Abstract
Toxin-antitoxin (TA) modules, composed of a toxic protein and a counteracting antitoxin, play important roles in bacterial physiology. We examined the experimental insertion of 1.5 million genes from 388 microbial genomes into an Escherichia coli host using over 8.5 million random clones. This revealed hundreds of genes (toxins) that could only be cloned when the neighboring gene (antitoxin) was present on the same clone. Clustering of these genes revealed novel TA families widespread in bacterial genomes, some of which deviate from the classical characteristics previously described for such modules. Introduction of these genes into E. coli validated that the toxin toxicity is mitigated by the antitoxin. Infection experiments with T7 phage showed that two of the new modules can provide resistance against phage. Moreover, our experiments revealed an 'anti-defense' protein in phage T7 that neutralizes phage resistance. Our results expose active fronts in the arms race between bacteria and phage.
Introduction
In many ecosystems phages are tenfold more abundant than bacterial cells, posing significant predation pressure on bacterial populations (Bergh et al., 1989; Chibani-Chennoufi et al., 2004). To survive in the face of perpetual phage attacks, bacteria have developed a variety of anti-phage defense systems (Labrie et al., 2010; Stern and Sorek, 2011). These systems include restriction enzymes that recognize and cleave foreign DNA (King and Murray, 1994), abortive infection (Abi) mechanisms that lead the bacterial cell, upon phage invasion, to commit "suicide", thus protecting the colony against phage spread (Chopin et al., 2005); and the recently identified adaptive defense system called CRISPR/Cas, which uses small RNAs to target invading phage DNA (Deveau et al., 2010; Horvath and Barrangou, 2010; Sorek et al., 2008; van der Oost et al., 2009). Due to the rapid evolution and elaborated biological novelty associated with the bacteria-phage arms race, it is estimated that many additional, yet uncharacterized anti-phage defense systems are encoded by bacteria and archaea (Makarova et al., 2011; Stern and Sorek, 2011). As part of this continuous arms race, successful phages had also developed numerous counter-resistance mechanisms to overcome bacterial defense (Labrie et al., 2010; Stern and Sorek, 2011).
The growing availability of genomic sequences has elucidated the vast dispersion of toxin-antitoxin (TA) systems in prokaryotic genomes (Shao et al., 2011). These modules, composed of a toxic gene and a neutralizing gene, were first suggested to function as plasmid ‘addiction molecules’ (Van Melderen and Saavedra De Bast, 2009; Wozniak and Waldor, 2009), but their prevalent existence on chromosomes (Aizenman et al., 1996; Makarova et al., 2009; Shao et al., 2011) has led to the understanding that this is unlikely their major role. Accumulating evidence suggest that TA modules play pivotal roles in prokaryotic cellular biology including programmed cell death (Hazan et al., 2004), stress response (Christensen et al., 2001), generation of persister cells (Schumacher et al., 2009), biofilm formation (Kim et al., 2009) and phage defense via abortive infection (Fineran et al., 2009; Hazan and Engelberg-Kulka, 2004; Koga et al., 2011; Pecota and Wood, 1996).
The most prevalent kind of TA systems is type II systems, where both toxin and antitoxin are proteins (as opposed to types I and III where the antitoxin is a non-coding RNA (Fineran et al., 2009; Fozo et al., 2010)). The two genes, which reside on the same operon, code for small proteins and inhibition of the toxin is carried out through protein-protein interaction. As a rule, the toxin is a stable protein and the antitoxin is unstable and is degraded rapidly by one of the housekeeping bacterial proteases, usually Lon or ClpP (Aizenman et al., 1996; Cherny and Gazit, 2004; Christensen et al., 2004; Christensen et al., 2001; Christensen et al., 2003; Lehnherr and Yarmolinsky, 1995; Roberts et al., 1994; Van Melderen et al., 1996). As a result, continuous production of the antitoxin is required to prevent the toxin’s deleterious effects (Van Melderen and Saavedra De Bast, 2009). Most toxins target the translation process by cleaving cellular mRNA (Amitai et al., 2009; Daines et al., 2007; Hurley and Woychik, 2009; Jorgensen et al., 2009; Koga et al., 2011; Neubauer et al., 2009). Other toxin types exert their toxicity by phosphorylating the elongation factor EF-Tu (Schumacher et al., 2009), associating with the ribosome (Liu et al., 2008), inhibiting DNA replication (Bernard and Couturier, 1992; Jiang et al., 2002) or targeting cytoskeletal proteins (Masuda et al., 2012; Tan et al., 2010). Although several major families of type II TA systems have been described to date (Leplae et al., 2011; Makarova et al., 2009; Masuda et al., 2012; Shao et al., 2011), the extent and roles of such systems in bacterial genomes is far from fully understood.
In numerous cases where TA systems were studied experimentally, cloning of the toxin was nearly impossible in the absence of the cognate antitoxin (Fico and Mahillon, 2006; Goulard et al., 2010; Zhang et al., 2006). Based on this concept we reasoned that data derived from Sanger-based whole genome shotgun sequencing can experimentally and systematically reveal active TA pairs. In such a genome sequencing process, randomly fragmented DNA pieces of the genome are serially cloned and propagated within E. coli prior to sequencing. The ends of the cloned fragments are then sequenced, and overlapping sequences are used for genome assembly (Fig 1A). We have previously shown that analysis of clone distribution patterns can reveal genes toxic to bacteria, which are uncloneable and cause gaps (Kimelman et al., 2012; Sorek et al., 2007). However, the toxin in a TA pair is not expected to cause a gap, since the adjacent antitoxin will sometimes be found on the same clone, neutralizing the toxic effect. Nevertheless, random fragments that contain the toxin but not the antitoxin will cause cell death and will be absent from the set of clones covering the genome (Fig 1B).
Figure 1. Data derived from whole-genome shotgun sequencing exposes toxin-antitoxin pairs.
(A) The "Sanger" based process of DNA sequencing involves random genome fragmentation and transformation of DNA fragments into E. coli. (B) In a DNA locus spanning a toxin/antitoxin (TA) gene pair, random fragmentation leaving the toxin detached from its cognate antitoxin leads to E. coli growth arrest, whereas a fragment containing both genes, or the antitoxin alone, will be propagated and sequenced. (C) A known family of toxin/antitoxin gene pairs, of the VapBC type, was found in 11 of the analyzed genomes. In 7/11 cases the gene pair follows the "TA cloning pattern", significantly higher than the number expected by chance (p=4×10−3). (D) The vapBC locus in Haemophilus somnus 129T (accession NC_008309, locus tags HS_1769-HS_1770). Shown are clones (brown) mapped to the reference genome at that locus. Clones covering the antitoxin but not the toxin are numbered. Only five of the 22 clones covering both toxin and antitoxin are shown.
Here we took advantage of this typical biased cloning pattern to systematically detect toxin-antitoxin gene pairs within hundreds of microbial genomes. Our analyses, while retrieving many known TA systems, have also exposed several novel families of TA modules widespread in numerous bacterial species. These systems were subsequently experimentally validated as TA systems. Infection experiments with wild-type and multiple T7 phage deletion mutants further showed that two of these new TA pairs provide resistance against T7, and also revealed a general anti-TA mechanism encoded by the phage.
Results
Systematic discovery of TA families based on large-scale cloning experiments
We analyzed 360 bacterial and 28 archaeal genomes that were sequenced using the clone-based Sanger approach, and for which the raw sequencing data was accessible and mapped to the assembled genome (Kimelman et al., 2012). For the sequencing of each genome, an average of 22,313 different randomly fragmented clones (typically sized between 3kb–8kb, thus typically spanning ~3 to ~8 genes) were inserted into E. coli. Cumulatively, the analyzed genomes span over 1.5 million genes that were sequenced using over 8.5 million clones. We recorded for each gene, and for each pair of consecutive genes, the number of cloned DNA fragments that fully contain it.
To detect families of gene pairs in which one of the genes (putative toxin) is absent from clones unless the adjacent gene (putative antitoxin) is also present, we first searched for homologous gene pairs that repeatedly appear adjacent to each other in multiple genomes (at least 7 appearances) and clustered them into families of pairs (Methods). To avoid the analysis of housekeeping genes that appear in conserved operons (e.g., ribosomal protein genes), we focused on families showing high tendency to undergo horizontal gene transfer (Methods). Each pair of genes (X and Y) in each family was considered as following the "TA cloning pattern" if the number of clones covering gene X (toxin) but not Y (antitoxin) was 0, the number of clones covering gene Y but not X was >0 and the number of clones covering both X and Y was >0 (Fig 1B–C).
Not all toxins are expected to manifest their toxicity when cloned in E. coli, because their expression depends on the ability of the E. coli host to recognize their native promoters and translate them using available tRNA pool (Sorek et al., 2007). Therefore, for a given toxin-antitoxin family of gene pairs, we expect a significant fraction of pairs, but not necessarily all pairs, to follow the TA cloning pattern (Fig 1C). To assign a statistical significance for a given family as possibly coding for a bona fide TA family, we performed, for each pair in each family, 1000 random simulations, where the clones used for sequencing of the relevant genome were randomly shuffled on the genome (Methods; Fig S1). The results were then used to assign an empirical p-value per family, revealing families in which the fraction of pairs that follow the TA cloning pattern is significantly above the fraction expected by chance (p <= 0.05). This yielded 188 candidate families (Table S1, Fig 2).
