Abstract
Objective
To isolate, evaluate and characterize potential antibiotic producing actinomycetes from water and sediments of Lake Tana, Ethiopia.
Methods
A total of 31 strains of actinomycetes were isolated and tested against Gram positive and Gram negative bacterial strains by primary screening. In the primary screening, 11 promising isolates were identified and subjected to solid state and submerged state fermentation methods to produce crude extracts. The fermented biomass was extracted by organic solvent extraction method and tested against bacterial strains by disc and agar well diffusion methods. The isolates were characterized by using morphological, physiological and biochemical methods.
Results
The result obtained from agar well diffusion method was better than disc diffusion method. The crude extract showed higher inhibition zone against Gram positive bacteria than Gram negative bacteria. One-way analysis of variance confirmed most of the crude extracts were statistically significant at 95% confidence interval. The minimum inhibitory concentration and minimum bactericidal concentration of crude extracts were 1.65 mg/mL and 3.30 mg/mL against Staphylococcus aureus, and 1.84 mg/mL and 3.80 mg/mL against Escherichia coli respectively. The growth of aerial and substrate mycelium varied in different culture media used. Most of the isolates were able to hydrolysis starch and urea; able to survive at 5% concentration of sodium chloride; optimum temperature for their growth was 30 °C.
Conclusions
The results of the present study revealed that freshwater actinomycetes of Lake Tana appear to have immense potential as a source of antibacterial compounds.
Keywords: Actinomycetes, Solid state fermentation, Submerged state fermentation, Disc diffusion method, Agar well diffusion method, Crude extracts, Antibiotics, Antibacterial activity
1. Introduction
Actinomycetes are filamentous, antibiotics producing bacteria. They are found in freshwater and marine water habitats[1]–[3]. The dominant actinomycetes Micromonospora can be isolated from aquatic habitats such as streams, rivers, lake mud, river sediments, beach sands, sponge and marine sediments[4],[5]. Several novel bioactive compounds were discovered from aquatic actinomycetes, for example rifamycin from Micromonospora[6]; salinosporamide-A, an anticancer metabolite from a Salinispora strain[7]; marinomycins from Marinophilus sp.[8]; abyssomicin-C from Verrucosispora sp. and marinopyrroles from Streptomyces sp[9],[10].
Out of 22 500 biologically active compounds obtained from microbes, 45% are from actinomycetes, 38% from fungi and 17% from other bacteria[11]. Over 5 000 antibiotics have been identified from the culture of Gram positive, Gram negative bacteria and filamentous fungi[12]. Species of Streptomyces, account for more than 70% of the total antibiotic production and Micromonospora was less than one-tenth as many as Streptomyces[13]. Twenty seven actinomycetes were isolated from Mount Everest region soil samples and reported to have antibacterial activity against at least one tested bacteria among the two Gram positive and nine Gram negative bacteria[14]. Narendra Kumar et al. isolated 117 antibiotic producing actinomycetes from non agricultural wasteland alkaline soils and compost rich garden soil in which most of the isolates inhibits Gram negative bacterial growth[15]. Four potential antibacterial actinomycetes were isolated from the aquatic environment[16]. Valli et al. isolated 21 potential actinomycetes from marine environment and reported that all the isolates were promising against at least one tested organisms[17]. Kalyani et al. isolated 20 species from marine soil samples in which three showed significant antimicrobial activity against Staphylococcus aureus and Escherichia coli (E. coli)[18].
Antimicrobial drugs used for prophylactic or therapeutic purposes in human, veterinary and agricultural purposes were favoring the survival and spread of resistant organisms[19]. The appearances of multidrug resistant pathogenic strains caused substantial morbidity and mortality especially among the elderly and immuno-compromised patients. To overcome this situation, there is an interest to improve or discover novel class antibiotics that have different mechanisms of action worldwide[20],[21]. The continuous screening of secondary microbial products produced from potential bacterial taxa was important to discover novel chemicals for the development of new therapeutic agents[22]. Recently, many scientists are searching new antibiotics from different untouched habitats to find out for their productions of antibiotics[23]. In Ethiopia, no significant studies have been conducted so far to isolate and evaluate actinomycetes from different freshwater habitats that could produce useful antibiotics. Therefore, present study is intended to isolate, screen and characterize antibiotic producing actinomycetes from water and sediments of Lake Tana, Ethiopia.
2. Materials and methods
2.1. Sampling area
The samples of water and sediments were collected from Lake Tana, Amhara regional state, Ethiopia. The sampling area was located at a latitude of 12 (12° 0′ 0 N) and a longitude of 37.33 (37° 19′ 60 E). The lake was the source of Blue Nile with a total surface area of 3 600 km2, a volume of 28.00 km3 and an average elevation of 1 911 m above the sea level[24].
2.2. Sample collection
Totally 12 water and 12 sediment samples were collected from Gorgora site of Lake Tana which was located at 65 km North West direction of Gondar town. The water samples were collected in 500 mL sterile screw capped bottles and sufficient space was provided for aeration and thorough mixing. The sediments were collected by sterilized spatula and transferred to wide mouth sterilized bottle. All samples were labeled and transported to Microbiology Laboratory, Department of Biology, University of Gondar and stored at 4 °C for further studies.
2.3. Pretreatment of samples
The samples were subjected to various physical and chemical pretreatment methods in order to facilitate isolation of actinomycetes[25]. The sediment samples were air dried; heated aseptically, which stimulates the isolation of actinomycetes by eliminating most unwanted Gram negative bacteria. Appropriate selective media such as starch casein agar, glycerol yeast extract agar and antifungal antibiotics (amphotericin B) at 25 µg/mL were used for actinomycetes growth promotion and also for prevention of fungal contamination[26],[27].
2.4. Actinomycetes isolation and maintenance
Actinomycetes were isolated by serial dilution method from sediments[28]. Stock solution was prepared by diluting 1 g of sediment in 9 mL of sterile saline water and shaken well by using vortex mixer. From the stock solution, 1 mL was used to prepare the final volume of 10−1, 10−2, 10−3, 10−4 and 10−5 by serial dilution method. Finally, 0.1 mL of suspension from 10−1, 10−2, 10−3, 10−4 and 10−5 were used to spread on starch-casein agar medium aseptically. In water samples, 1 mL of aliquot was taken and spread evenly over the sterilized starch casein agar plates by using L-shaped glass rod. For each sample three plates were used and incubated at 30 °C for 7 to 14 d. The plates were observed periodically for the growth of actinomycetes. The pure colonies were selected, isolated and maintained in starch casein agar slants at 4 °C for subsequent studies.
2.5. Bacterial strains
Pathogenic bacterial strains such as E. coli (ATCC2592), Salmonella typhi (ATCC9289) (S. typhi), Staphylococcus aureus (ATCC2923) (S. aureus), Pseudomonas aeruginosa (ATCC27853) (P. aeruginosa) and Klebsiella pneumonia (ATCC7000603) (K. pneumonia) were obtained from Gondar College of Medical Science, University of Gondar.
