Abstract
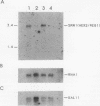
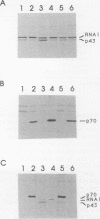
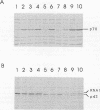
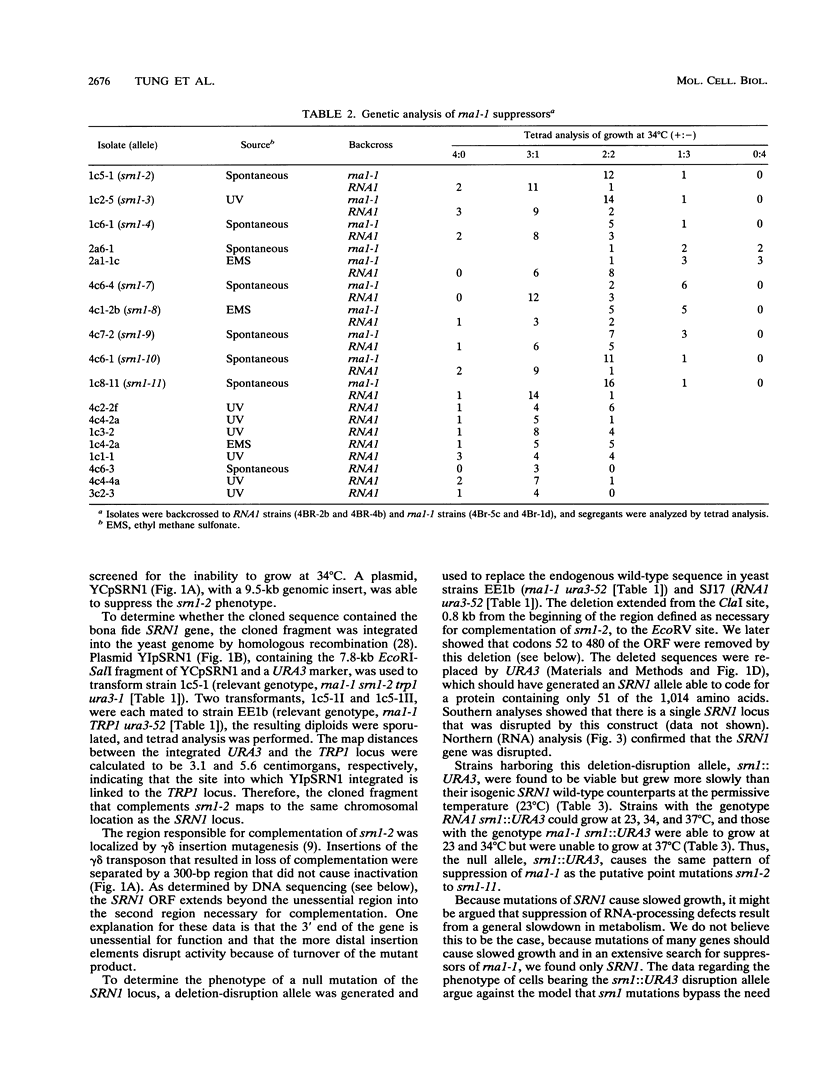
The yeast RNA1 gene encodes a cytosolic protein that affects pre-tRNA splicing, pre-rRNA processing, the production of mRNA, and the export of RNA from the nucleus to the cytosol. In an attempt to understand how the RNA1 protein affects such a diverse set of processes, we sought second-site suppressors of a mutation, rna1-1, of the RNA1 locus. Mutations in a single complementation group were obtained. These lesions proved to be in the same gene, SRN1, identified previously in a search for second-site suppressors of mutations that affect the removal of intervening sequences from pre-mRNAs. The SRN1 gene was mapped, cloned, and sequenced. DNA sequence analysis and the phenotype of disruption mutations showed that, surprisingly, SRN1 is identical to HEX2/REG1, a gene that negatively regulates glucose-repressible genes. Interestingly, SRN1 is not a negative regulator of RNA1 at the transcriptional, translational, or protein stability level. However, SRN1 does regulate the level of two newly discovered antigens, p43 and p70, one of which is not glucose repressible. These studies for the first time link RNA processing and carbon catabolite repression.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Atkinson N. S., Dunst R. W., Hopper A. K. Characterization of an essential Saccharomyces cerevisiae gene related to RNA processing: cloning of RNA1 and generation of a new allele with a novel phenotype. Mol Cell Biol. 1985 May;5(5):907–915. doi: 10.1128/mcb.5.5.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson M., Osmond B. C., Botstein D. Mutants of yeast defective in sucrose utilization. Genetics. 1981 May;98(1):25–40. doi: 10.1093/genetics/98.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson M. Regulation of sugar utilization in Saccharomyces species. J Bacteriol. 1987 Nov;169(11):4873–4877. doi: 10.1128/jb.169.11.4873-4877.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culbertson M. R., Winey M. Split tRNA genes and their products: a paradigm for the study of cell function and evolution. Yeast. 1989 Nov-Dec;5(6):405–427. doi: 10.1002/yea.320050602. [DOI] [PubMed] [Google Scholar]
- Entian K. D., Zimmermann F. K. Glycolytic enzymes and intermediates in carbon catabolite repression mutants of Saccharomyces cerevisiae. Mol Gen Genet. 1980 Jan;177(2):345–350. doi: 10.1007/BF00267449. [DOI] [PubMed] [Google Scholar]
- Entian K. D., Zimmermann F. K. New genes involved in carbon catabolite repression and derepression in the yeast Saccharomyces cerevisiae. J Bacteriol. 1982 Sep;151(3):1123–1128. doi: 10.1128/jb.151.3.1123-1128.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillman E. C., Slusher L. B., Martin N. C., Hopper A. K. MOD5 translation initiation sites determine N6-isopentenyladenosine modification of mitochondrial and cytoplasmic tRNA. Mol Cell Biol. 1991 May;11(5):2382–2390. doi: 10.1128/mcb.11.5.2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guyer M. S. Uses of the transposon gamma delta in the analysis of cloned genes. Methods Enzymol. 1983;101:362–369. doi: 10.1016/0076-6879(83)01027-7. [DOI] [PubMed] [Google Scholar]