Figure 2. Workflow for systematic discovery of families of toxin/antitoxin associated with anti-phage defense.
Families are divided to "known", representing families that were previously experimentally established as TA systems; "not previously validated", representing families for which there was so far no experimental support of being TA systems; and "complex genomic environment", representing families of genes pairs embedded in a larger operon. Cumulative number of gene pairs belonging to each subset of families is indicated.
The identified families may include genes that follow the TA cloning pattern as a by-product of their functions, and not for reasons associated with classical TA systems. For example, a metabolic enzyme whose expression results in accumulation of toxic intermediates might be neutralized by an accompanying transcriptional repressor. This may be the case for the argininosuccinate synthase and the ArgR repressor of the arginine regulon that obey the TA cloning patterns in 4 out of 12 homologous pairs (Table S1). Additional aspects of the clones distribution may also lead to false positive predictions (see Discussion). To identify TA systems more likely to play phage defense-related roles, we focused on those families that had high tendency to appear within bacterial "defense islands". It was recently shown that bacterial anti-phage immune systems such as restriction enzymes, CRISPR and Abi genes aggregate in such defense island loci in bacterial genomes (Makarova et al., 2011). We therefore selected those families in which the genomic neighborhood was enriched for defense genes (Methods). This analysis resulted in a set of 24 putative families of TA gene pairs, overall containing 400 pairs from 176 genomes (Fig 2; Tables 1–2; Table S2).
Table 1.
Families of previously known toxin-antitoxin systems retrieved by the TA discovery algorithm
| # pairs in family |
Antitoxin superfamily |
Toxin superfamily |
COGs associated with antitoxin |
COGs associated with toxin |
P value for TA cloning pattern |
|
|---|---|---|---|---|---|---|
| 1 | 16 | RelB | RelE | COG3905 | COG3668 | 5.0×10−3 |
| 2 | 25 | RelB | RelE | COG3077 | COG3041 | 3.0×10−3 |
| 3 | 13 | RelB | RelE | n/a | n/a | 1.1×10−2 |
| 4 | 33 | Xre | RelE | COG3620 | COG4679 | 2.5×10−2 |
| 5 | 28 | Xre | RelE | n/a | COG2944 | 2.5×10−2 |
| 6 | 32 | Xre | RelE | COG5499 | COG4680 | 7.0×10−3 |
| 7 | 27 | Phd | RelE | COG2161 | COG2026 | 3.4×10−2 |
| 8 | 9 | HicB | HicA | COG1598 | COG1724 | 9.0×10−3 |
| 9 | 11 | HicB | HicA | n/a | COG1724 | 3.4×10−2 |
| 10 | 12 | VapB | VapC | COG4456 | COG1487 | 4.0×10−3 |
| 11 | 10 | HigA | HigB | COG3093 | COG3549 | 1.0×10−3 |
| 12 | 22 | MosA | MosT | n/a | COG2253 | 0 |
Table 2.
Families of toxin-antitoxin systems retrieved by the TA discovery algorithm that were not validated previously.
| # pairs in fam. |
Antitoxin | Toxin | Antitoxin annotation |
Toxin annotation | Domains associated with antitoxin |
Domains associated with toxin |
P value for TA cloning pattern |
Exp a |
|---|---|---|---|---|---|---|---|---|
| 11 | psyrA | psyrT | Nucleotide-binding protein | RecQ-family DNA helicase | COG0758 | COG0514 | 0 | Yes |
| 10 | sanaA | sanaT | Hypothetical protein | Hypothetical protein | n/a | DUF1814 | 0 | Yes |
| 10 | pmenA | pmenT | ADP-ribose binding domain protein | Hypothetical protein | COG2110 | n/a | 0 | Yes |
| 20 | rlegA | rlegT | Predicted transcriptional regulator | Hypothetical protein | COG5340 | DUF1814 | 0 | Yes |
| 7 | sdenA | sdenT | Hypothetical protein | Hypothetical protein | n/a | DUF1814 | 3×10−3 | Yes |
| 44 | hhalA | hhalT | Nucleotidyl-transferase family protein (MNT domain) | Hypothetical protein (HEPN domain) | COG1669 | COG2361 | 2×10−3 | Yes |
| 7 | DNA polymerase, beta-like region (MNT domain) | HEPN domain-containing protein | n/a | COG1895 | 4×10−3 | |||
| 14 | Hypothetical protein (RHH domain) | Hypothetical protein | COG5304 | COG2929 | 2×10−3 | |||
| 8 | Hypothetical protein (associated with cas/cmr genes) | RAMP domain protein (Cmr6-like) | n/a | DUF324 | 2.9×10−2 | * | ||
| 15 | Uncharacterized membrane protein | Membrane protein, TraG-like N-terminal domain | n/a | PF07916 | 2×10−3 | * | ||
| 9 | Hypothetical protein | Adenine specific DNA methylase | n/a | COG2189 | 0 | * | ||
| 7 | T/G mismatch-specific endonuclease | Type II restriction endonuclease | COG3727 | PF09019 | 0 | * |
Validated experimentally in this study
TA system that is part of a larger operon putatively involved in bacterial defense (Fig S2).
Of the 24 identified families, 12 families (50%) were already previously experimentally described as TA systems, providing strong validation to our cloning-based approach for TA discovery (Table 1). Although a diverse set of known TA families is represented in the set retrieved by our algorithm, due to limitations of our approach not all known families were represented. For example, in the family consisting of HipAB gene pairs, 6 out of 19 pairs were found to conform with the TA cloning pattern, but since the toxin is relatively large and the antitoxin is a short gene, such a pattern has high probability to occur by chance in the random simulations, and hence this family did not pass our statistical threshold (p=0.43; see Discussion).
Experimental validation of novel TA families
Our analysis retrieved 8 putative novel families of binary TA systems for which no study has so far showed experimental support (although some of these systems have previously been predicted bioinformatically to function as TA systems, see below) (Table 2). Six of these families were selected for further experimental characterization (Table 3). A representative pair was selected from each family and co-transformed into E. coli BL21(DE3) on a compatible two-vector system, so that the putative toxin was under the control of an IPTG-induced promoter, and the antitoxin was under the control of an arabinose-induced promoter (Fig 3A). E. coli bacteria carrying these two plasmids were plated on agar plates containing IPTG, arabinose, or both IPTG and arabinose. In all six tested pairs, induction of toxin expression inhibited bacterial growth, while co-induction of the toxin and antitoxin resulted in bacterial survival (Fig 3A). Experiments in batch cultures, where bacteria were grown with or without toxin/antitoxin induction, confirmed the plate-based experiments (Fig 3B). These results validate our genome-wide approach for discovery of novel TA systems.
Table 3.
Gene pairs selected for experimental verification in a dual-plasmid arabinose/IPTG expression induction system
| Family | Antitoxin locus tag |
A size (aa) |
Toxin locus tag |
T size (aa) |
Organism | # clones that cover antitoxin |
# clones that cover toxin |
# clones that cover both |
Reversible toxicity? |
|---|---|---|---|---|---|---|---|---|---|
| sdenTA | Sden_0299 | 174 | Sden_0300 | 295 | Shewanella denitrificans OS217 | 8 | 0 | 31 | Yes |
| psyrTA | Psyr_3805 | 455 | Psyr_3804 | 698 | Pseudomonas syringae pv. syringae B728a | 13 | 0 | 1 | Partial |
| sanaTA | Shewana3_4160 | 136 | Shewana3_4161 | 313 | Shewanella sp. ANA-3 | 5 | 0 | 29 | Yes |
| pmenTA | Pmen_0566 | 360 | Pmen_0565 | 217 | Pseudomonas mendocina ymp | 3 | 0 | 10 | Partial |
| rlegTA | Rleg_6340 | 205 | Rleg_6339 | 289 | Rhizobium leguminosarum bv. trifolii WSM1325 | 11 | 0 | 31 | No |
| hhalTA | Hhal_0686 | 96 | Hhal_0685 | 119 | Halorhodospira halophila SL1 | 2 | 0 | 23 | Yes |
Figure 3. Properties of novel, experimentally verified TA systems.
(A) growth of bacteria when only antitoxin is induced (left), only toxin is induced (middle) and both are induced together (right). Toxin and antitoxin were cloned on pRSF (IPTG inducible promoter) and pBAD (arabinose inducible), respectively, in E. coli BL21(DE3)pLysS. C, control bacteria with empty plasmids; 1-pmen system; 2-sana system; 3-psyr system; 4-rleg system; 5-sden system; 6-hhal system. (B) Kinetics of E. coli BL21(DE3)pLysS growth when toxin and antitoxin are co-expressed simultaneously (purple), alone (red and blue for toxin and antitoxin, respectively) or when antitoxin is induced 2.5 hr following toxin induction (green). Kinetic measurements were performed on biological triplicates in technical duplicates. Error bars represent standard deviation. (C) Viability assays for cells following exposure to toxin. Transcription of toxin was induced by 100µM IPTG. At increasing time points following toxin induction (30, 60, 120, 180, 240 and 300 mins) cells were plated on LB-plates containing 0.3% arabinose and no IPTG, to activate antitoxin expression. Colony forming units (CFUs) were determined by colony counting.
