Abstract
The [FeFe] hydrogenase from Clostridium pasteurianum (CpI) harbors four Fe–S clusters that facilitate electron transfer to the H-cluster, a ligand-coordinated six-iron prosthetic group that catalyzes the redox interconversion of protons and H2. Here, we have used 57Fe nuclear resonance vibrational spectroscopy (NRVS) to study the iron centers in CpI, and we compare our data to that for a [4Fe–4S] ferredoxin as well as a model complex resembling the [2Fe]H catalytic domain of the H-cluster. In order to enrich the hydrogenase with 57Fe nuclei, we used cell-free methods to post-translationally mature the enzyme. Specifically, inactive CpI apoprotein with 56Fe-labeled Fe–S clusters was activated in vitro using 57Fe-enriched maturation proteins. This approach enabled us to selectively label the [2Fe]H subcluster with 57Fe, which NRVS confirms by detecting 57Fe–CO and 57Fe–CN normal modes from the H-cluster nonprotein ligands. The NRVS and iron quantification results also suggest that the hydrogenase contains a second 57Fe–S cluster. EPR spectroscopy indicates that this 57Fe-enriched metal center is not the [4Fe– 4S]H subcluster of the H-cluster. This finding demonstrates that the CpI hydrogenase retained an 56Fe-enriched [4Fe–4S]H cluster during in vitro maturation, providing unambiguous evidence for stepwise assembly of the H-cluster. In addition, this work represents the first NRVS characterization of [FeFe] hydrogenases.
Keywords: NRVS, EPR spectroscopy, 57Fe, synchrotron radiation, CpI, hydrogenase, H-cluster, Fe–S cluster, metalloenzyme, maturase, cell-free
Hydrogenases are metalloproteins that have unique iron-based cofactors capable of catalyzing the redox interconversion of H2 and protons. These enzymes have attracted interest not only for elucidating microbial evolution and physiology,1-3 but also for engineering renewable H2 production technologies.4,5 Furthermore, hydrogenases are appealing for their potential application as enzymes themselves6,7 and as inspirational targets toward designing biomimetic H2 catalysts.8,9
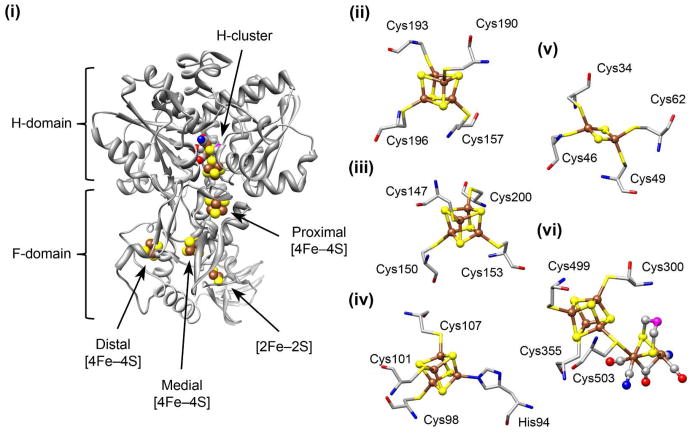
In the class of [FeFe] hydrogenases, the H2 redox chemistry occurs at the H-cluster. As shown in Figure 1, the H-cluster is located within the H-domain of the enzyme, and it consists of a catalytic [FeFe] subunit (termed [2Fe]H) linked via a cysteine ligand to a [4Fe–4S] cluster (termed [4Fe–4S]H).10,11 The [2Fe]H subcluster has multiple CO and CN– nonprotein ligands as well as a bridging dithiolate substituent (DTMX). Most [FeFe] hydrogenases also have ancillary Fe–S clusters that are involved in electron transfer to and from the H-cluster.12 In the particular case of the monomeric HydA hydrogenase from Clostridium pasteurianum – commonly referred to as CpI– the F-domain electron transport chain consists of three [4Fe–4S] clusters as well as a [2Fe–2S] cluster (Figure 1).11
Figure 1.
Structures of the iron cluster sites in the C. pasteurianum HydA hydrogenase (CpI). (i) Crystal structure of the CpI holoenzyme (Protein Data Bank ID code: 3C8Y) depicting the overall view of the electron transport chain within the F-domain. (ii) The proximal [4Fe–4S] accessory cluster. (iii) The medial [4Fe–4S] accessory cluster. (iv) The distal [4Fe–4S] accessory cluster, which is coordinated by three cysteine ligands and one histidine ligand. (v) The [2Fe–2S] ferredoxin-like cluster. (vi) The active site H-cluster metal center, which consists of the [4Fe–4S]H subcluster joined to the [2Fe]H subcluster via the Cys503 residue. The H-cluster is shown in the Hred state with an open site at the distal Fe nucleus. Nonprotein ligands coordinated to the [2Fe]H subcluster include three CO, two CN–, and a dithiol bridging molecule (DTMX) with an unknown central atom presumed to be either oxygen or nitrogen. The atoms shown are Fe (brown), S (yellow), C (gray), O (red), N (blue), and unknown (magenta). Molecular graphics were generated using the UCSF Chimera package.56
Unlike [4Fe–4S] and [2Fe–2S] clusters that are generally synthesized by host cell iron-sulfur cluster machinery, the [2Fe]H subcluster is made by three Fe–S accessory proteins called HydE, HydF, and HydG.12,13 Currently, it is not entirely understood how the [2Fe]H subcluster is assembled by these three Hyd maturases. Recent reports have shed light on the biosynthetic reaction sequence, mainly from in vitro studies focusing on characterizing the biochemistry of the Hyd proteins.14-17 For example, it has been suggested that HydF transfers the [2Fe]H subcluster to the [4Fe–4S]H-containing hydrogenase apoprotein, thus producing the active enzyme.18
Equally important to unraveling how the H-cluster is assembled will be to understand its structural and electronic transformations among the different redox configurations, which is crucial for establishing the catalytic cycle of the H-cluster. [FeFe] hydrogenases have been extensively characterized by X-ray crystallography and various advanced spectroscopies, and at least 7 different H-cluster substates have been observed.19-21 Quantum and molecular mechanics (QM/MM) calculations combined with density functional theory (DFT) have provided further insights into the catalytic cycle between various redox configurations such as the oxidized (Hox) and reduced (Hred) states.22 Yet, it has been challenging for researchers to develop synthetic catalysts that function similar to and as effectively as [FeFe] hydrogenases, due in part to a poor mechanistic understanding of how the H-cluster evolves H2.9 Certainly, elucidating the biosynthetic pathway of the H-cluster along with developing functional biomimetic catalysts are areas of active research that would benefit from new approaches to studying [FeFe] hydrogenases.12,15,23
Nuclear resonance vibrational spectroscopy (NRVS) is an exciting and novel approach for investigating the vibrational dynamics of Fe in metalloproteins.24-26 The fundamental physics of NRVS have been described in detail elsewhere.27-30 Measurements involve scanning an extremely monochromatic (< 1 meV) X-ray beam through a nuclear resonance and recording the vibrational structure associated with an 57Fe nuclear transition via the subsequent Fe Kα emission. While the resulting spectrum is similar to an infrared (IR) or resonance Raman (RR) spectrum,31 different intensity mechanisms apply. Specifically, the NRVS intensity for a particular normal mode is related to the movement of the resonant nucleus (in this case, 57Fe) along the direction of the incident X-ray beam,30,32 and results are generally reported as an 57Fe-centered partial vibrational density of states (PVDOS).32-34 With its particular elemental and isotopic specificity, NRVS provides a valuable complement to more conventional spectroscopies. Unlike IR spectroscopy, NRVS does not incur interference from water or protein-based modes. In contrast to RR spectroscopy, NRVS can probe any oxidation state and is impervious to sample fluorescence.
