Abstract
Epigenetic regulation can mediate long-lasting changes in gene expression, which makes it an attractive mechanism for the stable behavioral abnormalities that characterize drug addiction. Recent research has unveiled numerous types of epigenetic modifications within the brain’s reward circuitry in animal models of drug addiction. In this review, we summarize the latest advances in the field, focusing on histone modifications, DNA methylation, and non-coding RNAs. We also highlight several areas for future research. Unraveling the highly complex epigenetic mechanisms of addiction is adding to our understanding of this syndrome and has the potential to trigger novel approaches for better diagnosis and therapy.
Introduction
Epigenetics describes diverse mechanisms that regulate gene transcription without modifying underlying DNA sequences, and includes numerous types of histone modifications, DNA methylation, and non-coding RNAs. Such mechanisms play a key role in encoding environmental stimuli into cellular fate during development and behavioral adaptations throughout an individual’s lifetime [1].
Drug addiction can be viewed as maladaptive neural plasticity to drugs of abuse. Once formed, it can drive life-long behavioral abnormalities. While the RNA and protein molecules that presumably mediate these long-term effects are normally turned over on the order of days, it is speculated that epigenetic mechanisms might alter gene expression and thus the intrinsic properties of the brain over a much longer time course [2,3]. Epigenetic modifications, by causing long-lasting changes in the steady state levels of expression of a gene, in the inducibility of that gene in response to some subsequent stimulus, or in the splicing isoforms of a gene that are expressed, are ideally suited for mediating addiction-associated neural plasticity.
Virtually all principles of epigenetic regulation have come from studies of cultured cells in vitro and non-neuronal systems. Thus, elaborating the epigenetic mechanisms of drug addiction will contribute not only to our understanding of this syndrome, but also far more generally to the epigenetic basis of brain function and plasticity.
Histone modifications
Chromatin is composed of nucleosomes, DNA wrapped around histone octomers containing two copies each of H2A, H2B, H3, and H4 (Figure 1). Histones undergo many types of posttranslational modifications (PTMs) that alter their structure and interaction with neighboring DNA [4]. The N-terminal tails of histones protrude from the nucleosome and can be covalently modified at numerous residues by acetylation, methylation, phosphorylation, ubiquitination, SUMOylation, citrullination, and ADP-ribosylation. These histone modifications are formed and removed by large families of enzymes, which make them reversible, labile epigenetic “marks.” By far, histone acetylation and methylation are most studied in drug addiction.
Figure 1. Histone posttranslational modifications.
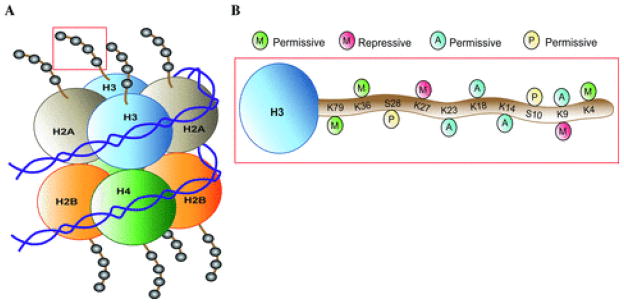
(A) The nucleosome core particle composed of 147 bp of DNA wrapped around an octamer of histone proteins (two copies each of H2A, H2B, H3, and H4). (B) Histone modifications on histone H3 tail. Permissive gene expression correlates with modifications that weaken the interaction between histones and DNA or that promote the recruitment of transcriptional activating complexes (e.g., histone acetylation at K23, K18, K14, and K9, as well as methylation at K79, K36, and K4 or phosphorylation at S28 and S10). Repressive transcription correlates with histone deacetylation (which compacts nucleosomes), histone methylation (e.g., on H3K27 or H3K9, which recruits repressive complexes to chromatin), or DNA methylation (not shown).