We named the 6 novel validated families based on the species from which the validated system was taken: pmenTA (P. mendocina); sanaTA (S. sp. ana-3); rlegTA (R. leguminosarum); psyrTA (P. syringae); sdenTA (S. denitrificans), and hhalTA (H. halophila)(Table 2). We further attempted to test whether the toxins in these new systems have a bacteriocidal (cell-killing) or bacteriostatic (growth-inhibiting) effect. For this, we used IPTG to induce toxin expression for different time intervals (ranging from 30 to 300 mins), and then plated the cells on agar plates containing arabinose to activate antitoxin expression (Fig 3C). For the rlegTA system, colony forming units dropped by 5 orders of magnitude following 120 minutes of toxin induction, implying that the rleg toxin has a bactericidal effect on the cells. These results were also supported by the kinetic assays, where induction of antitoxin expression 2.5 hours after toxin induction did not result in cell regrowth for rlegTA (Fig 3B). A milder effect was observed for the remaining TA systems, with sdenTA and hhalTA showing almost no reduction in colony forming units following toxin induction, suggestive of a bacteriostatic effect for these systems (Fig 3C). We note, however, that there is a general debate whether TA systems are bacteriostatic or bacteriocidal, and factors such as inducer concentrations of toxin and antitoxin and duration of toxin induction prior to antitoxin induction (‘point of no return’) may affect our interpretations.
Characteristics of detected TA families
Most type II TA modules described to date share several typical characteristics: the antitoxin appears upstream of the toxin; both the toxin and antitoxin are small proteins (typically ~100aa); and the antitoxin contains a DNA binding domain (Makarova et al., 2009). Since our approach does not rely on such attributes for TA modules discovery it has the potential to expand the premises of TA modules properties. Indeed, some of the new families we experimentally validated deviate significantly from the previously described characteristics. For example, the sizes of many new toxins and antitoxins are significantly larger than 100aa, with a maximum of 698aa in the toxin of the psyrTA system; in two families the toxin is located upstream of the antitoxin (Fig 4A); and several antitoxins do not contain a known DNA binding domain (although we cannot rule out the possibility that some antitoxins code for yet uncharacterized such domains)(Fig 4A).
Figure 4. Properties of families experimentally validated in this study.
(A) Operon and domain organization of the validated families. Representative pair of each family is shown. For each pair, red and blue genes denote toxin and antitoxin, respectively. (B) Distribution of TA families among different bacterial phyla and (C) human associated bacteria. Number of instances of each system within a phylum/bacterial species is indicated, with darker colors indicating higher number of instances (the phylogenetic distribution of the hhalTA family does not appear here as it was described previously by Makarova et al (2009)).
Although in most of the known TA systems the toxin is a ribonuclease, diverse domains within the new toxins we detected, including DNA helicase, phosphoribosyl-transferase and nucleotidyl-transferase suggest novel mechanisms of toxicity (Fig 4A). Similarly, the presence of an ADP-ribose-binding and nucletotide-binding domains in some of the antitoxins suggest that these antitoxins perform a more complex function than simply masking the activity of the toxin by protein-protein interactions (Fig 4A; Table 2).
Finally, in four additional families that we detected, it seems that the putative TA system is part of a larger operon that is co-horizontally transferred between genomes in the context of defense islands, suggesting their involvement in more complex defense mechanisms (Table 2; Fig S2). As such, these are probably not bona fide TA systems, but might represent false positive predictions of our approach (see Discussion).
Previous analyses have shown that toxin-antitoxin systems can be modular, such that members of one toxin family may be associated with several different types of antitoxin, and vice versa (Leplae et al., 2011; Makarova et al., 2009). Indeed, the toxins of three of our new families (rlegTA, sdenTA and sanaTA) carry the same domain, DUF1814, with a different antitoxin associated with this domain in each of the three families (Table 2; Fig 4A). One of these families, rlegTA, where the DUF1814 toxin is accompanied by COG5340 as an antitoxin, was previously shown to be enriched in defense islands in bacterial genomes and was suggested as a new TA system based on its two-gene nature (Makarova et al., 2011). Although the COG5340 domain was not identified in the sden and sana antitoxins, HHpred comparisons suggest that the antitoxins of the three families share distant homology, and hence may belong to a single superfamily. The DUF1814 domain was recently classified as a nucleotidyl-transferase domain based on structural information, but its specific substrates are yet unknown (Kuchta et al., 2009). Interestingly, the DUF1814 domain was also documented in AbiG, a two-gene system involved in abortive infection in Lactococcus lactis via an unknown mechanism (Makarova et al., 2011; O'Connor et al., 1996), although there is no direct homology between the AbiG system and any of the genes in the new TA systems we detected. Therefore, our results point to DUF1814 domain-containing proteins as a widespread superfamily of toxins that might be involved in anti-phage defense (see below).
Overall, members of the novel systems we detected appeared in 21% of the genomes analyzed, and in the vast majority of cases (93%) they appeared on chromosomal DNA rather than on plasmids. These results suggest that such systems have important roles in bacterial physiology/defense rather than functioning as plasmid addiction molecules. The distribution of most families is spread across multiple bacterial phyla including several important human pathogens (Table S3; Fig 4B–C). For example, the psyrTA, sanaTA, and pmenTA systems are abundant in enteropathogenic and uropathogenic E. coli strains; the pmenTA system also exists in many Mycobacterium tuberculosis isolates; psyrTA exists in Shigella and Pseudomonas aeruginosa strains; and the rlegTA and sanaTA systems exist in several pathogenic Legionella species. In addition, many resident bacteria of the human gut carry one or more of these TA modules, including bacteria belonging to the Bifidobacterium and Prevotella genera (Table S3, Fig 4C). This underscores the novel toxin-antitoxin families as potentially contributing to persistence, phage defense and stress responses of clinically important bacteria.
Novel TA systems provide phage resistance
We next set out to explore whether any of the TA systems we detected can provide defense against phage. For this, efficiency-of-plating (EOP) assays of T7 phage on E. coli hosts were performed (Fig. 5A). Since these new TA systems are widespread in E. coli strains (but are not found in the lab strains E. coli K-12 and E. coli BL21), we hypothesized that a successful coliphage, such as T7, might hold anti-defense mechanisms that mitigate the defense conferred by the TA systems. We therefore tested, in addition to the wild-type (WT) T7 phage, 12 additional T7 mutants lacking genes that are not-essential for infection of E. coli K-12 (Table S4). Each of these T7 mutants was used to infect E. coli K-12 expressing the verified new TA systems, as well as control clones expressing only the antitoxin of each system (Fig 5A). One of the tested systems, sanaTA, was found to provide E. coli with resistance against T7Δ4.5 and T7Δ4.3Δ4.5Δ4.7, reducing sensitivity to these phage strains by about 3 orders of magnitude (Fig 5B). A second system, rlegTA, resulted in opaque plaques with plaque diameters reduced almost fourfold for the WT T7 strain (diameters of 0.47mm ± 0.06 for E. coli expressing both the toxin and the antitoxin, as compared to 1.77mm ± 0.03 for bacteria expressing the antitoxin only).
