Abstract
Nuclear receptor (NR) dependent transcriptional action requires recruitment of diverse factors characterized as coregulators. PNRC (proline-rich nuclear receptor coregulatory protein) is a member of coregulators that are capable of potentiating the transcriptional activity of NRs. Here we identified three human PNRC splicing variants designated PNRC1c, PNRC1d and PNRC1f. PNRC1c and PNRC1f are generated through alternative recognition of the 3'-splice site in exon 1, leading to in-frame deletion of 79 amino acids aa) and an altered reading frame, respectively. PNRC1d is generated through the alternate promoter usage and forms a truncated protein containing C-terminus 142 aa of full-length PNRC. These isoforms differ in their abilities to bind NRs and potentiate NR mediated transcriptions. Moreover, PNRC1d can modulate the activity of full-length PNRC in enhancing ER mediated transcription. Our results suggest PNRC exists as functionally distinct isoforms and alternative splicing serves as a regulatory mechanism of PNRC coactivator activity.
Keywords: PNRC, alternative splicing, isoform, nuclear receptor, coactivator, estrogen receptor
1. Introduction
Nuclear receptors (NRs) comprise a superfamily of ligand-dependent transcription factors involved in growth, differentiation, development, and maintenance of cellular homeostasis. The NRs regulate transcription by binding to response elements in the promoters of target genes and acting as scaffolds for the assembly of large coactivator and corepressor complexes (Glass and Rosenfeld, 2000). Ligand-dependent dynamic exchange of corepressors for coactivators serves as a well-regulated switch from gene repression to gene activation (Xu et al., 1999; Baek and Rosenfeld, 2004).
PNRC (proline-rich nuclear receptor coregulatory protein) is one member of NR coactivators. To date, PNRC has been found to interact in a ligand-dependent manner with the ligand-binding domain of NRs including ERα, ERβ, PR, GR, TR, RARα, RARβ, RXR, RORα, RORβ, HNF4α, HNF4γ and LRH1, and in a ligand-independent manner with the orphan receptors SF1, ERRα1 and ERRγ (Zhou et al., 2000; Albers et al., 2005). The distinctive features of PNRC as a coactivator are significantly smaller (327aa, 35.2kD) than most of the previously identified coregulators, and proline-rich (14 %). Moreover, it interacts with NRs through a proline-rich Src homology domain-3 (SH3)-binding motif, S-D (E)-P-P-S-P-S, not through LXXLL motif (NR box) which mediates the interaction with NRs for most coactivators.
In addition to enhancing NR mediated transcriptions, PNRC has been found to down regulate the activation of Ras and MAP kinase through its interaction with Grb2, an important adapter protein involved in growth factor/Ras signaling pathway, and overexpression of PNRC inhibits the growth of HeLa cells (Zhou et al., 2004). Furthermore, PNRC has recently been demonstrated to stimulate RNA pol III transcription through its interaction with the subunit RPC39 of RNA pol III (Zhou et al., 2007). PNRC is widely expressed in human tissues, and its expression is significantly down-regulated in stomach, colorectal, and hepatocellular carcinomas and in breast cancer, implying a possibility of PNRC as a tumor-related gene and a role in carcinogenesis (Chen et al., 1995; Kanai et al., 2001; Zhou et al., 2004; Zucchi et al., 2004)
NRs and coregulators, like many other proteins, are often subjected to alternative splicing, which generates multiple isoforms that show different activities and play distinct biological roles. In the case of ER, some isoforms lose estrogen-dependent transactivation ability and show various effects on estrogen signaling, such as inhibitory effects on the wild-type ER (Wang and Miksicek, 1991). The nuclear receptor coactivator amplified in breast cancer 1 (AIB1) isoform AIB1-Δ3 shows more activity to increases hormone and growth factor sensitivity, leading to an increase in cell proliferation and carcinogenesis (Reiter et al., 2001; Reiter et al., 2004; Tilli et al., 2005). Coactivator activator (CoAA) isoform CoAM has no activity to enhance TRBP and CBP action, but represses the activity of CoAA. During retinoic-acid-induced P19 stem cell differentiation, the expression of CoAA undergoes a rapid switch to CoAM in the cavity of the embryoid body (Iwasaki et al., 2001; Auboeuf et al., 2004). Moreover, some isoforms of coregulators even exert the opposite function (Meng et al., 2006). For example, testicular zinc-finger protein (TZF) is a corepressor of androgen receptor (AR), but one of its isoforms, TZF-L, becomes a coactivator of AR (Tao et al., 2006a; Tao et al., 2006b).
Thus far, no PNRC isoform has been identified. We asked whether PNRC is alternatively spliced and what biological roles the isoforms play in regulating the activity of PNRC. In this study, we report the identification of three splice variants of PNRC. Functional analysis shows that these isoforms differ in their activities to bind NRs and potentiate NR mediated transcriptions. Furthermore, one of the isoforms designated PNRC1d has been found to modulate the activity of full-length PNRC.
2. Materials and Methods
2.1. RT-PCR and TA cloning
Total RNAs were extracted from human tissues and cell lines using TRIzol reagent (Invitrogen) according to the manufacturer’s guideline. Approximately 4 µg DNaseI-treated RNA was reverse transcribed to cDNA by superscript III (Invitrogen) using random hexamers and 2 µl cDNA was used as a template for PCR reaction. PCR was performed using Hotstar Taq polymerase (Qiagen) and the cycling was as follows: 95°C for 15 min; 94°C for 45 s, 55°C for 45 s and 72°C for 60 s, 40 cycles; 72°C for 10 min. The sequences of oligonucleotide primers in PCR are as follows: P1: 5’-ATGACTGTCGTCTCCGTCC -3’; P2: 5’-CTAAGTTTGAACTTTGAGGAGCGTT-3’; P3: 5’-TAAGGCCCACTTAAGTTGGGAAAG-3’; P4: 5’–CGGCGAAAGAAGAAGGTGCG-3’; P5: 5’-TTAAAAGGCTTTTTTTCCTGCAGAG–3’. Primers P1 and P2 were used to amplify simultaneously the coding sequence (CDS) of full-length PNRC (984bp), CDS of PNRC1c (747bp), and CDS plus part of 3’-UTR (untranslated sequence) of PNRC1f (523bp). Primes P3 and P2 were used to amplify part of 5’-UTR plus CDS of PNRC1d (474bp), and P4 and P5 for part of CDS of PNRC1e (414bp). The positions of the primers in each transcript are shown in Fig 1A. PCR products were purified using gel purification kit (Qiagen), and ligated into T vector pCR2.1, then transformed into DH5α competent cells. Colonies were screened by PCR and verified by restriction digestion and DNA sequencing.
