycan alterations in disease [11, 163]. While genetically engineered mouse models for GBM will also be useful, the ability to manipulate the genetics of the tumor and of the non-neoplastic cells independently will be critical to dissect the contribution of each to the oncogenic niche.
Figure 6. Tumor heterogeneity in human GBM and human GBM xenografts.
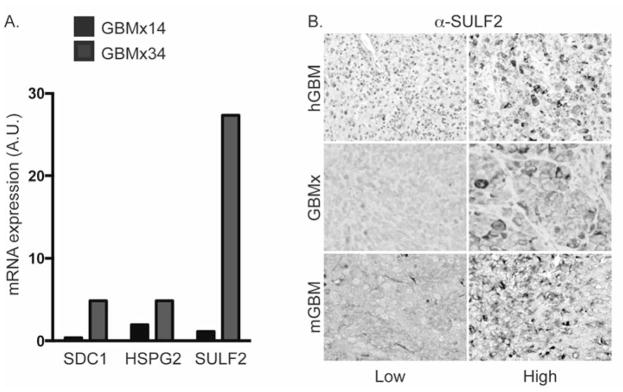
Two human GBM xenografts (GBMx14 and GBMx34) display divergent expression of SDC1, HSPG2, and SULF2 by quantitative RT-PCR (A). Immunohistochemistry (B) demonstrates differential expression of SULF2 protein in human GBM (hGBM), human GBM xenografts (GBMx), and in murine NSC-derived models for GBM (mGBM). High SULF2 expression (left) and low/no detectable expression (right). For qRT-PCR, primers: SDC1 (ID 55749479b1); HSPG2 (ID 140972288b2); SULF2 (ID 240255477b1). Immunohistochemistry performed as described previously [11].
Potential clinical applications
As extracellular proteins, HSPGs and the extracellular enzymes that modify them, such as SULF2 and HPSE, are amenable to therapeutic targeting. Heparan sulfate mimetics, highly sulfated oligosaccharides, inhibit both SULF and HPSE functions, and sequester HS-binding ligands, making them attractive candidates for GBM therapy across tumor subtypes [164–166]. In preclinical studies, heparan mimetics have effectively targeted multiple HSPG-dependent phenotypes, as indicated by their ability to decrease tumor growth, invasion, metastasis and angiogenesis [167, 168]. A phase II clinical trial for a HS mimetic in recurrent hepatocellular carcinoma has demonstrated safety and preliminary efficacy [169], and recent pre-clinical studies of a new rationally engineered HS mimetic, M402, suggest its potential as a therapeutic agent [168]. Indeed, M402 is now in a Phase I/II clinical trial for metastatic pancreatic cancer [www.clinicaltrials.gov NCT01621243]. In addition to potential direct anti-tumor effects, therapeutic targeting of proteoglycans could also modulate the tumor-associated immune response. This may be particularly useful in GBM as tumor associated macrophages are abundant and are likely to contribute to tumor growth [145, 170]. Indeed, therapeutic targeting of HSPGs with heparan sulfate mimetics decreased myeloid-derived suppressor cell levels in a murine mammary carcinoma model [168]. As a primary constituent of the brain extracellular environment, proteoglycans may impede diffusion of therapeutic agents in the brain and their disruption may improve therapeutic delivery to cancer cells [171]. Consistent with this concept, enzymatic removal of CS improved delivery of an oncolytic virus and increased its anti-tumor efficacy in an intracranial tumor model [172]. Importantly, this treatment did not increase tumor cell invasion. While heparan sulfate mimetics have not yet been tested in GBM, our data suggest that HS mimetics or more selective inhibitors of proteoglycan function may have anti-tumor efficacy in GBM. Proteoglycan function is diverse and selective inhibitors may be essential as suggested by a recent study in which single-chain variable fragment antibodies directed against endothelial HS elicited a pro-angiogenic response in primary human endothelial cells [173]. This illustrates the critical importance of conducting pre-clinical in vivo studies to test the therapeutic efficacy and safety of agents that target proteoglycans in human cancer.
Shed or secreted proteoglycans and their extracellular modifying enzymes can often be detected in the blood [174, 175]. As these are often altered in cancer, changes in their blood levels may be useful as biomarkers of disease. In lung cancer patients, plasma levels of SDC1 are significantly higher than in controls [174]. Moreover, in blood-based proteomic screens for cancer biomarkers extracellular and cell-surface proteins, including proteoglycans and growth factors, are often identified [175]. Proteoglycans and their modifying enzymes are present extracellularly, promote oncogenic signaling, and are altered in GBM. If these alterations can be detected in the blood of patients, then they may be useful as tumor biomarkers.
Conclusions
Proteoglycans are a major constituent of the brain extracellular environment and regulate cell signaling and cell migration. In this report we summarize the data supporting a functional role for proteoglycans in brain cancer and we demonstrate that proteoglycans and their modifying enzymes are altered in human GBM. Furthermore, we associate specific proteoglycan alterations with alterations in EGFR or PDGFRα signaling pathways. A mechanistic understanding of proteoglycan function in oncogenic signaling and tumor-microenvironment interactions in GBM is critical, and will likely lead to the identification of novel tumor biomarkers and druggable therapeutic targets.
Acknowledgments
We thank Noemi Andor for her helpful advice. We apologize to the many investigators whose articles we did not cite due to space constraints. This work was supported by the National Institutes of Health (U01CA168878 to JJP; R01 NS081117 to JJP; R01 CA164746 to CP; and R01 NS080619 to CDJ and CP).
Abbreviations
- GBM
glioblastoma
- HSPG
heparan sulfate proteoglycan
- CSPG
chondroitin sulfate proteoglycan
- RTK
receptor tyrosine kinase
- EGF
epidermal growth factor
- EGFR
epidermal growth factor receptor
- PDGF
platelet-derived growth factor
- PDGFRA
platelet-derived growth factor receptor alpha
- FGF
fibroblast growth factor
- VCAN
versican
- ACAN
aggrecan
- CSPG4/NG2
chondroitin sulfate proteoglycan 4
- SULFs
extracellular sulfatases
- ECM
extracellular matrix
- HPSE
heparanase
Footnotes
Conflict of interest
The authors declare they have no conflict of interest.