In this work, we present the NRVS of the CpI hydrogenase selectively enriched with 57Fe, which we refer to as CpI[57Fe]H. In order to produce the CpI[57Fe]H hydrogenase, a cell-free approach was used to post-translationally mature the apoenzyme. The inactive apoprotein was first expressed in Escherichia coli grown in 56Fe-supplemented medium, and the hydrogenase was then activated in vitro using 57Fe-enriched Hyd maturases. NRVS confirms the incorporation of 57Fe nuclei into the CpI[57Fe]H holoenzyme, and we compare our results to NRVS data for a model complex inspired by the [2Fe]H subcluster. In doing so, we confirm the presence of 57Fe–CO and 57Fe–CN normal modes. The NRVS data also show that a second Fe–S cluster contains 57Fe nuclei. However, EPR spectroscopy indicates that this iron center is not the [4Fe–4S]H subcluster.
Materials and Methods
Preparation of aqueous 57Fe
All steps were performed in a fume hood. Solutions were constantly stirred and chilled using ice baths. To prepare ∼80 mL of 65 mM 57Fe (final pH ∼1), metallic 57Fe was first dissolved using aqua regia. 5 mL of nitric acid (> 65%) was added to 300 mg of metallic 57Fe in a 150 mL glass beaker. Next, 12 mL of 12 M hydrochloric acid was added drop-wise. After the 57Fe was completely dissolved, 40 mL of 5 M sodium hydroxide was slowly added for partial neutralization, followed by 15 mL of 1.5 M sodium citrate and 15 mL of 1 M ammonium hydroxide.
In Vitro Preparation of 57Fe-enriched [FeFe] Hydrogenase for NRVS
Both the CpI apoprotein and the Shewanella oneidensis HydE, HydF, and HydG maturases were anaerobically produced in Escherichia coli strain BL21(DE3) ΔiscR∷kan using methods previously described for high-yield production of metalloproteins that contain stoichiometric amounts of Fe–S clusters.15,35 Inactive CpI–Strep-tag II apoprotein was produced in E. coli grown in complex medium with natural abundance Fe (250 mg·L–1 ferric ammonium citrate). HydE, HydF, and HydG were separately expressed in E. coli grown in complex medium supplemented with 200 μM 57Fe instead of natural abundance Fe. By using a colorimetric assay described by Fish,36 we estimated that the growth medium prepared from commercial LB Broth Miller (EMD Chemicals) contained less than 2 μM natural abundance Fe. Each maturase was individually expressed in E. coli in order to avoid the in vivo assembly of a HydF-bound [2Fe]H precursor that results when HydF is co-expressed with HydE and HydG.17,37 Cell lysates with the 57Fe-enriched maturases (HydE[57Fe], HydF[57Fe], and HydG[57Fe]) were prepared using BugBuster® Master Mix lysis solution (4 mL per gram wet-cell paste) supplemented with 100 mM HEPES (pH 8.2).
The CpI enzyme was post-translationally activated using previously described methods for cell-free synthesis of the H-cluster and hydrogenase maturation.15 In general terms, the in vitro maturation reaction is a combination of inactive CpI apoprotein, extrinsic low molecular weight substrates, and three E. coli lysates containing the HydE, HydF, and HydG maturases. Specifically, in vitro CpI maturation reaction mixtures (60 mL) contained 3 mL of HydE[57Fe] lysate, 10 mL of HydF[57Fe] lysate, 30 mL of HydG[57Fe] lysate, 1 mM aqueous 57Fe as described above, 1 mM sodium sulfide, 1 mM DTT, 2 mM S-adenosyl methionine, 2 mM L-cysteine, 2 mM L-tyrosine, 15 mM GTP, 1 mM pyridoxal-5′-phosphate, 2 mM sodium dithionite, and 300 mg·L–1 purified and desalted CpI apoenzyme (5 μM). After 24 hr of anaerobic incubation at 23°C, reaction mixtures were clarified at 20,000×g for 10 min to remove precipitate that formed during the reaction. Prior to the purification step, the pH of the clarified reaction mixture was adjusted to 7.5 using 1 M HEPES buffer (pH 8.2). Active CpI was isolated using Strep-Tactin® Superflow® high capacity resin (IBA GmbH), and the purified holoenzyme was concentrated to 25–50 μM using a stirred cell with a 5 kDa membrane (Amicon). Active CpI was further concentrated to ∼3 mM and a final volume of 75 μL using 30 kDa centrifugal filters (Amicon). The concentrated CpI[57Fe]H was loaded into 3 mm × 7 mm × 1 mm (interior dimensions) Lucite cuvettes and stored in liquid N2.
Biochemical Characterization of CpI[57Fe]H
CpI specific activities were measured using a methyl viologen activity assay.15 Protein concentrations were determined using the Bradford method38 along with a conversion factor specific to the CpI enzyme for dye-based assays.39 A ferrozine-based colorimetric assay was used to measure iron content.36
NRVS Measurements
NRVS measurements were recorded following described methods.30 The raw NRVS data were analyzed by the PHOENIX software program40 to extract the single-phonon 57Fe-centered partial vibrational densities of states (PVDOS). Each 57Fe PVDOS spectrum represents the average of the total number of scans, which was determined by adding scans and normalizing to the intensity of the incident beam.
The CpI[57Fe]H samples were measured at Beamline 09-XU at SPring-8 in Japan (energy resolution ∼0.8 meV, beam flux ∼1.4×109 photon·s–1).41 During NRVS measurements, samples were maintained at low temperatures using a liquid He cryostat. Actual temperatures for the CpI[57Fe]H samples were ∼50 K as obtained from the spectral imbalance analysis. Delayed nuclear fluorescence and Fe Kα fluorescence (from internal conversion) emitted by 57Fe atoms were recorded with a 4-channel avalanche photodiode array detector array. Scans took 45–60 min (varying from scan to scan), and measurements were taken with 0.27 meV steps. Scans of the CpI[57Fe]H samples were separated into multiple sections, and different scan times were used for each section. Specifically, points were measured for 1–3 s between 0–350 cm–1 and for 10–25 s between 350–800 cm–1. The resonant peak intensity for the CpI[57Fe]H measurements was 340 counts·s–1.
The [57Fe2(S2C3H6)(CN)2(CO)4]2– model complexes were measured at Beamline 3-ID at the Advanced Photon Source (APS) (resolution ∼1.0 meV, beam flux ∼3.2×109 photon·s–1).42 The samples were measured at low temperatures using a similar liquid helium cryostat. The packed and sealed powder samples were mounted onto the cryostat base with 2–4 screws. Actual sample temperatures were ∼75 K as obtained from the spectral imbalance analysis. The Fe emission was recorded with a single 1 cm2 square APD, and measurements were taken with ∼0.25 meV steps. The resonance peak intensities were 1100 and 1700 counts·s–1 for the natural abundance CN and 13CN-labeled model compounds, respectively.