Fig 1. Schematic representation of PNRC alternative splicing products.
(A). Schematic representation of human PNRC genomic structure, PNRC alternatively spliced transcript variants, and the position of the primers used in RT-PCR for identification of splicing variants. PNRC gene locates on chromosome 6 at 6q15 and contains two exons. Alternative splicing at exon 1 (termed 1a) or 2 of PNRC gene generates splicing variants PNRC1c, PNRC1f and PNRC1e. Alternative promoter usage generates splicing variant PNRC1d. The open boxes represent the putative amino acid-coding region of each transcript, the broken lines represent the part alternatively spliced out in the transcript, and the arrows P1–P5 indicate the positions of the primers used in RT-PCR for identification of PNRC splicing variants. (B). Schematic representation of deduced protein structure of PNRC isoforms. Full-length PNRC (fl PNRC) is composed of 327 amino acids (aa), containing two proline-rich Src homology domain-3 (SH3) binding motifs (aa 38–44, aa 285–291), one LXXLL motif (NR box, aa 319–323) and one putative nuclear localization signal (NLS, aa 94–101). PNRC1c lacks the middle 79 aa of fl PNRC (aa 103–181), PNRC1d is the carboxyl terminus 142 aa of fl PNRC (aa 186–327) due to noncoding of exon1b, whereas PNRC1e lacks 99 aa of fl PNRC (aa 229–327) but has 5 unique aa at the carboxyl terminus, and PNRC1f lacks 300 aa of fl PNRC (aa 28–327) but has 25 unique aa at the carboxyl terminus due to an altered reading frame and a stop codon resulting from alterative splicing.
2.2. Plasmid construction
All recombinant constructions were verified by DNA sequencing. The amino acid-coding sequences of PNRC splicing variants except PNRC1e were contained in the respective recombinants of pCR2.1-PNRC splicing variant generated by TA clone described above. The amino acid-coding sequence of PNRC1e was generated by PCR using pCR2.1-fl PNRC as the template and the following sequences as primers (forward primer: 5’-ATGACTGTCGTCTCCGTCCCG-3’; Reverse primer: 5’-TTAAAAGGCTTTTTTTCCTGCAGAGGTGATCTTGGTTAG -3’). The CDS of each PNRC isoforms was subcloned into the mammalian expression vector pCI (Promega) using NheI–XhoI restriction sites, respectively. To generate Gal4 AD-PNRC isoform fusion expression vectors for yeast two-hybrid, the CDS of each PNRC isoform was subcloned in-frame in the yeast expression vector pACT2 (Clontech) using BamHI–Xho I or EcoRI–XhoI restriction sites. To generate Gal4DBD-PNRC isoform fusion expression vectors for yeast two-hybrid, the CDS of each PNRC isoform was subcloned in-frame in the yeast expression vector pGBKT7 (Clontech) using only BamHI restriction site. To generate GFP-tagged PNRC isoform expression vectors, the CDS of each PNRC isoform was subcloned in-frame in the pEGFP-C1 vector using Xho I–BamHI or Xho I–EcoRI restriction sites. The construction of pGBT9-SF1, pGBT9-ERRa/HBD, pGBT9-ERα/HBD, pCI-ERα, pSG5-SF1, pGL3(SF1)3-SV40 and pGL3(ERE)3-SV40 were previously described (Zhou et al., 2000; Zhou and Chen, 2001).
2.3. Real-time PCR
Real-time quantitative PCR assay was performed using an Applied Biosystems 7700 sequence detector. Total RNA extraction and RT reaction were performed as described above. Real-time PCR was performed using QuantiTect SYBR Green PCR kit (Qiagen), according to the manufacturer's instructions. Samples were analyzed simultaneously for β-actin expression. Quantitative expression values were extrapolated from separate standard curves. Each sample was assayed in triplicate and normalized to β-actin. Real-time PCR was carried out using an isoform specific forward primer and a common reverse primer. The common reverse prime was 5’-GGCTTTTTTTCCTGCTTCTG-3’, and each isoform specific forward primer was as follows, full-length PNRC: 5’-CAAACCCCCCTCAGGAAAG -3’; PNRC1c: 5’-GCGAAAGAAGAAGGTTTTAAAAT-3’; PNRC1d: 5’-CCATCCTTTCAAGACTTCCTAT-3’; PNRC1f: 5’-CTTTTCCTCCCGAGGTTTTAAAAT-3’. The sizes of real-time PCR products are 236 bp (full-length PNRC), 228 bp (PNRC1c), 242 bp (PNRC1d) and 229 bp (PNRC1f), respectively.
2.4. Fluorescence microscopy
HeLa cells were maintained in Eagle’s MEM (Hyclone) supplemented with 10 % FBS, 200 IU/ml penicillin, 200 IU/ml streptomycin, and nonessential amino acid and sodium pyruvate at 37 °C in a 5 % CO2 incubator. The cells were seeded in 4-well chamber and cultured for 18–24 h, then transfected with 0.5µg GFP fusion expression vectors (pEGFP-C1-PNRC isoform). After 24h, the cells were fixed in cold methanol/acetone (1:1) for 10 min at −20 °C. Slides were mounted in DAPI solution (Vector) and analyzed under a confocal fluorescence microscope.