Histone acetylation is associated with transcriptional activation; it negates the positive charge of lysine (Lys) residues in histone tails and increases spacing between nucleosomes. It is controlled by histone acetyltransferases (HATs) and histone deacetylases (HDACs) [5]. Acute or repeated exposure to cocaine or other stimulant drugs of abuse increases global levels of histone acetylation in the nucleus accumbens (NAc), a key brain reward region [6] (Figure 2). Short-term increases in histone acetylation promote behavioral responses to cocaine, while sustained increases generally oppose cocaine’s effects, based on the actions of systemic or intra-NAc administration of HDAC inhibitors or NAc-specific deletions of HDAC genes [e.g., [6–9]. Altered histone acetylation has been demonstrated at several candidate genes in the NAc in response to stimulants, and these changes correlate with their altered expression. For example, H4 acetylation is increased at the c-Fos promoter acutely, with no changes seen chronically, consistent with desensitization of c-Fos expression after chronic drug exposure [6,10]. In contrast, the BDNF and Cdk5 promoters show H3 acetylation only after chronic cocaine, consistent with induction of these genes by chronic drug exposure [6]. A genome-wide study utilizing ChIP-chip—chromatin immunoprecipitation (ChIP) with antibodies against pan-acetylated H3 or H4 followed by promoter microarrays—has provided a more complete map of genes in NAc that display altered histone acetylation after chronic cocaine [11]. Numerous gene promoters were found to be hyper- or hypoacetylated; interestingly, there was minimal overlap between genes that display alterations in H3 versus H4. While many of the genes that showed altered histone acetylation in response to cocaine exhibit commensurate changes in mRNA expression—with hyperacetylation associated with increased expression and hypoacetylation decreased expression most— genes did not follow this pattern. These observations indicate that the “histone code” for gene regulation [4] is likely to be very complex, with histone acetylation contributing just a fraction of all epigenetic information that determines a gene’s activity. It will be important to repeat these genome-wide determinations for each of the many individual sites of histone acetylation, and in combination with other histone modifications, to better understand the role of each in gene regulation.
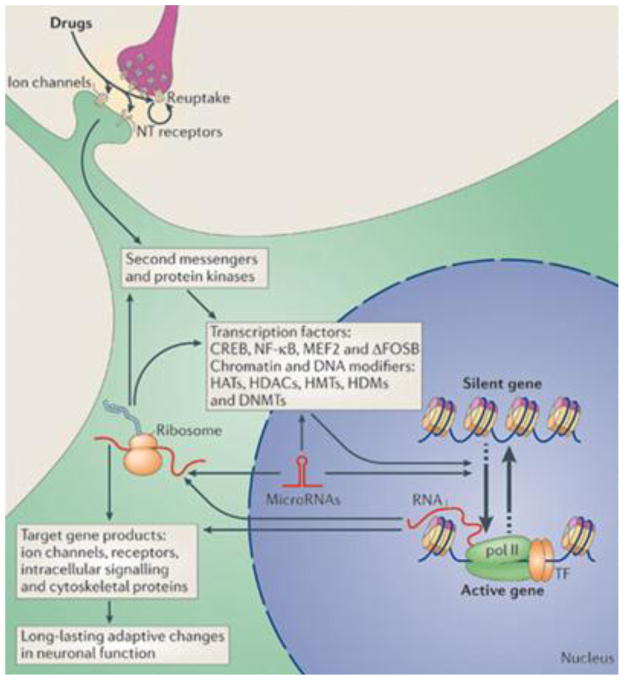
Figure 2. Mechanisms of transcriptional and epigenetic regulation by drugs of abuse.
Drugs of abuse act through synaptic targets (reuptake mechanisms, ion channels, and neurotransmitter [NT] receptors) to alter intracellular signalling cascades. This leads to the activation or inhibition of transcription factors and of many other nuclear targets, including chromatin-regulatory proteins (shown by thick arrows). These processes result in the induction or repression of particular genes, which can in turn further regulate gene transcription. It is proposed that some of these drug-induced changes at the chromatin level are extremely stable and thereby underlie the long-lasting behaviours that define addiction.
CREB, cAMP-response element binding protein; DNMTs, DNA methyltransferases; HATs, histone acetyltransferases; HDACs, histone deacetylases; HDMs, histone demethylases; HMTs, histone methyltransferases; MEF2, myocyte-specific enhancer factor 2; NF-κB, nuclear factor-κB; pol II, RNA polymerase II. Figure is reproduced with permission from Ref. 3
Repressive histone methylation has also been implicated in drug addiction [12–14]. G9a and GLP (G9a-like protein), two histone methyltransferases that catalyze the di-methylated state of Lys9 of H3 (H3K9me2), are downregulated in NAc by chronic cocaine [12] or opiate [14] administration, along with decreases in global levels of this mark. A similar downregulation is seen in NAc of animals that self-administer these drugs, as well as in NAc of human drug addicts. In contrast to G9a and GLP, numerous other types of histone methyltransferases and demethylases are not affected by drug exposure[12,14]. Genetic or pharmacological blockade of G9a in this brain region potentiates behavioral responses to both drugs of abuse, whereas increasing G9a function exerts the opposite effect [12,14]. Downregulation of G9a also increases the dendritic arborization of NAc neurons[12], which directly connects altered H3K9me2 in the synaptic plasticity associated with addiction.
