Abstract
Transcription factor SOX10 plays a role in the maintenance of progenitor cell multipotency, lineage specification, and cell differentiation and is a major actor in the development of the neural crest. It has been implicated in Waardenburg syndrome (WS), a rare disorder characterized by the association between pigmentation abnormalities and deafness, but SOX10 mutations cause a variable phenotype that spreads over the initial limits of the syndrome definition. On the basis of recent findings of olfactory-bulb agenesis in WS individuals, we suspected SOX10 was also involved in Kallmann syndrome (KS). KS is defined by the association between anosmia and hypogonadotropic hypogonadism due to incomplete migration of neuroendocrine gonadotropin-releasing hormone (GnRH) cells along the olfactory, vomeronasal, and terminal nerves. Mutations in any of the nine genes identified to date account for only 30% of the KS cases. KS can be either isolated or associated with a variety of other symptoms, including deafness. This study reports SOX10 loss-of-function mutations in approximately one-third of KS individuals with deafness, indicating a substantial involvement in this clinical condition. Study of SOX10-null mutant mice revealed a developmental role of SOX10 in a subpopulation of glial cells called olfactory ensheathing cells. These mice indeed showed an almost complete absence of these cells along the olfactory nerve pathway, as well as defasciculation and misrouting of the nerve fibers, impaired migration of GnRH cells, and disorganization of the olfactory nerve layer of the olfactory bulbs.
Introduction
SOX10 belongs to the SOX transcription-factor family, whose members are involved in a multitude of developmental and cellular processes.1 First identified as a glial cell transcription factor, it was soon revealed as a major player in the development of neural crest (NC) cells. NC cells are a population of multipotent precursor cells that emerge at the borders of the neural tube, migrate extensively throughout the embryo, and differentiate into a variety of cell types, including skin pigment cells and neurons and glia of the peripheral and enteric nervous systems.2 SOX10 plays a role in the maintenance of progenitor multipotency, specification, and differentiation of numerous cell types through the regulation of several transcriptional targets.1,3–5 Its involvement in Waardenburg syndrome (WS) contributed significantly to the understanding of its function in NC cells in general and in the melanocytic and enteric lineages in particular.6
WS is a clinically and genetically heterogeneous condition that manifests with sensorineural congenital deafness and abnormal pigmentation of the hair, skin, and iris. Four subtypes (WS1–WS4 [MIM 193500, 193510, 148820, 277580, respectively]) and a neurological variant (peripheral demyelinating neuropathy, central dysmyelinating leukodystrophy, Waardenburg syndrome, Hirschsprung disease [PCWH (MIM 609136)]) have been described.6 Since 1998, approximately 100 heterozygous point mutations or deletions of SOX10 (MIM 602229) have been reported, first in WS4 (WS with Hirschsprung disease, also called megacolon),7 then in its neurological variant,8 and finally in WS2 (WS without Hirschsprung disease)9 (see also the WS gene mutation database at LOVD [Leiden Open Variation Database]).6
To delineate the range of temporal-bone abnormalities associated with impaired SOX10 function, we recently performed a study of MRI and/or computed–tomography (CT) scans of inner ears from 15 WS individuals. Incidentally, this study also revealed an unexpectedly high frequency of olfactory-bulb agenesis (88% of the cases who could be analyzed),10 a finding that had been described only once before in association with a SOX10 mutation.11 Notably, one of the males included in our radiological study had previously been reported as having anosmia and hypogonadism.9
The association between anosmia and hypogonadotropic hypogonadism is known as Kallmann syndrome (KS [MIM 147950, 244200, 308700, 610628, 612370, and 612702]) and is explained by a pathological sequence in embryonic development. Premature interruption of the vomeronasal and terminal nerve fibers in the peripheral olfactory system have been shown to result in incomplete migration of the neuroendocrine gonadotropin-releasing hormone (GnRH) cells along these nerves and to thus preclude them from penetrating into the forebrain and reaching their final destination in the preoptic and hypothalamic region.12 The prevalence of KS has been estimated at 1 in 8,000 males and 1 in 40,000 females. To date, nine genes—namely KAL1, FGFR1, FGF8, PROKR2, PROK2, WDR11, HS6ST1, CHD7, and SEMA3A (MIM 300836, 136350, 600483, 607123, 607002, 606417, 604846, 608892, and 603961, respectively)—have been implicated, but mutations in any of these genes have been identified in only approximately 30% of KS individuals.13 KS can be associated with a variety of nonolfactory, nonreproductive symptoms, including deafness, but the association between KS and deafness has so far received little genetic explanation.
Here, we asked whether persons affected by KS carry mutations in SOX10. We found a notable prevalence (about 38%) of SOX10 mutations in a group of individuals presenting with the clinical association between KS and hearing impairment, thereby shedding new light on this clinical association. We showed that these mutations affect SOX10 function in vitro. Homozygous Sox10 mutant mice show an almost complete absence of olfactory ensheathing cells (OECs) along the olfactory, vomeronasal, and terminal nerves, abnormal fasciculation and axonal misrouting of the olfactory neurons, impaired GnRH cell migration, and disorganization of the olfactory-nerve layer of the olfactory bulbs.
Subjects and Methods
Subjects
This study was approved by the French national research-ethics committee (Agence de Biomedicine, Paris, France). Seventeen cases (nine males and eight females) presenting with KS and at least one WS-like feature were investigated, as well as 86 “random” cases (67 males and 19 females), 20 of whom had various nonolfactory, nonreproductive associated anomalies (including eight with cleft lip or palate), who were addressed for genetic exploration of KS. Hypogonadism was diagnosed on the basis of clinical and hormonal evaluation, whereas anosmia and hyposmia were diagnosed on the basis of the individual’s interview and were confirmed by olfactory tests with increasing concentrations of odorant molecules (olfactometry) and/or MRI showing agenesis of the olfactory bulbs. Informed consent for genetic testing was obtained. Genomic DNA was extracted from peripheral-blood leukocytes according to standard protocols. These individuals did not carry mutations in the coding sequences of previously analyzed genes, specifically, KAL1, FGFR1, FGF8, PROKR2, and PROK2, all involved in KS.
Mutation Screening
The coding exons of SOX10 were analyzed by Sanger sequencing of the PCR products as previously described.9 Mutations were named according to the international nomenclature based on RefSeq accession number NM_006941.3 for SOX10 cDNA. SOX10 deletions or rearrangements were sought with the use of quantitative multiplex fluorescent PCR with a method slightly modified from that of Bondurand et al.9 The three coding exons were amplified in five amplicons sorted in two different reaction mixes. The sequences of the primers used are available upon request. Individuals carrying a SOX10 mutation were further tested for the presence of mutations in the other genes involved in KS or in nonsyndromic congenital hypogonadotropic hypogonadism; specifically, these genes were CHD7, WDR11, HS6ST1, SEMA3A, GNRHR, GNRH1, TACR3, TAC3, KISS1R, and KISS1. To confirm the de novo occurrence of the mutation, we conducted a comparison with the parental DNA (when available) through analysis of six microsatellites located on different chromosomes by using the linkage mapping set (Applied Biosystems) according to the manufacturer’s instructions.
Animals and Genotyping
The generation and genotyping of Sox10lacZ mice (Sox10tm1Weg) have been described previously.14 Experiments were performed in accordance with the ethical guidelines of the Institut National de la Santé et de la Recherche Médicale. Embryos at embryonic day (E) 12.5 and E14.5 were obtained from staged pregnancies, fixed in 4% paraformaldehyde (PFA) at 4°C, and frozen in an optimal-cutting-temperature compound. Head sections (16 μm thick) were cut with a Leica CM3050S cryostat. Alternatively, embryo heads were fixed in 1% PFA and 0.2% glutaraldehyde and used for X-Gal staining.
Immunostaining and X-Gal Staining
X-Gal staining followed standard procedures. Immunostaining of mouse embryo sections was performed as described.15 The primary antibodies used were anti-SOX10 (N-20 or D-20) (goat, 1/50, Santa Cruz), anti-TUJ1 (mouse, 1/1,000, Eurogentec), anti-BLBP/BFABP (rabbit, 1/5,000, kindly provided by T. Müller),16 anti-P75 (rabbit, 1/500, Promega), anti-S100 (rabbit, 1/400, Dako Cytomation), anti-GnRH (rabbit, 1/500, Abcam), and anti-β-galactosidase (chicken, 1/500, Abcam). Secondary antibodies were anti-goat-FITC, anti-rabbit-Cy3, anti-mouse-Cy3, anti-goat-Alexa Fluor 555, anti-rabbit-Alexa Fluor 488, anti-mouse-Alexa Fluor 647, anti-guinea-pig-Cy3, and anti-chicken-Alexa Fluor 488 (1/200, Invitrogen or Jackson ImmunoResearch). TUNEL staining was performed with the In Situ Cell Death Detection kit, fluorescein (Roche) according to the manufacturer’s instructions. We estimated the total number of GnRH cells by counting the GnRH-positive cell bodies in more than two-thirds of the sections, from the vomeronasal organ to the end of their migration pathway. The number of GnRH cells on the missing slides was scored as the mean of the previous and following ones. Preparations were mounted in Vectashield and examined with an Olympus SZH10 stereo-microscope coupled to the Visilog capture program or a Zeiss Axioplan 2 confocal microscope coupled to the Metamorph software package.