EPR Spectroscopy
EPR measurements were performed at the CalEPR Center in the Department of Chemistry at the University of California, Davis. Continuous-wave (CW) spectra were collected at an X-band frequency (9.39 GHz) with either a Bruker Biospin ECS106 spectrometer or a Bruker Biospin Elexsys E500 spectrometer equipped with a cylindrical TE011-mode resonator (SHQE-W), an ESR-900 liquid helium cryostat, and an ITC-5 temperature controller (Oxford Instruments). Unless otherwise noted, measurement parameters were 15 K, 62 μW microwave power, 0.5 mT modulation amplitude, and 81.9 ms for the time constant and conversion time. Intensities were normalized to the temperature, time constant, receiver gain, number of scans, modulation amplitude, and square root of microwave power. Simulations were performed using MatLab™ and EasySpin software.43
Results
In Vitro Activation of the CpI[57Fe]H Hydrogenase
Two independent CpI[57Fe]H samples were prepared from separate cell-free maturation reactions that contained exogenous substrates including 57Fe, non-purified 57Fe-enriched Hyd maturases, and 56Fe-enriched CpI apoprotein. It is currently unclear which maturase harbors the Fe cluster onto which the CO, CN–, and DTMX ligands are coordinated. One hypothesis for [2Fe]H subcluster assembly involves the transfer of HydG-derived CO and CN– molecules to a HydF-bound [2Fe–2S] scaffold cluster.44 A second hypothesis states that HydG synthesizes an iron compound precursor with ligated CO and CN– that is shuttled to the hydrogenase via HydF.14 However, neither reaction sequence has been experimentally demonstrated. Therefore, all three maturases used in this study were enriched with 57Fe by producing them with growth medium supplemented with exogenous 57Fe.
Both the biochemical characterization and the measured NRVS spectra were similar for the two CpI[57Fe]H samples (Supporting Information, Figure S4). Based on activity measurements (558 ± 43 μmol H2 consumed·min–1·mg–1 of CpI), we estimated that 85% of the CpI apoprotein was matured during the in vitro reaction. Roughly 80% of the active enzyme was recovered during the purification step and prepared for NRVS measurements (3 mM CpI[57Fe]H). Iron quantification data from a ferrozine-based colorimetric assay indicate that the CpI apoenzyme and CpI[57Fe]H holoenzyme contained 12.7 ± 2.4 and 17.5 ± 1.7 total Fe per protein, respectively.
NRVS
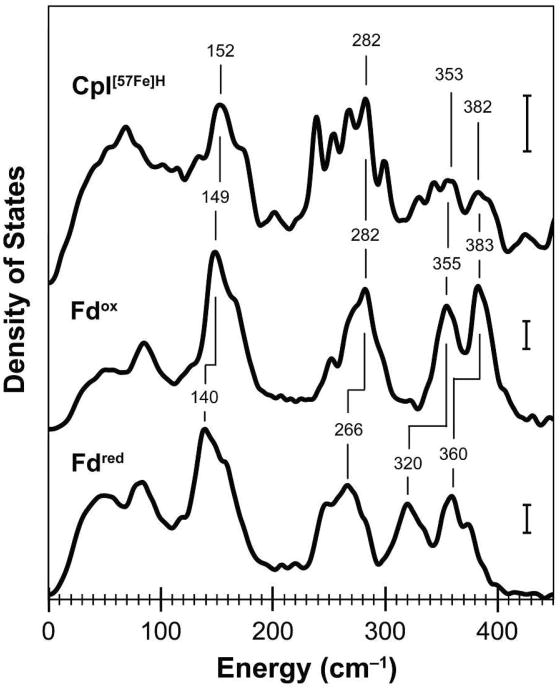
In Figure 2, the 57Fe PVDOS from 0–450 cm–1 for CpI[57Fe]H is compared to previously reported data for an 57Fe-enriched mutant of the Pyrococcus furiosus [4Fe–4S] ferredoxin (the Pf D14C Fd variant).25 Within this region, the main features include large-scale protein movements and Fe–S cluster torsional modes below 100 cm–1, strong Fe–S bending and breathing modes from 100–200 cm–1, and bands characteristic of Fe–S stretching modes (ν(FeS)) from 200–400 cm–1. The similar peak positions and line shapes between the 57Fe density of states for the CpI[57Fe]H hydrogenase and the oxidized Pf D14C Fd indicate the presence of an 57Fe-labeled [4Fe–4S]2+ cluster in the hydrogenase. Among the useful indicators, oxidized [4Fe–4S]2+ clusters have ν(FeS) stretches near 282, 355, and 383 cm–1,25 and similar strong bands were observed for the CpI[57Fe]H hydrogenase. In addition, the CpI spectrum shows an intense Fe bending mode around 150 cm–1 as well as ν(FeS) stretching modes between 230–290 cm–1 that are noticeably absent in a previously reported NRVS study of the Rhodobacter capsulatus [2Fe–2S] ferredoxin,45 which supports our assignment of these peaks to an 57Fe-enriched [4Fe–4S] cluster.
Figure 2.
NRVS data within the 0–450 cm–1 range of energies for the CpI[57Fe]H hydrogenase and the Pf D14C [4Fe–4S] ferredoxin (Fd). The 57Fe vibrational density of states are shown for CpI[57Fe]H (59 scans; top curve) and previously published data by Mitra and coworkers for both oxidized Fd (middle curve) and the reduced Fd (bottom curve).25 Energies for selected Fe–S vibrational modes are indicated, and vertical scale bars represent 30 cm for the 57Fe PVDOS.
The higher energy region of the NRVS spectrum is more chemically interesting since this region is where we expect vibrational modes from Fe–CO and Fe–CN moieties that should arise from the [FeFe] hydrogenase [2Fe]H subcluster. Figure 3 presents the spectrum for CpI[57Fe]H from 350–750 cm–1 and compares it to two NRVS spectra from the [57Fe2(S2C3H6)(CN)2(CO)4]2– model compound that has either CN– or 13CN– ligands. Our Fe–CO and Fe–CN mode assignments for the CpI[57Fe]H hydrogenase are based on prior NRVS of the iron-sulfur cluster-free hydrogenase (Hmd)24 as well as our NRVS results for the model complex. In order to assign the Fe–CO and Fe–CN vibrations for the model compound, we performed both DFT calculations and FTIR spectroscopy in the far-IR region (300–700 cm–1) and the mid-IR region (1700–2200 cm–1), and a more detailed analysis of the model complex is provided in the Supporting Information.
Figure 3.