G9a appears to be a critical control point for epigenetic regulation in NAc, as we know it functions in two negative feedback loops. It opposes the induction of ΔFosB, a long-lasting transcription factor important for drug addiction [2,3], while ΔFosB in turn suppresses G9a expression[12,14]. Also, G9a is induced in NAc upon prolonged HDAC inhibition, which explains the paradoxical attenuation of cocaine’s behavioral effects seen under these conditions, as noted above[9]. Genome-wide maps, utilizing ChIP-chip or the more powerful ChIP-seq (ChIP followed by deep sequencing), have been obtained for altered H3K9me2 binding in NAc after chronic cocaine or opiates[11,14]. As with histone acetylation, regulation of H3K9me2 is associated with altered gene expression, but is not in itself deterministic.
One surprising feature of early work in this field is that drug exposure alters global (or total cellular) levels of histone PTMs, such as increased histone acetylation or decreased H3K9me2 and H3K9me3 in NAc [6,12–14]. While genome-wide studies confirmed that a greater number of genomic sites show increased acetylation or reduced H3K9 methylation, hundreds of genes show opposite changes in these marks, and most genes show no alterations after drug exposure. This raises the crucial question of what determines whether a specific gene is modified in the face of a global change in a histone-modifying enzyme and its mark. These findings also raise the possible importance of histone modifications outside of gene promoters. This is particularly important, since the majority (>90%) of the genome has recently been proven to be transcribed and have regulatory roles [15]. Indeed, we have found using ChIP-seq close to 3,000 cocaine-induced H3K9me3 differential sites and more than 9,000 morphine-induced H3K9me2 differential sites in NAc, most of which are located at repetitive genomic sequences [13,14]. Cocaine-mediated decreases in H3K9me3 at specific repeats, such as LINE1, is associated with their increased expression [13]. This suggests that, while changes at specific genes are not tightly governed by global histone changes, such global patterns might reflect general genomic destabilization [16]after repeated drug exposure.
DNA Methylation
DNA methylation occurs with the addition of a methyl group to the C5 position of cytosine (5-mC) predominantly at CpG sites. It plays a pivotal role in cell differentiation/reprogramming, imprinting, X chromosome inactivation, repetitive element silencing, and tumor formation [1,17]. DNA methylation generally exerts a repressive effect on gene transcription. It can either prevent the association of DNA-binding factors with their target sequence or bind to methyl-CpG-binding proteins to recruit transcription co-repressors to modify the surrounding chromatin into a silencing state [18]. Compared with histone tail modifications, which are considered readily reversible, DNA methylation is viewed as a more stable epigenetic change.
DNMT3a, the only de novo DNA methyltransferase expressed in postnatal brain [19], is increased in NAc after prolonged (28 days) withdrawal from either repeated non-contingent cocaine administration or self-administration [20]. Local knockout of DNMT3a from the NAc, or local infusion of the DNMT inhibitor RG108, increased behavioral responses to cocaine, whereas DNMT3a overexpression in NAc had the opposite effect. DNMT3a likewise regulates dendritic arborizations of NAc neurons [20]. In addition, NAc knockout of MeCP2 (methyl CpG binding protein 2), an important modulator of neural plasticity [21,22], enhances amphetamine reward [23]. These findings suggest that DNMT3a and MeCP2 act to blunt drug action.
However, such actions of DNMT3a or MeCP2 do not necessarily indicate a role for altered DNA methylation, and information on the regulation of DNA methylation in addiction models remains limited. A small number of studies have investigated DNA methylation changes at particular genes of interest [24,25], but there has not yet been a genome-wide mapping of such regulation. Genome-wide measures of DNA methylation are challenging technically: most existing methods focus on gene promoter regions only and do not distinguish between several forms of DNA methylation that may have opposite effects on transcription (see next paragraph). True genome-wide maps of DNA methylation, with single base resolution based on deep sequencing, are still too expensive to be practical. Nevertheless, such maps are critically needed to understand how dynamic are alterations in DNA methylation at particular genomic sites over a broad time course of drug exposure, and to what extent such changes are reversible during drug abstinence.
Recent research has suggested, in fact, that DNA methylation in the adult brain is much more dynamic than previously recognized. Members of ten-eleven translocation (TET) family of proteins oxidize 5-mC into 5-hydroxymethylcytosine (5-hmC) [26,27], and subsequently into 5-formylcytosine and 5-carboxylcytosine [28,29]. Through deamination, glycosylation, and base excision repair, these newly discovered forms of cytosine modification can then be converted back into an unmethylated state [30,31]. These findings have attracted considerable interest, since they provide a mechanism by which 5-mC oxidation mediates active DNA demethylation in brain [32], a possibility that has long been debated [33]. Moreover, 5mC oxidation derivatives are expressed at highest levels in neurons [34]. In contrast to the repressive effect of 5-mC on gene expression, 5-hmC is more correlated with transactivation. In preliminary work, we have obtained genome-wide maps of 5-hmc in NAc after cocaine exposure and found that changes in 5-hmc correlate with altered gene expression (Feng et al, unpublished data). These findings further underscore the importance of obtaining genome-wide maps of several forms of DNA methylation in drug abuse models.