References
- 1.Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A, Scheithauer BW, Kleihues P. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114:97–109. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fan QW, Cheng C, Hackett C, Feldman M, Houseman BT, Nicolaides T, Haas-Kogan D, James CD, Oakes SA, Debnath J, Shokat KM, Weiss WA. Akt and autophagy cooperate to promote survival of drug-resistant glioma. Sci Signal. 2010;3:ra81. doi: 10.1126/scisignal.2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stommel JM, Kimmelman AC, Ying H, Nabioullin R, Ponugoti AH, Wiedemeyer R, Stegh AH, Bradner JE, Ligon KL, Brennan C, Chin L, DePinho RA. Coactivation of receptor tyrosine kinases affects the response of tumor cells to targeted therapies. Science. 2007;318:287–90. doi: 10.1126/science.1142946. [DOI] [PubMed] [Google Scholar]
- 4.Snuderl M, Fazlollahi L, Le LP, Nitta M, Zhelyazkova BH, Davidson CJ, Akhavanfard S, Cahill DP, Aldape KD, Betensky RA, Louis DN, Iafrate AJ. Mosaic amplification of multiple receptor tyrosine kinase genes in glioblastoma. Cancer Cell. 2011;20:810–7. doi: 10.1016/j.ccr.2011.11.005. [DOI] [PubMed] [Google Scholar]
- 5.Szerlip NJ, Pedraza A, Chakravarty D, Azim M, McGuire J, Fang Y, Ozawa T, Holland EC, Huse JT, Jhanwar S, Leversha MA, Mikkelsen T, Brennan CW. Intratumoral heterogeneity of receptor tyrosine kinases EGFR and PDGFRA amplification in glioblastoma defines subpopulations with distinct growth factor response. Proc Natl Acad Sci U S A. 2012;109:3041–6. doi: 10.1073/pnas.1114033109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fuster MM, Esko JD. The sweet and sour of cancer: glycans as novel therapeutic targets. Nat Rev Cancer. 2005;5:526–42. doi: 10.1038/nrc1649. [DOI] [PubMed] [Google Scholar]
- 7.Iozzo RV, Schaefer L. Proteoglycans in health and disease: novel regulatory signaling mechanisms evoked by the small leucine-rich proteoglycans. FEBS J. 2010;277:3864–75. doi: 10.1111/j.1742-4658.2010.07797.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rosen SD, Lemjabbar-Alaoui H. Sulf-2: an extracellular modulator of cell signaling and a cancer target candidate. Expert Opin Ther Targets. 2010;14:935–49. doi: 10.1517/14728222.2010.504718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Phillips JJ. Novel therapeutic targets in the brain tumor microenvironment. Oncotarget. 2012;3:568–75. doi: 10.18632/oncotarget.526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cecchi F, Pajalunga D, Fowler CA, Uren A, Rabe DC, Peruzzi B, Macdonald NJ, Blackman DK, Stahl SJ, Byrd RA, Bottaro DP. Targeted disruption of heparan sulfate interaction with hepatocyte and vascular endothelial growth factors blocks normal and oncogenic signaling. Cancer Cell. 22:250–62. doi: 10.1016/j.ccr.2012.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Phillips JJ, Huillard E, Robinson AE, Ward A, Lum DH, Polley MY, Rosen SD, Rowitch DH, Werb Z. Heparan sulfate sulfatase SULF2 regulates PDGFRalpha signaling and growth in human and mouse malignant glioma. xClin Invest. 2012;122:911–22. doi: 10.1172/JCI58215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sugiarto S, Persson AI, Munoz EG, Waldhuber M, Lamagna C, Andor N, Hanecker P, Ayers-Ringler J, Phillips J, Siu J, Lim DA, Vandenberg S, Stallcup W, Berger MS, Bergers G, Weiss WA, Petritsch C. Asymmetry-defective oligodendrocyte progenitors are glioma precursors. Cancer Cell. 2011;20:328–40. doi: 10.1016/j.ccr.2011.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Svendsen A, Verhoeff JJ, Immervoll H, Brogger JC, Kmiecik J, Poli A, Netland IA, Prestegarden L, Planaguma J, Torsvik A, Kjersem AB, Sakariassen PO, Heggdal JI, Van Furth WR, Bjerkvig R, Lund-Johansen M, Enger PO, Felsberg J, Brons NH, Tronstad KJ, Waha A, Chekenya M. Expression of the progenitor marker NG2/CSPG4 predicts poor survival and resistance to ionising radiation in glioblastoma. Acta Neuropathol. 2011;122:495–510. doi: 10.1007/s00401-011-0867-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chekenya M, Krakstad C, Svendsen A, Netland IA, Staalesen V, Tysnes BB, Selheim F, Wang J, Sakariassen PO, Sandal T, Lonning PE, Flatmark T, Enger PO, Bjerkvig R, Sioud M, Stallcup WB. The progenitor cell marker NG2/MPG promotes chemoresistance by activation of integrin-dependent PI3K/Akt signaling. Oncogene. 2008;27:5182–94. doi: 10.1038/onc.2008.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stallcup WB, Huang FJ. A role for the NG2 proteoglycan in glioma progression. Cell Adh Migr. 2008;2:192–201. doi: 10.4161/cam.2.3.6279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Su G, Meyer K, Nandini CD, Qiao D, Salamat S, Friedl A. Glypican-1 is frequently overexpressed in human gliomas and enhances FGF-2 signaling in glioma cells. Am J Pathol. 2006;168:2014–26. doi: 10.2353/ajpath.2006.050800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Viapiano MS, Matthews RT, Hockfield S. A novel membrane-associated glycovariant of BEHAB/brevican is up-regulated during rat brain development and in a rat model of invasive glioma. J Biol Chem. 2003;278:33239–47. doi: 10.1074/jbc.M303480200. [DOI] [PubMed] [Google Scholar]
- 18.Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, Miller CR, Ding L, Golub T, Mesirov JP, Alexe G, Lawrence M, O’Kelly M, Tamayo P, Weir BA, Gabriel S, Winckler W, Gupta S, Jakkula L, Feiler HS, Hodgson JG, James CD, Sarkaria JN, Brennan C, Kahn A, Spellman PT, Wilson RK, Speed TP, Gray JW, Meyerson M, Getz G, Perou CM, Hayes DN. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brennan C, Momota H, Hambardzumyan D, Ozawa T, Tandon A, Pedraza A, Holland E. Glioblastoma subclasses can be defined by activity among signal transduction pathways and associated genomic alterations. PLoS One. 2009;4:e7752. doi: 10.1371/journal.pone.0007752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Phillips HS, Kharbanda S, Chen R, Forrest WF, Soriano RH, Wu TD, Misra A, Nigro JM, Colman H, Soroceanu L, Williams PM, Modrusan Z, Feuerstein BG, Aldape K. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9:157–73. doi: 10.1016/j.ccr.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 21.Zimmermann DR, Dours-Zimmermann MT. Extracellular matrix of the central nervous system: from neglect to challenge. Histochem Cell Biol. 2008;130:635–53. doi: 10.1007/s00418-008-0485-9. [DOI] [PubMed] [Google Scholar]
- 22.Maeda N, Ishii M, Nishimura K, Kamimura K. Functions of chondroitin sulfate and heparan sulfate in the developing brain. Neurochem Res. 2011;36:1228–40. doi: 10.1007/s11064-010-0324-y. [DOI] [PubMed] [Google Scholar]
- 23.Abaskharoun M, Bellemare M, Lau E, Margolis RU. Glypican-1, phosphacan/receptor protein-tyrosine phosphatase-zeta/beta and its ligand, tenascin-C, are expressed by neural stem cells and neural cells derived from embryonic stem cells. ASN neuro. 2010;2:e00039. doi: 10.1042/AN20100001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Esko JD, Lindahl U. Molecular diversity of heparan sulfate. J Clin Invest. 2001;108:169–73. doi: 10.1172/JCI13530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kwok JC, Warren P, Fawcett JW. Chondroitin sulfate: a key molecule in the brain matrix. Int J Biochem Cell Biol. 2012;44:582–6. doi: 10.1016/j.biocel.2012.01.004. [DOI] [PubMed] [Google Scholar]
- 26.Greene DK, Tumova S, Couchman JR, Woods A. Syndecan-4 associates with alpha-actinin. J Biol Chem. 2003;278:7617–23. doi: 10.1074/jbc.M207123200. [DOI] [PubMed] [Google Scholar]