2.5. Yeast Two-Hybrid
Yeast two-hybrid assay was used to examine protein-protein interaction in vivo. The yeast two-hybrid assays were performed as previously described (Zhou et al., 2000; Zhou and Chen, 2001; Zhou et al., 2007). Briefly, the yeast strain Y187 was co-transformed with Gal4DBD-target fusion expression vector (pGBT9-SF1, pGBT9-ER/HBD, pGBT9-ERR1/HBD, or pGBKT7- PNRC isoform) and Gal4AD-PNRC isoform fusion expression vector (pACT2-PNRC isoform). The transformants that grew on SD/-Leu/-Trp agar plates were cultured overnight in liquid SD/-Leu/-Trp medium at 30°C, 250rpm. The overnight culture was diluted in liquid YPD medium containing appropriate amount of ligand or DMSO (vehicle control) and continued to culture for 4–6 h. Then a liquid culture β –galactosidase activity assay was performed using ONPG as substrate according to the manufacturer’s protocol (Clontech).
2.6. Transfection and luciferase assay
COS-7 cells were maintained in DMEM (Hyclone) supplemented with 10 % FBS, sodium pyruvate, 200 IU/ml penicillin, 200 IU/ml streptomycin, or in phenol-red-free DMEM supplemented with 10 % charcoal stripped FBS, sodium pyruvate, 200 IU/ml penicillin, 200 IU/ml streptomycin. COS-7 cells (1×105 cells per well in twelve-well plates) were transiently transfected using Lipofectamine2000 (Invitrogen) according to the manufacturer’s protocol. Generally, l.25 µg of plasmid DNA per well (0.2 µg reporter plasmid pGL3(ERE)3-SV40 or pGL3(SF1)3-SV40, 25 ng pCI-ERα or pSG5-SF1, and 0.2–1.0 µg of pCI-PNRC isoform) was used for the transfection, and the total amount of transfected DNA was kept constant by adding the responding empty plasmids. Five hours after transfection, the cells were changed to culture in medium containing 10 % FBS or 10 % charcoal stripped FBS with or without (in DMSO) 17 β-estrodial (10nM) for an additional 24 h. Subsequently, the cells were harvested and assayed for the protein concentrations and their luciferase activities using the Luciferase Reporter Assay System (Promega).
3. Results
3.1. Analysis of the human PNRC splicing variants by bioinformatics
Bioinformatics analysis, based on the blast of the expressed sequence tags (EST) of PNRC to human genome, revealed that there may exist six PNRC splicing variants named aAug05, bAug05, cAug05, dAug05, eAug05 and fAug05 (http://www.ncbi.nlm.nih.gov/IEB/Research/Acembly/. Their cDNA clone accession numbers are as follows: CR618392, BX463399, AY303779, BC044919, CR607268 and BG563008. For clarity, we refer to the original isoform as full-length PNRC or fl PNRC and the new six putative isoforms as PNRC1a, PNRC1b, PNRC1c, PNRC1d, PNRC1e and PNRC1f, respectively. Compared to fl PNRC (NM_006813), PNRC1a (CR618392) has196 bp longer 5’-UTR and additional 3’- UTR, it is supposed to code the same protein as fl PNRC although it may be an alternatively spliced transcript variant; PNRC1b (BX463399) is partial clone of fl PNRC because it just has incomplete 3’end compared to fl PNRC cDNA. Therefore, we discarded PNRC1a and PNRC1b for further study. Comparison of PNRC1c~PNRC1f sequences to the publicly available genomic sequence of human chromosome 6 (www.genome.ucsc.edu) revealed that PNRC1c and PNRC1f are produced through alternative recognition of the 3'-splice site in exon 1 (termed 1a) leading to the removal of part of exon 1 (237 bp and 461bp, respectively), and PNRC1e is yielded through the removal of part of exon 2 (59bp) resulting from alternative splicing in exon 2, whereas PNRC1d is generated by differential promoter usage. Details of the genomic organization of the human PNRC gene and the origin of various PNRC transcript variants are shown in Fig. 1A. The deduced amino acid sequences of four PNRC isoforms are shown in Figure1B. The fl PNRC contains 327 amino acids (aa) with a size of 35.2 kDa. By contrast, PNRC1c deletes in frame the middle 79 aa of fl PNRC (aa103-181), and has 248 aa with the predicted size of 27.3 kDa. PNRC1d is a truncated protein which has just the C-terminus 142 aa of fl PNRC (16.2 kDa) due to noncoding of exon 1b, while PNRC1e lacks 99 aa of fl PNRC but has 5 unique aa at the carboxyl terminus, and PNRC1f lacks 300 aa of fl PNRC but has 27 unique aa at the carboxyl terminus due to the reading frame shift and the introduction of a premature stop codon resulting from alterative splicing. The predicted size of PNRC1e (233aa) and PNRC1f (52aa) are 24.4 kDa and 5.6 kDa, respectively.
3.2. Identification and cloning of PNRC splicing variants
To test the real existence of PNRC splicing variants, a RT-PCR plus sequencing technique was used. We designed oligo-nucleotide primers targeted to the amino acid-coding sequence of the reference mRNA or isoform specific sequences based on these cDNA clone sequence (the positions of the primers in each transcript are shown in Fig 1A). RT-PCR was performed on five transformed cell lines derived from various tissues (MCF-7, HepG2, HeLa, Lovo and Raji cells) and three human tissues (breast, liver and brain). The PCR products were cloned in TA cloning vector pCR2.1 and sequenced for verification. Splicing variants PNRC1c, PNRC1d and PNRC1f as well as fl PNRC were detected in all the tested cell lines except that PNRC1c was not detected in HeLa cells (Fig 2). PNRC1d and PNRC1f were also detected in human breast, liver and brain tissues (data not shown). We failed to detect the splicing variant PNRC1e in all these cell lines and tissues. Sequence analysis of the cloned cDNA fragments verified that all the RT-PCR products were the exact corresponding splicing variants including PNRC1c, PNRC1d and PNRC1f.