Non-coding RNAs
The complete sequencing of the mammalian genome and its transcriptional products has revealed a surprisingly large number of expressed RNAs that are not translated into proteins. Such non-coding RNAs have been shown to play crucial regulatory roles in cell function [15,35].
Numerous types of microRNAs (miRNAs), a class of small non-coding RNAs, have been investigated in addiction models [36,37]. miRNAs exert a repressive role on gene expression by binding to specific mRNAs and thereby blocking their translation or inducing their degradation. Multiple miRNAs are reported to be up- or downregulated by drugs of abuse. For instance, cocaine increases levels of miR-181a and decreases miR-124 and let-7d in rat striatum [38,39], and mimicking the direction of each of these changes enhances cocaine reward. Since miRNAs function via base-pairing with complementary sequences within mRNA molecules, it is possible to infer target mRNAs of drug-altered miRNAs through computational predictions, although such methods can yield false positive and negative results. Recent work is encouraging. miR-212 is induced in rat dorsal striatum after cocaine self-administration, and serves to inhibit cocaine intake [40]. This action was attributed to the ability of miR-212 to indirectly lead to the activation of CREB [41], a transcription factor that antagonizes cocaine reward [2]. Several additional genes implicated in addiction models, such as ΔFosB, dopamine transporter, and glutamate receptor subunits, have also been related to drug-triggered alterations in specific miRNAs [39,41]. Next generation sequencing has recently been used to capture the repertoire of miRNAs that are altered in NAc whole extracts and purified striatal post-synaptic densities after chronic cocaine [42]. It was found that tens of miRNAs are regulated by cocaine. Future work is needed to identify the definitive mRNA targets for each regulated miRNA as well as to explore still un-annotated, novel miRNAs that are altered by drugs of abuse using available next generation sequencing datasets.
Lately, long non-coding RNAs (lncRNAs) are emerging as key regulators of gene transcription [35,36,43,44]. Such non-coding RNAs, defined as having a length >200 bp, are highly abundant and highly regulated. They appear to form RNA-protein interactions to carry out their functions by modulating chromatin-modifying complexes and interacting with transcription factors, among other actions. Though the role of lncRNAs in drug addiction has not yet been characterized, mining microarray data revealed that several lncRNAs are altered in the brains of addicted humans [45]. This is therefore an area ripe for future investigation.
Conclusion and future directions
Although still in relatively early stages, work to date has demonstrated that many forms of epigenetic regulation are altered in brain reward regions by drugs of abuse and in turn serve to regulate drug action. Already these initial studies have raised several key questions that will need to be addressed moving forward.
One complex question is how epigenetic regulation is translated into transcription change. As noted earlier, no single modification examined to date is deterministic for a change in gene expression, consistent with the required involvement of numerous modifications that work in concert. Deciphering such a code is an important goal for future research. A technical challenge in this effort is the heterogeneous cell population of even brain micronuclei, which makes it impossible to derive data as clear-cut as for cell culture systems. Methodologies are underway to isolate specific cell types from brain [46] and to perform genome-wide ChIP-seq, RNA-seq, and DNA methylation assays on much less starting material [47,48]. In the meantime, nearly all bioinformatics tools for genome-wide analysis have been developed based on clean cell culture data, which are not optimal to detect the more subtle signals from terminally differentiated neurons, particularly with the high background noise unavoidable with in vivo studies. Improved analytical tools will require creative collaborations between biologists and bioinformaticians.
An important part of this effort of defining an epigenetic code will be to decipher the cross-talk among histone modifications, DNA methylation, and non-coding RNAs. For example, certain histone methylation events are preferentially enriched at methylated DNA sites [49]. MeCP2 has been shown to interact with miRNAs to influence cocaine reward [50]. Long non-coding RNAs can serve as a scaffold for multiple chromatin modification enzymes such as DNMTs and G9a [43]. Studying these and many other potential interactions across epigenetic mechanisms in drug addiction models is a high priority of current research.