- Hartwell L. H. Macromolecule synthesis in temperature-sensitive mutants of yeast. J Bacteriol. 1967 May;93(5):1662–1670. doi: 10.1128/jb.93.5.1662-1670.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopper A. K. Genetic methods for study of trans-acting genes involved in processing of precursors to yeast cytoplasmic transfer RNAs. Methods Enzymol. 1990;181:400–421. doi: 10.1016/0076-6879(90)81139-l. [DOI] [PubMed] [Google Scholar]
- Hopper A. K., Traglia H. M., Dunst R. W. The yeast RNA1 gene product necessary for RNA processing is located in the cytosol and apparently excluded from the nucleus. J Cell Biol. 1990 Aug;111(2):309–321. doi: 10.1083/jcb.111.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurt D. J., Wang S. S., Lin Y. H., Hopper A. K. Cloning and characterization of LOS1, a Saccharomyces cerevisiae gene that affects tRNA splicing. Mol Cell Biol. 1987 Mar;7(3):1208–1216. doi: 10.1128/mcb.7.3.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison H. T., Hartwell L. H., McLaughlin C. S. Temperature-sensitive yeast mutant defective in ribonucleic acid production. J Bacteriol. 1969 Sep;99(3):807–814. doi: 10.1128/jb.99.3.807-814.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knapp G., Beckmann J. S., Johnson P. F., Fuhrman S. A., Abelson J. Transcription and processing of intervening sequences in yeast tRNA genes. Cell. 1978 Jun;14(2):221–236. doi: 10.1016/0092-8674(78)90109-5. [DOI] [PubMed] [Google Scholar]
- Kuo C. L., Campbell J. L. Cloning of Saccharomyces cerevisiae DNA replication genes: isolation of the CDC8 gene and two genes that compensate for the cdc8-1 mutation. Mol Cell Biol. 1983 Oct;3(10):1730–1737. doi: 10.1128/mcb.3.10.1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long R. M., Mylin L. M., Hopper J. E. GAL11 (SPT13), a transcriptional regulator of diverse yeast genes, affects the phosphorylation state of GAL4, a highly specific transcriptional activator. Mol Cell Biol. 1991 Apr;11(4):2311–2314. doi: 10.1128/mcb.11.4.2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto K., Yoshimatsu T., Oshima Y. Recessive mutations conferring resistance to carbon catabolite repression of galactokinase synthesis in Saccharomyces cerevisiae. J Bacteriol. 1983 Mar;153(3):1405–1414. doi: 10.1128/jb.153.3.1405-1414.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niederacher D., Entian K. D. Characterization of Hex2 protein, a negative regulatory element necessary for glucose repression in yeast. Eur J Biochem. 1991 Sep 1;200(2):311–319. doi: 10.1111/j.1432-1033.1991.tb16187.x. [DOI] [PubMed] [Google Scholar]
- Niederacher D., Entian K. D. Isolation and characterization of the regulatory HEX2 gene necessary for glucose repression in yeast. Mol Gen Genet. 1987 Mar;206(3):505–509. doi: 10.1007/BF00428892. [DOI] [PubMed] [Google Scholar]