Human Tissues
A human fetus was obtained from a medically terminated pregnancy at 8 weeks of embryonic development (the parents provided written informed consent). After fixation in a 4% formaldehyde solution, the head was embedded in paraffin. Serial sagittal sections (4 μm thick) were cut with a Microm HM340E microtome and collected on Superfrost Plus slides (Thermo Scientific). Immunochemistry analysis was conducted as described17 with the SOX10, TUJ1, and S100 antibodies cited above. Because of the limited number of slides, double SOX10/S100 staining was performed on the nasal mesenchyme only.
Plasmids
The pECE-SOX10, pCMV-SOX10Myc, pECE-PAX3, pECE-EGR2, pGL3-MITFdel1718, and pGL3-Cx32 vectors have been described previously.18–20 The mutations identified were introduced into the pECE-SOX10 (in the case of the c.2T>G mutation) or pCMV-SOX10Myc constructs by site-directed mutagenesis with the use of the QuikChange mutagenesis kit (Stratagene). The pGL3-MPZ construct was kindly provided by J. Svaren.21
Cell Culture, Transfection, and Reporter Assays
HeLa cells were grown in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum and transfected with Lipofectamine Plus reagent (Invitrogen). Cells were plated on 12-well plates and transfected 1 day later with 0.175 μg of each effector and reporter plasmid for the MITF (microphthalmia-associated transcription factor) and GJB1 (also known as Cx32) promoter study. In the MPZ enhancer study, 0.250 μg of reporter plasmid was used. As far as the competition assays are concerned, increasing amounts of altered SOX10 plasmids (0.175, 0.350, or 0.525 μg) were mixed with a fixed amount of wild-type SOX10 plasmid (0.175 μg) and reporter constructs. In each case, the total amount of plasmid was kept constant by the addition of empty pECE or pCMV-Myc vectors. Twenty-four or 48 hr after transfection, cells were washed twice with PBS and lysed, and the extracts were assayed for luciferase activity with the Luciferase Assay System (Promega) as described previously.9,19,22 Production of SOX10 was assessed by immunoblot analysis according to standard protocols.23 Quantification was performed with GeneTools v.3.05 software. Alternatively, cultures were fixed in 4% PFA for 10 min at room temperature, and immunocytochemistry was performed as described above with the use of anti-SOX10 (N-20 or D-20).
In Silico Analysis of the Mutations
Mutations were analyzed with PolyPhen-2, SIFT, Alamut 2.0.2 software for splicing, dbSNP, the 1000 Genomes Project, and the National Heart, Lung, and Blood Institute (NHLBI) Exome Sequencing Project Exome Variant Server. The conformation files for DNA-bound SOX17 were imported from the Protein Data Bank (accession number 3F27) and visualized with Swiss-PdbViewer software.24,25
Results
SOX10 Mutations Are Frequent in Individuals with KS and Deafness
We first searched for SOX10 point mutations and deletions in individuals who had been diagnosed with KS and who also presented with one or several of the known features of SOX10-linked WS. We screened 17 cases (9 males and 8 females) previously found negative for mutations in the main genes involved in KS; 13 of them presented with deafness (unilateral in three cases), two had pigmentation defects (hair and skin hypopigmentation; early graying in one, who was deaf), two had intellectual disability, and one showed psychomotor delay (both of these latter two features can be found in the neurological variant of WS [PCWH]). Eleven cases also had additional symptoms unrelated to WS.
Six new SOX10 nucleotide changes were identified in this cohort: a mutation of the initiation codon (c.2T>G [p.?]), two nucleotide changes predicted to cause missense substitutions within the HMG domain (c.331T>G [p.Phe111Val] and c.424T>C [p.Trp142Arg]), a splice-site mutation (c.698-1G>C), a 1 bp deletion predicted to result in a frameshift (c.1290del [p.Ser431Argfs*71]), and finally a nucleotide change predicted to result in a missense substitution located in the transactivation domain (c.1298G>A [p.Arg433Gln]). These changes were not found in at least 50 control individuals (100 chromosomes) of the same origin, and the relevant databases (dbSNP, 1000 Genomes Project, NHLBI Exome Variant Server) did not provide evidence that any of these variations are found in control populations.
Clinical and molecular findings in persons carrying SOX10 mutations are summarized in Table 1 (cases A–F). All six individuals had anosmia and either absent or delayed spontaneous puberty. Two males had cryptorchidism, and one also had a micropenis. One of them demonstrated early graying of the hair. Interestingly, all but one were deaf. Pedigrees of the two familial cases are represented in Figure S1A, available online. A sample from the clinically affected mother of case D (with anosmia and deafness) (see Table 1 and Figure S1A) was obtained, and cosegregation of the mutation and the disease was confirmed. Parents could also be analyzed for two of the sporadic cases (cases B and E), and the mutation was proved to be de novo in both cases.
Table 1.
Phenotypes of the KS Individuals Carrying SOX10 Mutations
| Case | Age (Years) | Gender | Occurrence | FSH (UI/l) Basal Peak | LH (UI/l) Basal Peak | E2 (pg/ml) | T (ng/ml) | Sponta -neous Puberty | Sense of Smell | Olfactory Bulb (MRI) | Hearing | Other Clinical Signs | DNA Mutation | Protein Alteration | In Silico Evidence | In Vitro Evidence |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Clinically Selected KS Individuals | ||||||||||||||||
| A | 26 | M | familial | 0.2–2.0 | 0.4–2.1 | - | <1.7 | no | anosmia | agenesis | unilateral deafness | micropenis, cryptorchidism, white hair (22 years old) | c.2T>G | p.? | pathogenic | pathogenic |
| B | 18 | F | sporadic | 2.8–ND | 0.3–ND | <10 | - | no | anosmia | agenesis | prelingual deafness | ptosis | c.331T>G (de novo) | p.Phe111Val | pathogenic | pathogenic |
| C | 39 | F | sporadic | <0.5–ND | 0.87–ND | 42 | - | no | anosmia | ND | prelingual deafness | obesity (BMI = 51) | c.424T>C | p.Trp142Arg | pathogenic | pathogenic |
| D | 25 | F | familial (anosmia and deafness in mother) | 2.1–7.9 | 0.8–8.0 | 20 | - | delayed | anosmia | agenesis | prelingual deafness | - | c.698-1G>C | p.spl? | pathogenic | - |
| E | 20 | M | sporadic | NA | NA | - | NA | delayed | anosmia | ND | prelingual deafness | cryptorchidism, SEMA3A c.1303G>A (p.Val435Ile) (+/−) | c.1290del (de novo) | p.Ser431Argfs*71 | pathogenic | pathogenic |
| F | 20 | M | sporadic | NA | NA | - | NA | - | anosmia | ND | normal | intellecual disability, dysmorphy, polymalformation | c.1298G>A | p.Arg433Gln | pathogenic | not pathogenic |
| Random KS Individuals | ||||||||||||||||
| G | 33 | M | familial? (a brother is most likely anosmic) | 0.8–0.9 | 0.41–0.6 | - | 0.23 | no | anosmia | ND | hypoacusis | ptosis | c.323T>C | p.Met108Thr | pathogenic | pathogenic |
| H | 19 | F | sporadic | <1.0–4.4 | 0.5–8.0 | <0.1 | - | no | anosmia | agenesis | normal | macroscelia | c.451C>T | p.Arg151Cys | pathogenic | pathogenic |
The age indicated was at the time of DNA sampling, usually age of diagnosis. Normal ranges are as follows: FSH/LH, 2–20 UI/l; E2, >20 pg/ml; and T, 2–10 ng/ml. The following abbreviations are used: M, male; F, female; BMI, body mass index; E2, estradiol; T, testosterone; FSH, follicle-stimulating hormone; LH, luteinizing hormone; ND, not determined; and NA, not available.
Five Mutations Result in Impaired SOX10 Function
In silico prediction suggested likely pathogenic consequences of all six mutations. Several programs predicted the splice-site mutation, c.698-1G>C, to result in complete loss of the last splice acceptor site and to have consequences similar to those of previously identified splice-site mutations at the same location (c.698-2A>C and c.698-2A>T).26,27 PolyPhen-2 and SIFT predicted the three missense mutations to be “likely damaging” and “not tolerated,” respectively. Functional effects of the mutations were tested through in vitro analysis.