2.6. Preparation of 0.5 McFarland standard
McFarland standard was prepared by adding 0.5 mL of 0.048 mol/L BaCl2 (1.17% w/v BaCl2.2H2O) in to 99.5 mL of 0.18 mol/L H2SO4 (1% w/v) with constant stirring. The equal volume of standard solution was distributed into same sized screw capped test tubes. These test tubes were tightly sealed and stored at room temperature to prevent from evaporation and light. Before use, turbidity standard was vigorously agitated by using vortex mixer[29].
2.7. Procedures for inoculum preparation and inoculation
A pure colony of test bacteria was taken by using a sterile wire loop and transferred into test tubes having a sterile nutrient broth. The test tubes were incubated at 37 °C for 24 h until the visible turbidity and density equal to that of 0.5 McFarland standard. After adjusting the turbidity, sterile cotton swab was dipped into the suspension and streaked over the entire surface of the plate medium for three times by rotating the plate approximately at 60 °C to ensure uniform distribution of the inoculum.
2.8. Primary screening of isolates
During the primary screening, isolates were screened against selected bacterial strains by using perpendicular streak and agar overlay methods. Antagonistic activity of isolates against Gram negative and Gram positive bacteria was primarily screened by using perpendicular streak method and later evaluated by agar overlay method[28],[30]. In perpendicular streak method, nutrient agar media was used and each plate was streaked with individual isolates at the center/diameter of the plate and incubated at 30 °C for 7 d. Later, 24 h fresh sub-cultured test bacteria were prepared and streaked perpendicular to the isolates and incubated at 37 °C for 24 h[30]. In agar overlay method, 5-7 d old colony of actinomycetes was spot inoculated on nutrient agar plates and incubated for 7 d at 30 °C. Then overlaid with 5 mL of semisolid nutrient agar with standardized suspension of test bacteria was inoculated and incubated at 37 °C for 24 h. The zone of inhibition (in mm) around the colonies was measured.
2.9. Production of crude extracts
The isolates showing potential antibacterial activities from the primary screening was subjected to solid state and submerged state fermentation methods to produce crude extracts.
2.9.1. Solid state fermentation method
In this method, 100 g of the substrate (rice grain) was taken in to 500 mL conical flask and 100 mL of distilled water was added and boiled until the rice grains become softened. The mineral salt solution such as K2HPO4 (2.00 g/L), NaCl (1.00 g/L), MgSO4.7H2O (0.10 g/L) and CaCl2 (0.05 g/L) was added for optimization and autoclaved at 121 °C for 15 minutes. Then it was allowed to cool and inoculated with 2 mL of culture suspension from 7 d old actinomycetes culture grown on starch casein agar. After inoculation, flasks were incubated at 30 °C for two weeks. The completely fermented rice grain was ground with mortar and pestle and allowed to dry for 24 h at room temperature. Later, 100 mL of ethyl acetate and 100 mL of methanol was added to the extract and placed on rotary shaker at 120 r/min for 12 h. Finally, the extract was filtered with Whatman No.1 filter paper and the solvent phase was removed by using rotary vacuum evaporator set at water bath temperature of 60 °C[31].
2.9.2. Submerged state fermentation method
In this method, 1 000 mL of yeast and malt extract broth was prepared in which 100 mL was dispensed into 500 mL Erlenmeyer flask, sterilized and cooled. At room temperature broth was inoculated with 2 mL suspension of isolates and kept for 14 d in a rotary shaker at 200 r/min. Then the culture broth was harvested by centrifugation at 4 000 r/min for 15 min. The supernatant was collected and added equal volumes of ethyl acetate (1:1 v/v) then shaken vigorously for 1 h and repeated twice. The solvent phase was separated from aqueous phase by using a separating funnel and subjected to rotary vacuum evaporator at a water bath temperature of 60 °C at 100 r/min to remove solvent and to get crude extracts[32].
2.10. Secondary screening
2.10.1. Disc diffusion method
Antibacterial activities of the crude extracts were tested by using agar disc diffusion method as described by Kirby-Bauer with modification[33]. The inoculum was prepared by mixing a few bacterial colonies (1 mL) from exponential phase with 9 mL of sterile nutrient broth and compared the turbidity with that of the standard 0.5 McFarland solution which is equivalent to 106-108 CFU/mL. The sterile swab was dipped into properly adjusted inoculum and excess liquid was removed by gentle rotation of the cotton swab against the inner surface of the test tube. To obtain even growth, the entire Mueller Hinton agar (MHA) surface was swabbed uniformLy by the cotton swab. The inoculated plates were left at room temperature for 3-5 minutes to allow for any surface moisture to be absorbed before applying the extract. Ten mg/mL of each crude extracts was impregnated with filter paper (6 mm diameter disc) and placed on the surface of MHA medium. Standard antibiotic (tetracycline) disc was used as a positive control and filter paper disc soaked with methanol as a negative control. The selected standard antibiotic and the crude extracts were applied on the MHA in triplicate and left for 15 min to allow the extracts to diffuse. Then the plates were incubated at 37 °C for 18-24 h and the zone of inhibition (mm) was measured.
2.10.2. Agar well diffusion method
In this method inoculum was prepared as that of disc diffusion method. The well was prepared in the plate by using sterile cork borer (6 mm in diameter). A volumn of 100 µL of 10 mg/mL of crude extracts was carefully dispensed into each well and allowed to diffuse for 2 h and incubated at 37 °C for 24 h. The sterilized methanol was filtered and used as negative control. After 24 h of incubation, zone of inhibition around each well was recorded and the experiment was repeated for three times[34].
2.11. Determination of minimum inhibitory concentration (MIC) and minimum bactericidal concentration (MBC)
MIC and MBC was determined by broth two fold serial dilution method[35]. In this method one Gram positive and one Gram negative bacterial strain was used. For determination of MIC and MBC, 12 sterile screw capped test tubes were used. A volume of 1 mL of nutrient broth was dispensed into test tubes 1-10 and 2 mL into test tube 11 (broth control). 1 mL of the crude extract solution was added into test tubes 1 and 2 and 2 mL to test tube 12 (crude extract control). One milliliter of well mixed solution was transferred from test tube 2 to 3 and this process was continued serially up to the test tube 10 by mixing and changing the micropipette tips at each dilution. Finally 1 mL was discarded from test tube 10 and 0.1 mL of standardized inoculum was added into test tubes 1-10 and incubated at 37 °C for 18-24 h. After incubation, by observing the growth of bacteria in the test tube MIC value was determined. From the above test tubes with no growth (no turbidity), 0.1 mL was spreaded over the surface of Mueller Hinton agar plates. After incubation at 37 °C for 24 h, the colonies were observed and MBC was determined.
2.12. Characterization of actinomycetes
The potential actinomycetes selected from the primary and secondary screening was characterized by morphological, biochemical and physiological methods[36].