- Orr-Weaver T. L., Szostak J. W., Rothstein R. J. Genetic applications of yeast transformation with linear and gapped plasmids. Methods Enzymol. 1983;101:228–245. doi: 10.1016/0076-6879(83)01017-4. [DOI] [PubMed] [Google Scholar]
- Orr-Weaver T. L., Szostak J. W., Rothstein R. J. Yeast transformation: a model system for the study of recombination. Proc Natl Acad Sci U S A. 1981 Oct;78(10):6354–6358. doi: 10.1073/pnas.78.10.6354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson N. J., Thorburn P. C., Haber J. E. A suppressor of temperature-sensitive rna mutations that affect mRNA metabolism in Saccharomyces cerevisiae. Mol Cell Biol. 1982 May;2(5):571–577. doi: 10.1128/mcb.2.5.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson W. R., Lipman D. J. Improved tools for biological sequence comparison. Proc Natl Acad Sci U S A. 1988 Apr;85(8):2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson G. L. A simplification of the protein assay method of Lowry et al. which is more generally applicable. Anal Biochem. 1977 Dec;83(2):346–356. doi: 10.1016/0003-2697(77)90043-4. [DOI] [PubMed] [Google Scholar]
- Piper P. W., Aamand J. L. Yeast mutation thought to arrest mRNA transport markedly increases the length of the 3' poly(A) on polyadenylated RNA. J Mol Biol. 1989 Aug 20;208(4):697–700. doi: 10.1016/0022-2836(89)90159-9. [DOI] [PubMed] [Google Scholar]
- Rosbash M., Harris P. K., Woolford J. L., Jr, Teem J. L. The effect of temperature-sensitive RNA mutants on the transcription products from cloned ribosomal protein genes of yeast. Cell. 1981 Jun;24(3):679–686. doi: 10.1016/0092-8674(81)90094-5. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiokawa K., Pogo A. O. The role of cytoplasmic membranes in controlling the transport of nuclear messenger RNA and initiation of protein synthesis. Proc Natl Acad Sci U S A. 1974 Jul;71(7):2658–2662. doi: 10.1073/pnas.71.7.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slusher L. B., Gillman E. C., Martin N. C., Hopper A. K. mRNA leader length and initiation codon context determine alternative AUG selection for the yeast gene MOD5. Proc Natl Acad Sci U S A. 1991 Nov 1;88(21):9789–9793. doi: 10.1073/pnas.88.21.9789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St John T. P., Davis R. W. The organization and transcription of the galactose gene cluster of Saccharomyces. J Mol Biol. 1981 Oct 25;152(2):285–315. doi: 10.1016/0022-2836(81)90244-8. [DOI] [PubMed] [Google Scholar]
- Towbin H., Staehelin T., Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traglia H. M., Atkinson N. S., Hopper A. K. Structural and functional analyses of Saccharomyces cerevisiae wild-type and mutant RNA1 genes. Mol Cell Biol. 1989 Jul;9(7):2989–2999. doi: 10.1128/mcb.9.7.2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolford J. L., Jr Nuclear pre-mRNA splicing in yeast. Yeast. 1989 Nov-Dec;5(6):439–457. doi: 10.1002/yea.320050604. [DOI] [PubMed] [Google Scholar]
- Zimmermann F. K., Scheel I. Mutants of Saccharomyces cerevisiae resistant to carbon catabolite repression. Mol Gen Genet. 1977 Jul 7;154(1):75–82. doi: 10.1007/BF00265579. [DOI] [PubMed] [Google Scholar]