The p.Phe111Val, p.Trp142Arg, p.Ser431Argfs*71, and p.Arg433Gln alterations were tested with a construction allowing production of SOX10 with a Myc tag at its amino terminus. After confirmation of the protein production by immunoblot analysis (Figure 1A), we first compared the transactivation capacities of the altered proteins on the MITF promoter and MPZ (myelin protein zero; P0) enhancer reporter constructs either alone or in synergy with known SOX10 partner transcription factors, PAX3 (paired box 3) and EGR2 (early growth response 2), respectively (Figures 1B and 1C).19,21 The p.Phe111Val, p.Trp142Arg, and p.Ser431Argfs*71 altered SOX10 were unable to transactivate the MITF reporter construct alone or in synergy with PAX3, whereas the p.Arg433Gln altered SOX10 behaved like the wild-type protein. On the MPZ enhancer reporter construct, the p.Phe111Val, p.Trp142Arg, and p.Ser431Argfs*71 altered SOX10 also lost their transactivation capacities alone and demonstrated variable synergistic activity with EGR2 (specifically, it was conserved for p.Phe111Val and p.Trp142Arg but lost for p.Ser431Argfs*71). Again, the p.Arg433Gln altered protein showed no change when compared with wild-type SOX10. Together, these findings indicated loss of function of the first three alterations and no pathogenic effect of the p.Arg433Gln substitution in vitro. These findings are consistent with the observation that Arg433 is located in the transactivation domain, where no WS-causing missense substitutions have been characterized thus far, whereas Phe111 and Trp142 are located within the highly conserved HMG domain, as are the amino acid residues modified by the previously identified SOX10 missense substitutions.24
Figure 1.
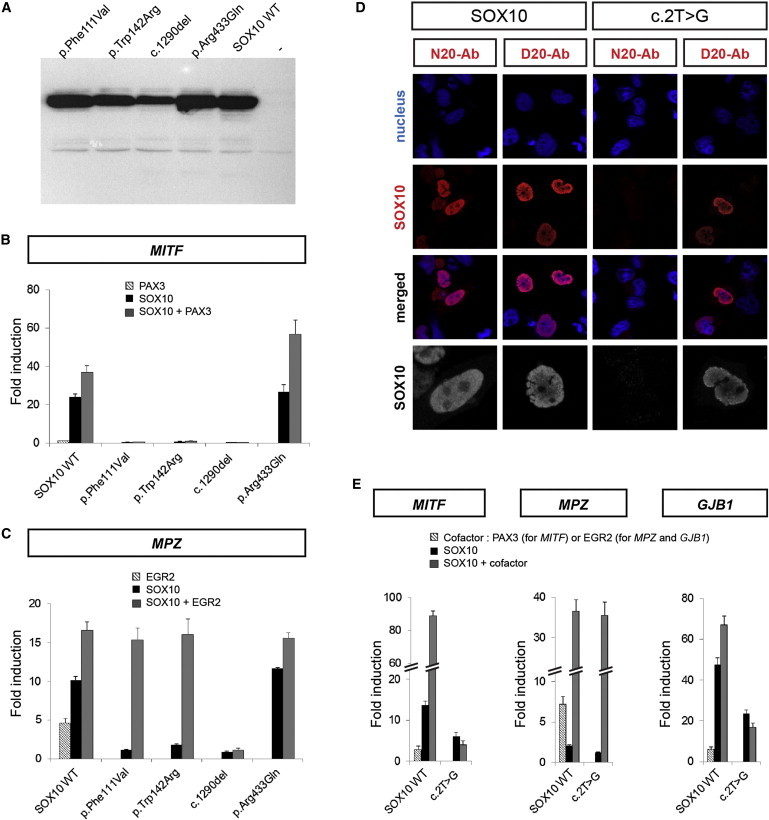
Functional Analysis of the Altered SOX10 Proteins
(A) Immunoblott analysis showing wild-type (WT) or altered SOX10 proteins with the use of an antibody directed against the carboxy terminal portion of the protein.
(B and C) Luciferase reporter-gene analysis. HeLa cells were cotransfected with the wild-type (WT) or altered SOX10 expression vector and a reporter construct containing the MITF promoter (B) or MPZ intronic enhancer (C) and known SOX10 cofactor expression vectors, i.e., PAX3 (B) or EGR2 (C). Reporter-gene activation is presented as luciferase fold induction relative to the empty vector. Results are the mean ± SEM of at least three different experiments, each performed in duplicate.
(D) Detection and localization of the c.2T>G (p.?) altered protein. HeLa cells were transfected with wild-type (left panels) or altered c.2T>G (p.?) (right panels) SOX10 constructs. Nuclei were counterstained with TO-PRO-3 iodide (blue). Transfected cells were immunostained with anti-SOX10 (red) directed against either the amino terminus (N20) or the carboxy terminus (D20) of the protein, as indicated. The merged images are presented below. A higher magnification of SOX10 labeling is also shown in gray (bottom panels).
(E) Luciferase reporter-gene analysis. HeLa cells were cotransfected with the wild-type (WT) or altered c.2T>G (p.?) SOX10 expression vectors and a reporter construct containing the MITF promoter (left panel), the MPZ intronic enhancer (central panel), or the GJB1 promoter (right panel) and the PAX3 (left panel) or EGR2 (central and right panels) expression vector. Reporter-gene activation is presented as fold induction relative to the empty vector. Results are the mean ± SEM of at least three different experiments, each performed in duplicate.
Because of its location in the translation initiation codon, the c.2T>G mutation was tested with a SOX10 construction allowing production of an untagged protein. Although not detected in immunoblot analysis performed under standard conditions (data not shown), the altered protein was visualized by immunocytochemistry. Use of an antibody directed against the carboxy-terminal portion of SOX10 revealed that an altered protein was produced and localized within the nucleus, as was the wild-type protein (Figure 1D, D20-Ab). Notably, we failed to detect the altered protein with another antibody directed against the amino terminus of SOX10 (Figure 1D, N20-Ab), which supports the use of a downstream methionine as an alternative initiation codon. This altered protein showed either absent or reduced transactivation capacity on the MITF, MPZ, or GJB1 (another SOX10 target gene, encoding connexin 32) reporter constructs (Figure 1E). Synergistic activity was lost with PAX3, whereas it was retained with EGR2 on MPZ, but not on GJB1 (Figure 1E).
The six mutations were also tested in a competition assay on the MITF promoter. They did not show any dominant-negative effect at a ratio of 1:1 and 2:1 to the wild-type protein (Figure S1B). Only at a ratio of 3:1 did the alterations (except p.Arg433Gln) induce a decrease of the luciferase induction.
Functional analysis of SOX10 missense mutations previously identified in the context of WS allowed us to show that amino acid substitutions in close contact with the main hydrophobic core of the HMG domain alter subcellular localization of the protein and thus lead to its accumulation in nuclear foci.24 The hydrophobic core is composed of four highly conserved residues, two tryptophans, and two phenylalanines, which are located at the intersection of the three α helices that compose the HMG domain and maintain their structural shape. The p.Phe111Val and p.Trp142Arg substitutions described here both remove one of the hydrophobic core components (Figure 2A). We therefore analyzed the effects of these substitutions on the subcellular localization of SOX10 by immunocytochemistry. Whereas the p.Ser431Argfs*71 and p.Arg433Gln altered proteins showed a nuclear localization pattern similar to that observed for the wild-type protein (Figure 2B), the p.Phe111Val and p.Trp142Arg altered proteins showed partial cytoplasmic relocalization (consistent with the location of Phe111 and Trp142 within the bipartite nuclear localization signals [NLSs] and the nuclear export signal [NES], respectively) and subnuclear accumulation in foci.
Figure 2.
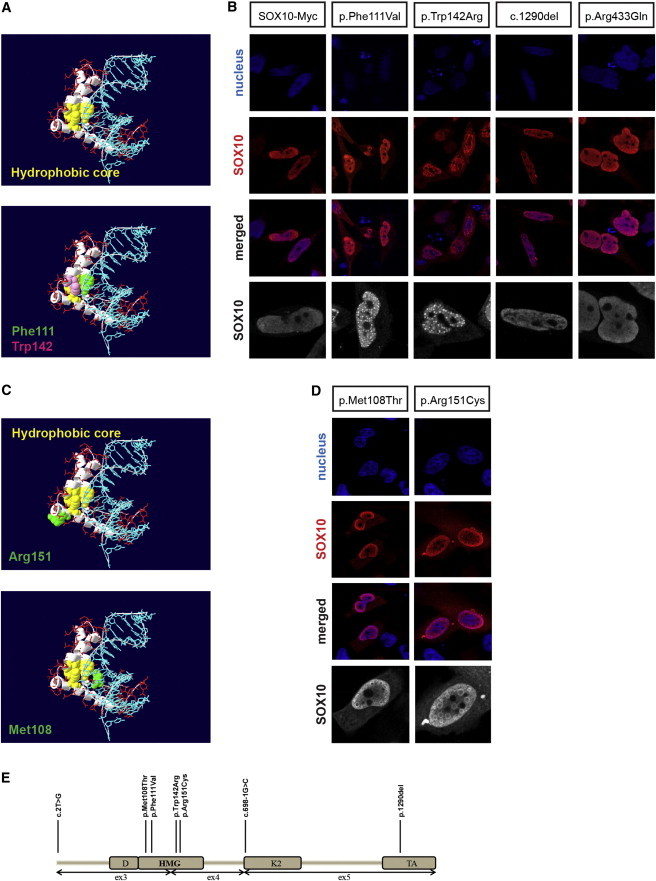
Subcellular Localization of the Altered SOX10 Proteins and Summary of All SOX10 Alterations Found in This Study
(A) Three-dimensional representation showing the location within the HMG-domain residues affected by the identified missense substitutions and a three-dimensional view of the SOX17 HMG domain (backbone in red) that forms three α helices (white ribbons) bound to its DNA target (blue). Upper panel: the four residues that form the hydrophobic core are shown in yellow. Lower panel: the residues corresponding to SOX10 Phe111 and Trp142, both belonging to the hydrophobic core, are indicated in green and pink, respectively.