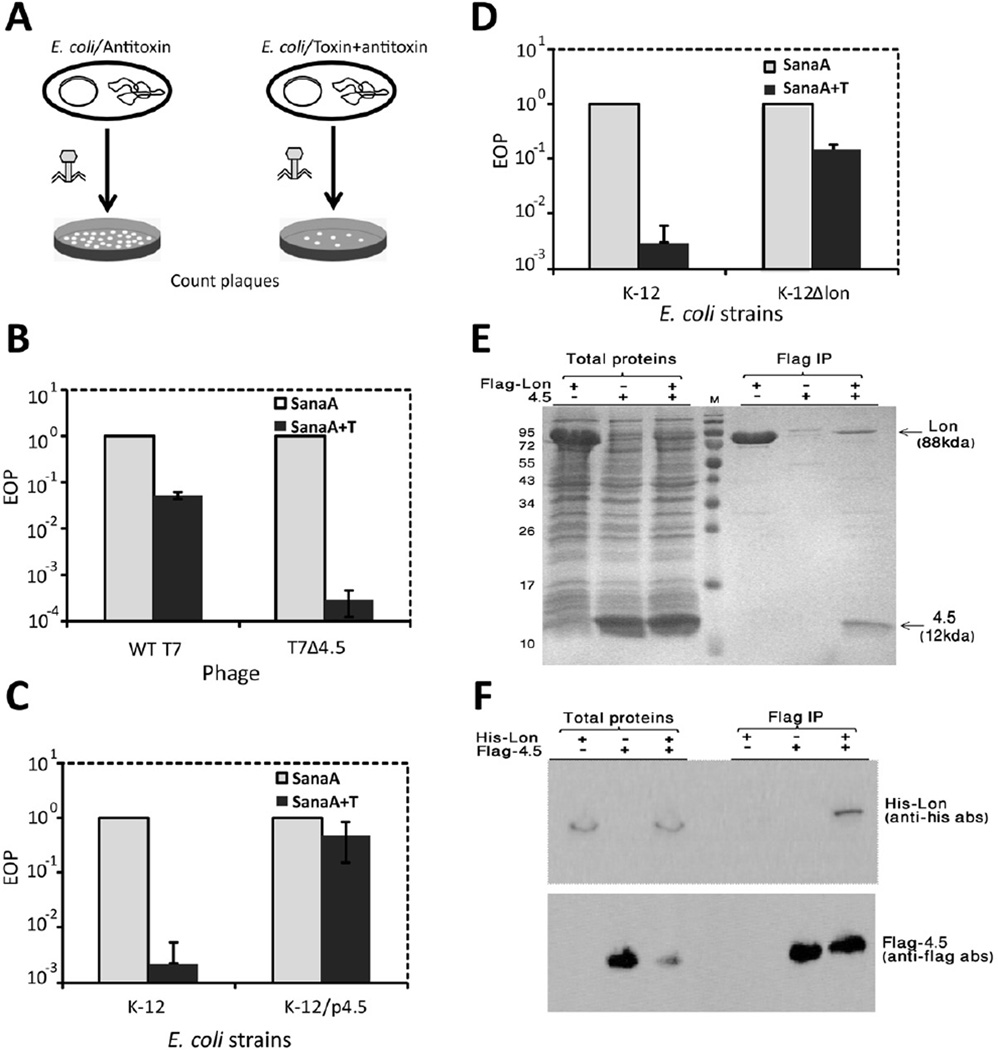
Figure 5. T7 Gp4.5 antagonizes abortive infection and interacts with Lon protease.
(A) Illustration of the plaque forming unit (PFU) assays on E. coli harboring toxin/antitoxin systems. Efficiency of plating (EOP) was calculated by dividing the number of PFUs obtained for a bacterial lawn expressing the toxin + antitoxin by the number of PFUs obtained on a lawn expressing the antitoxin alone. Bars in panels B, C and E represent average ± SD of three independent EOP experiments.
(B) EOP experiments with E. coli harboring the sanaTA system when infected by WT T7 (left) or by T7Δ4.5 (right). Error bars represent standard deviation between replicates.
(C) EOP experiments with WT E. coli (left) and E. coli expressing Gp4.5 (right), when infected by T7Δ4.3Δ4.5Δ4.7. Error bars represent standard deviation between replicates.
(D) EOP experiments with WT E. coli (left) and E. coli lacking lon (right), when infected by T7Δ4.3Δ4.5Δ4.7. Error bars represent standard deviation between replicates.
(E) Co-immunoprecipitation (co-IP) of Lon and Gp4.5. The E. coli Lon protease was Flag-tagged at the N-terminus and expressed within E. coli BL21(DE3) with or without co-expression of gene 4.5. Samples were analyzed by 15% SDS-PAGE. Three left lanes, total soluble proteins; three lanes on the right, following immunoprecipitation with anti-Flag antibody. Co-IP of Lon and Gp4.5 was validated by mass spectrometry analysis. Numbers on the left denote protein marker sizes in kDa.
(F) Western blot analysis on reciprocal co-IP with flag-tagged Gp4.5 and his-tagged Lon. Immunoprecipitation was done with anti-Flag antibody.
Since the sanaTA system provides resistance to the T7Δ4.5 mutant phage but not to the WT T7 phage, we hypothesized that the 4.5 gene codes for an anti-defense mechanism that overcomes the abortive infection imposed by the TA system. Indeed, complementation assay in E. coli K-12 expressing the 4.5 gene from a plasmid verified this gene as encoding the anti-Abi mechanism (Fig 5C). The 4.5 gene codes for a short protein (89aa) with no functional annotation.
We next asked whether the defense provided by the sanaTA system against the phage is Lon-dependent. The Lon protease is one of the major proteolytic machineries in the bacterial cell (Gottesman, 2003), and was implicated in degradation ("destabilization") of many types of antitoxins in E. coli, thus enabling toxin activity (Christensen et al., 2001; Van Melderen et al., 1996; Wang et al., 2011). Indeed, T7 mutant growth on E. coli containing the sanaTA system was restored by two orders of magnitude when the E. coli also lacked lon (Fig 5D), suggesting that the sanaTA protection from T7 phage depends on Lon activity.
We hypothesized that the phage gene product (Gp) 4.5 interacts with Lon to prevent antitoxin degradation and thus hinders the sanaTA abortive infection activity. To test this hypothesis we co-expressed Lon (Flag-tagged) and Gp4.5 within E. coli. Indeed, we found that Lon and Gp4.5 co-immuno-precipitate, indicating that 4.5 tightly binds Lon (Fig 5E). A reciprocal co-immuno-precipitation assay, where pull-down was performed on flag-tagged Gp4.5 protein, produced similar co-immuno-precipitation patterns (Fig 5F). Overall, these results imply that the T7 Gp4.5 neutralizes TA-system-mediated abortive infection by inhibiting the Lon protease activity, thus preventing antitoxin degradation and toxin activation.
Discussion
Due to the rapid evolution and the functional novelties frequently associated with TA modules, discovery of novel TA families is challenging and often depends on serendipity (Hayes and Van Melderen, 2011; Stern and Sorek, 2011). Even when a member of a known family is identified by sequence similarity, there is usually no experimental evidence to support functionality of that specific member. We used gene clonability in E. coli as an experimental readout for TA module functionality, and performed a massive scale analysis of millions of cloned genes to detect hundreds of functional TA modules within microbial genomes. Many functional members belonging to 12 known families were detected, as well as several novel families associated with defense islands, expanding the set of TA systems verified to date.
Three of the systems we identified (those containing MNT and RHH domains in their antitoxin) were previously predicted to be TA systems by a computational analysis based on comparative genomics and genomic context analyses (Makarova et al., 2009; Table 2). However, the cloning patterns we recorded for two of these systems that belong to the HEPN-MNT superfamily suggested that the HEPN is the toxin and the MNT is the antitoxin, and not the opposite as previously predicted (Makarova et al., 2009). Our experiments with one such system, hhalTA, verified that the gene with the HEPN domain (hhalT) is indeed the toxin and hhalA is the antitoxin, pointing to the strength of our cloning-based approach over purely computational approaches.
Some of the novel systems we detected contain domains that were not described in TA systems before. The antitoxin in the pmenTA system, for example, shares homology with the macro domain (COG2110), which was shown to bind the metabolite ADP-ribose (Han et al., 2011). It is therefore possible that this cofactor is important for the pmenA antitoxin activity. The psyrT shares homology with domains of the RecQ helicase, a family of proteins implicated in DNA repair (Bernstein et al., 2010); and the antitoxin of the same system, psyrA, has a nucleotide binding domain (COG0758) that was previously described in proteins involved in DNA uptake (Mortier-Barrière et al., 2007) and recently also in the shosT/shosA toxin/antitoxin system (Kimelman et al., 2012). The presence of these domains within the psyrTA system may suggest that the toxicity of this system is conferred by DNA manipulation of some sort.
Not all known TA families were retrieved by our approach. First, families represented in the studied genomes by less than 7 members were not analyzed due to lack of statistical power. Second, in families where the antitoxin is encoded by a very small gene, the occurrence of a randomly fragmented clone that contains the toxin but not the antitoxin has low probability, and this again reduces the statistical power for detection of such families. This has resulted in high p-values for several bona fide TA families, such as the HipAB family, and these did not pass our statistical thresholds. Our discovery method is therefore biased towards detection of systems characterized by larger genes than those found in most known families. It is therefore conceivable that additional novel families of TA systems, not detected by our algorithms, are yet to be discovered.
Our study had focused on type II TA systems, where both the toxin and antitoxin are protein entities. However, our general cloning-based approach can in principle detect other types of TA systems, where the antitoxin is a non-coding RNA molecule (type I and III). To detect such cases, one may search for genes (toxin) that cannot be cloned unless the nearby intergenic region (encoding the non-coding antitoxin) is also found on the same clone. However such an application is out of the scope of the current study.