- 27.Longley RL, Woods A, Fleetwood A, Cowling GJ, Gallagher JT, Couchman JR. Control of morphology, cytoskeleton and migration by syndecan-4. J Cell Sci. 1999;112(Pt 20):3421–31. doi: 10.1242/jcs.112.20.3421. [DOI] [PubMed] [Google Scholar]
- 28.Echtermeyer F, Baciu PC, Saoncella S, Ge Y, Goetinck PF. Syndecan-4 core protein is sufficient for the assembly of focal adhesions and actin stress fibers. J Cell Sci. 1999;112(Pt 20):3433–41. doi: 10.1242/jcs.112.20.3433. [DOI] [PubMed] [Google Scholar]
- 29.Beauvais DM, Rapraeger AC. Syndecan-1 couples the insulin-like growth factor-1 receptor to inside-out integrin activation. J Cell Sci. 2010;123:3796–807. doi: 10.1242/jcs.067645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shipp EL, Hsieh-Wilson LC. Profiling the sulfation specificities of glycosaminoglycan interactions with growth factors and chemotactic proteins using microarrays. Chem Biol. 2007;14:195–208. doi: 10.1016/j.chembiol.2006.12.009. [DOI] [PubMed] [Google Scholar]
- 31.Bespalov MM, Sidorova YA, Tumova S, Ahonen-Bishopp A, Magalhaes AC, Kulesskiy E, Paveliev M, Rivera C, Rauvala H, Saarma M. Heparan sulfate proteoglycan syndecan-3 is a novel receptor for GDNF, neurturin, and artemin. J Cell Biol. 2011;192:153–69. doi: 10.1083/jcb.201009136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ashikari-Hada S, Habuchi H, Kariya Y, Itoh N, Reddi AH, Kimata K. Characterization of growth factor-binding structures in heparin/heparan sulfate using an octasaccharide library. J Biol Chem. 2004;279:12346–54. doi: 10.1074/jbc.M313523200. [DOI] [PubMed] [Google Scholar]
- 33.Han C, Belenkaya TY, Khodoun M, Tauchi M, Lin X, Lin X. Distinct and collaborative roles of Drosophila EXT family proteins in morphogen signalling and gradient formation. Development. 2004;131:1563–75. doi: 10.1242/dev.01051. [DOI] [PubMed] [Google Scholar]
- 34.Han C, Yan D, Belenkaya TY, Lin X. Drosophila glypicans Dally and Dally-like shape the extracellular Wingless morphogen gradient in the wing disc. Development. 2005;132:667–79. doi: 10.1242/dev.01636. [DOI] [PubMed] [Google Scholar]
- 35.Rapraeger AC, Krufka A, Olwin BB. Requirement of heparan sulfate for bFGF-mediated fibroblast growth and myoblast differentiation. Science. 1991;252:1705–8. doi: 10.1126/science.1646484. [DOI] [PubMed] [Google Scholar]
- 36.Yayon A, Klagsbrun M, Esko JD, Leder P, Ornitz DM. Cell surface, heparin-like molecules are required for binding of basic fibroblast growth factor to its high affinity receptor. Cell. 1991;64:841–8. doi: 10.1016/0092-8674(91)90512-w. [DOI] [PubMed] [Google Scholar]
- 37.Kreuger J, Salmivirta M, Sturiale L, Gimenez-Gallego G, Lindahl U. Sequence analysis of heparan sulfate epitopes with graded affinities for fibroblast growth factors 1 and 2. J Biol Chem. 2001;276:30744–52. doi: 10.1074/jbc.M102628200. [DOI] [PubMed] [Google Scholar]
- 38.Ford-Perriss M, Guimond SE, Greferath U, Kita M, Grobe K, Habuchi H, Kimata K, Esko JD, Murphy M, Turnbull JE. Variant heparan sulfates synthesized in developing mouse brain differentially regulate FGF signaling. Glycobiology. 2002;12:721–7. doi: 10.1093/glycob/cwf072. [DOI] [PubMed] [Google Scholar]
- 39.Goodger SJ, Robinson CJ, Murphy KJ, Gasiunas N, Harmer NJ, Blundell TL, Pye DA, Gallagher JT. Evidence that heparin saccharides promote FGF2 mitogenesis through two distinct mechanisms. J Biol Chem. 2008;283:13001–8. doi: 10.1074/jbc.M704531200. [DOI] [PubMed] [Google Scholar]
- 40.Chuang CY, Lord MS, Melrose J, Rees MD, Knox SM, Freeman C, Iozzo RV, Whitelock JM. Heparan sulfate-dependent signaling of fibroblast growth factor 18 by chondrocyte-derived perlecan. Biochemistry. 2010;49:5524–32. doi: 10.1021/bi1005199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sarrazin S, Lamanna WC, Esko JD. Heparan sulfate proteoglycans. Cold Spring Harb Perspect Biol. 2011;3 doi: 10.1101/cshperspect.a004952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Haubst N, Georges-Labouesse E, De Arcangelis A, Mayer U, Gotz M. Basement membrane attachment is dispensable for radial glial cell fate and for proliferation, but affects positioning of neuronal subtypes. Development. 2006;133:3245–54. doi: 10.1242/dev.02486. [DOI] [PubMed] [Google Scholar]
- 43.Giros A, Morante J, Gil-Sanz C, Fairen A, Costell M. Perlecan controls neurogenesis in the developing telencephalon. BMC Dev Biol. 2007;7:29. doi: 10.1186/1471-213X-7-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Costell M, Gustafsson E, Aszodi A, Morgelin M, Bloch W, Hunziker E, Addicks K, Timpl R, Fassler R. Perlecan maintains the integrity of cartilage and some basement membranes. J Cell Biology. 1999;147:1109–22. doi: 10.1083/jcb.147.5.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pilia G, Hughes-Benzie RM, MacKenzie A, Baybayan P, Chen EY, Huber R, Neri G, Cao A, Forabosco A, Schlessinger D. Mutations in GPC3, a glypican gene, cause the Simpson-Golabi-Behmel overgrowth syndrome. Nat Genet. 1996;12:241–7. doi: 10.1038/ng0396-241. [DOI] [PubMed] [Google Scholar]
- 46.Kucharova K, Stallcup WB. The NG2 proteoglycan promotes oligodendrocyte progenitor proliferation and developmental myelination. Neuroscience. 2010;166:185–94. doi: 10.1016/j.neuroscience.2009.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chang Y, She ZG, Sakimura K, Roberts A, Kucharova K, Rowitch DH, Stallcup WB. Ablation of NG2 proteoglycan leads to deficits in brown fat function and to adult onset obesity. PLoS One. 2012;7:e30637. doi: 10.1371/journal.pone.0030637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Habuchi H, Habuchi O, Kimata K. Sulfation pattern in glycosaminoglycan: does it have a code? Glycoconj J. 2004;21:47–52. doi: 10.1023/B:GLYC.0000043747.87325.5e. [DOI] [PubMed] [Google Scholar]
- 49.Bink RJ, Habuchi H, Lele Z, Dolk E, Joore J, Rauch GJ, Geisler R, Wilson SW, den Hertog J, Kimata K, Zivkovic D. Heparan sulfate 6-o-sulfotransferase is essential for muscle development in zebrafish. J Biol Chem. 2003;278:31118–27. doi: 10.1074/jbc.M213124200. [DOI] [PubMed] [Google Scholar]
- 50.Bulow HE, Hobert O. Differential sulfations and epimerization define heparan sulfate specificity in nervous system development. Neuron. 2004;41:723–36. doi: 10.1016/s0896-6273(04)00084-4. [DOI] [PubMed] [Google Scholar]
- 51.Kamimura K, Fujise M, Villa F, Izumi S, Habuchi H, Kimata K, Nakato H. Drosophila heparan sulfate 6-O-sulfotransferase (dHS6ST) gene. Structure, expression, and function in the formation of the tracheal system. J Biol Chem. 2001;276:17014–21. doi: 10.1074/jbc.M011354200. [DOI] [PubMed] [Google Scholar]
- 52.Morimoto-Tomita M, Uchimura K, Werb Z, Hemmerich S, Rosen SD. Cloning and characterization of two extracellular heparin-degrading endosulfatases in mice and humans. J Biol Chem. 2002;277:49175–85. doi: 10.1074/jbc.M205131200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dhoot GK, Gustafsson MK, Ai X, Sun W, Standiford DM, Emerson CP., Jr Regulation of Wnt signaling and embryo patterning by an extracellular sulfatase. Science. 2001;293:1663–6. doi: 10.1126/science.293.5535.1663. [DOI] [PubMed] [Google Scholar]
- 54.Ai X, Do AT, Lozynska O, Kusche-Gullberg M, Lindahl U, Emerson CP., Jr QSulf1 remodels the 6-O sulfation states of cell surface heparan sulfate proteoglycans to promote Wnt signaling. J Cell Biol. 2003;162:341–51. doi: 10.1083/jcb.200212083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ai X, Kitazawa T, Do AT, Kusche-Gullberg M, Labosky PA, Emerson CP., Jr SULF1 and SULF2 regulate heparan sulfate-mediated GDNF signaling for esophageal innervation. Development. 2007;134:3327–38. doi: 10.1242/dev.007674. [DOI] [PubMed] [Google Scholar]
