Abstract
The Drosophila pair-rule gene, hairy (h), encodes a nuclear basic helix-loop-helix (bHLH) protein that regulates embryonic segmentation and adult bristle patterning. In both cases, the h protein behaves as a transcriptional repressor. In this study, we determined the molecular nature of 12 h alleles. One mutation maps within the HLH domain, consistent with h function requiring homodimerization or heterodimerization with other HLH proteins. A second mutation lies in the basic domain, suggesting that DNA binding is required for h activity. Several mutations show that the h C terminus, in particular the WRPW domain, is also required for h activity, perhaps by interacting with other proteins to mediate transcriptional repression. We show that the h protein in Drosophila virilis closely resembles that in D. melanogaster and includes completely conserved bHLH and WRPW domains.
Full text
PDF



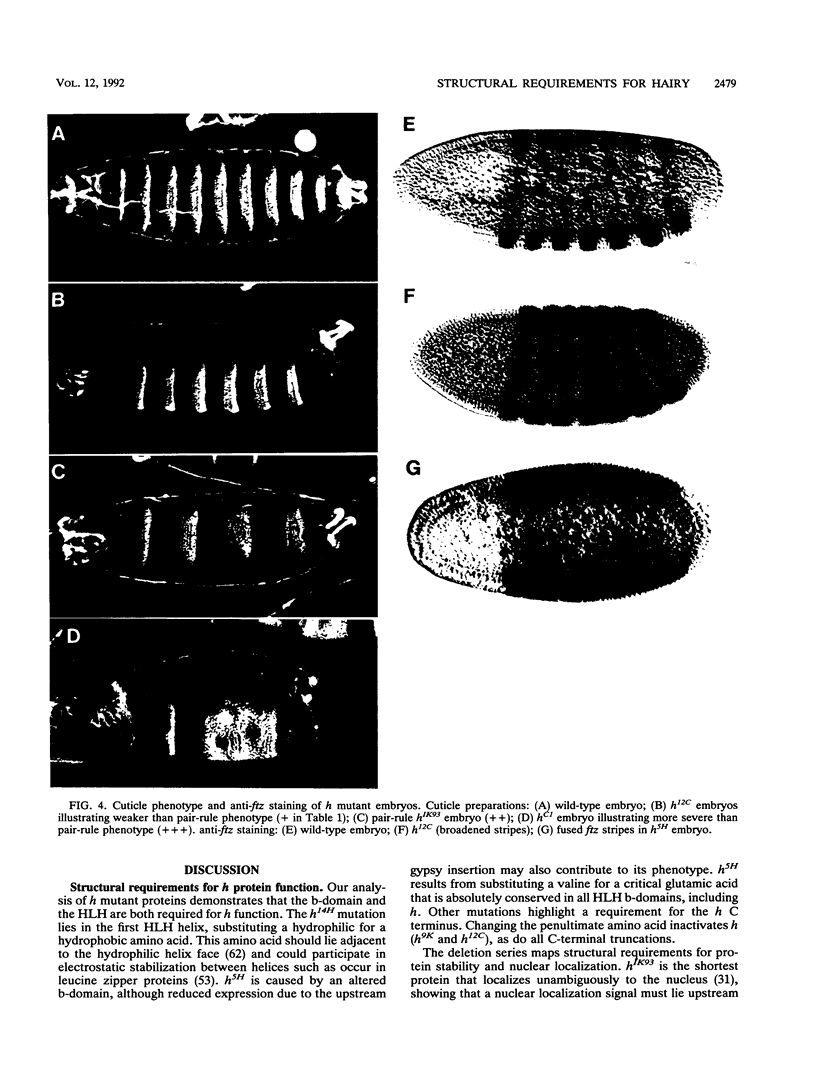




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alonso M. C., Cabrera C. V. The achaete-scute gene complex of Drosophila melanogaster comprises four homologous genes. EMBO J. 1988 Aug;7(8):2585–2591. doi: 10.1002/j.1460-2075.1988.tb03108.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artavanis-Tsakonas S., Simpson P. Choosing a cell fate: a view from the Notch locus. Trends Genet. 1991 Nov-Dec;7(11-12):403–408. doi: 10.1016/0168-9525(91)90264-q. [DOI] [PubMed] [Google Scholar]
- Benezra R., Davis R. L., Lockshon D., Turner D. L., Weintraub H. The protein Id: a negative regulator of helix-loop-helix DNA binding proteins. Cell. 1990 Apr 6;61(1):49–59. doi: 10.1016/0092-8674(90)90214-y. [DOI] [PubMed] [Google Scholar]
- Beverley S. M., Wilson A. C. Molecular evolution in Drosophila and the higher Diptera II. A time scale for fly evolution. J Mol Evol. 1984;21(1):1–13. doi: 10.1007/BF02100622. [DOI] [PubMed] [Google Scholar]
- Blackwell T. K., Weintraub H. Differences and similarities in DNA-binding preferences of MyoD and E2A protein complexes revealed by binding site selection. Science. 1990 Nov 23;250(4984):1104–1110. doi: 10.1126/science.2174572. [DOI] [PubMed] [Google Scholar]
- Blackwood E. M., Eisenman R. N. Max: a helix-loop-helix zipper protein that forms a sequence-specific DNA-binding complex with Myc. Science. 1991 Mar 8;251(4998):1211–1217. doi: 10.1126/science.2006410. [DOI] [PubMed] [Google Scholar]
- Botas J., Moscoso del Prado J., García-Bellido A. Gene-dose titration analysis in the search of trans-regulatory genes in Drosophila. EMBO J. 1982;1(3):307–310. doi: 10.1002/j.1460-2075.1982.tb01165.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan T. J., Chakraborty T., Olson E. N. Mutagenesis of the myogenin basic region identifies an ancient protein motif critical for activation of myogenesis. Proc Natl Acad Sci U S A. 1991 Jul 1;88(13):5675–5679. doi: 10.1073/pnas.88.13.5675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabrera C. V., Alonso M. C. Transcriptional activation by heterodimers of the achaete-scute and daughterless gene products of Drosophila. EMBO J. 1991 Oct;10(10):2965–2973. doi: 10.1002/j.1460-2075.1991.tb07847.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabrera C. V., Martinez-Arias A., Bate M. The expression of three members of the achaete-scute gene complex correlates with neuroblast segregation in Drosophila. Cell. 1987 Jul 31;50(3):425–433. doi: 10.1016/0092-8674(87)90496-x. [DOI] [PubMed] [Google Scholar]