Although we focused our validation and analyses on novel toxin-antitoxin families that preferentially appear in defense islands, other toxin-immunity modules may also be revealed by our algorithm. For example, secreted toxins that are targeted to other bacteria via the type VI secretion system in P. aeruginosa have an adjacent immunity gene to prevent the toxin from affecting the producing cell (Hood et al., 2010). Another example is of the contact dependent inhibition protein A (CdiA), which kills neighboring cells upon cell-cell contact; the toxicity of this protein is neutralized by the immunity protein CdiI, encoded by a small gene found immediately downstream to cdiA (Aoki et al., 2010; Poole et al., 2011). Since such antimicrobial toxin-immunity systems are expected to yield a cloning bias similar to that observed for TA systems, but are not expected to localize in phage defense islands, it is possible that the broader set of families retrieved by our algorithm (Table S1) may contain novel such systems related to antimicrobial activities rather than to phage defense. Nevertheless, some of these predicted families possibly represent false positive predictions. Such false positives may stem from long range effects of neighboring genes that change the clone-pattern distribution at the locus, leading to a distribution that may falsely appear as representing a TA system. Indeed, in 4 cases out of the 24 systems that our algorithm had retrieved, the genomic organization of the locus indicates on operons that are larger than 2 genes, which is not typical of TA systems. Therefore, single predictions need to be treated with caution until experimentally validated.
Our results show that the sanaTA system provides resistance against T7 phage lacking the non-essential 4.5 gene. We also found that expression of the rlegTA system results in significant reduction in T7 plaques diameter, although this result might be attributed to the partial toxicity of the rlegTA system when expressed in E. coli. To date, only two other type II TA systems were shown to provide phage resistance (Hazan et al., 2004; Koga et al., 2011). However, the distribution of numerous TA systems in bacterial defense islands (Makarova et al., 2011) implies that the involvement of TA systems in bacterial battle against phages might be underestimated. It is possible that the systems we tested which did not show anti-phage activity do indeed function in phage resistance in their genome of origin. Since the TA systems we tested were not originally derived from E. coli, the exogenous antitoxin might lack the sequence signals to be recognized and degraded by the E. coli proteases. In the absence of antitoxin destabilization, no anti-phage activity is possible. Another possibility is that, similar to the 4.5 gene reported here, T7 carries additional genes encoding anti-TA activities that were not tested in our study; these genes might have mitigated the defensive activity of the tested systems. Infection with additional mutant phages might expose anti-phage activity for additional systems, and might shed more light on the evolution of arms-race between bacteria and phage.
Two phage strategies to counteract bacterial activity of toxin-antitoxin systems have been described to date. One strategy is to bring an external antitoxin to substitute for the degraded antitoxin. This was shown for phage T4 which encodes a protein, Dmd, a suppressor of the RnlA toxin (Koga et al., 2011; Otsuka and Yonesaki, 2012). Another strategy is to inhibit the bacterial protease, thus overcoming the rapid destabilization of the antitoxin. This mode of action was demonstrated for T4 PinA protein, which blocks Lon protease (Skorupski et al., 1988) and for the λ RexB protein, which inhibits degradation of the Phd and MazE antitoxins by hindering activity of the ClpP protease (Engelberg-Kulka et al., 1998). Our results imply that the T7 Gp4.5 also resists TA-mediated Abi by inhibiting the Lon protease. This suggests that protease inhibition is a general anti-Abi mechanism used by phages. The complete lack of homology between the anti-protease phage proteins described to date might suggest that the protease inhibition strategy has been evolutionarily invented multiple times in parallel. Indeed, the bacterial proteases seem to be the weak link in the TA-mediated defense strategy, as a single protease usually dominates the destabilization of antitoxin belonging to multiple different TA systems in the same organism (Aizenman et al., 1996; Cherny and Gazit, 2004; Christensen et al., 2001; Lehnherr and Yarmolinsky, 1995; Roberts et al., 1994; Van Melderen et al., 1996).
Several studies suggest the involvement of TA systems and the Lon protease in generating bacterial persister cells (Maisonneuve et al., 2011; Rotem et al., 2010; Schumacher et al., 2009; Vazquez-Laslop et al., 2006). Persistence, in which bacteria enter a dormant, metabolically silent state, is a major obstacle in effective antibiotic treatment against pathogens such as Mycobacterium tuberculosis, Staphylococcus aureus, and Pseudomonas aeruginosa (Gomez and McKinney, 2004; Tuchscherr et al., 2011). Deletion of Lon in E. coli was shown to mitigate TA-system-mediated persistence and resulted in higher sensitivity to antibiotics (Maisonneuve et al., 2011). It appears that phages may be a rich source for molecules that inhibit bacterial proteases, and it is possible that with some molecular adaptations, such molecules could be used to treat bacterial persistence. One can envision that in the future such phage-derived compounds can enter clinical use as synergistic supplements to treatment with antibiotics. Thus, the arms race between bacteria and phage could be harnessed for the benefit of mankind.
Experimental procedures
Coverage analysis of pairs of genes
Mapping of sequencing clones on 360 bacteria and 28 archaea for which raw clone sequencing data was available (http://www.weizmann.ac.il/pandatox) was performed as described in (Kimelman et al., 2012). Gene positions and annotations were downloaded as described (Kimelman et al., 2012). For each consecutive pair of genes in each genome, three numbers were recorded:
x= [number of clones fully spanning the first gene but not the second gene]
y= [number of clones fully spanning the second gene but not the first gene]
z= [number of clones fully spanning both genes]
A pair of genes conforming with{x=0; y>0; z>0} or {x>0; y=0; z>0} was declared as "following the TA cloning pattern". Pairs in which the putative toxin was identified as a ‘hitchhiker’ (Kimelman et al., 2012) were eliminated from further counts in order to avoid cases in which this clonability pattern was a byproduct of a nearby single, standalone toxic gene. Pairs residing in replicons lacking sufficient clone coverage were also ignored in further counts.
Clustering of pairs into families
Clustering of individual genes based on sequence similarity was retrieved from IMG (http://img.jgi.doe.gov/cgi-bin/w/main.cgi) on November 2010 (based on the "IMG cluster" field). Cluster IDs were recorded for every consecutive pair of genes analyzed. Pairs containing a gene that lacked a cluster ID were discarded. All pairs having the same two cluster IDs (regardless of the order and the strand) were aggregated into a single "family of pairs". Families containing less than 7 pairs were ignored, to ensure statistical power in next steps of the analysis. This resulted in 21,417 families, containing at least 7 pairs of consecutive genes sharing the same two cluster IDs, which were further analyzed. The following "TA cloning fraction" (F) parameters were calculated for each family:
F1 = the fraction of family members that follow the TA cloning pattern {x=0; y>0; z>0}
F2 = the fraction of family members that follow the TA cloning pattern {x>0; y=0; z>0}
Directionality of putative family (i.e., determining whether gene "x" is the putative toxin or the putative antitoxin) was determined using Fmax= max{F1,F2}, such that:
Fmax = F1 → x is the putative toxin
Fmax = F2 → y is the putative toxin
For each family, the Genus names of all containing organisms were extracted. A "family diversity" (FD) measure was defined as the number of different genus names divided by the total number of family members (pairs). This measure was used to roughly assess the tendency of the family to undergo horizontal gene transfer (HGT) within a wide array of organisms, with higher FD corresponding to higher HGT tendency. For example, a 10-members family where all members are found in strains of Escherichia coli will receive a low FD of 0.1. Only families having FD>=0.5 were further analyzed, based on the empirical FD distribution among known families of TA systems (Fig S3A).
A statistical framework to detect TA families
Since a given pair of genes has the potential to follow the TA cloning pattern merely by chance (i.e., due to the random clone fragmentation) rather than reflecting a real toxin/antitoxin activity, clone distribution simulations were performed to asses statistical significance per family (Fig S1). For every pair of genes in each family, all clones covering its genome of origin were randomly distributed on the genome, shuffling clone positions but maintaining their number and sizes. Based on these random clone distributions, the x,y, and z values were measured for the gene pair, and a simulated "TA cloning fraction" (Fsim) was then calculated for the family. This procedure was repeated 1,000 times for each family, generating a distribution of Fsim(i) (i=1..1000). The real Fmax of the family was then compared to the distribution of Fsim values obtained from the simulations, generating an empirical p value for a family (Fig S1).
Since this procedure is computationally demanding to perform for >21000 families, only families where Fmax >=0.3 (i.e., at least 30% of family members followed the TA cloning pattern) were thus analyzed. Families presenting p-value <= 0.05 were considered as following the TA cloning pattern in a statistically significantly manner (Table S1).
To identify families significantly localized to Defense Islands (DIs)(Makarova et al., 2011), a value of ‘mean number of defense island genes in proximity’ (DIval) was calculated for each family based on a list of 132 COGs (DI genes) that were shown to be enriched in defense island regions (Makarova et al., 2011). For this, the average number of DI genes within a range of ±5 genes from each family member was calculated. Families having DIval > 0.5 were defined as DI associated (Fig 2), based on the empirical DIval distribution among known families of TA systems (Fig S3B; Fig S4). Families were further reviewed manually to remove transposon-containing families possibly associated with defense islands due to their transposition-related properties rather than being genuine TA systems.
Analysis of known TA systems within the identified set
The locus tags of all genes from the 24 final families (Fig 2) were checked against a previously compiled list of known TA systems (Makarova et al., 2009). For a given family, if at least one pair was found in the list of known TA systems, the family was declared as ‘known’. Families that were not declared as ‘known’ were further similarly checked against a list of predicted TA genes downloaded from TADB (http://bioinfomml.sjtu.edu.cn/TADB/) (Shao et al., 2011). Families in which at least one pair was found in this TADB list were declared as ‘predicted’.