2.12.1. Morphological characterization by macroscopic method
Morphological characters of the selected isolates were studied by inoculating into sterile media like, glycerol yeast extract agar, oatmeal agar, mineral agar, and starch-casein agar[37]. The media was sterilized and poured into sterile Petri dishes. After solidification, selected isolates were streaked aseptically and incubated at 30 °C for 7 d. Morphological characters such as colony characteristics, pigment production, absence or presence of aerial and substrate mycelium was observed.
2.12.2. Morphological characterization by microscopic cover slip culture method
The arrangement of spores and sporulating structures were examined microscopically by using cover slip culture method by inserting sterile cover slip at an angle of 45 °C in the starch casein agar medium[38],[39]. A loop full of isolates was taken from 7 d old culture media, inoculated at the insertion place of the cover slip and incubated at 30 °C for 7 d. The cover slip was carefully removed by using sterile forceps and placed upward on a clean glass slide. The bacterial growth on the cover slip was fixed with few drops of absolute methanol for 15 min and washed with tap water then flooded with crystal violet reagent for 1 min followed by washing and blot drying. Finally, the cover slip was examined under the microscope by using oil immersion lens (100×).
2.12.3. Biochemical and physiological characterization
In this test, isolates was streaked on starch agar plates and incubated at 30 °C for 7 d. After incubation, iodine solution was poured on the agar and examined for hydrolysis of starch by the production of clear zone around the microbial growth.
Isolates was streaked on gelatin agar plates and incubated at 30 °C for 7 d to test for gelatin hydrolysis. After incubation, plates were flooded with 1 mL of mercuric chloride solution and observed for zone of hydrolysis.
Isolates was streaked on skimmed milk agar medium and incubated at 30 °C for 7 d and observed for zone of casein hydrolysis.
In the urea hydrolysis test, isolates was inoculated in to sterile urea agar slants and incubated at 30 °C for 7 d and a change in colour was observed.
Isolates was streaked on Simon's citrate slant agar and incubated at 30 °C for 7 d and a change in colour was observed.
To test sodium chloride resistance, starch casein agar was prepared in three batches and supplemented with 5%, 7% and 10% sodium chloride. The medium was autoclaved, poured in to the plates and allowed for solidification. The plates were streaked with isolates and incubated at 30 °C for 7 d. The visual observations was made to record the growth of isolates at the highest salt concentration.
The isolates was streaked on starch casein agar plates and incubated at 25 °C, 30 °C, 35 °C and 40 °C for 7 d. The optimum temperature for maximum growth was determined by visual examination of the growth[40].
2.13. Data analysis
The data collected from the secondary screening method was subjected to one way ANOVA to compare the level of significance within the isolates by using SPSS version 16 and the results were interpreted.
3. Results
3.1. Actinomycetes isolation
Actinomycetes were isolated and the morphological appearance of isolates is shown in Figure 1.
Figure 1. Morphological appearance of isolates.
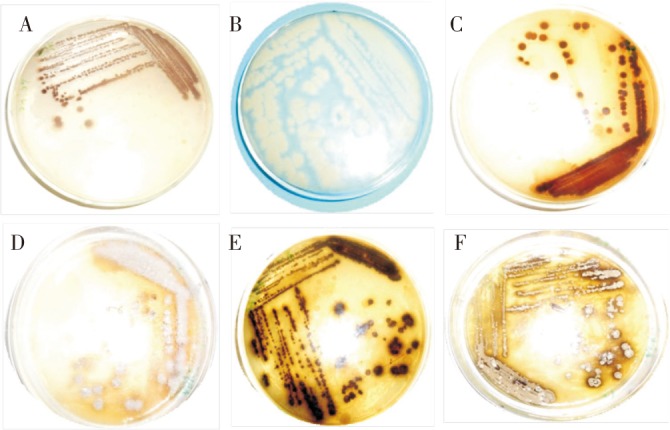
(A) LT004 (B) LT008 (C) LT001 (D) LT005 (E) LT009 and (F) LT010.
3.2. Primary screening
Among the 31 actinomycetes isolated from water and sediments of Lake Tana, 13 isolates showed antibacterial activities against at least one of the tested bacteria. In perpendicular streak plate method, results revealed that the isolates, LT001 and LT009 exhibited broad spectrum activities against tested bacteria. The isolate LT002 showed potential activity against E. coli, P. aeruginosa, S. aureus and LT015 has shown a wide range zone of inhibition against E. coli, S. typhi and S. aureus. Isolates LT004 and LT007 was active only against P. aeruginosa. The isolates LT015 showed MIC (33 mm diameter) followed by LT009 (21 mm) and LT 002 (18 mm) against E. coli (Table 1).
Table 1. Zone of inhibition (mm) of the Isolates against tested bacteria using perpendicular streak method.
| Isolates | Gram negative |
Gram positive |
|||
| E. coli | P. aeruginosa | S. typhi | K. pneumonia | S. aureus | |
| LT001 | 12.06±0.45 | 13.00±0.10 | 13.10±0.51 | 11.10±0.31 | 10.40±0.96 |
| LT002 | 18.03±0.45 | 19.00±0.10 | - | - | 16.03±0.35 |
| LT003 | 9.00±0.20 | 19.60±0.65 | - | 10.00±0.50 | 19.60±0.77 |
| LT004 | - | 11.00±0.20 | - | - | - |
| LT005 | 8.00±0.60 | 10.00±0.20 | 6.00±0.60 | 6.00±0.36 | - |
| LT006 | 7.00±0.20 | - | - | - | - |
| LT007 | - | 4.00±0.20 | - | - | 10.00±0.85 |
| LT008 | 14.00±0.30 | 8.00±0.40 | 11.00±0.20 | - | 18.00±0.40 |
| LT009 | 21.00±0.65 | 12.00±0.60 | 15.00±0.80 | 9.00±0.20 | 22.00±0.40 |
| LT010 | 13.00±0.40 | 7.00±0.80 | 12.00±0.30 | - | 21.00±0.60 |
| LT015 | 33.00±0.26 | - | 35.00±0.80 | - | 39.00±0.60 |
Values are mean±SD of three replications; -: No zone of inhibition.
In agar overlay method, isolates of LT003, LT005 and LT010 was most effective against E. coli, P. aeruginosa, S. typhi and S. aureus. The isolate LT015 was active against E. coli, S. typhi, and S. aureus. The isolates of LT003, LT010 and LT005 showed maximum zone of inhibition i.e., 24 mm, 22 mm and 20mm respectively against P. aeruginosa. The isolate LT005 was observed with maximum zone of inhibition (30 mm) against S. typhi (Table 2).