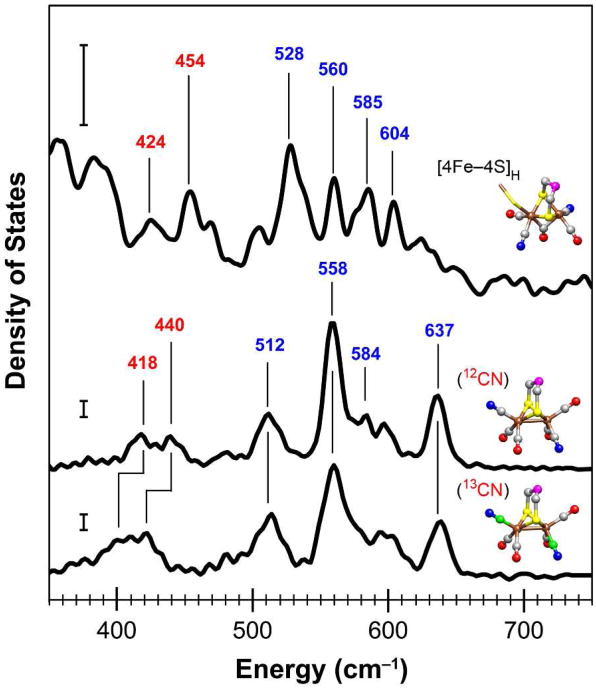
NRVS data within the 350–750 cm–1 range of energies for the CpI[57Fe]H hydrogenase and the [57Fe2(S2C3H6)(CN)2(CO)4]2– model complex. The 57Fe vibrational density of states are shown for CpI[57Fe]H (59 scans; top curve) as well as the model compound containing either CN– ligands (5 scans; middle curve) or isotopically labeled 13CN– ligands (5 scans; bottom curve). Selected energies are indicated for Fe–CN modes (red font type) and Fe–CO modes (blue font type). The vertical scale bars represent 30 cm for the 57Fe PVDOS. Structures are shown for the [2Fe]H subcluster (PDB ID code 3C8Y) and the model complex (CCDC code ODEDUW) with the following color scheme: Fe (brown), S (yellow), C (gray), 13C (green), O (red), N (blue), and unknown (magenta). Molecular graphics were generated using the UCSF Chimera package.56
In the NRVS spectrum for the [57Fe2(S2C3H6)(CN)2(CO)4]2– model compound, two peaks at 418 and 440 cm–1 are shown within the 375–500 cm–1 region, which shift downward ∼18 cm–1 upon 13CN substitution. We assign these features as mainly the combination of Fe–CN stretching modes (ν(Fe–CN)) and Fe–CN bending modes (δ(Fe– CN)). In the 500–700 cm–1 region, the model compound exhibits a complex density of states pattern that we assign to both Fe–CO stretching modes (ν(Fe–CO)) and Fe–CO bending modes (δ(Fe–CO)), which are generated by two pairs of cis-CO ligands in inequivalent geometries. Similar patterns of ν(Fe–CO) stretches and δ(Fe–CO) bends were observed from 450–700 cm–1 for the Hmd hydrogenase as well as a cis-(CO)2 ligated model complex, Fe(S2C2H4)(CO)2(PMe3)2.24 It is known that there are significant differences in the dynamics of Fe–CO and Fe–CN moieties. For example, strong back-bonding in the Fe–CO moiety favors a linear Fe–C–O geometry and pushes the δ(Fe– CO) bends to higher frequencies than the ν(Fe–CO) stretches.46 Our DFT calculations for the model complex support this finding (Supporting Information). Specifically, the 512 and 558 cm–1 ν(Fe–CO) bands are mainly from symmetric and asymmetric stretching motions, respectively, while the weak 584 cm–1 shoulder and strong 637 cm–1 band are mainly from in-plane and out-of-plane δ(Fe–CO) bending motions, respectively.
Turning to the 57Fe PVDOS for the CpI[57Fe]H hydrogenase (Figure 3), a complex pattern of at least 9 bands can be seen from 390–700 cm–1, a region where no significant intensities are observed for either [4Fe–4S] or [2Fe–2S] clusters.25,45 In the higher energy portion (500–700 cm–1), strong bands are shown at 528, 560, 585, and 604 cm–1. Based on our analysis for the model compound, we assign these four strong bands as Fe–CO vibrations. We further propose the assignment of the 528 and 560 cm–1 peaks as mainly ν(Fe–CO) stretches, and the 585 and 604 cm–1 pair of peaks as mainly δ(Fe–CO) bends. Contrary to the Fe–CO moieties, weak back-bonding interactions exist with the Fe–CN moieties, resulting in a more nuanced situation in which either Fe–CN stretching or bending motions can be at the higher frequency, and in which there is likely extensive mixing.47 Therefore, in the lower energy portion of the 390–700 cm–1 region for the CpI[57Fe] hydrogenase, we assign the two bands at 425 and 454 cm–1 as either ν(Fe–CN) stretches or δ(Fe–CN) bends. Relatively weaker bands are present from 460–510 cm–1 as well as 610–670 cm–1, and additional studies will help further characterize these possible Fe–CO and Fe–CN features.
EPR Spectroscopy
The iron analysis and NRVS data clearly indicate that the CpI[57Fe]H hydrogenase has two 57Fe-enriched Fe–S clusters, one of which is the [2Fe]H subcluster. A key question is whether the second 57Fe–S cluster is the [4Fe–4S]H subcluster or one of the F-domain accessory clusters. EPR spectroscopy offers a route to characterize the 57Fe content within the H-cluster by quantifying the amount of broadening associated with the Hox signal. It has been shown that this broadening arises from 57Fe hyperfine interactions and increases with the number of 57Fe nuclei in the six-iron H-cluster.48 Considering that we already established the presence of an 57Fe-labeled [2Fe]H subcluster from NRVS, we then used continuous-wave EPR to determine if the [4Fe–4S]H associated with the CpI[57Fe]H hydrogenase was the second Fe–S cluster labeled with 57Fe nuclei.
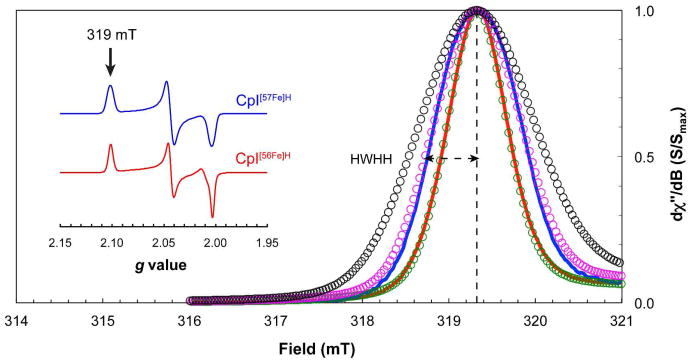
Since the anaerobically purified CpI[57Fe]H was prepared under a H2 atmosphere (97% N2, 3% H2), the NRVS sample was a mixture of both oxidized and H2-reduced enzyme (Supporting Information, Figure S5). Following the NRVS experiments, the CpI[57Fe]H sample was thawed, diluted, and treated with excess thionin acetate under a 100% N2 atmosphere. This generated a completely oxidized hydrogenase sample with an S = ½ rhombic signal (Figure 4, inset; g1 = 2.100, g2 = 2.042, g3 = 2.002) nearly identical to the Hox spectrum for CpI activated in vitro using natural abundance Fe (CpI[56Fe]H), and for wild-type CpI produced in the native organism.49,50 The main difference is that, as expected, the lines in the Hox spectrum for the CpI[57Fe]H hydrogenase were broader as a result of the hyperfine couplings that derives from the integrated 57Fe nuclei. Specifically, the half-width at the half-height (HWHH) at the g1 tensor is 0.58 mT (Figure 4). Based on previous EPR spectroscopy of the Desulfovibrio desulfuricans [FeFe] hydrogenase,48 this amount of broadening indicates that the CpI[57Fe]H contains an H-cluster with approximately two 57Fe nuclei. These results suggests that CpI[57Fe]H hydrogenase has an H-cluster in which the [2Fe]H subcluster is enriched with 57Fe, whereas the [4Fe–4S]H subcluster contains natural abundance 56Fe.
Figure 4.