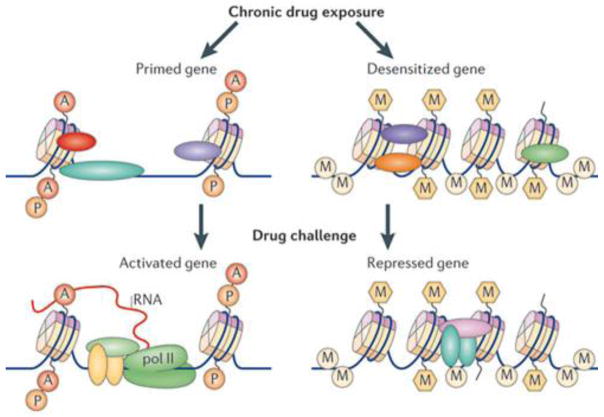
Another goal is to better understand the influence of different forms of epigenetic regulation on a gene’s activity. As noted at the outset, some epigenetic modifications likely alter the steady-state level of a gene’s transcription, while others poise genes for greater (gene priming) or lesser (gene desensitization) induction in response to some subsequent stimulus in the absence of a steady state change in expression[3]. There are several precedents for such priming and desensitization in addiction models, and such regulation needs to be examined more globally [10,12,51](Figure 3). Recent work has indicated that epigenetic modifications also control pre-mRNA alternative splicing [52]. Accordingly, an epigenetic change will correlate with altered expression of a particular isoform of a gene without detectable regulation of total transcript levels. As well, we know that many epigenetic modifications occur in non-genic regions. Each of these consequences of epigenetic regulation, beyond alterations in steady state levels of expressed RNAs, is far beyond the coverage of traditional microarrays. We expect that next generation sequencing technologies (e.g., ChIP-seq, RNA-seq) [53,54] will soon provide more complete answers to these questions.
Figure 3. Gene priming and desensitization.
Epigenetic mechanisms are important in mediating gene priming and desensitization, which are not reflected by stable changes in steady-state mRNA levels. Instead, a later drug challenge induces a given gene to a greater (primed) or lesser (desensitized) extent based on the epigenetic modifications that are induced by previous chronic drug exposure. A, acetylation; M, methylation; P, phosphorylation; pol II, RNA polymerase II. Figure is reproduced with permission from Ref. 3.
We also need to better understand the intracellular signaling pathways through which synaptic transmission is translated into epigenetic modifications. We still know very little about these steps, with only a small number of examples reported to date [e.g., [55–57] (see Figure 2). The next step is to learn how a particular epigenetic modification is targeted to a given gene, as stated earlier, and to decipher why some, perhaps even most, epigenetic changes are highly labile, while a smaller subset may persist for longer times.
Finally, we need to ascertain whether any drug-induced epigenetic modifications are transferred to offspring to influence their susceptibility to drug abuse or other conditions. Such trans-generational transmission would require drug-induced epigenetic changes in sperm or ova to persist in the fertilized embryo and to influence adult brain function. There are early reports that this may be the case [58], however, much further work is needed to demonstrate definitively an epigenetic basis of such transmission and to understand the underlying mechanisms involved.
Highlights.
Role of histone acetylation and methylation in drug addiction.
Role of DNA methylation, 5-methylcytosine and 5-hydroxymethylcytosine, in addiction.
Role of microRNAs and long non-coding RNAs in drug addiction.
Acknowledgments
Preparation of this review was supported by grants from the National Institute on Drug Abuse. The authors apologize for work not cited in this review due to space limitations.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
• of special interest
•• of outstanding interest
- 1.Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33(Suppl):245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 2.Nestler EJ. Molecular basis of long-term plasticity underlying addiction. Nat Rev Neurosci. 2001;2:119–128. doi: 10.1038/35053570. [DOI] [PubMed] [Google Scholar]
- 3.Robison AJ, Nestler EJ. Transcriptional and epigenetic mechanisms of addiction. Nat Rev Neurosci. 2011;12:623–637. doi: 10.1038/nrn3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 5.Borrelli E, Nestler EJ, Allis CD, Sassone-Corsi P. Decoding the epigenetic language of neuronal plasticity. Neuron. 2008;60:961–974. doi: 10.1016/j.neuron.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kumar A, Choi KH, Renthal W, Tsankova NM, Theobald DE, Truong HT, Russo SJ, Laplant Q, Sasaki TS, Whistler KN, et al. Chromatin remodeling is a key mechanism underlying cocaine-induced plasticity in striatum. Neuron. 2005;48:303–314. doi: 10.1016/j.neuron.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 7.Rogge GA, Wood MA. The role of histone acetylation in cocaine-induced neural plasticity and behavior. Neuropsychopharmacology: official publication of the American College of Neuropsychopharmacology. 2013;38:94–110. doi: 10.1038/npp.2012.