Domain analysis of genes in the identified families was performed by searching their sequences against the Conserved Domain Database (CDD)(Marchler-Bauer et al., 2011) using rpsblast (ftp://ftp.ncbi.nih.gov/blast/documents/rpsblast.html) with e-value threshold of 0.05.
For the Phylogenetic distribution analysis of families (Fig 4B–C), the IMG cluster ids (http://img.jgi.doe.gov/cgi-bin/w/main.cgi) of the toxin and antitoxin for each family were used to retrieve all adjacent pairs of genes in IMG that have the same two cluster ids. For each family, number and identity of organisms carrying members of the family were extracted (Table S3; Fig 4B–C).
Experimental evaluation of ‘TA cloning pattern’
Toxin and antitoxin were amplified from their genomes of origin (in the case of sanaTA, psyrTA, sdenTA and hhalTA) or synthesized (GenScript) for the families pmenTA and rlegTA. The toxin was then directionally ligated into the pRSFDuet-1 vector (EMD Chemicals Inc.) and the antitoxin ligated into the pBAD/HisA plasmid (Invitrogen). Since transformation of the toxin gene alone resulted in mutations in the toxin due to toxicity, the two plasmids (carrying the toxin and antitoxin) were co-transformed into E. coli BL21(DE3)pLysS (Stratagene) in the presence of 0.3% arabinose to induce the antitoxin. The clones were verified by direct sequencing with primers on the pRSFduet-1 and pBAD/HisA vectors.
For the toxicity assay on plates, clones were cultured in LB medium with 100 µg/ml ampicillin, 50 µg/ml kanamycin, 34 µg/ml chloramphenicol and 0.3% arabinose overnight. The next day, a portion of each overnight culture was inoculated into fresh medium (10-fold dilution) and 10µl were spotted on LB plates supplemented with 100 µg/ml ampicillin, 50 µg/ml kanamycin and 34 µg/ml chloramphenicol. Toxin, antitoxin or both were induced by 100µM IPTG and 0.3% arabinose, respectively.
For the growth kinetics experiments 3 different colonies of each system were cultured in LB medium with 100 µg/ml ampicillin, 50 µg/ml kanamycin, 34 µg/ml chloramphenicol and 0.3% arabinose overnight. The next day cells were diluted 1:20 and measured for OD using 1cm path cuvetts. Samples were equilibrated to the same OD and 5 µl of these samples were added to 175 µl LB medium supplemented with 100 µg/ml ampicillin, 50 µg/ml kanamycin, 34 µg/ml chloramphenicol in a 96-wells microplate. Cells were placed in a microplate reader (Infinite M200) in a script employing OD measurement every ~5 minutes. Measurements were done using 486ex/516em bandwidth (Gain 95, 105 and 110) and OD at 600nm overnight. For each of the colonies the following treatments were applied: No induction, induction of 100 µM IPTG and/or 0.3% arabinose after ~4h, and induction of 100 µM IPTG after ~4h and of 0.3% arabinose after another ~2.5 hours. The overnight growth replicate values were averaged and the measured OD values were plotted against time.
The viability assay (Fig 3C) was performed as described by Pedersen et al (2002). Briefly, each strain was grown overnight at 37°C in LB medium containing 100 µg/ml ampicillin, 50 µg/ml kanamycin, and 34 µg/ml chloramphenicol. In the next morning, cells were then diluted 1:1000 in the same medium as above and grown for 3 hours. At time zero cells were then washed once with LB, and then transcription of toxin was induced by 100µM IPTG. At increasing time points after toxin induction (30, 60, 120, 180, 240 and 300 mins) cells were plated in several dilutions on LB-plates containing 100 µg/ml ampicillin, 50 µg/ml kanamycin, 34 µg/ml chloramphenicol and 0.3% arabinose. CFUs were determined in the next morning by colony counting.
Plaque assays
E. coli strains harboring the antitoxin only or the toxin-antitoxin of psyrTA, sanaTA, pmenTA, rlegTA, and sdenTA binary toxin-antitoxin systems were grown overnight in LB liquid medium supplemented with 35 µg/ml chloramphenicol, 100 µg/ml ampicillin and 0.3% L-arabinose with or without 50 µg/ml kanamycin, respectively. Overnight cultures were diluted 1:100 in fresh LB medium supplemented with inducers and antibiotics as above and aerated with shaking at 37°C until reaching OD 600≈0.5. Cultures were then centrifuged for 10 min and re-suspended in LB to an OD 600 of exactly 0.5. Volumes of 200 µl of culture and 10 µl of the indicated T7 phages were mixed in 3 ml of warm 0.8% agar LB supplemented with 35 µg/ml chloramphenicol, 100 µg/ml ampicillin and 0.3% L-arabinose or 35 µg/ml chloramphenicol, 100 µg/ml ampicillin, 50 µg/ml kanamycin, 0.3% L-arabinose, and 50–100 µM IPTG for the antitoxin-only- or toxin-antitoxin-harboring cultures, respectively. The mixtures were immediately overlaid on LB plates supplemented with the above indicated inducers and antibiotics. Overlaid plates were incubated at 37 °C for 3 hr and plaques were then counted. For a given TA pair, efficiency of plating (EOP) was calculated by dividing the number of plaque forming units (PFU) obtained for bacterial lawn expressing the toxin + antitoxin by the number of PFU obtained on the bacterial lawn expressing the antitoxin alone.
Expression of Lon and Gp4.5 proteins and co-immunoprecipitation assay
Expression of Lon and Gp4.5 was performed using the expression vector pRSFDuet-1 (EMD Chemicals, Inc.). For co-expression experiments the 4.5 gene (3’ His-tagged) and full-length Lon protease gene (5’ Flag-tagged) were cloned into the 1st and 2nd expression cassettes, respectively. For reciprocal Co-IP experiments the 4.5 gene (5’ Flag-tagged) and the full-length Lon protease (5’ His-tagged) were cloned into pRSFDuet-1 as described above. Induction was carried out at 37°C for 3 hr by addition of 200 µM of IPTG. Cell pellets were lysed by sonication in a buffer containing 50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1%(v/v) Triton X-100, 1 mM phenylmethylsulfonyl fluoride (PMSF) and 1 µl/mL protease inhibitor cocktail (Set IV, EMD Chemicals, Inc.). Cell debris were removed by centrifugation at 4°C for 15 minutes at 18,000 g. Clear supernatants were transferred to 1.5 ml tubes and incubated on a rotator shaker at 4°C for 1 hr with 80 µl pre-washed anti-Flag M2-agarose beads (Sigma, # A2220). The beads were washed three times with the buffer described above and the Flag-tagged protein or protein complex were eluted using Flag-peptide (Sigma, #F3290) using the manufacturers’ recommendations. Anti Flag-tag and anti-His tags Abs were purchased from Sigma.
Supplementary Material
Highlights.