Fig 2. RT-PCR analysis of PNRC splicing variants in various human cell lines.
Total RNAs from the indicated cell lines were analyzed by RT-PCR using primer pairs of P1 and P2 (upper panel) or P3 and P2 (lower panel). P1 and P2 were used to amplify simultaneously the coding sequence (CDS) of fl PNRC (984bp), CDS of PNRC1c (747bp), and CDS plus part of 3’-UTR of PNRC1f (523bp). P3 and P2 were used to amplify part of 5’-UTR plus CDS of PNRC1d (474bp). Arrows indicate fl PNRC and its splicing variants PNRC1c, PNRC1f and PNRC1d, respectively.
Our results demonstrated that splicing variants PNRC1c, PNRC1d and PNRC1f are truly expressed in the cells or tissues. Although the expression of PNRC1e in the tested cell lines and tissues was not detected, we could not rule out the possibility that PNRC1e could be a natural occurring isoform of PNRC. Therefore, we generated the amino acid-coding region of PNRC1e by PCR (detail see the material and method) and also characterized PNRC1e in this paper when analyzing other identified PNRC splicing variants.
3.3. The expression of PNRC splicing variants in different cell lines
As reverse transcription PCR cannot be used reliably to compare the levels of mRNA expression in a sample (Noton et al., 2006). We next used real-time PCR to provide accurate quantity of mRNA expression of these variants in the cell line MCF-7 and MCF-10A. As shown in Fig 3A, real-time PCR revealed that fl PNRC was the most abundant form, followed by PNRC1d and PNRC1f respectively, and PNRC1c was the least abundant in both cell lines.
Fig 3. Differential expression of PNRC splicing variants in human cell lines.
Total RNAs were isolated from the indicated cell lines and subjected to reverse transcription quantitative PCR. Real-time PCR was carried out using an isoform specific forward primer and a common reverse primer for each of the isoforms. Each sample was assayed in triplicate and normalized to β-actin. (A). Real-time quantitative PCR analysis of human PNRC splicing variants in MCF-10A and MCF-7. Expression of each PNRC splicing variant was plotted relative to β-actin levels. (B). Relative expression of each PNRC splicing variant in various cell lines as indicated.
Given that PNRC was firstly identified as a coactivator from mammary gland tissue and PNRC was found to be down-regulated in breast cancer (Zhou et al., 2000; Zhou et al., 2004; Zucchi et al., 2004), we compared the relative expression level of PNRC splicing variants in six different transformed cell lines derived from breast tissues using real-time quantitative PCR. As shown in Fig 3, the expression level of PNRC splicing variants varied in these cell lines, and the ratios of these splicing variants to fl PNRC also varied in these transformed cell lines (data not shown). It is noteworthy that the expression level of fl PNRC was much higher in the MCF-10A cell line than any other cell lines, whereas other splicing variants were expressed at a lower level in the MCF-10A cell line compared to other cell lines.
3.4. Subcellular localization of PNRC isoforms
To investigate subcellular localization of PNRC isoforms, GFP-tagged PNRC isoform expression vectors were generated and transfected into HeLa cells. The subcellular localization of GFP-tagged PNRC isoforms was examined under confocal microscope. As shown in Fig 4, GFP-tagged fl PNRC, PNRC1c or PNRC1e was predominantly localized in the nucleus, while GFP-tagged PNRC1d or PNRC1f distributed in both the cytoplasm and nucleus.
Fig 4. Subcellular localization of GFP-tagged PNRC isoforms.
HeLa cells were transfected with plasmids expressing GFP-tagged PNRC isoform, respectively. Twenty-four hours after transfection, cells were fixed and stained with DAPI. Subcellular localization of GFP-tagged PNRC isoforms was examined under confocal fluorescence microscopy. GFP-tagged PNRC isoforms are shown in green, and nuclei stained with DAPI are shown in blue.