X-band CW EPR spectroscopy of the CpI hydrogenase. Spectra near the g1 tensor of the Hox signal (g1 = 2.101; ∼319 mT) were measured at 50 K and 50 μW for CpI[56Fe]H (red curve) and CpI[57Fe]H (blue curve) following oxidation with excess thionin acetate. Simulations for the Hox signal around 319 mT include hyperfine interactions from zero 57Fe nuclei (green circles), two 57Fe nuclei (magenta circles), and six 57Fe nuclei (black circles), as determined using the A1ox and A2ox hyperfine values from the D. desulfuricans HydA hydrogenase.48 Dashed lines are shown for the half-width at the half-height (HWHH) for the CpI[57Fe]H sample. The spectra shown in the inset represent the full Hox signal for each hydrogenase sample as measured at 15 K and 62 μW.
Discussion
Enriching proteins with rare isotopes such as 13C and 15N commonly involves growing a microorganism on growth medium supplemented with a substrate containing the particular isotopic element.48 For the case of labeling Fe–S clusters, exogenous 57Fe can be added to the cell culture, and the host cell iron-sulfur cluster machinery will ubiquitously assemble 57Fe–S clusters onto proteins that coordinate these metal centers. Nevertheless, global enrichment of a single protein is often undesirable if numerous isotopic nuclei will lead to confounding experimental results. Targeted isotopic enrichment is far more challenging and, ultimately, it requires a method in which there is precise control over the biochemical process that involves the incorporation of the isotopic element.
In vitro systems offer unique opportunities to label proteins with a variety of non-natural or isotopic elements. For example, cell-free protein synthesis platforms can be used to incorporate amino acid isotopologs,51,52 or even non-natural amino acids,53 either globally or at selected sites within a polypeptide chain. Yet, unlike protein translation with its generalized machinery, post-translational processes commonly require specialized accessory proteins. Incorporating isotopically enriched metal clusters and other cofactors during a particular post-translational reaction likely necessitates an in vitro system in which the biochemical pathway has been reconstituted. Interestingly, we have developed a cell-free system in which the [2Fe]H subcluster can be synthesized and transferred to the hydrogenase in a reaction sequence that is distinctly separated from protein translation.15
As we have shown in this study, precise control of both the maturases and the low molecular weight substrates allowed us to design a cell-free reaction mixture that synthesized an 57Fe-labeled [2Fe]H subcluster onto a hydrogenase apoprotein containing 56Fe-labeled Fe–S clusters. We then utilized two spectroscopic techniques to probe and characterize the [FeFe] hydrogenase, and our results confirm the incorporation of 57Fe into the enzyme's active site metal center. Specifically, we used NRVS to assign various normal modes for the hydrogenase by comparing the 57Fe vibrational density of states to those of a [4Fe–4S] ferredoxin and a [2Fe]H subcluster model complex, revealing 57Fe– CO and 57Fe–CN moieties associated with the CpI enzyme (Figure 3) and confirming the cell-free assembly of an 57Fe-enriched [2Fe]H catalytic subunit. By complementing the NRVS with EPR spectroscopy, we observed distinct broadening of the Hox EPR signal for the CpI[57Fe]H enzyme (Figure 4), which further exemplifies that the hydrogenase contains an 57Fe-labeled [2Fe]H subcluster.
The region in which we would expect Fe–CO and Fe–CN vibrational modes (400– 700 cm–1) is distinct from the region with Fe–S vibrational modes. This fortuitous circumstance simplifies the characterization of modes that come from the nonprotein ligands of the H-cluster. The clear observation of Fe–CO and Fe-CN normal modes in this work demonstrates that NRVS is another capable approach to studying [FeFe] hydrogenases. More interestingly, our results establish precedence for future NRVS studies to probe bound hydrides (i.e. Fe–H and Fe–2H modes). This will be crucial for understanding the mechanism of hydrogen activation and for developing synthetic catalysts that function similar to the H-cluster. In addition, spectral simulations along with isotopic labeling of the [2Fe]H subcluster using 13C, 15N, 18O, and 36S could help definitively assign the Fe–S, Fe–CO, and Fe–CN normal modes. The in vitro system used in this work has proven effective for the synthesis of the [2Fe]H subcluster with isotopically labeled substituents.15 In our previous study, we demonstrated the cell-free production of 13CO and 13CN–/13C15N– ligands from various tyrosine isotopologs, conclusively showing that all five CO and CN– adducts derive from this amino acid.
Regarding the Fe–CO modes, CO behaves like a single, more massive atom, resulting in larger amplitudes in Fe–CO vibrations, and thus bands at higher energies relative to Fe–CN vibrations. Our general characterization of the Fe–CO modes for the CpI[57Fe]H hydrogenase and the model complex is supported by our DFT calculations for the model compound as well as from previous NRVS of the Hmd hydrogenase where multiple peaks from 480–700 cm–1 were attributed to Fe–CO modes.24
With respect to the [2Fe]H subcluster CN– ligands, we have assigned peaks from 410– 450 cm–1 to Fe–CN vibrational modes. In reasonable agreement with our assignments, and based on isotope shifts along with normal mode analysis for the model complex, Fiedler and Brunold assigned a resonance Raman band at 390 cm–1 to a δ(Fe–CN) bending mode, and bands at 432 and 450 cm–1 to ν(Fe–CN) stretching modes.54 We also note that the [2Fe]H subcluster Fe–CN modes have modestly lower frequencies than those observed in spectroscopic studies of heme-related Fe–CN moieties. For example, ferrous cytochrome a3 oxidases have FeII–CN modes with energies from 470–510 cm–1.47 Conversely, the [57Fe2(S2C3H6)(CN)2(CO)4]2– model compound, which is an FeI/FeI complex, has FeI–CN modes at lower energies from 410–450 cm–1. Regarding the electronic structures of the [2Fe]H subcluster, conflicting results have been reported. Mössbauer studies have indicated the electronic structures to be FepII/FedIII (Hox) and FepII/FedII (Hred).55 EPR and FTIR spectroscopy have pointed to structures of FepI/FedII (Hox) and FepI/FedI (Hred).48 In this study, the NRVS spectrum for the CpI[57Fe]H hydrogenase shows peaks at 425 and 454 cm–1, which are likely attributed to FeI–CN modes, supporting the FepI/FedII (Hox) and FepI/FedI (Hred) electronic structures assigned by Silakov and coworkers.
Analysis of the density of states within the Fe–S region (Figure 2) suggests that the CpI[57Fe]H hydrogenase has an additional 57Fe–S cluster besides the [2Fe]H subcluster that was assembled during the in vitro activation reaction. The three peaks at 282, 353, and 382 cm–1 together with NRVS data for the oxidized ferredoxin indicate the presence of a [4Fe–4S]2+ cluster. We also compared the relative amounts of total NRVS intensity within the Fe–S and the Fe–CO/Fe–CN regions (Supporting Information, Table S2). Roughly twice as much intensity derives from 57Fe–S modes for CpI[57Fe]H relative to that from the 57Fe–CO/57Fe–CN modes. This result, together with the difference in the iron content between the apoprotein (∼13 Fe per CpI) and the holoenzyme (∼18 Fe per CpI), further support our assignment of an 57Fe–enriched [4Fe–4S] cluster associated with the CpI[57Fe]H hydrogenase.