Table 2. Primary screening of the isolates by using agar overlay method. Zone of inhibition (mm) against tested bacteria.
| Isolates | Gram negative |
Gram positive |
|||
| E. coli | P. aeruginosa | S. typhi | K. pneumonia | S. aureus | |
| LT003 | 20.00±0.40 | 24.00±0.36 | 16.00±0.17 | - | 23.00±0.34 |
| LT004 | - | - | - | 16.00±0.10 | - |
| LT005 | 16.00±0.30 | 20.00±0.43 | 30.00±0.40 | - | 10.00±0.40 |
| LT008 | - | - | 15.00±0.46 | - | 15.00±0.10 |
| LT009 | - | - | - | - | - |
| LT010 | 20.00±0.83 | 22.00±0.20 | 20.00±0.20 | - | 19.00±0.40 |
| LT015 | 17.00±0.40 | - | 21.00±0.40 | - | 25.00±0.20 |
| LT022 | 7.00±0.40 | 9.00±0.87 | - | - | 6.00±0.20 |
| LT027 | 8.00±0.40 | - | - | 6.00±0.80 | - |
Values are mean±SD of three replications; (-) denotes no zone of inhibition.
3.3. Secondary screening of crude extracts
The crude extracts prepared from 11 potential isolates by using solid state and submerged state fermentation methods was subjected to secondary screening by disc and agar well diffusion methods. In disc diffusion method, isolates LT001, LT002, LT004, LT005 and LT008 showed promising results against S. typhi and the results were statistically significant (df 5,12; F=62.90 P=3.44E-08). The isolates LT002 and LT004 was active against E. coli and the results was statistically significant within the tested isolates (df 2, 6; F=183.87; P= 4.14E-06). The inhibition zone of LT007 and LT010 was at the maximum of 11 mm against P. aeruginosa which was comparatively greater than positive control (10 mm) and the result was statistically significant within the crude extracts (df 4, 10; F=7.66; P=0.004 2) (Table 3). The isolates showing promising results against K. pneumonia was statistically significant (df 2, 6; F=6.81; P=0.028 5). Within the isolates tested against S. aureus showed statistically significant results (df 4, 10; F=275.40; P= 3.51E-10).
Table 3. Zone of inhibition (mm) in secondary screening of crude extracts (10 mg/mL) produced from solid state fermentation by using disc diffusion method.
| Isolates | Gram negative |
Gram positive |
|||
| S. typhi | E. coli | P. aeruginosa | K. pneumonia | S. aureus | |
| LT001 | 8.00±0.40 | - | - | 10.00±0.60 | - |
| LT002 | 9.00±0.20 | 8.00±0.40 | - | 9.00±0.20 | 8.00±0.34 |
| LT003 | - | - | - | - | - |
| LT004 | 9.00±0.20 | 10.00±0.34 | - | - | - |
| LT005 | 11.00±0.50 | - | - | - | - |
| LT006 | - | - | - | - | - |
| LT007 | - | - | 11.00±0.20 | 10.00±0.20 | 9.00±0.20 |
| LT008 | 10.00±0.20 | - | 10.00±0.72 | - | - |
| LT009 | - | - | 9.00±0.40 | - | 10.00±0.20 |
| LT010 | - | - | 11.00±0.40 | - | 8.00±0.52 |
| LT015 | - | - | - | - | - |
| Positive control | 12.00±0.30 | 13.00±0.17 | 10.00±0.70 | - | 16.00±0.36 |
| Negative control | - | - | - | - | - |
Values are mean±SD of three replications; (-) denotes inactive; positive control: 25 µg/mL of standard tetracycline; Negative control: 50% of methanol.
In agar well diffusion method, maximum zone of inhibition (13 mm) was recorded from LT001 and LT008 against S. typhi and the results were statistically significant (df 3, 8; F=25.67; P=0.000 18). The inhibition zones of crude extracts from LT001 and LT002 were at the maximum of 12 mm and 10 mm respectively against E. coli and the results were statistically significant (df 2, 6; F=285.36; P=1.13E-06) (Table 4). The extracts of LT003, LT007 and LT009 showed statistically significant results against P. aeruginosa (df 3, 8; F=27.53; P=0.000 14).
Table 4. Zone of inhibition (mm) in secondary screening of crude extracts (10 mg/mL) produced from solid state fermentation by using well diffusion method.
| Extracts | Gram negative |
Gram positive |
|||
| S. typhi | E. coli | P. aeruginosa | K. pneumonia | S. aureus | |
| LT001 | 13.00±0.20 | 12.00±0.40 | - | - | - |
| LT002 | - | 10.00±0.34 | - | - | - |
| LT003 | - | - | 12.00±0.52 | - | - |
| LT004 | - | - | - | - | - |
| LT005 | 12.00±0.70 | - | - | - | 16.00±0.20 |
| LT006 | - | - | - | - | - |
| LT007 | - | - | 12.00±0.20 | - | - |
| LT008 | 13.00±0.30 | - | - | - | - |
| LT009 | - | - | 10.00±0.40 | - | - |
| LT010 | - | - | - | - | - |
| LT015 | - | - | - | - | - |
| Positive control | 15.00±0.34 | 17.00±0.36 | 13.00±0.45 | - | 21.00±0.40 |
| Negative control | - | - | - | - | - |
Values are mean±SD of three replications; -: denotes inactive; Positive control: 25 µg/mL of standard tetracycline; Negative control: 50% of methanol.
The crude extracts obtained from submerged state fermentation, results revealed that LT003, LT006 and LT008 were active against S. typhi; LT003, LT005 and LT008 against E. coli; LT003 and LT009 against P. aeruginosa; LT005 and LT008 against K. pneumonia and LT003, LT005, LT008 and LT009 against S. aureus. The maximum inhibition zone of 11 mm was observed from LT008 against S. typhi which was comparatively similar to positive control (11 mm). The zone of inhibition was greater in all the tested extracts against E. coli compared to positive control. However, maximum inhibition zone of 15 mm was recorded from LT003 against E. coli which was comparatively greater than the positive control (10 mm) (Table 5). Within the isolates the results showed statistically significant (S. typhi, df 3.8; F=10.576; P=0.0037; E. coli, df 3,8; F=49.166; P=1.69E-05; P. aeruginosa, df 2, 8; F=8.25; P=0.0189; K. pneumonia, df 1, 4; F=9.375; P=0.0375 and S. aureus, df 4, 10; F=51.88; P=1.18E-06).
Table 5. Zone of inhibition (mm) in secondary screening of crude extracts (10 mg/mL) produced from submerged fermentation by using disc diffusion method.
| Extracts | Gram negative |
Gram positive |
|||
| S. typhi | E. coli | P. aeruginosa | K. pneumonia | S. aureus | |
| LT001 | - | - | - | - | - |
| LT002 | - | - | - | - | - |
| LT003 | 10.00±0.62 | 15.00±0.36 | 11.00±0.45 | - | 10.00±0.70 |
| LT004 | - | - | - | - | - |
| LT005 | - | 13.00±0.91 | - | 10.00±0.20 | 10.00±0.61 |
| LT006 | 9.00±0.72 | - | - | - | - |
| LT007 | - | - | - | - | - |
| LT008 | 11.00±0.30 | 11.00±0.43 | - | 11.00±0.53 | 11.00±0.69 |
| LT009 | - | - | 10.00±0.70 | - | 9.00±0.36 |
| LT010 | - | - | - | - | - |
| LT015 | - | - | - | - | - |
| Positive control | 11.00±0.20 | 10.00±0.20 | 12.00±0.62 | - | 15.00±0.35 |
| Negative control | - | - | - | - | - |
Values are mean±SD of three replications; -: denotes inactive; Positive control: 25 µg/mL of standard tetracycline; Negative control: 50% of methanol.