(B) Subcellular localization of wild-type and altered proteins by immunostaining. HeLa cells transfected with wild-type (left panels) or altered (other panels) SOX10 constructs. Nuclei were counterstained with TO-PRO-3 iodide (blue). Transfected cells were immunostained with anti-SOX10 (red). Images shown are with D20, but similar results were obtained with N20 directed against the amino terminus of the protein. The merged images are presented below. A higher magnification of SOX10 labeling is also shown in gray (bottom panels).
(C) Three-dimensional representation of the location of the HMG-domain residues affected by the missense substitutions identified in “random” KS individuals, as in (A). Upper panel: the residue corresponding to Arg151, in contact with the hydrophobic core, is indicated in green. Lower panel: the residue corresponding to Met108, in contact with the DNA target, is indicated in green.
(D) Subcellular localization of the p.Arg151Cys and p.Met108Thr altered proteins by immunostaining, as in (B).
(E) Schematic representation of the SOX10 protein and the alterations found in this study. Abbreviations are as follows: D, dimerization domain; K2, K2 domain; HMG, HMG domain; and TA, transactivation domain.
Together, these results strongly indicate that five of the six identified mutations have a pathogenic effect and that all are found in individuals presenting with the clinical association between KS and deafness.
SOX10 Mutations Are Rare in KS Individuals without Hearing Impairment
We then aimed to determine whether SOX10 is also involved in other clinical forms of KS, particularly in KS without deafness. We screened 86 random KS individuals, including 66 KS cases without associated features and 20 cases with nonolfactory, nonreproductive additional symptoms, for SOX10 point mutations or deletions. We identified two additional mutations (c.323T>C [p.Met108Thr] and c.451C>T [p.Arg151Cys]) (Table 1, cases G and H; see Figure S1C for case G pedigree) affecting amino acid residues of the HMG domain. Notably, the individual who carried the p.Met108Thr substitution showed hypoacusis, whereas the absence of hearing impairment was confirmed in the individual carrying the p.Arg151Cys substitution.
Arg151 is located close to the hydrophobic core of the protein (Figure 2C, upper panel), whereas the Met108 residue contacts both the hydrophobic core and the DNA target (Figure 2C, lower panel) and is located in the bipartite NLS of SOX10. Its substitution could thus affect the structure, DNA binding, and/or nuclear localization of SOX10 as the primary defect. We performed immunocytochemistry experiments to analyze the subcellular location of the altered proteins. Both p.Met108Thr and p.Arg151Cys missense substitutions resulted in partial cytoplasmic relocalization of SOX10 and subnuclear accumulation in foci (Figure 2D). Luciferase assays confirmed the loss-of-function effect of these substitutions on the MITF promoter both alone and in the presence of the cofactor PAX3 (Figure S1D).
The seven likely pathogenic alterations found in this study are shown along with the main functional domains of SOX10 (Figure 2E). Additional screening for mutations in the genes involved in KS or congenital nonsyndromic forms of hypogonadotropic hypogonadism was performed in the individuals with a SOX10 mutation (A–H, Table 1) and only yielded a SEMA3A heterozygous missense variant (c.1303G>A [p.Val435Ile]) in case E. This variant has been found at similar frequencies in KS individuals and control subjects, although it has been reported to have some deleterious functional consequences in vitro.28
These results indicate that SOX10 mutations are rare in KS individuals without associated features and confirm that the clinical association between KS and hearing impairment is more specific. We then used a mouse model to explore the embryonic defect leading to KS as a result of impaired SOX10 function.
SOX10 Is Expressed in Embryonic OECs in Mice and Humans
OECs, peculiar glial cells involved in the growth and guidance of the olfactory axons, have recently been shown to derive from NC cells in mice and chicks. SOX10 was used as a marker of NC-derived cells in these studies, and its pattern of expression indeed suggested that it is expressed by OECs.29–31 Given that OECs are heterogeneous in the pattern of markers they express,32 we fully characterized the SOX10 expression profile during development of the olfactory structures by using antibodies directed against SOX10 and several markers of OECs.
In the E12.5 mouse embryo, the olfactory, vomeronasal, and terminal nerve fibers can be seen in the mesenchyme surrounding the olfactory epithelium and converge on the migratory mass, a heterogeneous group of cells located in the frontonasal mesenchyme beneath the presumptive olfactory bulbs. OECs are detected all along the nerve pathway, where their cytoplasms ensheathe the axon bundles, and begin to invade the olfactory nerve layer (ONL) of the developing olfactory bulb at this stage (Figure 3A). Immunostaining on coronal sections of the E12.5 embryo head (Figure 3A) revealed that SOX10 is coexpressed with the OEC marker BLBP (also known as BFABP), whereas it showed mutually exclusive expression with the neuronal marker TUJ1 (β3-tubulin). More specifically, SOX10 (nuclear staining, red) and BLBP were coexpressed along the olfactory, vomeronasal, and terminal nerve pathways, in the frontonasal mesenchyme, and in the migratory mass below the presumptive olfactory bulb (Figure 3B and data not shown). SOX10-positive cells also expressed the OEC markers P75 (also known as NGFR; note that this marker is not specific to OECs and also labels most cells in the nasal mesenchyme) and S100 along the nerve fibers and in the nasal mesenchyme (Figure 3B). By contrast, they did not express P75 and showed variable S100 immunostaining within the migratory mass (Figure 3B).
Figure 3.
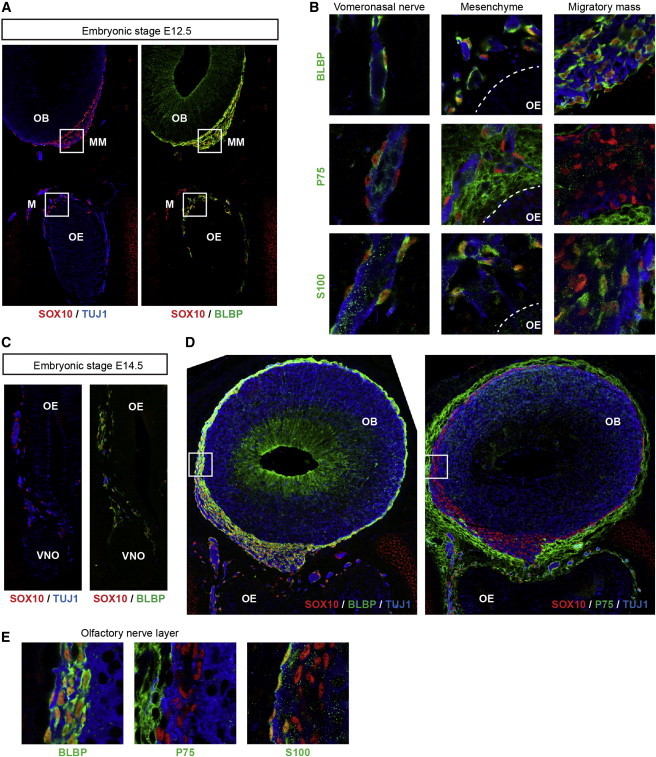
SOX10 Expression in the Mouse Embryonic Olfactory System
(A) Double immunolabeling of the E12.5 embryo head (coronal sections) for SOX10 (red), the neuronal marker (TUJ1; blue), or the OEC marker (BLBP; green). The boxed regions are the migratory mass and the nasal mesenchyme magnified in (B).
(B) Higher magnification of the vomeronasal nerve (left panel), nasal mesenchyme (middle panel), and migratory mass (right panel). Triple labeling for SOX10 (red), the neuronal marker (TUJ1; blue), and an OEC marker (BLBP, P75, or S100 as indicated; green) is shown. The dotted lines represent the limit between the olfactory epithelium and the mesenchyme.
(C) Double labeling of the E14.5 embryo olfactory epithelium and mesenchyme (coronal sections) for SOX10 (red), the neuronal marker (TUJ1; blue), or the OEC marker (BLBP; green).
(D) Triple labeling of E14.5 embryo olfactory bulbs (coronal sections) for SOX10 (red), the neuronal marker (TUJ1; blue), or the OEC marker (BLBP or P75 as indicated; green). The boxed regions in the medial ONL are magnified in (E).
(E) Higher magnification at the level of the medial ONL. Triple labeling for SOX10 (red), the neuronal marker (TUJ1; blue), and an OEC marker (BLBP, P75, or S100 as indicated; green) is shown.
Abbreviations are as follows: M, mesenchyme; MM, migratory mass; OB, olfactory bulb; OE, olfactory epithelium; and VNO, vomeronasal organ.