- 56.Dai Y, Yang Y, MacLeod V, Yue X, Rapraeger AC, Shriver Z, Venkataraman G, Sasisekharan R, Sanderson RD. HSulf-1 and HSulf-2 are potent inhibitors of myeloma tumor growth in vivo. J Biol Chem. 2005;280:40066–73. doi: 10.1074/jbc.M508136200. [DOI] [PubMed] [Google Scholar]
- 57.Viviano BL, Paine-Saunders S, Gasiunas N, Gallagher J, Saunders S. Domain-specific modification of heparan sulfate by Qsulf1 modulates the binding of the bone morphogenetic protein antagonist Noggin. J Biol Chem. 2004;279:5604–11. doi: 10.1074/jbc.M310691200. [DOI] [PubMed] [Google Scholar]
- 58.Danesin C, Agius E, Escalas N, Ai X, Emerson C, Cochard P, Soula C. Ventral neural progenitors switch toward an oligodendroglial fate in response to increased Sonic hedgehog (Shh) activity: involvement of Sulfatase 1 in modulating Shh signaling in the ventral spinal cord. J Neurosci. 2006;26:5037–48. doi: 10.1523/JNEUROSCI.0715-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Freeman SD, Moore WM, Guiral EC, Holme AD, Turnbull JE, Pownall ME. Extracellular regulation of developmental cell signaling by XtSulf1. Dev Biol. 2008;320:436–45. doi: 10.1016/j.ydbio.2008.05.554. [DOI] [PubMed] [Google Scholar]
- 60.Lamanna WC, Frese MA, Balleininger M, Dierks T. Sulf loss influences N-, 2-O-, and 6-O-sulfation of multiple heparan sulfate proteoglycans and modulates fibroblast growth factor signaling. J Biol Chem. 2008;283:27724–35. doi: 10.1074/jbc.M802130200. [DOI] [PubMed] [Google Scholar]
- 61.Fujita K, Takechi E, Sakamoto N, Sumiyoshi N, Izumi S, Miyamoto T, Matsuura S, Tsurugaya T, Akasaka K, Yamamoto T. HpSulf, a heparan sulfate 6-O-endosulfatase, is involved in the regulation of VEGF signaling during sea urchin development. Mech Dev. 2010;127:235–45. doi: 10.1016/j.mod.2009.12.001. [DOI] [PubMed] [Google Scholar]
- 62.Uchimura K, Morimoto-Tomita M, Bistrup A, Li J, Lyon M, Gallagher J, Werb Z, Rosen SD. HSulf-2, an extracellular endoglucosamine-6-sulfatase, selectively mobilizes heparin-bound growth factors and chemokines: effects on VEGF, FGF-1, and SDF-1. BMC Biochem. 2006;7:2. doi: 10.1186/1471-2091-7-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Habuchi H, Nagai N, Sugaya N, Atsumi F, Stevens RL, Kimata K. Mice deficient in heparan sulfate 6-O-sulfotransferase-1 exhibit defective heparan sulfate biosynthesis, abnormal placentation, and late embryonic lethality. J Biol Chem. 2007;282:15578–88. doi: 10.1074/jbc.M607434200. [DOI] [PubMed] [Google Scholar]
- 64.Sugaya N, Habuchi H, Nagai N, Ashikari-Hada S, Kimata K. 6-O-sulfation of heparan sulfate differentially regulates various fibroblast growth factor-dependent signalings in culture. J Biol Chem. 2008;283:10366–76. doi: 10.1074/jbc.M705948200. [DOI] [PubMed] [Google Scholar]
- 65.Hayano S, Kurosaka H, Yanagita T, Kalus I, Milz F, Ishihara Y, Islam MN, Kawanabe N, Saito M, Kamioka H, Adachi T, Dierks T, Yamashiro T. Roles of heparan sulfate sulfation in dentinogenesis. J Biol Chem. 2012;287:12217–29. doi: 10.1074/jbc.M111.332924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kalus I, Salmen B, Viebahn C, von Figura K, Schmitz D, D’Hooge R, Dierks T. Differential involvement of the extracellular 6-O-endosulfatases Sulf1 and Sulf2 in brain development and neuronal and behavioural plasticity. J Cell Mol Med. 2009;13:4505–21. doi: 10.1111/j.1582-4934.2008.00558.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ratzka A, Kalus I, Moser M, Dierks T, Mundlos S, Vortkamp A. Redundant function of the heparan sulfate 6-O-endosulfatases Sulf1 and Sulf2 during skeletal development. Dev Dyn. 2008;237:339–53. doi: 10.1002/dvdy.21423. [DOI] [PubMed] [Google Scholar]
- 68.Ida M, Shuo T, Hirano K, Tokita Y, Nakanishi K, Matsui F, Aono S, Fujita H, Fujiwara Y, Kaji T, Oohira A. Identification and functions of chondroitin sulfate in the milieu of neural stem cells. J Biol Chem. 2006;281:5982–91. doi: 10.1074/jbc.M507130200. [DOI] [PubMed] [Google Scholar]
- 69.Mercier F, Arikawa-Hirasawa E. Heparan sulfate niche for cell proliferation in the adult brain. Neurosci Lett. 2012;510:67–72. doi: 10.1016/j.neulet.2011.12.046. [DOI] [PubMed] [Google Scholar]
- 70.Wang Q, Yang L, Alexander C, Temple S. The niche factor syndecan-1 regulates the maintenance and proliferation of neural progenitor cells during mammalian cortical development. PLoS One. 2012;7:e42883. doi: 10.1371/journal.pone.0042883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sirko S, von Holst A, Weber A, Wizenmann A, Theocharidis U, Gotz M, Faissner A. Chondroitin sulfates are required for fibroblast growth factor-2-dependent proliferation and maintenance in neural stem cells and for epidermal growth factor-dependent migration of their progeny. Stem Cells. 2010;28:775–87. doi: 10.1002/stem.309. [DOI] [PubMed] [Google Scholar]
- 72.Trotter J, Karram K, Nishiyama A. NG2 cells: Properties, progeny and origin. Brain Res Rev. 2010;63:72–82. doi: 10.1016/j.brainresrev.2009.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Goretzki L, Burg MA, Grako KA, Stallcup WB. High-affinity binding of basic fibroblast growth factor and platelet-derived growth factor-AA to the core protein of the NG2 proteoglycan. J Biol Chem. 1999;274:16831–7. doi: 10.1074/jbc.274.24.16831. [DOI] [PubMed] [Google Scholar]
- 74.Fukushi J, Makagiansar IT, Stallcup WB. NG2 proteoglycan promotes endothelial cell motility and angiogenesis via engagement of galectin-3 and alpha3beta1 integrin. Mol Biol Cell. 2004;15:3580–90. doi: 10.1091/mbc.E04-03-0236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nishiyama A, Lin XH, Giese N, Heldin CH, Stallcup WB. Interaction between NG2 proteoglycan and PDGF alpha-receptor on O2A progenitor cells is required for optimal response to PDGF. J Neurosci Res. 1996;43:315–30. doi: 10.1002/(SICI)1097-4547(19960201)43:3<315::AID-JNR6>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 76.Cattaruzza S, Ozerdem U, Denzel M, Ranscht B, Bulian P, Cavallaro U, Zanocco D, Colombatti A, Stallcup WB, Perris R. Multivalent proteoglycan modulation of FGF mitogenic responses in perivascular cells. Angiogenesis. 2012 doi: 10.1007/s10456-012-9316-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Burg MA, Tillet E, Timpl R, Stallcup WB. Binding of the NG2 proteoglycan to type VI collagen and other extracellular matrix molecules. J Biol Chem. 1996;271:26110–6. doi: 10.1074/jbc.271.42.26110. [DOI] [PubMed] [Google Scholar]
- 78.Wen Y, Makagiansar IT, Fukushi J, Liu FT, Fukuda MN, Stallcup WB. Molecular basis of interaction between NG2 proteoglycan and galectin-3. J Cell Biochem. 2006;98:115–27. doi: 10.1002/jcb.20768. [DOI] [PubMed] [Google Scholar]
- 79.Nishiyama A, Komitova M, Suzuki R, Zhu X. Polydendrocytes (NG2 cells): multifunctional cells with lineage plasticity. Nat Rev Neurosci. 2009;10:9–22. doi: 10.1038/nrn2495. [DOI] [PubMed] [Google Scholar]
- 80.Barritt DS, Pearn MT, Zisch AH, Lee SS, Javier RT, Pasquale EB, Stallcup WB. The multi-PDZ domain protein MUPP1 is a cytoplasmic ligand for the membrane-spanning proteoglycan NG2. J Cell Biochem. 2000;79:213–24. doi: 10.1002/1097-4644(20001101)79:2<213::aid-jcb50>3.0.co;2-g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chatterjee N, Stegmuller J, Schatzle P, Karram K, Koroll M, Werner HB, Nave KA, Trotter J. Interaction of syntenin-1 and the NG2 proteoglycan in migratory oligodendrocyte precursor cells. J Biol Chem. 2008;283:8310–7. doi: 10.1074/jbc.M706074200. [DOI] [PubMed] [Google Scholar]