Data from shotgut sequencing projects can identify novel toxin/antitoxin systems
Toxin/antitoxin systems were identified by analyzing 8.5 million DNA clones
Some of the novel toxin/antitoxin families confer resistance against phage
Phage T7 contains a protein that mitigates toxin/antitoxin activity
Acknowledgements
We thank Shany Doron, Eyal Weinstock, Daniel Dar, Uri Gophna, Omri Wurtzel, Tal Dagan, Gil Amitai and Debbie Lindell for comments and stimulating discussions. We also thank Ada Dantes for excellent technical support. R.S. was supported, in part, by NIH grant R01AI082376-01, ISF-FIRST program (grant 1615/09), ISF (grant 1303/12) and ERC-StG program (grant 260432).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aizenman E, Engelberg-Kulka H, Glaser G. An Escherichia coli chromosomal "addiction module" regulated by guanosine [corrected] 3',5'-bispyrophosphate: a model for programmed bacterial cell death. Proc Natl Acad Sci U S A. 1996;93:6059–6063. doi: 10.1073/pnas.93.12.6059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amitai S, Kolodkin-Gal I, Hananya-Meltabashi M, Sacher A, Engelberg-Kulka H. Escherichia coli MazF leads to the simultaneous selective synthesis of both "death proteins" and "survival proteins". PLoS Genet. 2009;5:e1000390. doi: 10.1371/journal.pgen.1000390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki SK, Diner EJ, de Roodenbeke CT, Burgess BR, Poole SJ, Braaten BA, Jones AM, Webb JS, Hayes CS, Cotter PA, et al. A widespread family of polymorphic contact-dependent toxin delivery systems in bacteria. Nature. 2010;468:439–442. doi: 10.1038/nature09490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergh O, Borsheim KY, Bratbak G, Heldal M. High abundance of viruses found in aquatic environments. Nature. 1989;340:467–468. doi: 10.1038/340467a0. [DOI] [PubMed] [Google Scholar]
- Bernard P, Couturier M. Cell killing by the F plasmid CcdB protein involves poisoning of DNA-topoisomerase II complexes. J Mol Biol. 1992;226:735–745. doi: 10.1016/0022-2836(92)90629-x. [DOI] [PubMed] [Google Scholar]
- Bernstein KA, Gangloff S, Rothstein R. The RecQ DNA helicases in DNA repair. Annu Rev Genet. 2010;44:393–417. doi: 10.1146/annurev-genet-102209-163602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherny I, Gazit E. The YefM antitoxin defines a family of natively unfolded proteins: implications as a novel antibacterial target. J Biol Chem. 2004;279:8252–8261. doi: 10.1074/jbc.M308263200. [DOI] [PubMed] [Google Scholar]
- Chibani-Chennoufi S, Bruttin A, Dillmann ML, Brussow H. Phage-host interaction: an ecological perspective. J Bacteriol. 2004;186:3677–3686. doi: 10.1128/JB.186.12.3677-3686.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopin MC, Chopin A, Bidnenko E. Phage abortive infection in lactococci: variations on a theme. Curr Opin Microbiol. 2005;8:473–479. doi: 10.1016/j.mib.2005.06.006. [DOI] [PubMed] [Google Scholar]
- Christensen SK, Maenhaut-Michel G, Mine N, Gottesman S, Gerdes K, Van Melderen L. Overproduction of the Lon protease triggers inhibition of translation in Escherichia coli: involvement of the yefM-yoeB toxin-antitoxin system. Mol Microbiol. 2004;51:1705–1717. doi: 10.1046/j.1365-2958.2003.03941.x. [DOI] [PubMed] [Google Scholar]
- Christensen SK, Mikkelsen M, Pedersen K, Gerdes K. RelE, a global inhibitor of translation, is activated during nutritional stress. Proc Natl Acad Sci U S A. 2001;98:14328–14333. doi: 10.1073/pnas.251327898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen SK, Pedersen K, Hansen FG, Gerdes K. Toxin-antitoxin loci as stress-response-elements: ChpAK/MazF and ChpBK cleave translated RNAs and are counteracted by tmRNA. J Mol Biol. 2003;332:809–819. doi: 10.1016/s0022-2836(03)00922-7. [DOI] [PubMed] [Google Scholar]
- Daines DA, Wu MH, Yuan SY. VapC-1 of nontypeable Haemophilus influenzae is a ribonuclease. J Bacteriol. 2007;189:5041–5048. doi: 10.1128/JB.00290-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deveau H, Garneau JE, Moineau S. CRISPR/Cas system and its role in phage-bacteria interactions. Annu Rev Microbiol. 2010;64:475–493. doi: 10.1146/annurev.micro.112408.134123. [DOI] [PubMed] [Google Scholar]
- Duman RE, Lowe J. Crystal structures of Bacillus subtilis Lon protease. J Mol Biol. 2010;401:653–670. doi: 10.1016/j.jmb.2010.06.030. [DOI] [PubMed] [Google Scholar]
- Engelberg-Kulka H, Reches M, Narasimhan S, Schoulaker-Schwarz R, Klemes Y, Aizenman E, Glaser G. rexB of bacteriophage lambda is an anti-cell death gene. Proc Natl Acad Sci U S A. 1998;95:15481–15486. doi: 10.1073/pnas.95.26.15481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fico S, Mahillon J. TasA-tasB, a new putative toxin-antitoxin (TA) system from Bacillus thuringiensis pGI1 plasmid is a widely distributed composite mazE-doc TA system. BMC Genomics. 2006;7:259. doi: 10.1186/1471-2164-7-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fineran PC, Blower TR, Foulds IJ, Humphreys DP, Lilley KS, Salmond GP. The phage abortive infection system, ToxIN, functions as a protein-RNA toxin-antitoxin pair. Proc Natl Acad Sci U S A. 2009;106:894–899. doi: 10.1073/pnas.0808832106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fozo EM, Makarova KS, Shabalina SA, Yutin N, Koonin EV, Storz G. Abundance of type I toxin-antitoxin systems in bacteria: searches for new candidates and discovery of novel families. Nucleic Acids Res. 2010;38:3743–3759. doi: 10.1093/nar/gkq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg AL, Moerschell RP, Chung CH, Maurizi MR. ATP-dependent protease La (lon) from Escherichia coli. Methods Enzymol. 1994;244:350–375. doi: 10.1016/0076-6879(94)44027-1. [DOI] [PubMed] [Google Scholar]
- Gomez JE, McKinney JD. M. tuberculosis persistence, latency, and drug tolerance. Tuberculosis (Edinb) 2004;84:29–44. doi: 10.1016/j.tube.2003.08.003. [DOI] [PubMed] [Google Scholar]
- Gottesman S. Proteolysis in bacterial regulatory circuits. Annu Rev Cell Dev Biol. 2003;19:565–587. doi: 10.1146/annurev.cellbio.19.110701.153228. [DOI] [PubMed] [Google Scholar]
- Goulard C, Langrand S, Carniel E, Chauvaux S. The Yersinia pestis chromosome encodes active addiction toxins. J Bacteriol. 2010;192:3669–3677. doi: 10.1128/JB.00336-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han W, Li X, Fu X. The macro domain protein family: Structure, functions, and their potential therapeutic implications. Mutat Res. 2011;727:86–103. doi: 10.1016/j.mrrev.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes F, Van Melderen L. Toxins-antitoxins: diversity, evolution and function. Crit Rev Biochem Mol Biol. 2011;46:386–408. doi: 10.3109/10409238.2011.600437. [DOI] [PubMed] [Google Scholar]
- Hazan R, Engelberg-Kulka H. Escherichia coli mazEF-mediated cell death as a defense mechanism that inhibits the spread of phage P1. Mol Genet Genomics. 2004;272:227–234. doi: 10.1007/s00438-004-1048-y. [DOI] [PubMed] [Google Scholar]
- Hazan R, Sat B, Engelberg-Kulka H. Escherichia coli mazEF-mediated cell death is triggered by various stressful conditions. J Bacteriol. 2004;186:3663–3669. doi: 10.1128/JB.186.11.3663-3669.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood RD, Singh P, Hsu F, Guvener T, Carl MA, Trinidad RR, Silverman JM, Ohlson BB, Hicks KG, Plemel RL, et al. A type VI secretion system of Pseudomonas aeruginosa targets a toxin to bacteria. Cell Host Microbe. 2010;7:25–37. doi: 10.1016/j.chom.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath P, Barrangou R. CRISPR/Cas, the immune system of bacteria and archaea. Science. 2010;327:167–170. doi: 10.1126/science.1179555. [DOI] [PubMed] [Google Scholar]
- Hurley JM, Woychik NA. Bacterial toxin HigB associates with ribosomes and mediates translation-dependent mRNA cleavage at A-rich sites. J Biol Chem. 2009;284:18605–18613. doi: 10.1074/jbc.M109.008763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Pogliano J, Helinski DR, Konieczny I. ParE toxin encoded by the broad-host-range plasmid RK2 is an inhibitor of Escherichia coli gyrase. Mol Microbiol. 2002;44:971–979. doi: 10.1046/j.1365-2958.2002.02921.x. [DOI] [PubMed] [Google Scholar]
- Jorgensen MG, Pandey DP, Jaskolska M, Gerdes K. HicA of Escherichia coli defines a novel family of translation-independent mRNA interferases in bacteria and archaea. J Bacteriol. 2009;191:1191–1199. doi: 10.1128/JB.01013-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Wang X, Ma Q, Zhang XS, Wood TK. Toxin-antitoxin systems in Escherichia coli influence biofilm formation through YjgK (TabA) and fimbriae. J Bacteriol. 2009;191:1258–1267. doi: 10.1128/JB.01465-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimelman A, Levy A, Sberro H, Kidron S, Amitai G, Yoder-Himes D, Zhu Y, Wurtzel O, Rubin EM, Sorek R. A vast collection of microbial genes that are toxic to bacteria. Genome Research. 2012;22:802–809. doi: 10.1101/gr.133850.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King G, Murray NE. Restriction enzymes in cells, not eppendorfs. Trends Microbiol. 1994;2:465–469. doi: 10.1016/0966-842x(94)90649-1. [DOI] [PubMed] [Google Scholar]