At E14.5, the olfactory epithelium developed. SOX10 demonstrated the same pattern of expression in OECs (defined as BLBP+, P75+, S100+, TUJ1− cells) of the nasal mesenchyme and along the nerve pathway (Figure 3C and data not shown). At this stage, the olfactory bulb had formed from the rostral telencephalon, and a continuous outer layer of SOX10-expressing cells, corresponding to the ONL, was observed (Figure 3D). These cells were positive for BLBP and negative for P75, whereas only the outermost cells were positive for S100 (Figures 3D and 3E). Apart from observing OECs, we observed some SOX10 expression in the nasal glands (Figures S2A and S2B). We also confirmed the presence of a few nonneuronal SOX10-expressing cells in the olfactory epithelium at this stage (Figure S2C), as previously reported.29,31
Finally, we studied SOX10 expression in the peripheral olfactory system of a human embryo at 8 weeks of embryonic development (this stage is equivalent to mouse E14.5). Immunostaining experiments revealed that SOX10-expressing cells (nuclear, green) were located along the olfactory, vomeronasal, and terminal nerve trajectories (TUJ1 staining, red) in the nasal mesenchyme (Figure 4A), in the migratory mass, and in the olfactory bulb (Figure 4B). Double staining of SOX10 with the OEC marker S100 along the olfactory nerves confirmed that SOX10-expressing cells are OECs (Figure 4C).
Figure 4.
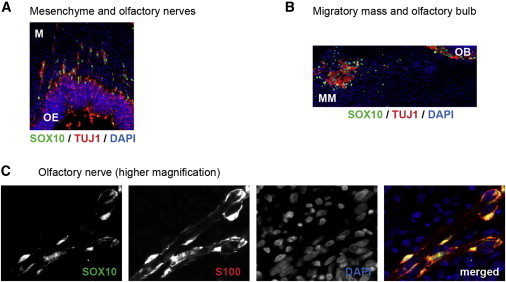
SOX10 Expression in the Human Embryonic Olfactory System
(A and B) Immunostaining of the head of a human fetus at 8 weeks of embryonic development (sagittal sections) at the level of the nasal mesenchyme (A) or migratory mass and olfactory bulb (B). Double labeling for SOX10 (green) and the neuronal marker (TUJ1; blue) and counterstaining with DAPI (blue) are shown. Abbreviations are as follows: M, mesenchyme; MM, migratory mass; OB, olfactory bulb; and OE, olfactory epithelium.
(C) Immunostaining showing an olfactory nerve at higher magnification. Double staining for SOX10 (green) and the OEC marker (S100; red) and counterstaining with DAPI are shown.
Together, these results confirm SOX10 expression in OECs during early development of the peripheral olfactory system in mice and humans.
The Sox10 Mutant Mouse Has OEC Defects
We then used mice bearing a Sox10 mutation (Sox10lacZ)14 to explore the embryonic defect leading to KS as a result of impaired SOX10 function. We first performed X-Gal staining to detect β-galactosidase (lacZ expression) in heterozygous and homozygous mutant E14.5 embryos. The olfactory bulbs had formed in both genotypes, but an abnormal colonization of the olfactory bulbs by X-Gal-stained (blue) OECs was apparent in homozygotes (Figure 5A). We then compared β-galactosidase expression with various markers by immunohistochemistry on coronal sections. A general overview of the olfactory system showed that the olfactory, vomeronasal, and terminal nerves had formed in the homozygous mutant embryos, but OECs (β-galactosidase-positive cells) were almost absent in the nasal mesenchyme and along the nerve pathway (Figure 5B and higher magnification in Figure 5C). Only a few OECs were observed in the upper part of the frontonasal mesenchyme, under the migratory mass. By contrast, numerous OECs were present in the migratory mass and the ONL, but they appeared to encircle the olfactory bulbs incompletely by forming a thinner, disorganized, and discontinued layer.
Figure 5.
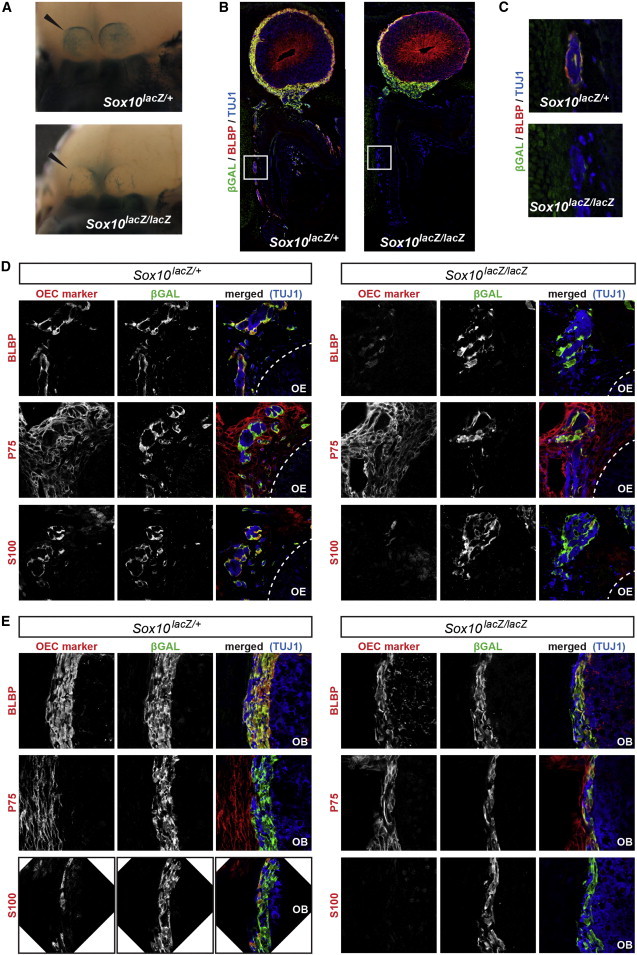
OEC Defect in the E14.5 Sox10 Mutant Mice
(A) Whole-mount X-Gal staining of the head (facial view) in heterozygous (Sox10lacZ/+, left panel) and homozygous (Sox10lacZ/lacZ, right panels) mutant embryos. The skin was removed for visualization of the olfactory bulbs, indicated by the black arrowheads.
(B) General overview of the olfactory system upon triple labeling for β-galactosidase (green), the neuronal marker (TUJ1; blue), and the OEC marker (BLBP; red), in Sox10 heterozygous (left panel) and homozygous (right panels) mutant embryos.
(C) Higher magnification of the regions boxed in (B) shows the presence of OECs ensheathing the vomeronasal nerve fibers in the heterozygous Sox10 mutant embryos and the absence of these cells in the homozygous embryos.
(D and E) Higher magnification of the nasal mesenchyme (D) or ONL (E) immunostained for an OEC marker (BLBP, P75, or S100; red), β-galactosidase (green), and the neuronal marker (TUJ1; blue), as indicated in the figure, in Sox10 heterozygous (left panels) and homozygous (right panels) mutant embryos.
Abbreviations are as follows: OB, olfactory bulb; and OE, olfactory epithelium.
We then determined the expression pattern of OEC markers in the absence of SOX10. In the upper part of the frontonasal mesenchyme, the marker profile of the few β-galactosidase-positive cells present in the mutant homozygotes differed from that of cells in heterozygotes: homozygotes had fainter BLBP and absent S100 labeling in most of these cells, whereas P75 staining was preserved (Figure 5D). In the ONL of the olfactory bulbs, BLBP staining was preserved, and S100 staining (normally found in the outermost cell layer of the ONL) was lost, but most, if not all, OECs were now positive for P75 (Figure 5E).
In summary, these results indicate an almost complete absence of OECs in the nasal mesenchyme of the Sox10 homozygous mutant embryos, defective colonization of the olfactory bulbs, and abnormal profiles of the remaining OECs in both the nasal mesenchyme and the ONL of the olfactory bulbs.
To explain these defects, we compared the development of OECs in heterozygous and homozygous Sox10 mutant embryos at an earlier stage. At E12.5, the absence of OECs in most of the nasal mesenchyme was already visible in the mutant homozygotes (Figure S3A). The migratory mass had formed; however, defective colonization of the olfactory-bulb anlage by OECs was already detectable on more rostral sections (Figure S3B). To determine whether the absence of OECs in the nasal mesenchyme was due to apoptosis of Sox10-defective cells, we counted the number of TUNEL-positive cells in the nasal mesenchyme. We did not find a significant difference in the number of apoptotic cells between heterozygous and homozygous Sox10lacZ embryos (Figure S3C), indicating that the defect responsible for the low number of OECs in the homozygous mutant embryos occurred earlier or by a nonapoptotic mechanism.
The OEC Defect Impacts the Development of Olfactory, Vomeronasal, and Terminal Nerves and the Migration of Neuroendocrine GnRH Cells
OECs ensheathe the olfactory, vomeronasal, and terminal nerve fibers and also contribute to axonal pathfinding, fasciculation, and defasciculation.32 We thus looked at the trajectory of these nerve fibers in the Sox10 mutant embryos. TUJ1 staining on E14.5 heads showed that the nerves had formed in mutant homozygotes despite the almost complete absence of OECs in the nasal mesenchyme (Figure 6A). However, axons were abnormally routed; some of them did not target the olfactory bulb and instead contacted axons from the other side, dorsally to the nasal septum (red arrowhead in Figure 6A). Furthermore, as a result of the absence of OECs, axons were not ensheathed along their most ventral trajectory, and their defasciculation could be observed near the nasal septum (red arrow in Figure 6A; defasciculation is also visible in Figure 5D).