We speculated that the 57Fe-labeled [4Fe–4S] cluster might be the companion [4Fe– 4S]H subcluster rather than one of the three accessory [4Fe–4S] cluster. As previously stated, EPR spectroscopy provides a way to probe as well as to quantitate protein-bound 57Fe atoms based on the hyperfine interactions resulting from these magnetic nuclei. When the CpI hydrogenase is in the oxidized and active state, the H-cluster (Hox) is EPR-active, whereas the accessory Fe–S clusters are EPR-silent. This fortunate situation means that the entire amount of line broadening associated with the Hox signal that we observed results only from 57Fe nuclei within the H-cluster. Based on previous results by Silakov and coworkers, the amount of broadening for CpI[57Fe]H (HWHH of 0.58 mT) suggests that the H-cluster has two 57Fe nuclei rather than the expected six if the H-cluster was completely enriched.48 This finding clearly indicates that only the [2Fe]H subcluster is 57Fe-labeled, whereas the [4Fe–4S]H subcluster is not enriched with 57Fe. It is noteworthy that in this study, the maturation reactions for producing the CpI[57Fe]H samples contained 57Fe-enriched Hyd maturases as well as exogenously added 57Fe. Therefore, the [2Fe]H subcluster is presumably homogeneously enriched with 57Fe, and all 56Fe-enriched Fe–S clusters associated with CpI[57Fe]H were likely assembled in vivo during heterologous expression of the hydrogenase apoprotein. It follows that the [4Fe– 4S]H subcluster was not synthesized in vitro during hydrogenase maturation.
By illustrating that the CpI[57Fe]H hydrogenase retained an 56Fe-labeled [4Fe–4S]H during the 57Fe-enriched cell-free maturation reaction, we provide unequivocal evidence that supports a stepwise maturation sequence postulated by Mulder and coworkers.18 In their study, X-ray crystallography of the Chlamydomonas reinhardtii HydA1 hydrogenase revealed a cationic channel associated with the [4Fe–4S]H-containing apoprotein. The authors suggested that HydF transfers the [2Fe]H subcluster through the channel to the H-domain, which is followed by a conformational change that leads to an active hydrogenase devoid of the channel.
It could be expected that the enclosed nature of the H-domain would necessitate the assembly of the [4Fe–4S]H subcluster before the activation reaction, perhaps prior to or during protein folding. In contrast to the H-cluster, the N-terminal domain ancillary clusters are more accessible to the intracellular environment. It seems possible that one of the iron centers is fairly unstable, and thus labile before or during the purification process. The similarities and differences amongst the three F-domain accessory [4Fe–4S] clusters are not well characterized, and NRVS of multiple protein variants would be required to elucidate which [4Fe–4S] cluster in labeled with 57Fe.
In summary, we have used NRVS to measure the 57Fe-based vibrational density of states of the [FeFe] hydrogenase from C. pasteurianum as well as a model complex resembling the [2Fe]H subcluster. This report not only presents the first NRVS of [FeFe] hydrogenases, but, to the best of our knowledge, it also provides the first example of isotopically enriching a specific protein-bound metal cluster amongst several other clusters for NRVS. Since the CpI enzyme contains multiple Fe–S centers, selective labeling of the [2Fe]H subcluster with 57Fe substantially improved the 57Fe PVDOS intensities within the Fe–CO/Fe–CN region, thus facilitating a more facile characterization of these relatively weak vibrational modes. Such increased signal intensity within the higher energy region will be crucial for probing another interesting H-cluster feature: the Fe–H vibration. Finally, the combination of NRVS with EPR spectroscopy allowed us to demonstrate the separate insertion of the [2Fe]H and [4Fe– 4S]H subclusters. In this way, we have shown how cell-free biochemistry combined with advanced spectroscopic techniques is a powerful combination capable of answering fundamental questions about the biological chemistry of a complex metalloenzyme and its unique maturation pathway.
Supplementary Material
Acknowledgments
We thank Thomas Rauchfuss and Saeed Kamali for insightful discussions pertaining to the manuscript.
Abbreviations
- NRVS
nuclear resonance vibrational spectroscopy
- PVDOS
partial vibrational density of states
- EPR
electron paramagnetic resonance
- IR
infrared
- RR
resonance Raman
- DFT
density functional theory
- Pf D14C Fd
Pyrococcus furiosus [4Fe–4S] ferredoxin D14C variant
- CpI
Clostridium pasteurianum HydA [FeFe] hydrogenase
- CO
carbon monoxide
- CN
cyanide
- DTMX
H-cluster bridging ligand that is either dithiomethylamine (DTMA) or dithiomethylether (DTMO)
- GTP
guanosine-5′-triphosphate
- DTT
dithiothreitol
Footnotes
This work was funded by the National Institutes of Health (GM-65440, SPC); the U.S. Department of Energy, Office of Biological and Environmental Research; and the U.S. Department of Energy, Office of Basic Energy Sciences (DE-FG02-09ER46632, JRS & SPC). Research at SPring-8 was funded by JASRI, and research at the APS was supported by the U.S. DOE, Office of Basic Energy Sciences.
Supporting Information – Synthesis details, DFT calculations, and IR spectroscopy for the (Et4N)2[57Fe2(S2C3H6)(CN)2(CO)4] model compound; figure comparing the full NRVS spectra for two CpI[57Fe]H samples, two model compounds (12CN-labeled and 13CN-labeled), and two Pf D14C Fd samples (oxidized and reduced); table with percentages of total 57Fe intensity within the Fe–S and Fe–CO/Fe–CN regions; EPR spectra of 57Fe-enriched CpI (pre- and post-NRVS) and 56Fe-enriched CpI. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Ghirardi ML, Dubini A, Yu J, Maness PC. Photobiological hydrogen-producing systems. Chem Soc Rev. 2009;38:52–61. doi: 10.1039/b718939g. [DOI] [PubMed] [Google Scholar]
- 2.Vignais PM, Billoud B. Occurrence, classification, and biological function of hydrogenases: an overview. Chem Rev. 2007;107:4206–4272. doi: 10.1021/cr050196r. [DOI] [PubMed] [Google Scholar]
- 3.Martin W, Müller M. The hydrogen hypothesis for the first eukaryote. Nature. 1998;392:37–41. doi: 10.1038/32096. [DOI] [PubMed] [Google Scholar]
- 4.Kruse O, Hankamer B. Microalgal hydrogen production. Curr Opin Biotechnol. 2010;21:1–6. doi: 10.1016/j.copbio.2010.03.012. [DOI] [PubMed] [Google Scholar]