- Koga M, Otsuka Y, Lemire S, Yonesaki T. Escherichia coli rnlA and rnlB compose a novel toxin-antitoxin system. Genetics. 2011;187:123–130. doi: 10.1534/genetics.110.121798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchta K, Knizewski L, Wyrwicz LS, Rychlewski L, Ginalski K. Comprehensive classification of nucleotidyltransferase fold proteins: identification of novel families and their representatives in human. Nucleic Acids Res. 2009;37:7701–7714. doi: 10.1093/nar/gkp854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labrie SJ, Samson JE, Moineau S. Bacteriophage resistance mechanisms. Nat Rev Microbiol. 2010;8:317–327. doi: 10.1038/nrmicro2315. [DOI] [PubMed] [Google Scholar]
- Lehnherr H, Yarmolinsky MB. Addiction protein Phd of plasmid prophage P1 is a substrate of the ClpXP serine protease of Escherichia coli. Proc Natl Acad Sci U S A. 1995;92:3274–3277. doi: 10.1073/pnas.92.8.3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leplae R, Geeraerts D, Hallez R, Guglielmini J, Dreze P, Van Melderen L. Diversity of bacterial type II toxin-antitoxin systems: a comprehensive search and functional analysis of novel families. Nucleic Acids Res. 2011;39:5513–5525. doi: 10.1093/nar/gkr131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Zhang Y, Inouye M, Woychik NA. Bacterial addiction module toxin Doc inhibits translation elongation through its association with the 30S ribosomal subunit. Proc Natl Acad Sci U S A. 2008;105:5885–5890. doi: 10.1073/pnas.0711949105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maisonneuve E, Shakespeare LJ, Jorgensen MG, Gerdes K. Bacterial persistence by RNA endonucleases. Proc Natl Acad Sci U S A. 2011;108:13206–13211. doi: 10.1073/pnas.1100186108. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Makarova KS, Wolf YI, Koonin EV. Comprehensive comparative-genomic analysis of type 2 toxin-antitoxin systems and related mobile stress response systems in prokaryotes. Biol Direct. 2009;4:19. doi: 10.1186/1745-6150-4-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Wolf YI, Snir S, Koonin EV. Defense islands in bacterial and archaeal genomes and prediction of novel defense systems. J Bacteriol. 2011;193:6039–6056. doi: 10.1128/JB.05535-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, Lu S, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, et al. CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 2011;39:D225–D229. doi: 10.1093/nar/gkq1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda H, Tan Q, Awano N, Yamaguchi Y, Inouye M. A novel membrane-bound toxin for cell division, CptA (YgfX), inhibits polymerization of cytoskeleton proteins, FtsZ and MreB, in Escherichia coli. FEMS Microbiol Lett. 2012;328:174–181. doi: 10.1111/j.1574-6968.2012.02496.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortier-Barrière I, Velten M, Dupaigne P, Mirouze N, Piétrement O, McGovern S, Fichant G, Martin B, Noirot P, Le Cam E, et al. A key presynaptic role in transformation for a widespread bacterial protein: DprA conveys incoming ssDNA to RecA. Cell. 2007;130:824–836. doi: 10.1016/j.cell.2007.07.038. [DOI] [PubMed] [Google Scholar]
- Neubauer C, Gao YG, Andersen KR, Dunham CM, Kelley AC, Hentschel J, Gerdes K, Ramakrishnan V, Brodersen DE. The structural basis for mRNA recognition and cleavage by the ribosome-dependent endonuclease RelE. Cell. 2009;139:1084–1095. doi: 10.1016/j.cell.2009.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor L, Coffey A, Daly C, Fitzgerald GF. AbiG, a genotypically novel abortive infection mechanism encoded by plasmid pCI750 of Lactococcus lactis subsp. cremoris UC653. Appl Environ Microbiol. 1996;62:3075–3082. doi: 10.1128/aem.62.9.3075-3082.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuka Y, Yonesaki T. Dmd of bacteriophage T4 functions as an antitoxin against Escherichia coli LsoA and RnlA toxins. Mol Microbiol. 2012;83:669–681. doi: 10.1111/j.1365-2958.2012.07975.x. [DOI] [PubMed] [Google Scholar]
- Pecota DC, Wood TK. Exclusion of T4 phage by the hok/sok killer locus from plasmid R1. J Bacteriol. 1996;178:2044–2050. doi: 10.1128/jb.178.7.2044-2050.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen K, Christensen SK, Gerdes K. Rapid induction and reversal of a bacteriostatic condition by controlled expression of toxins and antitoxins. Mol Microbiol. 2002;45:501–510. doi: 10.1046/j.1365-2958.2002.03027.x. [DOI] [PubMed] [Google Scholar]
- Poole SJ, Diner EJ, Aoki SK, Braaten BA, t'Kint de Roodenbeke C, Low DA, Hayes CS. Identification of functional toxin/immunity genes linked to contact-dependent growth inhibition (CDI) and rearrangement hotspot (Rhs) systems. PLoS Genet. 2011;7:e1002217. doi: 10.1371/journal.pgen.1002217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts RC, Strom AR, Helinski DR. The parDE operon of the broad-host-range plasmid RK2 specifies growth inhibition associated with plasmid loss. J Mol Biol. 1994;237:35–51. doi: 10.1006/jmbi.1994.1207. [DOI] [PubMed] [Google Scholar]
- Rotem E, Loinger A, Ronin I, Levin-Reisman I, Gabay C, Shoresh N, Biham O, Balaban NQ. Regulation of phenotypic variability by a threshold-based mechanism underlies bacterial persistence. Proc Natl Acad Sci U S A. 2010;107:12541–12546. doi: 10.1073/pnas.1004333107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher MA, Piro KM, Xu W, Hansen S, Lewis K, Brennan RG. Molecular mechanisms of HipA-mediated multidrug tolerance and its neutralization by HipB. Science. 2009;323:396–401. doi: 10.1126/science.1163806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Y, Harrison EM, Bi D, Tai C, He X, Ou HY, Rajakumar K, Deng Z. TADB: a web-based resource for Type 2 toxin-antitoxin loci in bacteria and archaea. Nucleic Acids Res. 2011;39:D606–D611. doi: 10.1093/nar/gkq908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skorupski K, Tomaschewski J, Ruger W, Simon LD. A bacteriophage T4 gene which functions to inhibit Escherichia coli Lon protease. J Bacteriol. 1988;170:3016–3024. doi: 10.1128/jb.170.7.3016-3024.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorek R, Kunin V, Hugenholtz P. CRISPR--a widespread system that provides acquired resistance against phages in bacteria and archaea. Nat Rev Microbiol. 2008;6:181–186. doi: 10.1038/nrmicro1793. [DOI] [PubMed] [Google Scholar]
- Sorek R, Zhu Y, Creevey CJ, Francino MP, Bork P, Rubin EM. Genome-wide experimental determination of barriers to horizontal gene transfer. Science. 2007;318:1449–1452. doi: 10.1126/science.1147112. [DOI] [PubMed] [Google Scholar]
- Stahlberg H, Kutejova E, Suda K, Wolpensinger B, Lustig A, Schatz G, Engel A, Suzuki CK. Mitochondrial Lon of Saccharomyces cerevisiae is a ring-shaped protease with seven flexible subunits. Proc Natl Acad Sci U S A. 1999;96:6787–6790. doi: 10.1073/pnas.96.12.6787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern A, Sorek R. The phage-host arms race: shaping the evolution of microbes. Bioessays. 2011;33:43–51. doi: 10.1002/bies.201000071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Q, Awano N, Inouye M. YeeV is an Escherichia coli toxin that inhibits cell division by targeting the cytoskeleton proteins, FtsZ and MreB. Mol Microbiol. 2010;79:109–118. doi: 10.1111/j.1365-2958.2010.07433.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuchscherr L, Medina E, Hussain M, Volker W, Heitmann V, Niemann S, Holzinger D, Roth J, Proctor RA, Becker K, et al. Staphylococcus aureus phenotype switching: an effective bacterial strategy to escape host immune response and establish a chronic infection. EMBO Mol Med. 2011;3:129–141. doi: 10.1002/emmm.201000115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Oost J, Jore MM, Westra ER, Lundgren M, Brouns SJ. CRISPR-based adaptive and heritable immunity in prokaryotes. Trends Biochem Sci. 2009;34:401–407. doi: 10.1016/j.tibs.2009.05.002. [DOI] [PubMed] [Google Scholar]
- Van Melderen L, Saavedra De Bast M. Bacterial toxin-antitoxin systems: more than selfish entities? PLoS Genet. 2009;5:e1000437. doi: 10.1371/journal.pgen.1000437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Melderen L, Thi MH, Lecchi P, Gottesman S, Couturier M, Maurizi MR. ATP-dependent degradation of CcdA by Lon protease. Effects of secondary structure and heterologous subunit interactions. J Biol Chem. 1996;271:27730–27738. doi: 10.1074/jbc.271.44.27730. [DOI] [PubMed] [Google Scholar]
- Vazquez-Laslop N, Lee H, Neyfakh AA. Increased persistence in Escherichia coli caused by controlled expression of toxins or other unrelated proteins. J Bacteriol. 2006;188:3494–3497. doi: 10.1128/JB.188.10.3494-3497.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Kim Y, Hong SH, Ma Q, Brown BL, Pu M, Tarone AM, Benedik MJ, Peti W, Page R, et al. Antitoxin MqsA helps mediate the bacterial general stress response. Nat Chem Biol. 2011;7:359–366. doi: 10.1038/nchembio.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wozniak RA, Waldor MK. A toxin-antitoxin system promotes the maintenance of an integrative conjugative element. PLoS Genet. 2009;5:e1000439. doi: 10.1371/journal.pgen.1000439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XZ, Yan X, Cui ZL, Hong Q, Li SP. mazF, a novel counter-selectable marker for unmarked chromosomal manipulation in Bacillus subtilis. Nucleic Acids Res. 2006;34:e71. doi: 10.1093/nar/gkl358. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.