Figure 6.
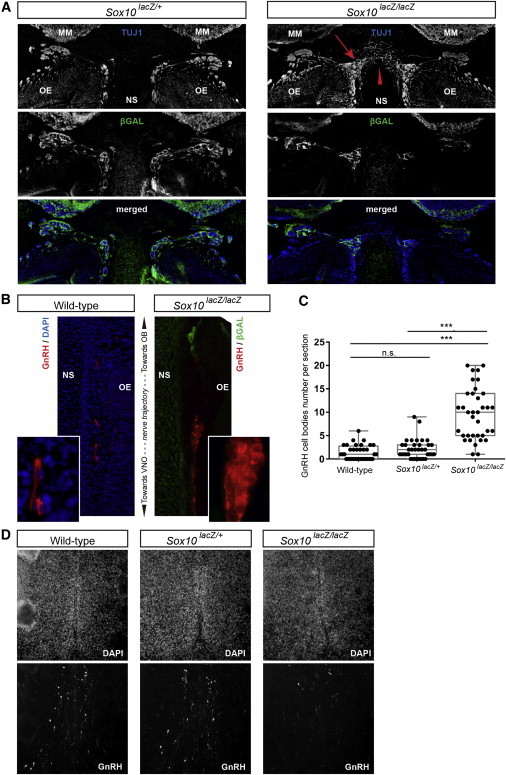
Abnormal Nerve Fasciculation, Axonal Pathfinding, and GnRH Cell Migration in Sox10 Mutant Mice
(A) TUJ1 (upper panel) and β-galactosidase (middle panel) immunostaining and a merged image (lower panel) over the nasal septum at a similar level of heterozygous (Sox10lacZ/+) and homozygous (Sox10lacZ/lacZ) mutant E14.5 embryos, as indicated. The arrow and arrowhead in the homozygote indicate the defasciculation of sensory axons and their misrouting over the nasal septum, respectively.
(B) GnRH cells along the vomeronasal nerve trajectory in wild-type (left panel) and homozygous Sox10 mutant (right panel) E14.5 embryos. The GnRH immunostaining is in red, DAPI staining is in blue, and β-galactosidase immunostaining is in green. Insets show higher magnifications.
(C) Box plots showing quantification of the number of GnRH cells along the trajectory of the vomeronasal nerve in wild-type, heterozygous, and homozygous mutant mouse embryos. Each dot corresponds to the number of GnRH-positive cell bodies counted on one side of a section. The top and bottom of each box represent the 25th and 75th percentiles, respectively. The middle line is the median. Statistical significance was tested with a Student’s t test. ***p < 0.0001. The following abbreviation is used: ns, not significant.
(D) DAPI (upper panel) and GnRH (lower panel) staining of sections at the level of the preoptic area in the wild-type (left), heterozygous (center) and homozygous (right) E14.5 embryos.
Abbreviations are as follows: OB, olfactory bulb; OE, olfactory epithelium; NS, nasal septum; and VNO, vomeronasal organ.
Hypogonadism in KS results from the so-called olfactogenital fetopathological sequence, whereby incomplete embryonic migration of the neuroendocrine GnRH cells from the nose to the brain arises from the disruption of vomeronasal and terminal nerve fibers.12 We therefore analyzed the migration of these cells in the Sox10 mutant embryo. In E14.5 wild-type and heterozygous embryos, most GnRH cells had already left the extracerebral nerve pathway and were migrating within the two cerebral hemispheres. In homozygous mutant embryos, GnRH cells were found to accumulate on the vomeronasal and terminal nerve trajectories in the nasal mesenchyme (Figure 6B and data not shown). GnRH cell count along the nerve pathway confirmed this observation in that there was a significantly larger number of GnRH cell bodies present in the Sox10laZ homozygotes than in wild-type and heterozygous embryos (Figure 6C). Although some GnRH cells did reach the migratory mass in the homozygous mutant embryos, few cells had penetrated into the forebrain en route to the hypothalamic region (Figure 6D). The overall number of GnRH cells appeared unchanged (estimated at 969 in wild-type, 949 in heterozygous mutant, and 937 in homozygous mutant embryos).
These results show that the Sox10 homozygous mutant mice, which have a complete lack of SOX10, also undergo an embryonic pathological sequence similar to that responsible for KS. The defects we observed in the Sox10 homozygous mutant mice are schematized in Figure 7.
Figure 7.
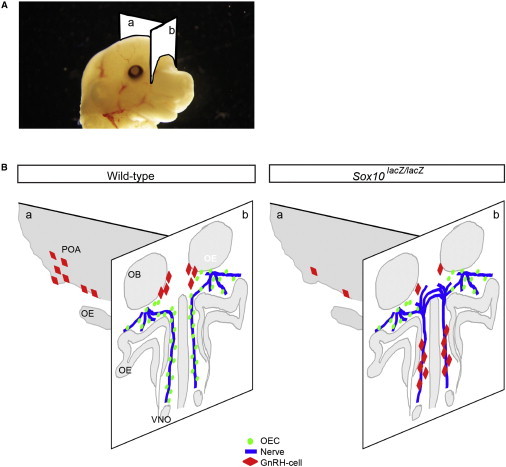
Summary of the Defects Observed in the Sox10 Mutant Embryo
(A) Schematization of the cut planes (a and b) drawn in (B) on the head of an E14.5 mouse embryo.
(B) Summary of the defects observed in the Sox10 homozygous mutant embryos (Sox10lacZ/lacZ, right panel) compared to wild-type embryos (left panel). The OECs are shown in green, the nerve fibers are in blue, and the GnRH-synthesizing cells are in red. Abbreviations are as follows: OE, olfactory epithelium; OB, olfactory bulb; POA, preoptic area; and VNO, vomeronasal organ.
Discussion
KS is genetically heterogeneous and has various modes of transmission: it can be autosomal recessive, autosomal dominant with incomplete penetrance, linked to the X chromosome, or oligogenic. However, only 30% of cases have mutations in any of the nine genes identified thus far.13,28 Deafness is one of various nonolfactory, nonreproductive anomalies that are sometimes found together with KS.13 Its prevalence in KS individuals has been estimated at approximately 5%.33 As our understanding of the molecular basis of KS has progressed, deafness has been reported in individuals with KAL1, FGFR1, FGF8, or CHD7 mutations, but these cases remain relatively rare.34–38 Given the high prevalence of hearing impairment in the general population, the association between KS and deafness might sometimes be coincidental, as was shown in a family in which deafness cosegregated neither with KS nor with the FGF8 mutation.39 Comparatively, the remarkably large penetrance of deafness in the KS individuals who carry mutations in SOX10 appears highly significant.
We characterized seven different SOX10 mutations in persons affected by KS. As in WS, the mutations were found in the heterozygous state together with a dominant mode of inheritance. Some phenotypic variation was observed. In family D, the mother, who also carries the mutation, suffered from deafness and anosmia but had normal puberty and a spontaneous pregnancy. Although mosaicism cannot be excluded, this finding most likely refers to the usual phenotypic variability that is frequently observed in developmental disorders, and particularly in KS or SOX10-linked WS. Stochastic events and modifier genes are often proposed to explain at least part of the phenotypic variability. In this respect, the SEMA3A sequence variant that was found in case E might influence the expression of the disease.
In the first part of our study, five mutations were found among 13 individuals with the clinical association between KS and deafness, corresponding to a SOX10 involvement of 38%. This rate might be slightly overestimated because a few KS individuals with deafness and already known mutations in other genes had been excluded. However, to date, SOX10 stands as the main gene involved in this specific clinical association.
The hearing loss was sensorineural, profound, or total in most cases. The audiograms of cases D and E are shown in Figure S4 and illustrate a total loss of hearing in the two ears at all frequencies. In contrast, case G showed a mild sensorineural hearing loss (30 dB at 1,000 and 2,000 Hz in the left ear and 40 dB from 250 to 1,000 Hz in the right ear). A striking finding is the high proportion of females among persons carrying SOX10 mutations in this series (four females and three males) given that KS has been estimated to be three to five times more frequent in males than in females.13 However, an approximately 1:1 sex ratio was also found in the group of individuals with KS and deafness (7 females out of 13 cases), an observation that needs to be confirmed by further studies.
Seven of the eight SOX10 sequence variations identified (four missense mutations, one frameshift, one splice-site mutation, and one mutation of the initiation codon) either result in or are predicted to result in an altered production or function of SOX10 (Figure 2E). We performed transactivation assays on two target genes of SOX10, namely MITF and MPZ.19,21 MITF was chosen because of its critical role in melanocyte development, whereas MPZ was selected on the basis of its known expression in OECs.40 Our results do not support a deleterious effect of the p.Arg433Gln substitution. The other seven alterations were found to alter subcellular localization and/or transactivation capacities, thus validating their deleterious effect. The four missense substitutions located in the HMG domain showed a peculiar aspect of relocalization to nuclear bodies, which we had previously observed for amino acid substitutions in close contact with the main hydrophobic core of the HMG domain.24 Whether these foci are a cause or a consequence of the pathogenic effect is still unclear.