- 5.Zhang YHP, Evans BR, Mielenz JR, Hopkins RC, Adams MWW. High-yield hydrogen production from starch and water by a synthetic enzymatic pathway. PLoS ONE. 2007;2:e456. doi: 10.1371/journal.pone.0000456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hambourger M, Gervaldo M, Svedruzic D, King PW, Gust D, Ghirardi M, Moore AL, Moore TA. [FeFe]-hydrogenase-catalyzed H2 production in a photoelectrochemical biofuel cell. J Am Chem Soc. 2008;130:2015–2022. doi: 10.1021/ja077691k. [DOI] [PubMed] [Google Scholar]
- 7.Iwuchukwu IJ, Vaughn M, Myers N, O'Neill H, Frymier P, Bruce BD. Self-organized photosynthetic nanoparticle for cell-free hydrogen production. Nat Nanotechnol. 2010;5:73–79. doi: 10.1038/nnano.2009.315. [DOI] [PubMed] [Google Scholar]
- 8.Le Goff A, Artero V, Jousselme B, Tran PD, Guillet N, Métayé R, Fihri A, Palacin S, Fontecave M. From hydrogenases to noble metal-free catalytic nanomaterials for H2 production and uptake. Science. 2009;326:1384–1387. doi: 10.1126/science.1179773. [DOI] [PubMed] [Google Scholar]
- 9.Tard C, Pickett CJ. Structural and functional analogues of the active sites of the [Fe]-, [NiFe]-, and [FeFe]-hydrogenases. Chem Rev. 2009;109:2245–2274. doi: 10.1021/cr800542q. [DOI] [PubMed] [Google Scholar]
- 10.Nicolet Y, Piras C, Legrand P, Hatchikian CE, Fontecilla-Camps JC. Desulfovibrio desulfuricans iron hydrogenase: the structure shows unusual coordination to an active site Fe binuclear center. Structure. 1999;7:13–23. doi: 10.1016/s0969-2126(99)80005-7. [DOI] [PubMed] [Google Scholar]
- 11.Peters JW, Lanzilotta WN, Lemon BJ, Seefeldt LC. X-ray crystal structure of the Fe-only hydrogenase (CpI) from Clostridium pasteurianum to 1.8 angstrom resolution. Science. 1998;282:1853–1858. doi: 10.1126/science.282.5395.1853. [DOI] [PubMed] [Google Scholar]
- 12.Mulder DW, Shepard EM, Meuser JE, Joshi N, King PW, Posewitz MC, Broderick JB, Peters JW. Insights into [FeFe]-hydrogenase structure, mechanism, and maturation. Structure. 2011;19:1038–1052. doi: 10.1016/j.str.2011.06.008. [DOI] [PubMed] [Google Scholar]
- 13.Posewitz MC, King PW, Smolinski SL, Zhang L, Seibert M, Ghirardi ML. Discovery of two novel radical S-adenosylmethionine proteins required for the assembly of an active [Fe] hydrogenase. J BiolChem. 2004;279:25711–25720. doi: 10.1074/jbc.M403206200. [DOI] [PubMed] [Google Scholar]
- 14.Kuchenreuther JM, Britt RD, Swartz JR. New insights into [FeFe] hydrogenase activation and maturase function. PLoS ONE. 2012;7:e45850. doi: 10.1371/journal.pone.0045850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuchenreuther JM, George SJ, Grady-Smith CS, Cramer SP, Swartz JR. Cell-free H-cluster synthesis and [FeFe] hydrogenase activation: all five CO and CN- ligands derive from tyrosine. PLoS ONE. 2011;6:e20346. doi: 10.1371/journal.pone.0020346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuchenreuther JM, Stapleton JA, Swartz JR. Tyrosine, cysteine, and S-adenosyl methionine stimulate in vitro [FeFe] hydrogenase activation. PLoS ONE. 2009;4:e7565. doi: 10.1371/journal.pone.0007565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McGlynn SE, Shepard EM, Winslow MA, Naumov AV, Duschene KS, Posewitz MC, Broderick WE, Broderick JB, Peters JW. HydF as a scaffold protein in [FeFe] hydrogenase H-cluster biosynthesis. FEBS Lett. 2008;582:2183–2187. doi: 10.1016/j.febslet.2008.04.063. [DOI] [PubMed] [Google Scholar]
- 18.Mulder DW, Boyd ES, Sarma R, Lange RK, Endrizzi JA, Broderick JB, Peters JW. Stepwise [FeFe]-hydrogenase H-cluster assembly revealed in the structure of HydA(DeltaEFG) Nature. 2010;465:248–252. doi: 10.1038/nature08993. [DOI] [PubMed] [Google Scholar]
- 19.Chen Z, Lemon BJ, Huang S, Swartz DJ, Peters JW, Bagley KA. Infrared studies of the CO-inhibited form of the Fe-only hydrogenase from Clostridium pasteurianum I: examination of its light sensitivity at cryogenic temperatures. Biochemistry. 2002;41:2036–2043. doi: 10.1021/bi011510o. [DOI] [PubMed] [Google Scholar]
- 20.Lubitz W, Reijerse E, van Gastel M. [NiFe] and [FeFe] hydrogenases studied by advanced magnetic resonance techniques. Chem Rev. 2007;107:4331–4365. doi: 10.1021/cr050186q. [DOI] [PubMed] [Google Scholar]
- 21.Roseboom W, de Lacey AL, Fernandez VM, Hatchikian EC, Albracht SPJ. The active site of the [FeFe]-hydrogenase from Desulfovibrio desulfuricans. II. Redox properties, light sensitivity and CO-ligand exchange as observed by infrared spectroscopy. J Biol Inorg Chem. 2006;11:102–118. doi: 10.1007/s00775-005-0040-2. [DOI] [PubMed] [Google Scholar]
- 22.Bruschi M, Greco C, Kaukonen M, Fantucci P, Ryde U, De Gioia L. Influence of the [2Fe]H subcluster environment on the properties of key intermediates in the catalytic cycle of [FeFe] hydrogenases: hints for the rational design of synthetic catalysts. Angew Chem Int Ed. 2009;48:3503–3506. doi: 10.1002/anie.200900494. [DOI] [PubMed] [Google Scholar]
- 23.Bingham AS, Smith PR, Swartz JR. Evolution of an [FeFe] hydrogenase with decreased oxygen sensitivity. Int J Hydrogen Energy. 2012;37:2965–2976. [Google Scholar]
- 24.Guo Y, Wang H, Xiao Y, Vogt S, Thauer RK, Shima S, Volkers PI, Rauchfuss TB, Pelmenschikov V, Case DA, Alp EE, Sturhahn W, Yoda Y, Cramer SP. Characterization of the Fe site in iron-sulfur cluster-free hydrogenase (Hmd) and of a model compound via nuclear resonance vibrational spectroscopy (NRVS) Inorg Chem. 2008;47:3969–3977. doi: 10.1021/ic701251j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mitra D, Pelmenschikov V, Guo Y, Case DA, Wang H, Dong W, Tan ML, Ichiye T, Jenney FE, Adams MWW, Yoda Y, Zhao J, Cramer SP. Dynamics of the [4Fe-4S] cluster in Pyrococcus furiosus D14C ferredoxin via nuclear resonance vibrational and resonance Raman spectroscopies, force field simulations, and density functional theory calculations. Biochemistry. 2011;50:5220–5235. doi: 10.1021/bi200046p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xiao Y, Fisher K, Smith MC, Newton WE, Case DA, George SJ, Wang H, Sturhahn W, Alp EE, Zhao J, Yoda Y, Cramer SP. How nitrogenase shakes--initial information about P-cluster and FeMo-cofactor normal modes from nuclear resonance vibrational spectroscopy (NRVS) J Am Chem Soc. 2006;128:7608–7612. doi: 10.1021/ja0603655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alp E, Sturhahn W, Toellner T, Zhao J, Hu M, Brown D. Vibrational Dynamics Studies by Nuclear Resonant Inelastic X-Ray Scattering. Hyperfine Interact. 2002;144-145:3–20. [Google Scholar]