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Levine A, Huang Y, Drisaldi B, Griffin EA, Jr, Pollak DD, Xu S, Yin D, Schaffran C, Kandel DB, Kandel ER. Molecular mechanism for a gateway drug: epigenetic changes initiated by nicotine prime gene expression by cocaine. Science translational medicine. 2011;3:107ra109. doi: 10.1126/scitranslmed.3003062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kennedy PJ, Robison AJ, Maze I, Feng J, Badimon A, Bassel-Duby R, Olson EN, Nestler EJ. HDAC1 inhibition blocks cocaine-induced plasticity through targeted changes in histone methylation. Nat Neuroscience. 2013 doi: 10.1038/nn.3354. provisionally accepted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Renthal W, Carle TL, Maze I, Covington HE, 3rd, Truong HT, Alibhai I, Kumar A, Montgomery RL, Olson EN, Nestler EJ. Delta FosB mediates epigenetic desensitization of the c-fos gene after chronic amphetamine exposure. J Neurosci. 2008;28:7344–7349. doi: 10.1523/JNEUROSCI.1043-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Renthal W, Kumar A, Xiao G, Wilkinson M, Covington HE, 3rd, Maze I, Sikder D, Robison AJ, LaPlant Q, Dietz DM, et al. Genome-wide analysis of chromatin regulation by cocaine reveals a role for sirtuins. Neuron. 2009;62:335–348. doi: 10.1016/j.neuron.2009.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12••.Maze I, Covington HE, 3rd, Dietz DM, LaPlant Q, Renthal W, Russo SJ, Mechanic M, Mouzon E, Neve RL, Haggarty SJ, et al. Essential role of the histone methyltransferase G9a in cocaine-induced plasticity. Science. 2010;327:213–216. doi: 10.1126/science.1179438. This study reports for the first time the regulatory role of repressive histone methylation in drug addiction. It also highlights a feedback loop between a histone methyltransferase and the transcription factor ΔFosB. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13•.Maze I, Feng J, Wilkinson MB, Sun H, Shen L, Nestler EJ. Cocaine dynamically regulates heterochromatin and repetitive element unsilencing in nucleus accumbens. Proc Natl Acad Sci U S A. 2011;108:3035–3040. doi: 10.1073/pnas.1015483108. By using next generation sequencing technology, this study deciphers a previously unrecognized dynamic regulation by cocaine of intergenic repetitive elements, which expands our understanding of the epigenetic regulation of drug addiction. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun H, Maze I, Dietz DM, Scobie KN, Kennedy PJ, Damez-Werno D, Neve RL, Zachariou V, Shen L, Nestler EJ. Morphine Epigenomically Regulates Behavior through Alterations in Histone H3 Lysine 9 Dimethylation in the Nucleus Accumbens. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2012;32:17454–17464. doi: 10.1523/JNEUROSCI.1357-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15•.Dunham I, Kundaje A, Aldred SF, Collins PJ, Davis CA, Doyle F, Epstein CB, Frietze S, Harrow J, Kaul R, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. This is a representative report of the Encyclopedia of DNA Elements (ENCODE) Consortium. A critical finding is that the majority of the human genome, while not transcribed, plays a regulatory role. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Muotri AR, Chu VT, Marchetto MC, Deng W, Moran JV, Gage FH. Somatic mosaicism in neuronal precursor cells mediated by L1 retrotransposition. Nature. 2005;435:903–910. doi: 10.1038/nature03663. [DOI] [PubMed] [Google Scholar]
- 17.Bird AP, Wolffe AP. Methylation-induced repression--belts, braces, and chromatin. Cell. 1999;99:451–454. doi: 10.1016/s0092-8674(00)81532-9. [DOI] [PubMed] [Google Scholar]
- 18.Goll MG, Bestor TH. Eukaryotic cytosine methyltransferases. Annu Rev Biochem. 2005;74:481–514. doi: 10.1146/annurev.biochem.74.010904.153721. [DOI] [PubMed] [Google Scholar]
- 19.Feng J, Chang H, Li E, Fan G. Dynamic expression of de novo DNA methyltransferases Dnmt3a and Dnmt3b in the central nervous system. J Neurosci Res. 2005;79:734–746. doi: 10.1002/jnr.20404. [DOI] [PubMed] [Google Scholar]
- 20•.LaPlant Q, Vialou V, Covington HE, 3rd, Dumitriu D, Feng J, Warren BL, Maze I, Dietz DM, Watts EL, Iniguez SD, et al. Dnmt3a regulates emotional behavior and spine plasticity in the nucleus accumbens. Nat Neurosci. 2010;13:1137–1143. doi: 10.1038/nn.2619. The authors first reported the functional role of DNA methyltransferases in addiction-related changes in neuronal morphology and behavior. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen WG, Chang Q, Lin Y, Meissner A, West AE, Griffith EC, Jaenisch R, Greenberg ME. Derepression of BDNF transcription involves calcium-dependent phosphorylation of MeCP2. Science. 2003;302:885–889. doi: 10.1126/science.1086446. [DOI] [PubMed] [Google Scholar]