- Carroll S. B., Scott M. P. Zygotically active genes that affect the spatial expression of the fushi tarazu segmentation gene during early Drosophila embryogenesis. Cell. 1986 Apr 11;45(1):113–126. doi: 10.1016/0092-8674(86)90543-x. [DOI] [PubMed] [Google Scholar]
- Caudy M., Grell E. H., Dambly-Chaudière C., Ghysen A., Jan L. Y., Jan Y. N. The maternal sex determination gene daughterless has zygotic activity necessary for the formation of peripheral neurons in Drosophila. Genes Dev. 1988 Jul;2(7):843–852. doi: 10.1101/gad.2.7.843. [DOI] [PubMed] [Google Scholar]
- Caudy M., Vässin H., Brand M., Tuma R., Jan L. Y., Jan Y. N. daughterless, a Drosophila gene essential for both neurogenesis and sex determination, has sequence similarities to myc and the achaete-scute complex. Cell. 1988 Dec 23;55(6):1061–1067. doi: 10.1016/0092-8674(88)90250-4. [DOI] [PubMed] [Google Scholar]
- Cline T. W. A female-specific lethal lesion in an X-linked positive regulator of the Drosophila sex determination gene, Sex-lethal. Genetics. 1986 Jul;113(3):641–663. doi: 10.1093/genetics/113.3.641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline T. W. A sex-specific, temperature-sensitive maternal effect of the daughterless mutation of Drosophila melanogaster. Genetics. 1976 Dec;84(4):723–742. doi: 10.1093/genetics/84.4.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline T. W. Evidence that sisterless-a and sisterless-b are two of several discrete "numerator elements" of the X/A sex determination signal in Drosophila that switch Sxl between two alternative stable expression states. Genetics. 1988 Aug;119(4):829–862. doi: 10.1093/genetics/119.4.829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubas P., de Celis J. F., Campuzano S., Modolell J. Proneural clusters of achaete-scute expression and the generation of sensory organs in the Drosophila imaginal wing disc. Genes Dev. 1991 Jun;5(6):996–1008. doi: 10.1101/gad.5.6.996. [DOI] [PubMed] [Google Scholar]
- Davis R. L., Cheng P. F., Lassar A. B., Weintraub H. The MyoD DNA binding domain contains a recognition code for muscle-specific gene activation. Cell. 1990 Mar 9;60(5):733–746. doi: 10.1016/0092-8674(90)90088-v. [DOI] [PubMed] [Google Scholar]
- Davis R. L., Weintraub H., Lassar A. B. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell. 1987 Dec 24;51(6):987–1000. doi: 10.1016/0092-8674(87)90585-x. [DOI] [PubMed] [Google Scholar]
- Edgar B. A., Odell G. M., Schubiger G. A genetic switch, based on negative regulation, sharpens stripes in Drosophila embryos. Dev Genet. 1989;10(3):124–142. doi: 10.1002/dvg.1020100303. [DOI] [PubMed] [Google Scholar]
- Ellis H. M., Spann D. R., Posakony J. W. extramacrochaetae, a negative regulator of sensory organ development in Drosophila, defines a new class of helix-loop-helix proteins. Cell. 1990 Apr 6;61(1):27–38. doi: 10.1016/0092-8674(90)90212-w. [DOI] [PubMed] [Google Scholar]
- Erickson J. W., Cline T. W. Molecular nature of the Drosophila sex determination signal and its link to neurogenesis. Science. 1991 Mar 1;251(4997):1071–1074. doi: 10.1126/science.1900130. [DOI] [PubMed] [Google Scholar]
- Garrell J., Modolell J. The Drosophila extramacrochaetae locus, an antagonist of proneural genes that, like these genes, encodes a helix-loop-helix protein. Cell. 1990 Apr 6;61(1):39–48. doi: 10.1016/0092-8674(90)90213-x. [DOI] [PubMed] [Google Scholar]