- 28.Petrenko T, Sturhahn W, Neese F. First-principles calculation of nuclear resonance vibrational spectra. Hyperfine Interact. 2007;175:165–174. [Google Scholar]
- 29.Scheidt WR, Durbin SM, Sage JT. Nuclear resonance vibrational spectroscopy--NRVS. J Inorg Biochem. 2005;99:60–71. doi: 10.1016/j.jinorgbio.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 30.Sturhahn W. Nuclear resonant spectroscopy. J Phys: Condens Matter. 2004;16:S497. [Google Scholar]
- 31.Nakamoto K. Infrared and Raman Spectra of Inorganic and Coordination Compounds∷ Theory and applications in inorganic chemistry (Part A) 5th. Wiley-Interscience; 1997. [Google Scholar]
- 32.Leu BM, Zgierski MZ, Wyllie GRA, Scheidt WR, Sturhahn W, Alp EE, Durbin SM, Sage JT. Quantitative Vibrational Dynamics of Iron in Nitrosyl Porphyrins. J Am Chem Soc. 2004;126:4211–4227. doi: 10.1021/ja038526h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sage J, Paxson C, Wyllie GRA, Sturhahn W, Durbin SM, Champion PM, Alp EE, Scheidt WR. Nuclear resonance vibrational spectroscopy of a protein active-site mimic. J Phys: Condens Matter. 2001;13:7707–7722. [Google Scholar]
- 34.Sturhahn W, Toellner TS, Alp EE, Zhang X, Ando M, Yoda Y, Kikuta S, Seto M, Kimball CW, Dabrowski B. Phonon density of states measured by inelastic nuclear resonant scattering. Phys Rev Lett. 1995;74:3832–3835. doi: 10.1103/PhysRevLett.74.3832. [DOI] [PubMed] [Google Scholar]
- 35.Kuchenreuther JM, Grady-Smith CS, Bingham AS, George SJ, Cramer SP, Swartz JR. High-yield expression of heterologous [FeFe] hydrogenases in Escherichia coli. PLoS ONE. 2010;5:e15491. doi: 10.1371/journal.pone.0015491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fish WW. Rapid colorimetric micromethod for the quantitation of complexed iron in biological samples. Method Enzymol. 1988;158:357–364. doi: 10.1016/0076-6879(88)58067-9. [DOI] [PubMed] [Google Scholar]
- 37.Czech I, Silakov A, Lubitz W, Happe T. The [FeFe]-hydrogenase maturase HydF from Clostridium acetobutylicum contains a CO and CN- ligated iron cofactor. FEBS Lett. 2009;584:638–642. doi: 10.1016/j.febslet.2009.12.016. [DOI] [PubMed] [Google Scholar]
- 38.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 39.Adams MW, Eccleston E, Howard JB. Iron-sulfur clusters of hydrogenase I and hydrogenase II of Clostridium pasteurianum. Proc Natl Acad Sci USA. 1989;86:4932–4936. doi: 10.1073/pnas.86.13.4932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sturhahn W. CONUSS and PHOENIX: evaluation of nuclear resonant scattering data. Hyperfine Interactions. 2000;125:149–172. [Google Scholar]
- 41.Yoda Y, Yabashi M, Izumi K, Zhang XW, Kishimoto S, Kitao S, Seto M, Mitsui T, Harami T, Imai Y, Kikuta S. Nuclear resonant scattering beamline at SPring-8. Nuclear Instruments & Methods in Physics Research Section A-Accelerators Spectrometers Detectors & Associated Equipment. 2001;467:715–718. [Google Scholar]
- 42.Toellner T. Monochromatization of synchrotron radiation for nuclear resonant scattering experiments. Hyperfine Interact. 2000;125:3–28. [Google Scholar]
- 43.Stoll S, Schweiger A. EasySpin, a comprehensive software package for spectral simulation and analysis in EPR. J Magn Reson. 2006;178:42–55. doi: 10.1016/j.jmr.2005.08.013. [DOI] [PubMed] [Google Scholar]
- 44.Shepard EM, McGlynn SE, Bueling AL, Grady-Smith CS, George SJ, Winslow MA, Cramer SP, Peters JW, Broderick JB. Synthesis of the 2Fe subcluster of the [FeFe]-hydrogenase H cluster on the HydF scaffold. Proc Natl Acad Sci USA. 2010;107:10448–10453. doi: 10.1073/pnas.1001937107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xiao Y, Tan ML, Ichiye T, Wang H, Guo Y, Smith MC, Meyer J, Sturhahn W, Alp EE, Zhao J, Yoda Y, Cramer SP. Dynamics of Rhodobacter capsulatus [2Fe-2S] ferredoxin VI and Aquifex aeolicus ferredoxin 5 via nuclear resonance vibrational spectroscopy (NRVS) and resonance Raman spectroscopy. Biochemistry. 2008;47:6612–6627. doi: 10.1021/bi701433m. [DOI] [PubMed] [Google Scholar]
- 46.Nakamoto K. Infrared and Raman Spectra of Inorganic and Coordination Compounds: Applications in coordination, organometallic, and bioinorganic chemistry (Part B) 5th. Wiley-Interscience; 1997. [Google Scholar]
- 47.Kim Y, Babcock GT, Surerus KK, Fee JA, Dyer RB, Woodruff WH, Oertling WA. Cyanide binding and active site structure in heme-copper oxidases: normal coordinate analysis of iron-cyanide vibrations of a3(2+)CN- complexes of cytochromes ba3 and aa3. Biospectroscopy. 1998;4:1–15. doi: 10.1002/(sici)1520-6343(1998)4:1<1::aid-bspy1>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 48.Silakov A, Reijerse EJ, Albracht SPJ, Hatchikian EC, Lubitz W. The electronic structure of the H-cluster in the [FeFe]-hydrogenase from Desulfovibrio desulfuricans: a Q-band 57Fe-ENDOR and HYSCORE study. J Am Chem Soc. 2007;129:11447–11458. doi: 10.1021/ja072592s. [DOI] [PubMed] [Google Scholar]
- 49.Adams MW. The mechanisms of H2 activation and CO binding by hydrogenase I and hydrogenase II of Clostridium pasteurianum. J Biol Chem. 1987;262:15054–15061. [PubMed] [Google Scholar]
- 50.Chen JS, Mortenson LE. In: In Iron and Copper Proteins. Yasunobu KT, Mower HF, Hayaishi O, editors. Plenum Press; New York: 1976. pp. 68–82. [Google Scholar]
- 51.Boyer ME, Stapleton JA, Kuchenreuther JM, Wang CW, Swartz JR. Cell-free synthesis and maturation of [FeFe] hydrogenases. Biotechnol Bioeng. 2008;99:59–67. doi: 10.1002/bit.21511. [DOI] [PubMed] [Google Scholar]
- 52.Boyer ME, Wang CW, Swartz JR. Simultaneous expression and maturation of the iron-sulfur protein ferredoxin in a cell-free system. Biotechnol Bioeng. 2006;94:128–138. doi: 10.1002/bit.20830. [DOI] [PubMed] [Google Scholar]
- 53.Patel KG, Ng PP, Kuo CC, Levy S, Levy R, Swartz JR. Cell-free production of Gaussia princeps luciferase--antibody fragment bioconjugates for ex vivo detection of tumor cells. Biochem Biophys Res Commun. 2009;390:971–976. doi: 10.1016/j.bbrc.2009.10.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fiedler AT, Brunold TC. Combined spectroscopic/computational study of binuclear Fe(I)-Fe(I) complexes: implications for the fully-reduced active-site cluster of Fe-only hydrogenases. Inorg Chem. 2005;44:1794–1809. doi: 10.1021/ic048739n. [DOI] [PubMed] [Google Scholar]
- 55.Popescu C, Münck E. Electronic structure of the H cluster in [Fe]-hydrogenases. J Am Chem Soc. 1999;121:7877–7884. [Google Scholar]
- 56.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.