- 22.Martinowich K, Hattori D, Wu H, Fouse S, He F, Hu Y, Fan G, Sun YE. DNA methylation-related chromatin remodeling in activity-dependent BDNF gene regulation. Science. 2003;302:890–893. doi: 10.1126/science.1090842. [DOI] [PubMed] [Google Scholar]
- 23.Deng JV, Rodriguiz RM, Hutchinson AN, Kim IH, Wetsel WC, West AE. MeCP2 in the nucleus accumbens contributes to neural and behavioral responses to psychostimulants. Nat Neurosci. 2010;13:1128–1136. doi: 10.1038/nn.2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Anier K, Malinovskaja K, Aonurm-Helm A, Zharkovsky A, Kalda A. DNA methylation regulates cocaine-induced behavioral sensitization in mice. Neuropsychopharmacology. 2010;35:2450–2461. doi: 10.1038/npp.2010.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nielsen DA, Yuferov V, Hamon S, Jackson C, Ho A, Ott J, Kreek MJ. Increased OPRM1 DNA methylation in lymphocytes of methadone-maintained former heroin addicts. Neuropsychopharmacology: official publication of the American College of Neuropsychopharmacology. 2009;34:867–873. doi: 10.1038/npp.2008.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.He YF, Li BZ, Li Z, Liu P, Wang Y, Tang Q, Ding J, Jia Y, Chen Z, Li L, et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333:1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, He C, Zhang Y. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu H, Zhang Y. Mechanisms and functions of Tet protein-mediated 5-methylcytosine oxidation. Genes & development. 2011;25:2436–2452. doi: 10.1101/gad.179184.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moore LD, Le T, Fan G. DNA methylation and its basic function. Neuropsychopharmacology: official publication of the American College of Neuropsychopharmacology. 2013;38:23–38. doi: 10.1038/npp.2012.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32••.Guo JU, Su Y, Zhong C, Ming GL, Song H. Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell. 2011;145:423–434. doi: 10.1016/j.cell.2011.03.022. By combining both in vitro and in vivo studies, the authors demonstrate that the 5-mC hydroxylase, TET1, promotes active DNA demethylation in adult hippocampal neurons through a process that requires base excision repair. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ooi SK, Bestor TH. The colorful history of active DNA demethylation. cell. 2008;133:1145–1148. doi: 10.1016/j.cell.2008.06.009. [DOI] [PubMed] [Google Scholar]
- 34•.Szulwach KE, Li X, Li Y, Song CX, Wu H, Dai Q, Irier H, Upadhyay AK, Gearing M, Levey AI, et al. 5-hmC-mediated epigenetic dynamics during postnatal neurodevelopment and aging. Nat Neurosci. 2011;14:1607–1616. doi: 10.1038/nn.2959. The authors profiled the genome-wide pattern of 5-hmC in both mouse hippocampus and cerebellum during development. They also proposed a role of 5-hmC in neurological disorders. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annual review of biochemistry. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sartor GC, St Laurent G, 3rd, Wahlestedt C. The Emerging Role of Non-Coding RNAs in Drug Addiction. Frontiers in genetics. 2012;3:106. doi: 10.3389/fgene.2012.00106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schaefer A, Im HI, Veno MT, Fowler CD, Min A, Intrator A, Kjems J, Kenny PJ, O’Carroll D, Greengard P. Argonaute 2 in dopamine 2 receptor-expressing neurons regulates cocaine addiction. J Exp Med. 2010;207:1843–1851. doi: 10.1084/jem.20100451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chandrasekar V, Dreyer JL. microRNAs miR-124, let-7d and miR-181a regulate cocaine-induced plasticity. Molecular and cellular neurosciences. 2009;42:350–362. doi: 10.1016/j.mcn.2009.08.009. [DOI] [PubMed] [Google Scholar]
- 39•.Chandrasekar V, Dreyer JL. Regulation of MiR-124, Let-7d, and MiR-181a in the accumbens affects the expression, extinction, and reinstatement of cocaine-induced conditioned place preference. Neuropsychopharmacology: official publication of the American College of Neuropsychopharmacology. 2011;36:1149–1164. doi: 10.1038/npp.2010.250. By using lentiviral vector-expressing miRNAs, the group addressed the behavioral role of certain miRNA candidates that are altered by cocaine exposure and explored the target gene networks affected by these miRNAs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40••.Hollander JA, Im HI, Amelio AL, Kocerha J, Bali P, Lu Q, Willoughby D, Wahlestedt C, Conkright MD, Kenny PJ. Striatal microRNA controls cocaine intake through CREB signalling. Nature. 2010;466:197–202. doi: 10.1038/nature09202. The study reports that miR-212 is upregulated in the dorsal striatum of rats after cocaine self-administration. miR-212 decreases responsiveness to the motivational properties of cocaine via indirect upregulation of CREB, an antagonist of cocaine reward. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Saba R, Storchel PH, Aksoy-Aksel A, Kepura F, Lippi G, Plant TD, Schratt GM. Dopamine-regulated microRNA MiR-181a controls GluA2 surface expression in hippocampal neurons. Mol Cell Biol. 2012;32:619–632. doi: 10.1128/MCB.05896-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42•.Eipper-Mains JE, Kiraly DD, Palakodeti D, Mains RE, Eipper BA, Graveley BR. microRNA-Seq reveals cocaine-regulated expression of striatal microRNAs. RNA. 2011;17:1529–1543. doi: 10.1261/rna.2775511. This is the first report of using next generation sequencing technology to profile miRNA alterations genome-wide in mouse brain after repeated cocaine administration. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Qureshi IA, Mehler MF. Emerging roles of non-coding RNAs in brain evolution, development, plasticity and disease. Nature reviews Neuroscience. 2012;13:528–541. doi: 10.1038/nrn3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guil S, Esteller M. Cis-acting noncoding RNAs: friends and foes. Nature structural & molecular biology. 2012;19:1068–1075. doi: 10.1038/nsmb.2428. [DOI] [PubMed] [Google Scholar]
- 45.Michelhaugh SK, Lipovich L, Blythe J, Jia H, Kapatos G, Bannon MJ. Mining Affymetrix microarray data for long non-coding RNAs: altered expression in the nucleus accumbens of heroin abusers. Journal of neurochemistry. 2011;116:459–466. doi: 10.1111/j.1471-4159.2010.07126.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jiang Y, Matevossian A, Huang HS, Straubhaar J, Akbarian S. Isolation of neuronal chromatin from brain tissue. BMC Neurosci. 2008;9:42. doi: 10.1186/1471-2202-9-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Adli M, Bernstein BE. Whole-genome chromatin profiling from limited numbers of cells using nano-ChIP-seq. Nature protocols. 2011;6:1656–1668. doi: 10.1038/nprot.2011.402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gu H, Smith ZD, Bock C, Boyle P, Gnirke A, Meissner A. Preparation of reduced representation bisulfite sequencing libraries for genome-scale DNA methylation profiling. Nature protocols. 2011;6:468–481. doi: 10.1038/nprot.2010.190. [DOI] [PubMed] [Google Scholar]
- 49.Cedar H, Bergman Y. Linking DNA methylation and histone modification: patterns and paradigms. Nature reviews Genetics. 2009;10:295–304. doi: 10.1038/nrg2540. [DOI] [PubMed] [Google Scholar]
- 50.Im HI, Hollander JA, Bali P, Kenny PJ. MeCP2 controls BDNF expression and cocaine intake through homeostatic interactions with microRNA-212. Nat Neurosci. 2010;13:1120–1127. doi: 10.1038/nn.2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Damez-Werno D, LaPlant Q, Sun H, Scobie KN, Dietz DM, Walker IM, Koo JW, Vialou VF, Mouzon E, Russo SJ, et al. Drug experience epigenetically primes Fosb gene inducibility in rat nucleus accumbens. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2012;32:10267–10272. doi: 10.1523/JNEUROSCI.1290-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Luco RF, Allo M, Schor IE, Kornblihtt AR, Misteli T. Epigenetics in alternative pre-mRNA splicing. Cell. 2011;144:16–26. doi: 10.1016/j.cell.2010.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nature reviews Genetics. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Park PJ. ChIP-seq: advantages and challenges of a maturing technology. Nature reviews Genetics. 2009;10:669–680. doi: 10.1038/nrg2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Renthal W, Maze I, Krishnan V, Covington HE, 3rd, Xiao G, Kumar A, Russo SJ, Graham A, Tsankova N, Kippin TE, et al. Histone deacetylase 5 epigenetically controls behavioral adaptations to chronic emotional stimuli. Neuron. 2007;56:517–529. doi: 10.1016/j.neuron.2007.09.032. [DOI] [PubMed] [Google Scholar]
- 56.Bertran-Gonzalez J, Hakansson K, Borgkvist A, Irinopoulou T, Brami-Cherrier K, Usiello A, Greengard P, Herve D, Girault JA, Valjent E, et al. Histone H3 phosphorylation is under the opposite tonic control of dopamine D2 and adenosine A2A receptors in striatopallidal neurons. Neuropsychopharmacology: official publication of the American College of Neuropsychopharmacology. 2009;34:1710–1720. doi: 10.1038/npp.2008.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Taniguchi M, Carreira MB, Smith LN, Zirlin BC, Neve RL, Cowan CW. Histone deacetylase 5 limits cocaine reward through cAMP-induced nuclear import. Neuron. 2012;73:108–120. doi: 10.1016/j.neuron.2011.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58•.Vassoler FM, White SL, Schmidt HD, Sadri-Vakili G, Pierce RC. Epigenetic inheritance of a cocaine-resistance phenotype. Nat Neurosci. 2013;16:42–47. doi: 10.1038/nn.3280. This is the first report of the trans-generational transmission of behavioral responses to cocaine. [DOI] [PMC free article] [PubMed] [Google Scholar]