The secondary screening of extracts produced from submerged state fermentation method was tested by agar well diffusion method and the results revealed that the zone of inhibition from LT008 was 14 mm; LT001 and LT003 was 13 mm against S. typhi. The activity of all tested extracts was comparatively greater than positive control (9 mm). Maximum zone of inhibition (17 mm) was recorded against E. coli from LT005. The zone of inhibition observed from LT005 and LT008 was superior to positive control (12 mm) (Table 6).
Table 6. Zone of inhibition (mm) in secondary screening of crude extracts (10 mg/mL) produced from submerged fermentation by using well diffusion method.
| Extracts | Gram negative |
Gram positive |
|||
| S. typhi | E. coli | P. aeruginosa | K. pneumonia | S. aureus | |
| LT001 | 13.00±0.72 | - | 11.00±0.80 | - | - |
| LT002 | - | 10.00±0.78 | - | - | - |
| LT003 | 13.00±0.79 | 12.00±0.70 | 18.00±0.87 | - | 13.00±0.20 |
| LT004 | 11.00±0.7 | - | - | - | - |
| LT005 | 11.00±0.79 | 17.00±0.86 | 14.00±0.87 | 13.00±0.35 | 25.00±0.20 |
| LT006 | - | - | - | - | 17.00±0.53 |
| LT007 | - | - | - | - | 12.00±0.40 |
| LT008 | 14.00±0.72 | 13.00±0.40 | 12.00±0.35 | - | 20.00±0.61 |
| LT009 | - | - | - | - | 15.00±0.40 |
| LT010 | - | - | 10.00±0.35 | 12.00±0.72 | 14.00±0.36 |
| LT015 | - | - | 11.00±0.86 | 12.00±0.70 | - |
| Positive control | 9.00±0.53 | 12.00±0.36 | 13.00±0.40 | - | 18.00±0.70 |
| Negative control | - | - | - | - | - |
Values are mean±SD of three replications; -: denotes inactive; Positive control: 25 µg/mL of standard tetracycline; Negative control: 50% of methanol.
Within the isolates tested the results showed statistical significant (S. typhi, df 5, 12; F=19.739; P=2.06E-05; E. coli, df 4, 10; F=46.962: P=1.9E-06; P. aeruginosa, df 6, 14 F=45.92; P=2.04E-08; K. pneumonia, df 2, 6; F=2.654; P=0.149 and S. aureus, df 6,14; F=287.41; 7.8E-14). Among the crude extracts tested against individual bacterial strains, all showed statistically significant result (P<0.05) except K. pneumonia.
3.4. Determination of MIC and MBC
MIC of crude extracts ranged from 1.46 mg/mL to 2.52 mg/mL against Gram positive bacterium (S. aureus). In Gram negative bacterium such as E. coli MIC ranged from 1.84 mg/mL to 2.82 mg/mL. Among the 11 crude extracts tested the results of MBC ranged from 2.92 to 7.56 mg/mL against S. aureus and for E. coli, MBC ranged from 3.80 to 8.46 mg/mL (Table 7).
Table 7. MIC and MBC of the crude extracts (mg/mL).
| Isolates | MIC |
MBC |
||
| S. aureus | E. coli | S. aureus | E. coli | |
| LT001 | 1.95 | 2.11 | 3.90 | 6.33 |
| LT002 | 2.13 | 2.16 | 4.26 | 4.32 |
| LT003 | 1.70 | 2.20 | 3.40 | 4.40 |
| LT004 | 2.25 | 2.35 | 6.75 | 6.75 |
| LT005 | 1.65 | 1.90 | 3.30 | 3.80 |
| LT006 | 2.30 | 2.65 | 6.90 | 7.95 |
| LT007 | 2.52 | 2.82 | 7.56 | 8.46 |
| LT008 | 1.46 | 1.84 | 2.92 | 3.86 |
| LT009 | 1.84 | 2.19 | 3.86 | 4.38 |
| LT010 | 2.01 | 2.52 | 4.02 | 5.04 |
| LT015 | 2.15 | 2.34 | 4.30 | 4.68 |
3.5. Morphological characteristics of selected isolates
Results of morphological characteristics of the selected isolates revealed that the growth of the isolates was excellent in starch casein agar. The isolates of LT005, LT007 and LT015 showed excellent growth in starch casein agar and glycerol yeast extract. In mineral agar and starch casein agar growth was excellent for LT009. The aerial and substrate mycelium colour varied among the isolates such as LT004 was observed with blackish pigment and LT008 was observed with yellowish pigment (Table 8).
Table 8. Morphological characteristics of isolates.