- Ghysen A., Dambly-Chaudiere C. Genesis of the Drosophila peripheral nervous system. Trends Genet. 1989 Aug;5(8):251–255. doi: 10.1016/0168-9525(89)90097-8. [DOI] [PubMed] [Google Scholar]
- Hodgkin J. Sex determination compared in Drosophila and Caenorhabditis. Nature. 1990 Apr 19;344(6268):721–728. doi: 10.1038/344721a0. [DOI] [PubMed] [Google Scholar]
- Holmgren R. Cloning sequences from the hairy gene of Drosophila. EMBO J. 1984 Mar;3(3):569–573. doi: 10.1002/j.1460-2075.1984.tb01849.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper K. L., Parkhurst S. M., Ish-Horowicz D. Spatial control of hairy protein expression during embryogenesis. Development. 1989 Nov;107(3):489–504. doi: 10.1242/dev.107.3.489. [DOI] [PubMed] [Google Scholar]
- Howard K., Ingham P. Regulatory interactions between the segmentation genes fushi tarazu, hairy, and engrailed in the Drosophila blastoderm. Cell. 1986 Mar 28;44(6):949–957. doi: 10.1016/0092-8674(86)90018-8. [DOI] [PubMed] [Google Scholar]
- Howard K. The blastoderm prepattern. Semin Cell Biol. 1990 Jun;1(3):161–172. [PubMed] [Google Scholar]
- Ingham P. W., Pinchin S. M., Howard K. R., Ish-Horowicz D. Genetic Analysis of the Hairy Locus in DROSOPHILA MELANOGASTER. Genetics. 1985 Nov;111(3):463–486. doi: 10.1093/genetics/111.3.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingham P. W. The molecular genetics of embryonic pattern formation in Drosophila. Nature. 1988 Sep 1;335(6185):25–34. doi: 10.1038/335025a0. [DOI] [PubMed] [Google Scholar]
- Ish-Horowicz D., Howard K. R., Pinchin S. M., Ingham P. W. Molecular and genetic analysis of the hairy locus in Drosophila. Cold Spring Harb Symp Quant Biol. 1985;50:135–144. doi: 10.1101/sqb.1985.050.01.019. [DOI] [PubMed] [Google Scholar]
- Ish-Horowicz D., Pinchin S. M. Pattern abnormalities induced by ectopic expression of the Drosophila gene hairy are associated with repression of ftz transcription. Cell. 1987 Nov 6;51(3):405–415. doi: 10.1016/0092-8674(87)90636-2. [DOI] [PubMed] [Google Scholar]
- Kassis J. A., Poole S. J., Wright D. K., O'Farrell P. H. Sequence conservation in the protein coding and intron regions of the engrailed transcription unit. EMBO J. 1986 Dec 20;5(13):3583–3589. doi: 10.1002/j.1460-2075.1986.tb04686.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellerman K. A., Mattson D. M., Duncan I. Mutations affecting the stability of the fushi tarazu protein of Drosophila. Genes Dev. 1990 Nov;4(11):1936–1950. doi: 10.1101/gad.4.11.1936. [DOI] [PubMed] [Google Scholar]
- Klämbt C., Knust E., Tietze K., Campos-Ortega J. A. Closely related transcripts encoded by the neurogenic gene complex enhancer of split of Drosophila melanogaster. EMBO J. 1989 Jan;8(1):203–210. doi: 10.1002/j.1460-2075.1989.tb03365.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knust E., Bremer K. A., Vässin H., Ziemer A., Tepass U., Campos-Ortega J. A. The enhancer of split locus and neurogenesis in Drosophila melanogaster. Dev Biol. 1987 Jul;122(1):262–273. doi: 10.1016/0012-1606(87)90351-4. [DOI] [PubMed] [Google Scholar]
- Kohl N. E., Legouy E., DePinho R. A., Nisen P. D., Smith R. K., Gee C. E., Alt F. W. Human N-myc is closely related in organization and nucleotide sequence to c-myc. Nature. 1986 Jan 2;319(6048):73–77. doi: 10.1038/319073a0. [DOI] [PubMed] [Google Scholar]