3.5. PNRC isoforms differ in their binding to nuclear receptors
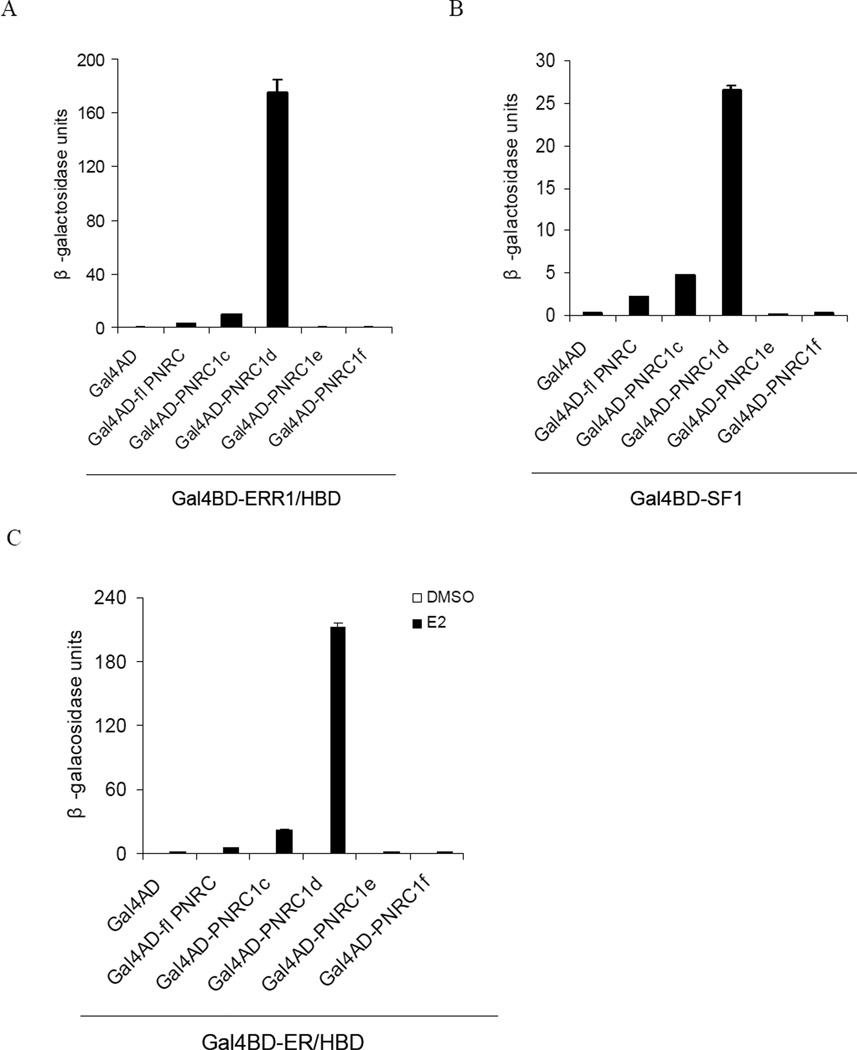
To test PNRC isoforms for their activity to bind NRs, a yeast two-hybrid assay was performed to examine the interactions between PNRC isoforms and SF1, the hormone-binding domain (HBD) of ERR1 or ERα. As shown in Fig 5, fl PNRC, PNRC1c and PNRC1d interacted with ERα/HBD in a ligand-dependent manner, and interacted with ERR1/HBD and SF1 in a ligand-independent manner, whereas PNRC1e and PNRC1f failed to interact with all these tested NRs. Compared to fl PNRC, both PNRC1c and PNRC1d had an increased affinity to bind the tested NRs, especially PNRC1d, displayed an over 10-fold increase of affinity in the yeast two-hybrid assay.
Fig 5. PNRC isoforms differ in their activity to interact with nuclear receptors in yeast.
Yeast strain Y187 was cotransformed with Gal4DBD fusion expression vector for nuclear receptor SF1, ERRα/HBD or ER/HBD (pGBT9-ERR1/HBD, pGBT9-SF1 or pGBT9-ER/HBD) and Gal4AD fusion expression vector for PNRC isoform (pACT2-PNRC isoform). The positive yeast clones were selected by growth on SD/-Trp/-Leu agar plate. The interactions between PNRC isoforms and ERR1/HBD (A), SF1 (B) or ERα/HBD (C) were analyzed by measuring β-galacosidase activity of liquid cultures of the positive yeast clones bearing both vectors. Relative β-galactosidase activities in liquid cultures were expressed in Miller units as mean ± SD of three independent assays.
3.6. PNRC isoforms differ in their ability to potentiate transcription by ER or SF1
To determine whether these PNRC isoforms also differ in their ability to potentiate the NR mediated transcriptions, the respective isoform expression vectors were co-transfected into Cos-7 cells with an expression plasmid encoding the ERα or SF1 and a luciferase reporter plasmid containing three copies of ERE or SF1 response element. As shown in Fig 6, PNRC1c and PNRC1d stimulated ERα and SF1 transactivation function, but both were less potent in their actions than fl PNRC, whereas PNRC1e and PNRC1f did not potentiate ERα or SF1 mediated transcription. It is noteworthy that fl PNRC, PNRC1c and PNRC1d also stimulated the transcriptional activity of ERα in the absence of ligand, which could result from their interaction with transactivation function 1 (AF1) of ERα because in our study they were found to interact with ERα/AF1 in the absence of ligand (data not shown). Our previous study also showed that fl PNRC could interact with ERα/AF1 in the absence of ligand (Zhou et al., 2006).
Fig 6. PNRC isoforms differ in their activity to stimulate nuclear receptor mediated transcription by ERα or SF1.
Cos-7 cells were maintained in phenol-red-free DMEM with 10 % charcoal stripped FBS (A and C), or in the regular DMEM with 10 % FBS (B). Cells were transiently co-transfected with 0.125µg reporter pGL3(ERE)3-SV40 or pGL3(SF1)3-SV40, 25ng pCI-hERα or 0.2µg pSG5-SF1, and 1µg pCI-PNRC isoform or pCI. The total amount of transfected DNA was kept constant by adding the corresponding empty vectors. The transfected cells were added with DMSO or the ligand. Twenty-four hours after transfection, the cells were harvested and assayed for the protein concentrations and their luciferase activities using the Luciferase Reporter Assay System. The activity of PNRC isoforms to stimulate ERα (A) or SF1 (B) meditated transcription was analyzed by measuring relative luciferase activity of 10 µg cell extracts. The data are expressed as Mean ± SD of triplicate transfections. (C), Comparison of the ability of PNRC isoforms to potentiate ERα mediated transcription in the presence of the ligand at different concentration. Cos-7 cells were transiently cotransfected with 0.2 µg reporter plasmid pGL3(ERE)3-SV40, 10ng pCI-hERα, and different amount of PNRC isoform expression vectors (0.05µg, 0.2µg and 0.5µg). The transfection and the luciferase assays were performed as described for (A) and (B).