| Isolates | Culture media | Growth | Aerial mycelium | Substrate mycelium | Pigments |
| LT001 | Mineral agar | Good | White | Red | None |
| Starch casein agar | Excellent | White | Red | None | |
| Glycerol yeast extract agar | Very good | White | Red | None | |
| Oatmeal agar | Good | white | Red | None | |
| LT002 | Mineral agar | Very good | White | Black | None |
| Starch casein agar | Excellent | Chocolate | Black | None | |
| Glycerol yeast extract agar | Very good | Chocolate | Black | None | |
| Oatmeal agar | Good | Grey | Black | None | |
| LT003 | Mineral agar | Good | White | Yellow | None |
| Starch casein agar | Excellent | Grey | Yellow | None | |
| Glycerol yeast extract agar | Very good | Grey | Yellow | None | |
| Oatmeal agar | Good | White | Yellow | None | |
| LT004 | Mineral agar | Very good | Grey | Black | Blackish |
| Starch casein agar | Excellent | Chocolate | Black | Blackish | |
| Glycerol yeast extract agar | Very good | Chocolate | Black | Blackish | |
| Oatmeal agar | Good | Grey | Black | Blackish | |
| LT005 | Mineral agar | Good | White-grey | Green | None |
| Starch casein agar | Excellent | White- grey | Green | None | |
| Glycerol yeast extract agar | Excellent | White- grey | Green | None | |
| Oatmeal agar | Good | White | Green | None | |
| LT006 | Mineral agar | Very good | Green | Yellow | None |
| Starch casein agar | Excellent | Green | Yellow | None | |
| Glycerol yeast extract agar | Very good | Green | Yellow | None | |
| Oatmeal agar | Good | Green | Yellow | None | |
| LT007 | Mineral agar | Very good | White grey | Lighter- black | None |
| Starch casein agar | Excellent | Grey | Black | None | |
| Glycerol yeast extract agar | Excellent | Grey | Black | None | |
| Oatmeal agar | Very good | White | Black | None | |
| LT008 | Mineral agar | Very good | White | Yellow | Yellowish |
| Starch casein agar | Excellent | Chocolate | Yellow | Yellowish | |
| Glycerol yeast extract agar | Very good | Grey | Yellow | Yellowish | |
| Oatmeal agar | Good | Grey | Yellow | Yellowish | |
| LT009 | Mineral agar | Excellent | Light-Yellow | Yellow | None |
| Starch casein agar | Excellent | Light-Yellow | Yellow | None | |
| Glycerol yeast extract agar | Very good | White-Yellow | Yellow | None | |
| Oatmeal agar | Good | Yellow | Yellow | None | |
| LT010 | Mineral agar | Very good | Grey | Yellow | None |
| Starch casein agar | Excellent | Bright-grey | Yellow | None | |
| Glycerol yeast extract agar | Very good | White-grey | Yellow | None | |
| Oatmeal agar | Good | White | Yellow | None | |
| LT015 | Mineral agar | Very good | White | White | None |
| Starch casein agar | Excellent | White-grey | White | None | |
| Glycerol yeast extract agar | Excellent | White | White | None | |
| Oatmeal agar | Very good | White | White | None |
3.6. Physiological and biochemical characteristics of isolates
Physiological and biochemical characteristics result indicates that all isolates showed the ability of starch and urea hydrolysis. The isolates from LT001 to LT005 were able to hydrolysis celatin; LT001 to LT003 and LT005 to LT008 were able to hydrolysis casein. The positive utilization of citrate was recorded in LT001, LT003, LT005, LT006 and LT009. The tested actinomycetes showed resistance capacity to grow in 5% concentration of sodium chloride. The optimum temperature for the growth of most isolates was between 25-30 °C and LT008 exceed up to 40 °C (Table 9).
Table 9. Physiological and biochemical characteristics of isolates.
| Types of test | Characteristics of isolates |
||||||||||
| LT001 | LT002 | LT003 | LT004 | LT005 | LT006 | LT007 | LT008 | LT009 | LT010 | LT015 | |
| Starch | + | + | + | + | + | + | + | + | + | + | + |
| Gelatin | + | + | + | + | + | - | - | - | - | - | - |
| Casein | + | + | + | - | + | + | + | + | - | - | - |
| Urea | + | + | + | + | + | + | + | + | + | + | + |
| Citrate utilization | + | - | + | - | + | + | - | - | + | - | - |
| Resistance to NaCl | 5% | 5% | 5%-7% | 5% | 7% | 5% | 5% | 5% | 5% | 5% | 5% |
| Optimum temperature | 30 °C | 25-35 °C | 30 °C | 25-30 °C | 30-35 °C | 30 °C | 25-35 °C | 30-40 °C | 30 °C | 25-30 °C | 30 °C |
-: Negative; +: Positive.
4. Discussion
Currently, the incidence of multidrug resistant organisms is increasing and compromising the treatment of a growing number of infectious diseases. As a result, there is an urgent need for developing new drugs which are effective against current antibiotic resistant pathogens. Actinomycetes have been proven as a potential source of bioactive compounds and richest source of secondary metabolites[41]. Isolation of antibacterial compounds from the freshwater environment is in advance interest in recent times to isolate novel bioactive actinomycetes. According to Sibanda et al., extracts from three freshwater actinomycetes showed inhibition to Gram negative bacterial strains compared to Gram positive bacterial strains[42]. These recent studies motivated to isolate antibiotic producing actinomycetes from fresh water and sediments of Lake Tana.
In the present study 31 actinomycetes were isolated from water and sediments and 13 showed wide range of zone of inhibition in the primary screening. More yield of crude extract was produced through solid state fermentation method compared to submerged state fermentation method when extracted with methanol and ethyl acetate solvents. The reason for the increased production of yield in solid state fermentation was due to lack of water and completely miscible in organic solvents (ethyl acetate and methanol) with the fermented biomass. The higher yields obtained by the solid state fermentation method was agreed with the previous reports[43],[44]. The lower yields obtained from submerged state fermentation method was attributed to the use of water immiscible solvent such as ethyl acetate during extraction. Similar findings were earlier reported by Subramaniyam and Vimala[45].
During secondary screening, 11 crude extracts showed wide range of inhibition zone against tested bacterial strains. Similar findings were reported earlier by Schimel and Hattenschwiler[46] and Anupama et al[47]. In general the antibacterial activity of crude extracts was fluctuated widely. This may be associated with disintegration of the crude extracts during extraction process. The bioactivity of the isolates was dissimilar between Gram positive and Gram negative bacterial strains. The results clearly demonstrate that a Gram positive bacterium was highly susceptible to the tested crude extracts compared to Gram negative bacteria. This result was in agreement with the report of Ilic et al[32]. They have suggested that different sensitivity between Gram positive and Gram negative bacteria could be ascribed to morphological differences such as, outer membrane of Gram negative bacteria having lipopolysaccharide which makes the cell wall impermeable to lipophilic extracts. But, a Gram positive bacterium was more susceptible because lack of outer membrane. There was a marked difference between the crude extracts and pure antibacterial drug (tetracycline). Some of the crude extracts showed higher or equivalent zone of inhibition compared to standard drug tetracycline. Rex et al. reported that significant difference was normally presented in crude (unpurified) extracts compared with pure drug that was already in clinical use[48].
The MIC and MBC varied among the tested isolates against S. aureus and E. coli. These results were similar with the report of Sibanda et al[42]. Therefore, crude extracts could be a potent source for antibiotic production, which leads to the development of novel drugs for the treatment of infectious diseases. The report of antibacterial activities of actinomycetes isolated from water and sediments of Lake Tana was first hand information as per our knowledge.
The aerial mycelium, substrate mycelium growth and pigmentation showed distinct variation based on the culture media in which the isolates were grown. Among the four culture media used, most of the isolates growth was excellent in starch casein agar and this may be due to sufficient amount of nutrient included in this media. Valli et al. also observed leathery, white powdery, creamy, pinpoint and powder colonies of actinomycetes[17]. All the potential isolates in this study have the ability to hydrolysis starch and urea. Most of the isolates can tolerate at 5% concentration of sodium chloride and the optimum temperature for the growth of isolates was ranged from 25 to 30 °C. Therefore, these results indicate that most of the isolates obtained from Lake Tana were grouped under the genera of Streptomyces.
Acknowledgments
The first author acknowledges Dr. Mulugeta Aemero, Head, Department of Biology for his kind cooperation and facilities extended in the Microbiology laboratory to complete this research work. The financial assistance received from University of Gondar under teaching and learning program (UoG/Budget code: 6417) was greatly acknowledged.