- 82.Stegmuller J, Werner H, Nave KA, Trotter J. The proteoglycan NG2 is complexed with alpha-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptors by the PDZ glutamate receptor interaction protein (GRIP) in glial progenitor cells. Implications for glial-neuronal signaling. J Biol Chem. 2003;278:3590–8. doi: 10.1074/jbc.M210010200. [DOI] [PubMed] [Google Scholar]
- 83.Makagiansar IT, Williams S, Dahlin-Huppe K, Fukushi J, Mustelin T, Stallcup WB. Phosphorylation of NG2 proteoglycan by protein kinase C-alpha regulates polarized membrane distribution and cell motility. J Biol Chem. 2004;279:55262–70. doi: 10.1074/jbc.M411045200. [DOI] [PubMed] [Google Scholar]
- 84.Makagiansar IT, Williams S, Mustelin T, Stallcup WB. Differential phosphorylation of NG2 proteoglycan by ERK and PKCalpha helps balance cell proliferation and migration. J Cell Biol. 2007;178:155–65. doi: 10.1083/jcb.200612084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Forsberg M, Holmborn K, Kundu S, Dagalv A, Kjellen L, Forsberg-Nilsson K. Undersulfation of heparan sulfate restricts differentiation potential of mouse embryonic stem cells. J Biol Chem. 2012;287:10853–62. doi: 10.1074/jbc.M111.337030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Braquart-Varnier C, Danesin C, Clouscard-Martinato C, Agius E, Escalas N, Benazeraf B, Ai X, Emerson C, Cochard P, Soula C. A subtractive approach to characterize genes with regionalized expression in the gliogenic ventral neuroepithelium: identification of chick sulfatase 1 as a new oligodendrocyte lineage gene. Mol Cell Neurosci. 2004;25:612–28. doi: 10.1016/j.mcn.2003.11.013. [DOI] [PubMed] [Google Scholar]
- 87.Touahri Y, Escalas N, Benazeraf B, Cochard P, Danesin C, Soula C. Sulfatase 1 Promotes the Motor Neuron-to-Oligodendrocyte Fate Switch by Activating Shh Signaling in Olig2 Progenitors of the Embryonic Ventral Spinal Cord. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2012;32:18018–34. doi: 10.1523/JNEUROSCI.3553-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ishii M, Maeda N. Oversulfated chondroitin sulfate plays critical roles in the neuronal migration in the cerebral cortex. J Biol Chem. 2008;283:32610–20. doi: 10.1074/jbc.M806331200. [DOI] [PubMed] [Google Scholar]
- 89.Asher RA, Morgenstern DA, Shearer MC, Adcock KH, Pesheva P, Fawcett JW. Versican is upregulated in CNS injury and is a product of oligodendrocyte lineage cells. J Neurosci. 2002;22:2225–36. doi: 10.1523/JNEUROSCI.22-06-02225.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lau LW, Keough MB, Haylock-Jacobs S, Cua R, Doring A, Sloka S, Stirling DP, Rivest S, Yong VW. Chondroitin sulfate proteoglycans in demyelinated lesions impair remyelination. Ann Neurol. 2012;72:419–32. doi: 10.1002/ana.23599. [DOI] [PubMed] [Google Scholar]
- 91.Bradbury EJ, Moon LD, Popat RJ, King VR, Bennett GS, Patel PN, Fawcett JW, McMahon SB. Chondroitinase ABC promotes functional recovery after spinal cord injury. Nature. 2002;416:636–40. doi: 10.1038/416636a. [DOI] [PubMed] [Google Scholar]
- 92.Sato Y, Nakanishi K, Tokita Y, Kakizawa H, Ida M, Maeda H, Matsui F, Aono S, Saito A, Kuroda Y, Hayakawa M, Kojima S, Oohira A. A highly sulfated chondroitin sulfate preparation, CS-E, prevents excitatory amino acid-induced neuronal cell death. J Neurochem. 2008;104:1565–76. doi: 10.1111/j.1471-4159.2007.05107.x. [DOI] [PubMed] [Google Scholar]
- 93.Kucharova K, Chang Y, Boor A, Yong VW, Stallcup WB. Reduced inflammation accompanies diminished myelin damage and repair in the NG2 null mouse spinal cord. J Neuroinflammation. 2011;8:158. doi: 10.1186/1742-2094-8-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Higginson JR, Thompson SM, Santos-Silva A, Guimond SE, Turnbull JE, Barnett SC. Differential sulfation remodelling of heparan sulfate by extracellular 6-o-sulfatases regulates fibroblast growth factor-induced boundary formation by glial cells: implications for glial cell transplantation. J Neurosci. 2012;32:15902–12. doi: 10.1523/JNEUROSCI.6340-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Iozzo RV, Sanderson RD. Proteoglycans in cancer biology, tumour microenvironment and angiogenesis. J Cell Mol Med. 2011;15:1013–31. doi: 10.1111/j.1582-4934.2010.01236.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Theocharis AD, Skandalis SS, Tzanakakis GN, Karamanos NK. Proteoglycans in health and disease: novel roles for proteoglycans in malignancy and their pharmacological targeting. FEBS J. 2010;277:3904–23. doi: 10.1111/j.1742-4658.2010.07800.x. [DOI] [PubMed] [Google Scholar]
- 97.Aikawa T, Whipple CA, Lopez ME, Gunn J, Young A, Lander AD, Korc M. Glypican-1 modulates the angiogenic and metastatic potential of human and mouse cancer cells. J Clin Invest. 2008;118:89–99. doi: 10.1172/JCI32412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kleeff J, Ishiwata T, Kumbasar A, Friess H, Buchler MW, Lander AD, Korc M. The cell-surface heparan sulfate proteoglycan glypican-1 regulates growth factor action in pancreatic carcinoma cells and is overexpressed in human pancreatic cancer. The Journal of clinical investigation. 1998;102:1662–73. doi: 10.1172/JCI4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Whipple CA, Young AL, Korc M. A Kras(G12D)-driven genetic mouse model of pancreatic cancer requires glypican-1 for efficient proliferation and angiogenesis. Oncogene. 2011;31:2535–44. doi: 10.1038/onc.2011.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Vlodavsky I, Beckhove P, Lerner I, Pisano C, Meirovitz A, Ilan N, Elkin M. Significance of heparanase in cancer and inflammation. Cancer Microenviron. 2012;5:115–32. doi: 10.1007/s12307-011-0082-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ramani VC, Yang Y, Ren Y, Nan L, Sanderson RD. Heparanase plays a dual role in driving hepatocyte growth factor (HGF) signaling by enhancing HGF expression and activity. J Biol Chem. 2011;286:6490–9. doi: 10.1074/jbc.M110.183277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Purushothaman A, Uyama T, Kobayashi F, Yamada S, Sugahara K, Rapraeger AC, Sanderson RD. Heparanase-enhanced shedding of syndecan-1 by myeloma cells promotes endothelial invasion and angiogenesis. Blood. 2010;115:2449–57. doi: 10.1182/blood-2009-07-234757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Purushothaman A, Babitz SK, Sanderson RD. Heparanase enhances the insulin receptor signaling pathway to activate ERK in multiple myeloma. J Biol Chem. 2012 doi: 10.1074/jbc.M112.391417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Yang Y, Macleod V, Miao HQ, Theus A, Zhan F, Shaughnessy JD, Jr, Sawyer J, Li JP, Zcharia E, Vlodavsky I, Sanderson RD. Heparanase enhances syndecan-1 shedding: a novel mechanism for stimulation of tumor growth and metastasis. J Biol Chem. 2007;282:13326–33. doi: 10.1074/jbc.M611259200. [DOI] [PubMed] [Google Scholar]
- 105.Morimoto-Tomita M, Uchimura K, Bistrup A, Lum DH, Egeblad M, Boudreau N, Werb Z, Rosen SD. Sulf-2, a proangiogenic heparan sulfate endosulfatase, is upregulated in breast cancer. Neoplasia. 2005;7:1001–10. doi: 10.1593/neo.05496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lemjabbar-Alaoui H, van Zante A, Singer MS, Xue Q, Wang YQ, Tsay D, He B, Jablons DM, Rosen SD. Sulf-2, a heparan sulfate endosulfatase, promotes human lung carcinogenesis. Oncogene. 2010;29:635–46. doi: 10.1038/onc.2009.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lai J, Chien J, Staub J, Avula R, Greene EL, Matthews TA, Smith DI, Kaufmann SH, Roberts LR, Shridhar V. Loss of HSulf-1 up-regulates heparin-binding growth factor signaling in cancer. J Biol Chem. 2003;278:23107–17. doi: 10.1074/jbc.M302203200. [DOI] [PubMed] [Google Scholar]