The c.2C>T mutation affects the SOX10 initiation codon. However, immunostaining of cells transfected with cDNA carrying the mutation, with the use of antibodies directed against the SOX10 amino or carboxy terminus, showed that an in-frame protein is produced. The first in-frame ATG codon corresponds to Met90 and, if used, would produce a protein conserving the HMG and transactivation domains but lacking most of the dimerization domain.41 Because Met90 is the only in-frame methionine more proximal to the two NLSs, this hypothesis is consistent with our observation that the protein produced did not relocalize to the cytoplasm.42
Significantly, one of the KS individuals had early graying and deafness and therefore fulfilled the diagnosis criteria for type 2 WS, but no pigmentation defects were reported for the other cases. For some persons, clinical reevaluation was possible after the identification of the SOX10 mutation. The absence of WS-like pigmentation disturbance was confirmed in case D and her mother, as well as in cases B and C, whereas case E had belatedly developed a frontal white forelock at approximately 25 years of age. No evidence at this point indicates what makes a given SOX10 mutation a KS- or WS-causing mutation. As a whole, the mutations we identified here were not typically different from the mutations previously identified in WS except for their relative frequency: we found mostly missense mutations causing protein changes in the HMG domain and a few protein-truncating mutations in our group of KS individuals (most SOX10 mutations are predicted to result in a truncated protein in WS), a finding that must be confirmed in additional studies. In regard to previous clinical findings in WS (very rare reports of anosmia and hypogonadism and/or cryptorchidism among almost 100 reported persons with different SOX10 mutations) and the recent observation that 88% of WS individuals with a SOX10 mutation and inner-ear morphological abnormalities also have olfactory-bulb agenesis, it remains possible that anosmia and hypogonadism have been underestimated in WS. This phenomenon could possibly be explained in part because people usually do not spontaneously complain of anosmia and by the fact that WS is often diagnosed in childhood.
The involvement of SOX10 in mouse olfactory development was not suspected until recently, when its expression was reported in OECs.29–31 Here, we show that SOX10 plays a major role in the development of these peculiar NC-derived glial cells. In Sox10-null mutant embryos, a large part of the peripheral embryonic olfactory system lacked OECs at the embryonic stages analyzed.
Our results show that the OEC deficiency occurs prior to E12.5 in mice. On the basis of current knowledge about SOX10 function in other tissues, the defect most likely occurs between the emergence of NC cells from the neural tube and their arrival at the olfactory placode. SOX10 is thought to play a role in sustaining the survival of multipotent NC cells. In the absence of SOX10, a dramatic increase in cell death has been reported for vagal NC cells prior to their entry into the foregut, as well as in various other NC cell derivatives.43–47
The remaining OECs were located in the uppermost part of the frontonasal mesenchyme, the migratory mass, and the ONL. They had an abnormal expression profile and formed a discontinued and disorganized layer around the olfactory bulbs, indicating that SOX10 is also probably involved in their identity and function at several locations. Several explanations can be proposed as to why some OECs persisted in the mutant mice. One possibility is that not all OECs are dependent on SOX10. Another hypothesis is a partial functional redundancy between SOX10 and another protein of the SOX family. SOX8, which together with SOX10 and SOX9 forms the E subgroup of SOX transcription factors, is a good candidate given its expression in the ONL during embryogenesis and the described redundancy between SOXE subfamily members in other tissues.48–50
Despite the markedly reduced number of OECs, the olfactory, vomeronasal, and terminal nerves had formed in the Sox10-null embryos, as had the olfactory bulbs. Given that OECs not only ensheathe and fasciculate the nerve fibers but also extend processes ahead of the pioneer olfactory axons they ensheathe, suggesting that they orchestrate their growth and guidance,32,51 this finding might be surprising. However, although the olfactory axons were formed and extended some processes toward the migratory mass and the olfactory bulb in the Sox10-null mutant embryos, we found that they were partially misrouted and defasciculated, thus confirming the role of OECs in these processes.
In mice, neuroendocrine GnRH cells begin to leave the epithelium of the olfactory pit at approximately E11.5. They migrate in close association with growing fibers of the vomeronasal and terminal nerves and then penetrate into the rostral forebrain and continue their migration toward the hypothalamus along the terminal nerve or a branch of the vomeronasal nerve.52 In E14.5 Sox10-null embryos, the latest embryonic stage we could examine, GnRH cells accumulated outside the brain, along the vomeronasal and terminal nerve pathways, in the regions devoid of OECs. Few GnRH cells were seen in the forebrain of these mice, which could reflect an impaired or delayed migration. To the extent that mice and humans are comparable, it remains to be determined whether the defective GnRH cell migration in this particular genetic form of KS is primarily due to the absence of OECs on the vomeronasal and terminal nerve trajectories or to the defective structure of these nerves, as previously suggested in other genetic forms.12
All the abnormalities we found in the Sox10 mutant mice were observed in homozygous embryos, and embryonic lethality of these mice precludes further testing of late olfactory development and fertility. In the normal ONL, OECs contribute to the defasciculation, sorting, and refasciculation of axons and are therefore crucial to the formation of the olfactory-bulb glomeruli and to the establishment of the olfactory topographic map.32 They are also involved in the renewal of olfactory receptor neurons and their axonal growth throughout life. Accordingly, it is tempting to speculate that the defect in the number and function of OECs in Sox10 mutant mice could have drastic consequences on the maturation of the olfactory bulbs and the ability of olfactory nerve fibers to renew throughout life. The use of a conditional knockout mouse model to avoid lethality would be of interest to explore the late consequences of the absence of SOX10 in the peripheral olfactory system.
SOX10 mutations have been found in the heterozygous state in both KS and WS individuals. No reproduction defect has been reported in Sox10 heterozygous mutant mice, suggesting that both olfaction and GnRH neurosecretion are not strongly affected, although it remains possible that subtle, still unrecognized defects in OECs have some functional consequences in adult mice. By contrast, depigmentation and megacolon are found in heterozygous mice as they are in WS4 individuals. Such differences in sensitivity to genetic diseases between mice and humans are not rare. Alternatively, oligogenicity has been described in KS, and mutations in unknown genes might contribute to the disease in some instances. Notably, the inner-ear morphological defects associated with SOX10 mutations are also quite marked and have a high penetrance in humans, whereas they are much less evident in the Sox10 homozygous mutant mice.10,53
Sox10 is expressed in the melanocytic intermediate cells of the cochlear stria vascularis; these cells produce the endocochlear electric potential essential to the hearing process.54 It is also widely expressed during early inner-ear development before being restricted to the cochlear and vestibular ganglia and to supporting cells of the sensory epithelium. In addition to having a specific role in glial development of the ganglia, SOX10 has been shown to promote the survival of cochlear progenitors during otocyst formation and differentiation of the organ of Corti.53–55 In the Sox10 homozygous mutant mice, the structure and cellular organization of the organ of Corti appear normal but the cochlear duct is shortened and there is no apparent malformation of the vestibule.53 In humans, apart from the cochlear degeneration thought to take place in WS in the absence of intermediate cells of the stria vascularis, a proportion of WS individuals with SOX10 mutations had an enlarged vestibule, agenesis or hypoplasia of semicircular canals, and an abnormally shaped cochlea.10 Together, these results suggest that the high penetrance of deafness among persons who carry SOX10 mutations is most likely the consequence of several different defects in the development of the inner ear.
MRI or CT scans of the temporal bone have been performed in four of the seven individuals. Abnormal images were not found in case B (not all the temporal bone structures could be analyzed, but the semicircular canals were present and normally shaped). A vestibulocochlear dysplasia was reported in case D. The temporal bone CT scan of case C showed a bilateral hypoplasia of the lateral and posterior semicircular canals, as well as a vesicular vestibule on one side and an enlargement of the vestibular aqueduct on the other side. The CT scan of case E showed bilateral enlargement of the vestibule and hypoplasia of the lateral semicircular canals. Interestingly, the association between semicircular canal hypoplasia or agenesis and olfactory-bulb agenesis and hypogonadotropic hypogonadism is also found in CHARGE (coloboma, heart defects, choanal atresia, retardation, genital and ear anomalies) syndrome, due to mutations in CHD7.56 It is not difficult to distinguish individuals affected by typical CHARGE syndrome from persons carrying SOX10 mutations, but in individuals who are affected by mild forms of CHARGE syndrome and who thus do not show all the cardinal signs of the disease, it might be difficult to differentiate this condition in the absence of a complete clinical description.56–58 From now on, the existence of semicircular canal hypoplasia or agenesis in a person affected by KS should be considered an indication for both CHD7 and SOX10 molecular analyses. Isolated or minor signs of CHARGE syndrome, such as coloboma and heart defects but also facial asymmetry, malformations of the middle or external ear, and a hypoplastic vestibule, might point toward a CHD7 mutation, whereas some depigmentation features or an enlarged vestibule might point to a SOX10 defect.
Our findings implicate NC-derived OECs in the pathogenesis of KS, thus defining the SOX10-related genetic form of the disease as a neurocristopathy. The peripheral olfactory nervous system is unique in that it renews throughout life, a property attributed to the presence of the OECs, which makes them strong candidates for cell-mediated repair of a variety of neural lesions.32,51 Despite this major medical relevance, the consequences of the absence or depletion of OECs in the peripheral olfactory system have been poorly characterized. In this respect, the Sox10-knockout mouse might prove to be a valuable model.