3.7. PNRC1d modulates the activity of full-length PNRC
Given that PNRC1d had much stronger interaction with NRs but less potent to enhance NR meditated transcriptions compared to fl PNRC, we hypothesized that PNRC1d competes for binding NRs and therefore modulates the activity of fl PNRC. To test this hypothesis, two cotransfection experiments were performed. Firstly, Cos-7 cells were transfected with two different equal amounts of fl PNRC and PNRC1d expression vectors (ratio 1:1) with the fixed concentration of reporter gene and ERα expression plasmid. As shown in Figure 7A, PNRC1d impaired fl PNRC activity of enhancing ERα mediated transcription at both concentrations. Secondly, when Cos-7 cells were transfected with increasing amounts of PNRC1d expression plasmid in the presence of fixed concentrations of fl PNRC expression plasmid, PNRC1d attenuated the activity of fl PNRC in a dose-dependent manner (Fig. 7B). These results suggest that PNRC1d could function as a modulator on fl PNRC in enhancing ERα mediated transcription.
Fig 7. PNRC1d modulates the activity of fl PNRC in enhancing ERα mediated transcription.
Cos-7 cells grown in phenol-red-free DMEM with 10 % charcoal stripped FBS were transiently cotransfected with reporter pGL3(ERE)3-SV40 and ERα expression plasmid and additional plasmids for fl PNRC and PNRC1d as indicated. Five hours after transfection, cells were changed to culture in the presence of 10 nM 17β-estrodail. Twenty-four hours after transfection, the cells were harvested and assayed for the protein concentrations and their luciferase activities using the Luciferase Reporter Assay System. The data are expressed as mean of triplicate transfections ± standard error. (A). Cells were cotransfected with different equal amounts of fl PNRC and PNRC1d expression vectors (ratio 1:1) as indicated in addition to reporter gene (0.2µg) and ERα expression plasmid (10 ng). (B). Cells were cotransfected with increasing amounts of PNRC1d expression vectors (0.05µg, 0.1µg, 0.2 µg and 0.5 µg) and a constant amount of fl PNRC expression vector (0.5 µg) in addition to reporter gene (0. 2 µg) and ERα expression plasmid (25 ng).
3.8. PNRC1d does not interact with full-length PNRC
Previously, we had isolated many cDNA clones encoding the various length fragments of PNRC C-terminus when using aa 270–327 of PNRC as the bait to screen PNRC-interacting proteins in a yeast two-hybrid (data not published), suggesting that PNRC and its isoforms containing aa 270–327 of PNRC may interact with each other. To test the possibility that PNRC1d inhibited fl PNRC activity through its direct interaction with fl PNRC, a yeast two-hybrid assay was performed. There was no detectable interaction between fl PNRC and PNRC1d or other isoforms, while a weak interaction between PNRC1d and itself was detected (Fig 8). This result suggests that PNRC1d repressed the activity of fl PNRC not through its direct interaction with fl PNRC.
Fig 8. Analysis of the interaction between PNRC isoforms by yeast two-hybrid.
Yeast strain Y187 was cotransformed with Gal4DBD fusion expression vector for fl PNRC or PNRC1d (pGBKT7-fl PNRC or pGBKT7-PNRC1d) and Gal4AD fusion expression vector for PNRC isoform (pACT2-PNRC isoform). The positive yeast clones were selected by growth on SD/-Trp/-Leu agar plate. The interactions between PNRC isoforms were analyzed by measuring β-galacosidase activity of liquid cultures of the yeast clones bearing both vectors. Relative β-galactosidase activities in liquid cultures were expressed in Miller units as mean ± SD of three independent assays.
4. Discussion
Alternative splicing is one of the most important mechanisms used to generate diversity and complexity of proteome from the limited human genes. It is estimated that up to 35–60 % human genes are alternatively spliced (Stamm et al., 2005). Unlike regulation of promoter activity, which just alters the amount of transcripts, alternative splicing can change the structure of transcripts and their encoded proteins, resulting in removing and/or inserting a specific domain or altering a few residues of protein. Through changing the protein primary structure, alternative splicing produces distinctly functional isoforms that are often critical for specific cellular functions. Alternative splicing is cell-type-specific and developmentally regulated, and altered alternative splicing patterns are often associated with cancer and other different kinds of diseases (Faustino and Cooper, 2003; Brinkman, 2004; Kalnina et al., 2005).
In this study, we have identified three PNRC isoforms designated PNRC1c, PNRC1d and PNRC1f, resulting from alternative splicing or alternate promoter usage. PNRC1c and PNRC1d both remove a few residues and are just part of fl PNRC in protein primary structure. PNRC1f misses 300 aa of fl PNRC but gains 25 unique aa at the carboxyl terminus due to an altered reading frame and a stop codon resulting from alternative recognition of the 3'-splice site in exon 1. Since no reliable antibodies recognizing PNRC and its isoforms are available, we could not detect these isoforms in protein level.
The four PNRC isoforms including fl PNRC are differentially expressed in the test cell lines derived from breast normal tissue or cancer, fl PNRC is expressed at higher levels in MCF-10A cells compared with other cell lines derived from breast cancer, whereas other isoforms are expressed at lower levels in MCF-10A. The significance of this observation remains to be explored. These isoforms also have different subcellular distributions. Unlike other isoforms that predominantly localize in the nucleus, PNRC1d and PNRC1f distribute in both the cytoplasm and nucleus, which may result from the removal of the nuclear localization signal sequence due to alternative splicing. This result implies these isoforms may function distinctively in the cell.