Comments
Background
Among the microorganism fungi, actinomycetes are best known for their outstanding ability to produce a great variety of medically and economically important secondary metabolites or compounds with antibacterial, antifungal and antiviral properties and encompass a large variety of antibiotics. Soil microbes represent an important source of biologically active compounds. These molecules present original and unexpected structure and are selective inhibitors of the molecular targets.
Research frontiers
Studies are being performed in order to determine the effective actinomycetes isolate from water and sediments of Lake Tana, Ethiopia.
Related Reports
At Bioresearch Italica, discovery of new bioactive molecules is mostly carried out through the exploitation of a proprietary strain collection of over 50 000 strains, mostly unusual genera of actinomycetes. Nearly 755 of all described antibiotics are produced by actinomycetes. Some isolate of Streptomyces species can produce more than 180 different secondary metabolites. Novel antibiotics and other bioactive secondary metabolite can still be discovered from microbial sources.
Innovations and breakthroughs
The results of the present study revealed that freshwater actinomycetes of Lake Tana, Ethiopia appear to have immense potential as a source of antibacterial compounds.
Applications
Currently the incidence of multidrug resistant organisms is increasing. As a result there is an urgent need for new drugs which are effective against current antibiotic resistant pathogens. Actinomycetes are one of the potential sources for isolation of bioactive antibiotics for multidrug resistance organisms.
Peer review
This is a good study in which the authors isolated and evaluated the actinomycetes which producing antibiotics from different freshwater habitats and tested against different Gram positive and Gram negative bacterial strains.
Footnotes
Foundation Project: Supported by University of Gondar under Teaching and Learning Program (UoG/Budget code: 6417).
Conflict of interest statement: We declare that we have no conflict of interest.
References
- 1.Fenical W, Jensen PR. Developing a new resource for drug discovery: marine actinomycete bacteria. Nat Chem Biol. 2006;2:666–673. doi: 10.1038/nchembio841. [DOI] [PubMed] [Google Scholar]
- 2.Singh B, Saikia R, Yadav M, Singh R, Chauhan VS, Dilip K. Molecular characterization of Fusarium oxysporum f. sp. ciceri causing wilt of chickpea. Afr J Biotechnol. 2006;5(6):497–502. [Google Scholar]
- 3.Weinstein MP, Litvin SY, Guida VG. Consideration of habit linkages, estuarine landscapes and the trophic spectrum in wetland restoration design. J Coastal Res. 2005;40:51–63. [Google Scholar]
- 4.Rifaat HM. The biodiversity of actinomycetes in the River Nile exhibiting antifungal activity. J Mediter Ecol. 2003;4:5–7. [Google Scholar]
- 5.Eccleston GP, Brooks PR, Kurtboke DI. The occurrence of bioactive micromonsoporae in aquatic habitats of the sunshine coast in Australia. Mar Drugs. 2008;6:243–261. doi: 10.3390/md20080012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang H, Lv J, Hu Y, Fang Z, Zhang K, Bao S. Micromonospora rifamycinia sp. nov., a novel actinomycetes from mangrove sediments. Int J Syst Evol Microbiol. 2008;58(1):17–20. doi: 10.1099/ijs.0.64484-0. [DOI] [PubMed] [Google Scholar]
- 7.Fehling RH, Buchanan GO, Mincer TJ, Kauffman CA, Jensen PR, Fenical WR. A highly cytotoxic proteasome inhibitor from a novel microbial source, a marine bacterium of the new genus Salinospora. Angew Chem. 2003;42:355–357. doi: 10.1002/anie.200390115. [DOI] [PubMed] [Google Scholar]
- 8.Jensen PR, Gontang E, Mafnas C, Mincer TJ, Fenical W. Culturable marine actinomycete diversity from tropical Pacific Ocean sediments. Environ Microbiol. 2005;7:1039–1048. doi: 10.1111/j.1462-2920.2005.00785.x. [DOI] [PubMed] [Google Scholar]
- 9.Riedlinger J, Reicke A, Zahner H, Krishmer B, Bull AT, Maldonado LA, et al. et al. Abyssomicins, inhibitors of the para aminobenzoic acid pathway produced by the marine Verrucosispora strain AB-18-032. J Antibiot. 2004;57:271–279. doi: 10.7164/antibiotics.57.271. [DOI] [PubMed] [Google Scholar]
- 10.Hughes CC, Prieto-Davo A, Jensen PR, Fenical W. The marinopyrroles, antibiotics of an unprecedented structure class from a marine Streptomyces sp. Org Lett. 2008;10:629–631. doi: 10.1021/ol702952n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berdy J. Bioactive microbial metabolites: a personal view. J Antibiot. 2005;58(1):1–26. doi: 10.1038/ja.2005.1. [DOI] [PubMed] [Google Scholar]
- 12.Hayakawa Y, Shirasaki S, Shiba S, Kawasaki T, Matsuo Y, Adachi K, et al. et al. Piericidins C7 and C8, new cytotoxic antibiotics produced by a marine Streptomyces sp. J Antibiot. 2007;60:196–200. doi: 10.1038/ja.2007.22. [DOI] [PubMed] [Google Scholar]
- 13.Lam KS. Discovery of novel metabolities from marine actinomycetes. Curr Opin Microbiol. 2006;9:245–251. doi: 10.1016/j.mib.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 14.Gurung TD, Sherpa C, Agrawal VP, Lekhak B. Isolation and characterization of antibacterial actinomycetes from soil samples of Kapapatthar, Mount Everest Region. Nepal J Sci Technol. 2009;10:173–182. [Google Scholar]
- 15.Kumar N, Singh RK, Mishra SK, Singh AK, Pachouri UC. Isolation and screening of soil actinomycetes as source of antibiotics active against bacteria. Int J Microbiol Res. 2010;2(2):12–16. [Google Scholar]
- 16.Cwala Z, Igbinosa EO, Okoh AI. Assessment of antibiotics production potential in four actinomycetes isolated from aquatic environments of the Eastern Cape Province of South Africa. Afr J Pharm Pharmacol. 2011;5(2):118–124. [Google Scholar]
- 17.Valli S, Suvathi Sugasini S, Aysha OS, Nirmala P, Vinot Kumar P, Reena A. Antimicrobial potential of actinomycetes species isolated from marine environment. Asian Pacific J Trop Biomed. 2012;9:416–473. doi: 10.1016/S2221-1691(12)60078-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kalyani ALT, Ramya Sravani KM, Annapurana J. Isolation and characterization of antibiotic producing actinomycetes from marine soils samples. Int J Curr Pharmaceu Res. 2012;4(2):109–112. [Google Scholar]