- 108.Nawroth R, van Zante A, Cervantes S, McManus M, Hebrok M, Rosen SD. Extracellular sulfatases, elements of the Wnt signaling pathway, positively regulate growth and tumorigenicity of human pancreatic cancer cells. PLoS One. 2007;2:e392. doi: 10.1371/journal.pone.0000392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hur K, Han TS, Jung EJ, Yu J, Lee HJ, Kim WH, Goel A, Yang HK. Up-regulated expression of sulfatases (SULF1 and SULF2) as prognostic and metastasis predictive markers in human gastric cancer. J Pathol. 2012;228:88–98. doi: 10.1002/path.4055. [DOI] [PubMed] [Google Scholar]
- 110.Yang JD, Sun Z, Hu C, Lai J, Dove R, Nakamura I, Lee JS, Thorgeirsson SS, Kang KJ, Chu IS, Roberts LR. Sulfatase 1 and sulfatase 2 in hepatocellular carcinoma: associated signaling pathways, tumor phenotypes, and survival. Genes Chromosomes Cancer. 2011;50:122–35. doi: 10.1002/gcc.20838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Johansson FK, Brodd J, Eklof C, Ferletta M, Hesselager G, Tiger CF, Uhrbom L, Westermark B. Identification of candidate cancer-causing genes in mouse brain tumors by retroviral tagging. Proc Natl Acad Sci U S A. 2004;101:11334–7. doi: 10.1073/pnas.0402716101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Fuster MM, Wang L, Castagnola J, Sikora L, Reddi K, Lee PH, Radek KA, Schuksz M, Bishop JR, Gallo RL, Sriramarao P, Esko JD. Genetic alteration of endothelial heparan sulfate selectively inhibits tumor angiogenesis. J Cell Biol. 2007;177:539–49. doi: 10.1083/jcb.200610086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ferreras C, Rushton G, Cole CL, Babur M, Telfer BA, van Kuppevelt TH, Gardiner JM, Williams KJ, Jayson GC, Avizienyte E. Endothelial heparan sulfate 6-o-sulfation levels regulate angiogenic responses of endothelial cells to fibroblast growth factor 2 and vascular endothelial growth factor. J Biol Chem. 2012;287:36132–46. doi: 10.1074/jbc.M112.384875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Huang FJ, You WK, Bonaldo P, Seyfried TN, Pasquale EB, Stallcup WB. Pericyte deficiencies lead to aberrant tumor vascularizaton in the brain of the NG2 null mouse. Dev Biol. 2010;344:1035–46. doi: 10.1016/j.ydbio.2010.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Parish CR. The role of heparan sulphate in inflammation. Nat Rev Immunol. 2006;6:633–43. doi: 10.1038/nri1918. [DOI] [PubMed] [Google Scholar]
- 116.Kim S, Takahashi H, Lin WW, Descargues P, Grivennikov S, Kim Y, Luo JL, Karin M. Carcinoma-produced factors activate myeloid cells through TLR2 to stimulate metastasis. Nature. 2009;457:102–6. doi: 10.1038/nature07623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lerner I, Hermano E, Zcharia E, Rodkin D, Bulvik R, Doviner V, Rubinstein AM, Ishai-Michaeli R, Atzmon R, Sherman Y, Meirovitz A, Peretz T, Vlodavsky I, Elkin M. Heparanase powers a chronic inflammatory circuit that promotes colitis-associated tumorigenesis in mice. J Clin Invest. 2011;121:1709–21. doi: 10.1172/JCI43792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Colman H, Zhang L, Sulman EP, McDonald JM, Shooshtari NL, Rivera A, Popoff S, Nutt CL, Louis DN, Cairncross JG, Gilbert MR, Phillips HS, Mehta MP, Chakravarti A, Pelloski CE, Bhat K, Feuerstein BG, Jenkins RB, Aldape K. A multigene predictor of outcome in glioblastoma. Neuro Oncol. 2010;12:49–57. doi: 10.1093/neuonc/nop007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Mischel PS, Shai R, Shi T, Horvath S, Lu KV, Choe G, Seligson D, Kremen TJ, Palotie A, Liau LM, Cloughesy TF, Nelson SF. Identification of molecular subtypes of glioblastoma by gene expression profiling. Oncogene. 2003;22:2361–73. doi: 10.1038/sj.onc.1206344. [DOI] [PubMed] [Google Scholar]
- 120.Sturm D, Witt H, Hovestadt V, Khuong-Quang DA, Jones DT, Konermann C, Pfaff E, Tonjes M, Sill M, Bender S, Kool M, Zapatka M, Becker N, Zucknick M, Hielscher T, Liu XY, Fontebasso AM, Ryzhova M, Albrecht S, Jacob K, Wolter M, Ebinger M, Schuhmann MU, van Meter T, Fruhwald MC, Hauch H, Pekrun A, Radlwimmer B, Niehues T, von Komorowski G, Durken M, Kulozik AE, Madden J, Donson A, Foreman NK, Drissi R, Fouladi M, Scheurlen W, von Deimling A, Monoranu C, Roggendorf W, Herold-Mende C, Unterberg A, Kramm CM, Felsberg J, Hartmann C, Wiestler B, Wick W, Milde T, Witt O, Lindroth AM, Schwartzentruber J, Faury D, Fleming A, Zakrzewska M, Liberski PP, Zakrzewski K, Hauser P, Garami M, Klekner A, Bognar L, Morrissy S, Cavalli F, Taylor MD, van Sluis P, Koster J, Versteeg R, Volckmann R, Mikkelsen T, Aldape K, Reifenberger G, Collins VP, Majewski J, Korshunov A, Lichter P, Plass C, Jabado N, Pfister SM. Hotspot Mutations in H3F3A and IDH1 Define Distinct Epigenetic and Biological Subgroups of Glioblastoma. Cancer Cell. 2012;22:425–37. doi: 10.1016/j.ccr.2012.08.024. [DOI] [PubMed] [Google Scholar]
- 121.Bhat KP, Salazar KL, Balasubramaniyan V, Wani K, Heathcock L, Hollingsworth F, James JD, Gumin J, Diefes KL, Kim SH, Turski A, Azodi Y, Yang Y, Doucette T, Colman H, Sulman EP, Lang FF, Rao G, Copray S, Vaillant BD, Aldape KD. The transcriptional coactivator TAZ regulates mesenchymal differentiation in malignant glioma. Genes Dev. 2011;25:2594–609. doi: 10.1101/gad.176800.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.The Cancer Genome Atlas TCGA Research Network . Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Datta MW, Hernandez AM, Schlicht MJ, Kahler AJ, DeGueme AM, Dhir R, Shah RB, Farach-Carson C, Barrett A, Datta S. Perlecan, a candidate gene for the CAPB locus, regulates prostate cancer cell growth via the Sonic Hedgehog pathway. Mol Cancer. 2006;5:9. doi: 10.1186/1476-4598-5-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Jijiwa M, Demir H, Gupta S, Leung C, Joshi K, Orozco N, Huang T, Yildiz VO, Shibahara I, de Jesus JA, Yong WH, Mischel PS, Fernandez S, Kornblum HI, Nakano I. CD44v6 regulates growth of brain tumor stem cells partially through the AKT-mediated pathway. PLoS One. 2011;6:e24217. doi: 10.1371/journal.pone.0024217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Zhou Z, Wang J, Cao R, Morita H, Soininen R, Chan KM, Liu B, Cao Y, Tryggvason K. Impaired angiogenesis, delayed wound healing and retarded tumor growth in perlecan heparan sulfate-deficient mice. Cancer Res. 2004;64:4699–702. doi: 10.1158/0008-5472.CAN-04-0810. [DOI] [PubMed] [Google Scholar]
- 126.Foehr ED, Lorente G, Kuo J, Ram R, Nikolich K, Urfer R. Targeting of the receptor protein tyrosine phosphatase beta with a monoclonal antibody delays tumor growth in a glioblastoma model. Cancer Res. 2006;66:2271–8. doi: 10.1158/0008-5472.CAN-05-1221. [DOI] [PubMed] [Google Scholar]