Due to the change in the primary protein structure resulting from alternative splicing, PNRC isoforms displayed great difference in their activities to bind NRs and stimulate NR meditated transcriptions. Previous studies showed that PNRC interacted with NRs mainly through the SH3-binding motif at C-terminal regions of PNRC (aa 285–291), and the region aa 270–327 of PNRC was essential and sufficient to interact with NRs (Zhou et al., 2000; Zhou et al., 2006). Consistent with these results, in the present study isoforms PNRC1c and PNRC1d both harboring the SH3-binding motif interacted with ERR1, SF1 and ERα, whereas PNRC1e and PNRC1f without the SH3 motif were unable to. In addition, the less sequences outside the region aa 270–327 of PNRC the isoform contains, the higher affinity it has to bind NRs. In contrast to fl PNRC, PNRC1c displayed about two-to threefold increase of the affinity to bind SF1, ERR1/HBD and ER/HBD, while PNRC1c exhibited over tenfold increase of affinity to bind these NRs in yeast two-hybrid assay. Although PNRC1c and PNRC1d displayed stronger interactions with NRs than fl PNRC, our study showed that they had much less activity to enhance NR meditated transcriptions. These results suggest that the domains that PNRC1c and PNRC1d miss compared to fl PNRC are necessary for PNRC optimized coactivator activity. Our study further showed that PNRC1d could repress coactivator activity of fl PNRC through competing for ER binding. In our previous study, the fragment of PNRC (aa 270–327) was found to be a dominant negative inhibitor of PNRC (data not published). Therefore, we could consider PNRC1d as a natural occurring inhibitor of PNRC in vivo to some extent.
Our previous study showed that PNRC interacts with ERα in a different manner from the p160 coactivators like SRC2/GRIP1 (Zhou et al., 2006). It is tempting to speculate that ERα could form a more active complex by interaction with multiple coactivators simultaneously including PNRC. Now more and more researches reveal that isoforms of NRs including ER exhibit differences in ligand selectivity and coactivators recruitment, and different coactivators recruitment is the molecular basis of context-specific transcriptional responses to nuclear receptor signaling (Baek and Rosenfeld, 2004; Smith and O'Malley, 2004; Zhao et al., 2005). Therefore, we believe that PNRC may play a role in some specific ERα signaling. Work is under way to investigate the role of PNRC in ERα signaling, and physiological significance of these novel isoforms of PNRC. In conclusion, our results reveal the existence of three alternative PNRC transcripts variants resulting from alternative splicing or alternate promoter usage, and these isoforms differ in their activities to bind NRs and to enhance NR meditated transcriptions. Moreover, PNRC1d modulates activity of fl PNRC. Our finds suggest alternative splicing may serve as a regulatory mechanism of PNRC coactivator activity.
ACKNOWLEDGEMENTS
We thank Sheryl Phung for English proof reading of the manuscript. This work was supported by grants from National Natural Science Foundation of P.R. China (DZ, No. 30671056, 30600299 and 30672055) and NIH grants CA44735 and DK60560 (SC).
Abbreviations
- aa
amino acid
- AF1
transactivation function 1
- AIB1
amplified in breast cancer 1
- AR
androgen receptor
- CDS
coding sequences
- CoAA
coactivator activator
- ER
estrogen receptor
- ERR
estrogen receptor-related receptor
- EST
expressed sequence tags
- fl
full-length
- HBD
the hormone-binding domain
- NR
nuclear receptor
- PNRC
proline-rich nuclear receptor coregulatory protein
- SF1
steroidogenic factor 1
- SH3
Src homology domain-3
- TZF
testicular zinc-finger protein
- UTR
untranslated region
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Albers M, Kranz H, Kober I, Kaiser C, Klink M, Suckow J, Kern R, Koegl M. Automated yeast two-hybrid screening for nuclear receptor-interacting proteins. Mol Cell Proteomics. 2005;4:205–213. doi: 10.1074/mcp.M400169-MCP200. [DOI] [PubMed] [Google Scholar]
- Auboeuf D, Dowhan DH, Li X, Larkin K, Ko L, Berget SM, O'Malley BW. CoAA, a nuclear receptor coactivator protein at the interface of transcriptional coactivation and RNA splicing. Mol Cell Biol. 2004;24:442–453. doi: 10.1128/MCB.24.1.442-453.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek SH, Rosenfeld MG. Nuclear receptor coregulators: their modification codes and regulatory mechanism by translocation. Biochem Biophys Res Commun. 2004;319:707–714. doi: 10.1016/j.bbrc.2004.04.169. [DOI] [PubMed] [Google Scholar]
- Brinkman BM. Splice variants as cancer biomarkers. Clin Biochem. 2004;37:584–594. doi: 10.1016/j.clinbiochem.2004.05.015. [DOI] [PubMed] [Google Scholar]
- Chen J, Liu L, Pohajdak B. Cloning a cDNA from human NK/T cells which codes for a protein with high proline content. Biochim Biophys Acta. 1995;1264:19–22. doi: 10.1016/0167-4781(95)00159-e. [DOI] [PubMed] [Google Scholar]
- Faustino NA, Cooper TA. Pre-mRNA splicing and human disease. Genes Dev. 2003;17:419–437. doi: 10.1101/gad.1048803. [DOI] [PubMed] [Google Scholar]
- Glass CK, Rosenfeld MG. The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev. 2000;14:121–141. [PubMed] [Google Scholar]
- Iwasaki T, Chin WW, Ko L. Identification and characterization of RRM-containing coactivator activator (CoAA) as TRBP-interacting protein, and its splice variant as a coactivator modulator (CoAM) J Biol Chem. 2001;276:33375–33383. doi: 10.1074/jbc.M101517200. [DOI] [PubMed] [Google Scholar]