- 19.Enright MC. The evolution of a resistant pathogen the case of MRSA. Curr Opin Pharmacol. 2003;3:474–479. doi: 10.1016/s1471-4892(03)00109-7. [DOI] [PubMed] [Google Scholar]
- 20.Barsby T, Kelly MT, Gagne SM, Andersen RJ. Bogorol A produced in culture by a marine Bacillus sp. reveals a novel template for cationic peptide antibiotics. Org Lett. 2001;3:437–440. doi: 10.1021/ol006942q. [DOI] [PubMed] [Google Scholar]
- 21.Parungao MM, Maceda EBG, Villano MAF. Screening of antibiotic-producing actinomycetes from marine, brackish and terrestrial sediment of Samal Island, Philippines. J Res Sci Comput Eng. 2007;4(3):29–38. [Google Scholar]
- 22.Lazzarini A, Cavaletti L, Toppo G, Marinelli F. Rare genera of actinomycetes as potential producers of new antibiotics. Anton Leeuw. 2000;78:388–405. [PubMed] [Google Scholar]
- 23.Oskay AM, Tamer U, Azeri C. Antibacterial activity of some actinomycetes isolated from farming soil of Turkey. Afr J Biotechnol. 2004;3(9):441–446. [Google Scholar]
- 24.LAKENET . USA: World Lakes Website; 2003. Lake profile. [Online] Available from: http://www.worldlakes.org/lakedetails.asp?lakeid=8568. [Accessed on 3 July, 2012]. [Google Scholar]
- 25.Hong K, Gao A, Xie Q, Gao H, Zhuang L, Lin H, et al. et al. Actinomycetes from marine drug discovery isolated from mangrove soils and plants in China. Mar Drugs. 2009;7:24–44. doi: 10.3390/md7010024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cuesta G, García-de-la-Fuente R, Abad M, Fornes F. Isolation and identification of actinomycetes from a compost-amended soil with potential as biocontrol agents. J Environ Manag. 2010;11:10–16. doi: 10.1016/j.jenvman.2010.11.023. [DOI] [PubMed] [Google Scholar]
- 27.Zhang L, Wang AR, Sun J, Zhung N, Hu J. Exploring novel bioactive compounds from marine microbes. Curr Opin Microbiol. 2005;8:276–281. doi: 10.1016/j.mib.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 28.Hayakawa M, Ishiwwa K, Nonomurha H. Distribution of rare actinomycetes in Japanese soils. J Fermt Technol. 2004;66:367–373. [Google Scholar]
- 29.Andrews M, Wise R. Susceptibility testing of Bacillus species. J Antimicrob Chemother. 2002;49:1040–1042. doi: 10.1093/jac/dkf063. [DOI] [PubMed] [Google Scholar]
- 30.Williams ST, Goodfellow M, Wellington EMH, Vickers JC, Alderson G, Sneath PHA, et al. et al. A probability matrix for identification of Streptomyces. J Gen Microbiol. 1983;129:1815–1830. doi: 10.1099/00221287-129-6-1815. [DOI] [PubMed] [Google Scholar]
- 31.Robinson T, Singh D, Nigam P. Solid state fermentation: a promising microbial technology for secondary metabolite production. App Microbiol Biotech. 2001;55:284–289. doi: 10.1007/s002530000565. [DOI] [PubMed] [Google Scholar]
- 32.Ilic SB, Konstantinovic SS, Todorovic ZB, Lazic ML, Veljkovic VB, Jokovic N, et al. et al. Characterization and antimicrobial activity of the bioactive metabolites in Streptomycetes isolates. Microbiol. 2007;76:421–428. [PubMed] [Google Scholar]
- 33.Kirby-Bauer Susceptibility test with single, high-concentration antimicrobial disks. Antimicrob Agents Chemother. 1979;3(3):418–424. doi: 10.1128/aac.3.3.418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pandey B, Ghimire P, Agrawal VP. Studies on the antibacterial activity of actinomycetes isolated from Khumbu region of Mount Everest. J App Mirobiol. 2004;20:45–54. [Google Scholar]
- 35.Andrews M. Determination of minimum inhibitory concentrations. J Antimicrob Chemother. 2001;48:5–16. doi: 10.1093/jac/48.suppl_1.5. [DOI] [PubMed] [Google Scholar]
- 36.Pandey B, Ghimire P, Agrawal VP. Studies on the antimicrobial activity of the actinomycetes isolated from the Khumbu region of Nepal. Appl Microbiol. 2008;5:235–261. [Google Scholar]
- 37.Yang KQ, Han L, Vining LC. Regulation of jadomycin-B production in Streptomyces venezuelae Isp5230-involvement of a repressor gene. J Bacteriol. 1995;177:6111–6117. doi: 10.1128/jb.177.21.6111-6117.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cross T, William ST. Method in microbiology. 4th ed. London: Academic press; 1971. Actinomycetes; pp. 29–65. [Google Scholar]
- 39.Tiwari KD. Protocol for actinomycetes study in RLABB 2008/9. Tridhtuven University Tikiriupr: Central Department of Microbiology, Institution of Science and technology; 2009. p. 21. [Google Scholar]
- 40.Muiru WM, Mutitu EW, Mukunya DM. Identification of selected actinomycetes isolates and characterization of their antibiotic metabolites. J Biol Sci. 2008;8:1021–1026. [Google Scholar]
- 41.Suthindhiran K, Kannabiran K. Cytotoxic and antimicrobial potential of actinomycete species Saccharopolyspora salina VITSDK4 isolated from the Bay of Bengal Coast of India. Amer J Infect Dis. 2009;5:90–98. [Google Scholar]
- 42.Sibanda T, Mabinya LV, Mazomba N, Akinpelu DA, Bernard K, Olaniran AO, et al. et al. Antibiotic producing potentials of three freshwater actinomycetes isolated from the Eastern Cape Province of South Africa. Int J Mol Sci. 2010;11:2612–2623. doi: 10.3390/ijms11072612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.El-Naggar MY, EL-Assar SA, Abdul Gawad SM. Solid state fermentation for the production of meroparamycin by Streptomyces sp. strain MAR01. J Microbiol Biotechnol. 2009;19(5):468–473. doi: 10.4014/jmb.0807.457. [DOI] [PubMed] [Google Scholar]
- 44.Tabaraie B, Ghasemian E, Tabaraie T, Parvizi E, Rezazarandi M. Comparative evaluation of cephalosphorin C production in solid state fermentation and submerged liquid culture. J Microbiol Biotechnol Food Sci. 2012;2(1):83–94. [Google Scholar]
- 45.Subramaniyam R, Vimala R. Solid state fermentation and submerged fermentation for the production of bioactive substances: a comparative study. Int J Sci Nat. 2012;3(3):480–486. [Google Scholar]
- 46.Schimel JP, Hattenschwiler S. Nitrogen transfer between decomposing leaves of different N status. Soil Biol Biochem. 2007;39:1428–1436. [Google Scholar]
- 47.Anupama M, Narayana KJP, Vijayalakshmi M. Screening of Streptomyces purpeofuscus for antimicrobial metabolites. Res J Microbiol. 2007;2:992–994. [Google Scholar]
- 48.Rex MA, Stuart CT, Etter RJ. Do deep-sea nematodes show a positive latitudinal gradient of species diversity? The potential role of depth. Mar Ecol Prog Ser. 2001;210:297–298. [Google Scholar]