- 127.Arslan F, Bosserhoff AK, Nickl-Jockschat T, Doerfelt A, Bogdahn U, Hau P. The role of versican isoforms V0/V1 in glioma migration mediated by transforming growth factor-beta2. Br J Cancer. 2007;96:1560–8. doi: 10.1038/sj.bjc.6603766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Khotskaya YB, Dai Y, Ritchie JP, MacLeod V, Yang Y, Zinn K, Sanderson RD. Syndecan-1 is required for robust growth, vascularization, and metastasis of myeloma tumors in vivo. J Biol Chem. 2009;284:26085–95. doi: 10.1074/jbc.M109.018473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Berman B, Ostrovsky O, Shlissel M, Lang T, Regan D, Vlodavsky I, Ishai-Michaeli R, Ron D. Similarities and differences between the effects of heparin and glypican-1 on the bioactivity of acidic fibroblast growth factor and the keratinocyte growth factor. J Biol Chem. 1999;274:36132–8. doi: 10.1074/jbc.274.51.36132. [DOI] [PubMed] [Google Scholar]
- 130.Gengrinovitch S, Berman B, David G, Witte L, Neufeld G, Ron D. Glypican-1 is a VEGF165 binding proteoglycan that acts as an extracellular chaperone for VEGF165. J Biol Chem. 1999;274:10816–22. doi: 10.1074/jbc.274.16.10816. [DOI] [PubMed] [Google Scholar]
- 131.Qiao D, Meyer K, Mundhenke C, Drew SA, Friedl A. Heparan sulfate proteoglycans as regulators of fibroblast growth factor-2 signaling in brain endothelial cells. Specific role for glypican-1 in glioma angiogenesis. J Biol Chem. 2003;278:16045–53. doi: 10.1074/jbc.M211259200. [DOI] [PubMed] [Google Scholar]
- 132.Wang J, Svendsen A, Kmiecik J, Immervoll H, Skaftnesmo KO, Planaguma J, Reed RK, Bjerkvig R, Miletic H, Enger PO, Rygh CB, Chekenya M. Targeting the NG2/CSPG4 proteoglycan retards tumour growth and angiogenesis in preclinical models of GBM and melanoma. PLoS One. 2011;6:e23062. doi: 10.1371/journal.pone.0023062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Ulbricht U, Eckerich C, Fillbrandt R, Westphal M, Lamszus K. RNA interference targeting protein tyrosine phosphatase zeta/receptor-type protein tyrosine phosphatase beta suppresses glioblastoma growth in vitro and in vivo. J Neurochem. 2006;98:1497–506. doi: 10.1111/j.1471-4159.2006.04022.x. [DOI] [PubMed] [Google Scholar]
- 134.McClain CR, Sim FJ, Goldman SA. Pleiotrophin Suppression of Receptor Protein Tyrosine Phosphatase-beta/zeta Maintains the Self-Renewal Competence of Fetal Human Oligodendrocyte Progenitor Cells. J Neurosci. 2012;32:15066–75. doi: 10.1523/JNEUROSCI.1320-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Hernandez D, Miquel-Serra L, Docampo MJ, Marco-Ramell A, Cabrera J, Fabra A, Bassols A. V3 versican isoform alters the behavior of human melanoma cells by interfering with CD44/ErbB-dependent signaling. J Biol Chem. 2011;286:1475–85. doi: 10.1074/jbc.M110.127522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Leygue E, Snell L, Dotzlaw H, Troup S, Hiller-Hitchcock T, Murphy LC, Roughley PJ, Watson PH. Lumican and decorin are differentially expressed in human breast carcinoma. J Pathol. 2000;192:313–20. doi: 10.1002/1096-9896(200011)192:3<313::AID-PATH694>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 137.Theocharis AD. Human colon adenocarcinoma is associated with specific post-translational modifications of versican and decorin. Biochim Biophys Acta. 2002;1588:165–72. doi: 10.1016/s0925-4439(02)00161-8. [DOI] [PubMed] [Google Scholar]
- 138.Li Y, Sheu CC, Ye Y, de Andrade M, Wang L, Chang SC, Aubry MC, Aakre JA, Allen MS, Chen F, Cunningham JM, Deschamps C, Jiang R, Lin J, Marks RS, Pankratz VS, Su L, Sun Z, Tang H, Vasmatzis G, Harris CC, Spitz MR, Jen J, Wang R, Zhang ZF, Christiani DC, Wu X, Yang P. Genetic variants and risk of lung cancer in never smokers: a genome-wide association study. Lancet Oncol. 2010;11:321–30. doi: 10.1016/S1470-2045(10)70042-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Porsch H, Bernert B, Mehic M, Theocharis AD, Heldin CH, Heldin P. Efficient TGFbeta-induced epithelial-mesenchymal transition depends on hyaluronan synthase HAS2. Oncogene. 2012 doi: 10.1038/onc.2012.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Misra S, Toole BP, Ghatak S. Hyaluronan constitutively regulates activation of multiple receptor tyrosine kinases in epithelial and carcinoma cells. J Biol Chem. 2006;281:34936–41. doi: 10.1074/jbc.C600138200. [DOI] [PubMed] [Google Scholar]
- 141.Itano N, Sawai T, Atsumi F, Miyaishi O, Taniguchi S, Kannagi R, Hamaguchi M, Kimata K. Selective expression and functional characteristics of three mammalian hyaluronan synthases in oncogenic malignant transformation. J Biol Chem. 2004;279:18679–87. doi: 10.1074/jbc.M313178200. [DOI] [PubMed] [Google Scholar]
- 142.Itano N, Zhuo L, Kimata K. Impact of the hyaluronan-rich tumor microenvironment on cancer initiation and progression. Cancer Sci. 2008;99:1720–5. doi: 10.1111/j.1349-7006.2008.00885.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Hermanson M, Funa K, Hartman M, Claesson-Welsh L, Heldin CH, Westermark B, Nister M. Platelet-derived growth factor and its receptors in human glioma tissue: expression of messenger RNA and protein suggests the presence of autocrine and paracrine loops. Cancer Res. 1992;52:3213–9. [PubMed] [Google Scholar]
- 144.Ozawa T, Brennan CW, Wang L, Squatrito M, Sasayama T, Nakada M, Huse JT, Pedraza A, Utsuki S, Yasui Y, Tandon A, Fomchenko EI, Oka H, Levine RL, Fujii K, Ladanyi M, Holland EC. PDGFRA gene rearrangements are frequent genetic events in PDGFRA-amplified glioblastomas. Genes Dev. 2010;24:2205–18. doi: 10.1101/gad.1972310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Engler JR, Robinson AE, Smirnov I, Hodgson JG, Berger MS, Gupta N, James CD, Molinaro A, Phillips JJ. Increased microglia/macrophage gene expression in a subset of adult and pediatric astrocytomas. PLoS One. 2012;7:e43339. doi: 10.1371/journal.pone.0043339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Watanabe A, Mabuchi T, Satoh E, Furuya K, Zhang L, Maeda S, Naganuma H. Expression of syndecans, a heparan sulfate proteoglycan, in malignant gliomas: participation of nuclear factor-kappaB in upregulation of syndecan-1 expression. J Neurooncol. 2006;77:25–32. doi: 10.1007/s11060-005-9010-3. [DOI] [PubMed] [Google Scholar]
- 147.Beauvais DM, Burbach BJ, Rapraeger AC. The syndecan-1 ectodomain regulates alphavbeta3 integrin activity in human mammary carcinoma cells. J Cell Biol. 2004;167:171–81. doi: 10.1083/jcb.200404171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Beauvais DM, Ell BJ, McWhorter AR, Rapraeger AC. Syndecan-1 regulates alphavbeta3 and alphavbeta5 integrin activation during angiogenesis and is blocked by synstatin, a novel peptide inhibitor. J Exp Med. 2009;206:691–705. doi: 10.1084/jem.20081278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Kharabi Masouleh B, Ten Dam GB, Wild MK, Seelige R, van der Vlag J, Rops AL, Echtermeyer FG, Vestweber D, van Kuppevelt TH, Kiesel L, Gotte M. Role of the heparan sulfate proteoglycan syndecan-1 (CD138) in delayed-type hypersensitivity. J Immunol. 2009;182:4985–93. doi: 10.4049/jimmunol.0800574. [DOI] [PubMed] [Google Scholar]
- 150.Xu J, Park PW, Kheradmand F, Corry DB. Endogenous attenuation of allergic lung inflammation by syndecan-1. J Immunol. 2005;174:5758–65. doi: 10.4049/jimmunol.174.9.5758. [DOI] [PubMed] [Google Scholar]
- 151.Hayashida K, Parks WC, Park PW. Syndecan-1 shedding facilitates the resolution of neutrophilic inflammation by removing sequestered CXC chemokines. Blood. 2009;114:3033–43. doi: 10.1182/blood-2009-02-204966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Nishiyama A, Lin XH, Giese N, Heldin CH, Stallcup WB. Co-localization of NG2 proteoglycan and PDGF alpha-receptor on O2A progenitor cells in the developing rat brain. J Neurosci Res. 1996;43:299–314. doi: 10.1002/(SICI)1097-4547(19960201)43:3<299::AID-JNR5>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 153.Giamanco KA, Matthews RT. Deconstructing the perineuronal net: cellular contributions and molecular composition of the neuronal extracellular matrix. Neuroscience. 2012;218:367–84. doi: 10.1016/j.neuroscience.2012.05.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Giamanco KA, Morawski M, Matthews RT. Perineuronal net formation and structure in aggrecan knockout mice. Neuroscience. 2010;170:1314–27. doi: 10.1016/j.neuroscience.2010.08.032. [DOI] [PubMed] [Google Scholar]