- Krause H. M., Klemenz R., Gehring W. J. Expression, modification, and localization of the fushi tarazu protein in Drosophila embryos. Genes Dev. 1988 Aug;2(8):1021–1036. doi: 10.1101/gad.2.8.1021. [DOI] [PubMed] [Google Scholar]
- Lassar A. B., Buskin J. N., Lockshon D., Davis R. L., Apone S., Hauschka S. D., Weintraub H. MyoD is a sequence-specific DNA binding protein requiring a region of myc homology to bind to the muscle creatine kinase enhancer. Cell. 1989 Sep 8;58(5):823–831. doi: 10.1016/0092-8674(89)90935-5. [DOI] [PubMed] [Google Scholar]
- Lewin B. Commitment and activation at pol II promoters: a tail of protein-protein interactions. Cell. 1990 Jun 29;61(7):1161–1164. doi: 10.1016/0092-8674(90)90675-5. [DOI] [PubMed] [Google Scholar]
- McKeon F. D., Kirschner M. W., Caput D. Homologies in both primary and secondary structure between nuclear envelope and intermediate filament proteins. Nature. 1986 Feb 6;319(6053):463–468. doi: 10.1038/319463a0. [DOI] [PubMed] [Google Scholar]
- Murre C., McCaw P. S., Baltimore D. A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and myc proteins. Cell. 1989 Mar 10;56(5):777–783. doi: 10.1016/0092-8674(89)90682-x. [DOI] [PubMed] [Google Scholar]
- Murre C., McCaw P. S., Vaessin H., Caudy M., Jan L. Y., Jan Y. N., Cabrera C. V., Buskin J. N., Hauschka S. D., Lassar A. B. Interactions between heterologous helix-loop-helix proteins generate complexes that bind specifically to a common DNA sequence. Cell. 1989 Aug 11;58(3):537–544. doi: 10.1016/0092-8674(89)90434-0. [DOI] [PubMed] [Google Scholar]
- Nüsslein-Volhard C., Wieschaus E. Mutations affecting segment number and polarity in Drosophila. Nature. 1980 Oct 30;287(5785):795–801. doi: 10.1038/287795a0. [DOI] [PubMed] [Google Scholar]
- O'Shea E. K., Rutkowski R., Stafford W. F., 3rd, Kim P. S. Preferential heterodimer formation by isolated leucine zippers from fos and jun. Science. 1989 Aug 11;245(4918):646–648. doi: 10.1126/science.2503872. [DOI] [PubMed] [Google Scholar]
- Parkhurst S. M., Bopp D., Ish-Horowicz D. X:A ratio, the primary sex-determining signal in Drosophila, is transduced by helix-loop-helix proteins. Cell. 1990 Dec 21;63(6):1179–1191. doi: 10.1016/0092-8674(90)90414-a. [DOI] [PubMed] [Google Scholar]
- Prendergast G. C., Lawe D., Ziff E. B. Association of Myn, the murine homolog of max, with c-Myc stimulates methylation-sensitive DNA binding and ras cotransformation. Cell. 1991 May 3;65(3):395–407. doi: 10.1016/0092-8674(91)90457-a. [DOI] [PubMed] [Google Scholar]
- Richardson J. S., Richardson D. C. Amino acid preferences for specific locations at the ends of alpha helices. Science. 1988 Jun 17;240(4859):1648–1652. doi: 10.1126/science.3381086. [DOI] [PubMed] [Google Scholar]
- Rogers S., Wells R., Rechsteiner M. Amino acid sequences common to rapidly degraded proteins: the PEST hypothesis. Science. 1986 Oct 17;234(4774):364–368. doi: 10.1126/science.2876518. [DOI] [PubMed] [Google Scholar]
- Romani S., Campuzano S., Macagno E. R., Modolell J. Expression of achaete and scute genes in Drosophila imaginal discs and their function in sensory organ development. Genes Dev. 1989 Jul;3(7):997–1007. doi: 10.1101/gad.3.7.997. [DOI] [PubMed] [Google Scholar]