- Kalnina Z, Zayakin P, Silina K, Line A. Alterations of pre-mRNA splicing in cancer. Genes Chromosomes Cancer. 2005;42:342–357. doi: 10.1002/gcc.20156. [DOI] [PubMed] [Google Scholar]
- Kanai Y, Ushijima S, Saito Y, Nakanishi Y, Sakamoto M, Hirohashi S. MRNA expression of genes altered by 5-azacytidine treatment in cancer cell lines is associated with clinicopathological parameters of human cancers. J Cancer Res Clin Oncol. 2001;127:697–706. doi: 10.1007/s004320100284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X, Arulsundaram VD, Yousef AF, Webb P, Baxter JD, Mymryk JS, Walfish PG. Corepressor/coactivator paradox: potential constitutive coactivation by corepressor splice variants. Nucl Recept Signal. 2006;4:e022. doi: 10.1621/nrs.04022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noton EA, Colnaghi R, Tate S, Starck C, Carvalho A, Ko Ferrigno P, Wheatley SP. Molecular analysis of survivin isoforms: evidence that alternatively spliced variants do not play a role in mitosis. J Biol Chem. 2006;281:1286–1295. doi: 10.1074/jbc.M508773200. [DOI] [PubMed] [Google Scholar]
- Reiter R, Oh AS, Wellstein A, Riegel AT. Impact of the nuclear receptor coactivator AIB1 isoform AIB1-Delta3 on estrogenic ligands with different intrinsic activity. Oncogene. 2004;23:403–409. doi: 10.1038/sj.onc.1207202. [DOI] [PubMed] [Google Scholar]
- Reiter R, Wellstein A, Riegel AT. An isoform of the coactivator AIB1 that increases hormone and growth factor sensitivity is overexpressed in breast cancer. J Biol Chem. 2001;276:39736–39741. doi: 10.1074/jbc.M104744200. [DOI] [PubMed] [Google Scholar]
- Smith CL, O'Malley BW. Coregulator function: a key to understanding tissue specificity of selective receptor modulators. Endocr Rev. 2004;25:45–71. doi: 10.1210/er.2003-0023. [DOI] [PubMed] [Google Scholar]
- Stamm S, Ben-Ari S, Rafalska I, Tang Y, Zhang Z, Toiber D, Thanaraj TA, Soreq H. Function of alternative splicing. Gene. 2005;344:1–20. doi: 10.1016/j.gene.2004.10.022. [DOI] [PubMed] [Google Scholar]
- Tao RH, Kawate H, Ohnaka K, Ishizuka M, Hagiwara H, Takayanagi R. Opposite effects of alternative TZF spliced variants on androgen receptor. Biochem Biophys Res Commun. 2006;341:515–521. doi: 10.1016/j.bbrc.2005.12.213. [DOI] [PubMed] [Google Scholar]
- Tao RH, Kawate H, Wu Y, Ohnaka K, Ishizuka M, Inoue A, Hagiwara H, Takayanagi R. Testicular zinc finger protein recruits histone deacetylase 2 and suppresses the transactivation function and intranuclear foci formation of agonist-bound androgen receptor competitively with TIF2. Mol Cell Endocrinol. 2006;247:150–165. doi: 10.1016/j.mce.2005.12.052. [DOI] [PubMed] [Google Scholar]
- Tilli MT, Reiter R, Oh AS, Henke RT, McDonnell K, Gallicano GI, Furth PA, Riegel AT. Overexpression of an N-terminally truncated isoform of the nuclear receptor coactivator amplified in breast cancer 1 leads to altered proliferation of mammary epithelial cells in transgenic mice. Mol Endocrinol. 2005;19:644–656. doi: 10.1210/me.2004-0106. [DOI] [PubMed] [Google Scholar]
- Wang Y, Miksicek RJ. Identification of a dominant negative form of the human estrogen receptor. Mol Endocrinol. 1991;5:1707–1715. doi: 10.1210/mend-5-11-1707. [DOI] [PubMed] [Google Scholar]
- Xu L, Glass CK, Rosenfeld MG. Coactivator and corepressor complexes in nuclear receptor function. Curr Opin Genet Dev. 1999;9:140–147. doi: 10.1016/S0959-437X(99)80021-5. [DOI] [PubMed] [Google Scholar]
- Zhao C, Toresson G, Xu L, Koehler KF, Gustafsson JA, Dahlman-Wright K. Mouse estrogen receptor beta isoforms exhibit differences in ligand selectivity and coactivator recruitment. Biochemistry. 2005;44:7936–7944. doi: 10.1021/bi047691m. [DOI] [PubMed] [Google Scholar]
- Zhou D, Chen B, Ye JJ, Chen S. A novel crosstalk mechanism between nuclear receptor-mediated and growth factor/Ras-mediated pathways through PNRC-Grb2 interaction. Oncogene. 2004;23:5394–5404. doi: 10.1038/sj.onc.1207695. [DOI] [PubMed] [Google Scholar]
- Zhou D, Chen S. PNRC2 is a 16 kDa coactivator that interacts with nuclear receptors through an SH3-binding motif. Nucleic Acids Res. 2001;29:3939–3948. doi: 10.1093/nar/29.19.3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D, Quach KM, Yang C, Lee SY, Pohajdak B, Chen S. PNRC: a proline-rich nuclear receptor coregulatory protein that modulates transcriptional activation of multiple nuclear receptors including orphan receptors SF1 (steroidogenic factor 1) and ERRalpha1 (estrogen related receptor alpha-1) Mol Endocrinol. 2000;14:986–998. doi: 10.1210/mend.14.7.0480. [DOI] [PubMed] [Google Scholar]
- Zhou D, Ye JJ, Li Y, Lui K, Chen S. The molecular basis of the interaction between the proline-rich SH3-binding motif of PNRC and estrogen receptor alpha. Nucleic Acids Res. 2006;34:5974–5986. doi: 10.1093/nar/gkl764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D, Zhong S, Ye JJ, Quach KM, Johnson DL, Chen S. PNRC is a unique nuclear receptor coactivator that stimulates RNA polymerase III-dependent transcription. J Mol Signal. 2007;2:5. doi: 10.1186/1750-2187-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zucchi I, Mento E, Kuznetsov VA, Scotti M, Valsecchi V, Simionati B, Vicinanza E, Valle G, Pilotti S, Reinbold R, Vezzoni P, Albertini A, Dulbecco R. Gene expression profiles of epithelial cells microscopically isolated from a breast-invasive ductal carcinoma and a nodal metastasis. Proc Natl Acad Sci U S A. 2004;101:18147–18152. doi: 10.1073/pnas.0408260101. [DOI] [PMC free article] [PubMed] [Google Scholar]