- 155.Hu B, Kong LL, Matthews RT, Viapiano MS. The proteoglycan brevican binds to fibronectin after proteolytic cleavage and promotes glioma cell motility. J Biol Chem. 2008;283:24848–59. doi: 10.1074/jbc.M801433200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Jaworski DM, Kelly GM, Piepmeier JM, Hockfield S. BEHAB (brain enriched hyaluronan binding) is expressed in surgical samples of glioma and in intracranial grafts of invasive glioma cell lines. Cancer Res. 1996;56:2293–8. [PubMed] [Google Scholar]
- 157.Zhang H, Kelly G, Zerillo C, Jaworski DM, Hockfield S. Expression of a cleaved brain-specific extracellular matrix protein mediates glioma cell invasion In vivo. J Neurosci. 1998;18:2370–6. doi: 10.1523/JNEUROSCI.18-07-02370.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Matthews RT, Gary SC, Zerillo C, Pratta M, Solomon K, Arner EC, Hockfield S. Brain-enriched hyaluronan binding (BEHAB)/brevican cleavage in a glioma cell line is mediated by a disintegrin and metalloproteinase with thrombospondin motifs (ADAMTS) family member. J Biol Chem. 2000;275:22695–703. doi: 10.1074/jbc.M909764199. [DOI] [PubMed] [Google Scholar]
- 159.Viapiano MS, Hockfield S, Matthews RT. BEHAB/brevican requires ADAMTS-mediated proteolytic cleavage to promote glioma invasion. J Neurooncol. 2008;88:261–72. doi: 10.1007/s11060-008-9575-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Lum DH, Tan J, Rosen SD, Werb Z. Gene trap disruption of the mouse heparan sulfate 6-O-endosulfatase gene, Sulf2. Mol Cell Biol. 2007;27:678–88. doi: 10.1128/MCB.01279-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Gill RB, Day A, Barstow A, Zaman G, Chenu C, Dhoot GK. Mammalian Sulf1 RNA alternative splicing and its significance to tumour growth regulation. Tumour Biol. 2012;33:1669–80. doi: 10.1007/s13277-012-0423-2. [DOI] [PubMed] [Google Scholar]
- 162.Giannini C, Sarkaria JN, Saito A, Uhm JH, Galanis E, Carlson BL, Schroeder MA, James CD. Patient tumor EGFR and PDGFRA gene amplifications retained in an invasive intracranial xenograft model of glioblastoma multiforme. Neuro Oncol. 2005;7:164–76. doi: 10.1215/S1152851704000821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Bachoo RM, Maher EA, Ligon KL, Sharpless NE, Chan SS, You MJ, Tang Y, DeFrances J, Stover E, Weissleder R, Rowitch DH, Louis DN, DePinho RA. Epidermal growth factor receptor and Ink4a/Arf: convergent mechanisms governing terminal differentiation and transformation along the neural stem cell to astrocyte axis. Cancer Cell. 2002;1:269–77. doi: 10.1016/s1535-6108(02)00046-6. [DOI] [PubMed] [Google Scholar]
- 164.Dredge K, Hammond E, Handley P, Gonda TJ, Smith MT, Vincent C, Brandt R, Ferro V, Bytheway I. PG545, a dual heparanase and angiogenesis inhibitor, induces potent anti-tumour and anti-metastatic efficacy in preclinical models. Br J Cancer. 2011;104:635–42. doi: 10.1038/bjc.2011.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Hossain MM, Hosono-Fukao T, Tang R, Sugaya N, van Kuppevelt TH, Jenniskens GJ, Kimata K, Rosen SD, Uchimura K. Direct detection of HSulf-1 and HSulf-2 activities on extracellular heparan sulfate and their inhibition by PI-88. Glycobiology. 2010;20:175–86. doi: 10.1093/glycob/cwp159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Johnstone KD, Karoli T, Liu L, Dredge K, Copeman E, Li CP, Davis K, Hammond E, Bytheway I, Kostewicz E, Chiu FC, Shackleford DM, Charman SA, Charman WN, Harenberg J, Gonda TJ, Ferro V. Synthesis and biological evaluation of polysulfated oligosaccharide glycosides as inhibitors of angiogenesis and tumor growth. J Med Chem. 2010;53:1686–99. doi: 10.1021/jm901449m. [DOI] [PubMed] [Google Scholar]
- 167.Joyce JA, Freeman C, Meyer-Morse N, Parish CR, Hanahan D. A functional heparan sulfate mimetic implicates both heparanase and heparan sulfate in tumor angiogenesis and invasion in a mouse model of multistage cancer. Oncogene. 2005;24:4037–51. doi: 10.1038/sj.onc.1208602. [DOI] [PubMed] [Google Scholar]
- 168.Zhou H, Roy S, Cochran E, Zouaoui R, Chu CL, Duffner J, Zhao G, Smith S, Galcheva-Gargova Z, Karlgren J, Dussault N, Kwan RY, Moy E, Barnes M, Long A, Honan C, Qi YW, Shriver Z, Ganguly T, Schultes B, Venkataraman G, Kishimoto TK. M402, a novel heparan sulfate mimetic, targets multiple pathways implicated in tumor progression and metastasis. PLoS One. 2011;6:e21106. doi: 10.1371/journal.pone.0021106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Liu CJ, Lee PH, Lin DY, Wu CC, Jeng LB, Lin PW, Mok KT, Lee WC, Yeh HZ, Ho MC, Yang SS, Lee CC, Yu MC, Hu RH, Peng CY, Lai KL, Chang SS, Chen PJ. Heparanase inhibitor PI-88 as adjuvant therapy for hepatocellular carcinoma after curative resection: a randomized phase II trial for safety and optimal dosage. J Hepatol. 2009;50:958–68. doi: 10.1016/j.jhep.2008.12.023. [DOI] [PubMed] [Google Scholar]
- 170.Yang I, Han SJ, Kaur G, Crane C, Parsa AT. The role of microglia in central nervous system immunity and glioma immunology. Journal of clinical neuroscience : official journal of the Neurosurgical Society of Australasia. 2010;17:6–10. doi: 10.1016/j.jocn.2009.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Thorne RG, Lakkaraju A, Rodriguez-Boulan E, Nicholson C. In vivo diffusion of lactoferrin in brain extracellular space is regulated by interactions with heparan sulfate. Proc Natl Acad Sci U S A. 2008;105:8416–21. doi: 10.1073/pnas.0711345105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Dmitrieva N, Yu L, Viapiano M, Cripe TP, Chiocca EA, Glorioso JC, Kaur B. Chondroitinase ABC I-mediated enhancement of oncolytic virus spread and antitumor efficacy. Clin Cancer Res. 2011;17:1362–72. doi: 10.1158/1078-0432.CCR-10-2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Christianson HC, van Kuppevelt TH, Belting M. ScFv Anti-Heparan Sulfate Antibodies Unexpectedly Activate Endothelial and Cancer Cells through p38 MAPK: Implications for Antibody-Based Targeting of Heparan Sulfate Proteoglycans in Cancer. PLoS One. 2012;7:e49092. doi: 10.1371/journal.pone.0049092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Joensuu H, Anttonen A, Eriksson M, Makitaro R, Alfthan H, Kinnula V, Leppa S. Soluble syndecan-1 and serum basic fibroblast growth factor are new prognostic factors in lung cancer. Cancer Res. 2002;62:5210–7. [PubMed] [Google Scholar]
- 175.Taguchi A, Politi K, Pitteri SJ, Lockwood WW, Faca VM, Kelly-Spratt K, Wong CH, Zhang Q, Chin A, Park KS, Goodman G, Gazdar AF, Sage J, Dinulescu DM, Kucherlapati R, Depinho RA, Kemp CJ, Varmus HE, Hanash SM. Lung cancer signatures in plasma based on proteome profiling of mouse tumor models. Cancer Cell. 2011;20:289–99. doi: 10.1016/j.ccr.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Esko JD, Kimata K, Lindahl U. Proteoglycans and Sulfated Glycosaminoglycans. In: Varki A, Cummings RD, Esko JD, Freeze HH, Stanley P, Bertozzi CR, Hart GW, Etzler ME, editors. Essentials of Glycobiology. Cold Spring Harbor (NY): 2009. [PubMed] [Google Scholar]