- Romani S., Campuzano S., Modolell J. The achaete-scute complex is expressed in neurogenic regions of Drosophila embryos. EMBO J. 1987 Jul;6(7):2085–2092. doi: 10.1002/j.1460-2075.1987.tb02474.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushlow C. A., Hogan A., Pinchin S. M., Howe K. M., Lardelli M., Ish-Horowicz D. The Drosophila hairy protein acts in both segmentation and bristle patterning and shows homology to N-myc. EMBO J. 1989 Oct;8(10):3095–3103. doi: 10.1002/j.1460-2075.1989.tb08461.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skeath J. B., Carroll S. B. Regulation of achaete-scute gene expression and sensory organ pattern formation in the Drosophila wing. Genes Dev. 1991 Jun;5(6):984–995. doi: 10.1101/gad.5.6.984. [DOI] [PubMed] [Google Scholar]
- Sun X. H., Baltimore D. An inhibitory domain of E12 transcription factor prevents DNA binding in E12 homodimers but not in E12 heterodimers. Cell. 1991 Jan 25;64(2):459–470. doi: 10.1016/0092-8674(91)90653-g. [DOI] [PubMed] [Google Scholar]
- Tautz D., Pfeifle C. A non-radioactive in situ hybridization method for the localization of specific RNAs in Drosophila embryos reveals translational control of the segmentation gene hunchback. Chromosoma. 1989 Aug;98(2):81–85. doi: 10.1007/BF00291041. [DOI] [PubMed] [Google Scholar]
- Torres M., Sánchez L. The scute (T4) gene acts as a numerator element of the X:a signal that determines the state of activity of sex-lethal in Drosophila. EMBO J. 1989 Oct;8(10):3079–3086. doi: 10.1002/j.1460-2075.1989.tb08459.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaessin H., Caudy M., Bier E., Jan L. Y., Jan Y. N. Role of helix-loop-helix proteins in Drosophila neurogenesis. Cold Spring Harb Symp Quant Biol. 1990;55:239–245. doi: 10.1101/sqb.1990.055.01.026. [DOI] [PubMed] [Google Scholar]
- Van Doren M., Ellis H. M., Posakony J. W. The Drosophila extramacrochaetae protein antagonizes sequence-specific DNA binding by daughterless/achaete-scute protein complexes. Development. 1991 Sep;113(1):245–255. doi: 10.1242/dev.113.1.245. [DOI] [PubMed] [Google Scholar]
- Villares R., Cabrera C. V. The achaete-scute gene complex of D. melanogaster: conserved domains in a subset of genes required for neurogenesis and their homology to myc. Cell. 1987 Jul 31;50(3):415–424. doi: 10.1016/0092-8674(87)90495-8. [DOI] [PubMed] [Google Scholar]
- Vinson C. R., Sigler P. B., McKnight S. L. Scissors-grip model for DNA recognition by a family of leucine zipper proteins. Science. 1989 Nov 17;246(4932):911–916. doi: 10.1126/science.2683088. [DOI] [PubMed] [Google Scholar]
- Voronova A., Baltimore D. Mutations that disrupt DNA binding and dimer formation in the E47 helix-loop-helix protein map to distinct domains. Proc Natl Acad Sci U S A. 1990 Jun;87(12):4722–4726. doi: 10.1073/pnas.87.12.4722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub H., Davis R., Tapscott S., Thayer M., Krause M., Benezra R., Blackwell T. K., Turner D., Rupp R., Hollenberg S. The myoD gene family: nodal point during specification of the muscle cell lineage. Science. 1991 Feb 15;251(4995):761–766. doi: 10.1126/science.1846704. [DOI] [PubMed] [Google Scholar]
- Weintraub H., Dwarki V. J., Verma I., Davis R., Hollenberg S., Snider L., Lassar A., Tapscott S. J. Muscle-specific transcriptional activation by MyoD. Genes Dev. 1991 Aug;5(8):1377–1386. doi: 10.1101/gad.5.8.1377. [DOI] [PubMed] [Google Scholar]
- Wharton K. A., Yedvobnick B., Finnerty V. G., Artavanis-Tsakonas S. opa: a novel family of transcribed repeats shared by the Notch locus and other developmentally regulated loci in D. melanogaster. Cell. 1985 Jan;40(1):55–62. doi: 10.1016/0092-8674(85)90308-3. [DOI] [PubMed] [Google Scholar]