In summary, we characterized an additional role for SOX10 in human pathology 15 years after the cloning and first implication of this gene in WS. We found SOX10 loss-of-function mutations in individuals demonstrating the clinical association between KS and hearing impairment and more rarely in individuals affected by KS without associated features. From now on, SOX10 should be the first gene to be tested for the presence of mutations in the association between KS and hearing impairment. On the basis of a mouse-model study, we suggest that this particular genetic form of KS results from a primary defect affecting OECs during early embryonic development of the peripheral olfactory system.
Acknowledgments
We thank the index cases and family members for their contribution to the study. We thank Fabien Guimiot for providing sections of human fetuses and Michael Wegner for providing us with the Sox10lacZ mice. We thank the Hôpital Henri Mondor sequencing facility, Marjorie Collery for animal husbandry, and Xavier Ducrouy for confocal imaging. This work was supported by the Institut de la Santé et de la Recherche Médicale and the Agence Nationale de la Recherche (ANR-JCJC-2010 to N.B. and ANR-2009-GENOPAT-017 to C.D.). A.C. is a recipient of a fellowship from the Fondation pour la Recherche Médicale. S.M. is receiving a salary from the ANR-2009-GENOPAT-017 grant.
Supplemental Data
Web Resources
The URLs for data presented herein are as follows:
1000 Genomes Project, http://www.1000genomes.org/
LOVD – Leiden Open Variation Database, http://grenada.lumc.nl/LOVD2/WS/home.php?select_db=SOX10
NHLBI Exome Sequencing Project (ESP) Exome Variant Server, http://evs.gs.washington.edu/EVS/
Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/omim
PolyPhen-2, http://genetics.bwh.harvard.edu/pph2/
Protein Data Bank, http://www.rcsb.org/pdb/home/home.do
SIFT, http://sift.jcvi.orgWS
References
- 1.Wegner M. Secrets to a healthy Sox life: lessons for melanocytes. Pigment Cell Res. 2005;18:74–85. doi: 10.1111/j.1600-0749.2005.00218.x. [DOI] [PubMed] [Google Scholar]
- 2.Le Douarin N.M., Kalcheim C. Cambridge University press; Cambridge: 1999. The neural crest. [Google Scholar]
- 3.Lang D., Epstein J.A. Sox10 and Pax3 physically interact to mediate activation of a conserved c-RET enhancer. Hum. Mol. Genet. 2003;12:937–945. doi: 10.1093/hmg/ddg107. [DOI] [PubMed] [Google Scholar]
- 4.Mollaaghababa R., Pavan W.J. The importance of having your SOX on: role of SOX10 in the development of neural crest-derived melanocytes and glia. Oncogene. 2003;22:3024–3034. doi: 10.1038/sj.onc.1206442. [DOI] [PubMed] [Google Scholar]
- 5.Zhu L., Lee H.O., Jordan C.S., Cantrell V.A., Southard-Smith E.M., Shin M.K. Spatiotemporal regulation of endothelin receptor-B by SOX10 in neural crest-derived enteric neuron precursors. Nat. Genet. 2004;36:732–737. doi: 10.1038/ng1371. [DOI] [PubMed] [Google Scholar]
- 6.Pingault V., Ente D., Dastot-Le Moal F., Goossens M., Marlin S., Bondurand N. Review and update of mutations causing Waardenburg syndrome. Hum. Mutat. 2010;31:391–406. doi: 10.1002/humu.21211. [DOI] [PubMed] [Google Scholar]
- 7.Pingault V., Bondurand N., Kuhlbrodt K., Goerich D.E., Préhu M.O., Puliti A., Herbarth B., Hermans-Borgmeyer I., Legius E., Matthijs G. SOX10 mutations in patients with Waardenburg-Hirschsprung disease. Nat. Genet. 1998;18:171–173. doi: 10.1038/ng0298-171. [DOI] [PubMed] [Google Scholar]
- 8.Inoue K., Khajavi M., Ohyama T., Hirabayashi S., Wilson J., Reggin J.D., Mancias P., Butler I.J., Wilkinson M.F., Wegner M., Lupski J.R. Molecular mechanism for distinct neurological phenotypes conveyed by allelic truncating mutations. Nat. Genet. 2004;36:361–369. doi: 10.1038/ng1322. [DOI] [PubMed] [Google Scholar]
- 9.Bondurand N., Dastot-Le Moal F., Stanchina L., Collot N., Baral V., Marlin S., Attie-Bitach T., Giurgea I., Skopinski L., Reardon W. Deletions at the SOX10 gene locus cause Waardenburg syndrome types 2 and 4. Am. J. Hum. Genet. 2007;81:1169–1185. doi: 10.1086/522090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elmaleh-Bergès M., Baumann C., Noël-Pétroff N., Sekkal A., Couloigner V., Devriendt K., Wilson M., Marlin S., Sebag G., Pingault V. Spectrum of temporal bone abnormalities in patients with Waardenburg syndrome and SOX10 mutations. AJNR Am. J. Neuroradiol. 2012 doi: 10.3174/ajnr.A3367. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barnett C.P., Mendoza-Londono R., Blaser S., Gillis J., Dupuis L., Levin A.V., Chiang P.W., Spector E., Reardon W. Aplasia of cochlear nerves and olfactory bulbs in association with SOX10 mutation. Am. J. Med. Genet. A. 2009;149A:431–436. doi: 10.1002/ajmg.a.32657. [DOI] [PubMed] [Google Scholar]
- 12.Teixeira L., Guimiot F., Dodé C., Fallet-Bianco C., Millar R.P., Delezoide A.L., Hardelin J.P. Defective migration of neuroendocrine GnRH cells in human arrhinencephalic conditions. J. Clin. Invest. 2010;120:3668–3672. doi: 10.1172/JCI43699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dodé C., Hardelin J.P. Kallmann syndrome. Eur. J. Hum. Genet. 2009;17:139–146. doi: 10.1038/ejhg.2008.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Britsch S., Goerich D.E., Riethmacher D., Peirano R.I., Rossner M., Nave K.A., Birchmeier C., Wegner M. The transcription factor Sox10 is a key regulator of peripheral glial development. Genes Dev. 2001;15:66–78. doi: 10.1101/gad.186601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bondurand N., Natarajan D., Thapar N., Atkins C., Pachnis V. Neuron and glia generating progenitors of the mammalian enteric nervous system isolated from foetal and postnatal gut cultures. Development. 2003;130:6387–6400. doi: 10.1242/dev.00857. [DOI] [PubMed] [Google Scholar]
- 16.Kurtz A., Zimmer A., Schnütgen F., Brüning G., Spener F., Müller T. The expression pattern of a novel gene encoding brain-fatty acid binding protein correlates with neuronal and glial cell development. Development. 1994;120:2637–2649. doi: 10.1242/dev.120.9.2637. [DOI] [PubMed] [Google Scholar]
- 17.Schwanzel-Fukuda M., Bick D., Pfaff D.W. Luteinizing hormone-releasing hormone (LHRH)-expressing cells do not migrate normally in an inherited hypogonadal (Kallmann) syndrome. Brain Res. Mol. Brain Res. 1989;6:311–326. doi: 10.1016/0169-328x(89)90076-4. [DOI] [PubMed] [Google Scholar]
- 18.Bondurand N., Girard M., Pingault V., Lemort N., Dubourg O., Goossens M. Human Connexin 32, a gap junction protein altered in the X-linked form of Charcot-Marie-Tooth disease, is directly regulated by the transcription factor SOX10. Hum. Mol. Genet. 2001;10:2783–2795. doi: 10.1093/hmg/10.24.2783. [DOI] [PubMed] [Google Scholar]
- 19.Bondurand N., Pingault V., Goerich D.E., Lemort N., Sock E., Le Caignec C., Wegner M., Goossens M. Interaction among SOX10, PAX3 and MITF, three genes altered in Waardenburg syndrome. Hum. Mol. Genet. 2000;9:1907–1917. doi: 10.1093/hmg/9.13.1907. [DOI] [PubMed] [Google Scholar]
- 20.Sánchez-Mejías A., Watanabe Y., M Fernández R., López-Alonso M., Antiñolo G., Bondurand N., Borrego S. Involvement of SOX10 in the pathogenesis of Hirschsprung disease: report of a truncating mutation in an isolated patient. J. Mol. Med. 2010;88:507–514. doi: 10.1007/s00109-010-0592-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.LeBlanc S.E., Jang S.W., Ward R.M., Wrabetz L., Svaren J. Direct regulation of myelin protein zero expression by the Egr2 transactivator. J. Biol. Chem. 2006;281:5453–5460. doi: 10.1074/jbc.M512159200. [DOI] [PubMed] [Google Scholar]
- 22.Zhang H., Chen H., Luo H., An J., Sun L., Mei L., He C., Jiang L., Jiang W., Xia K. Functional analysis of Waardenburg syndrome-associated PAX3 and SOX10 mutations: report of a dominant-negative SOX10 mutation in Waardenburg syndrome type II. Hum. Genet. 2012;131:491–503. doi: 10.1007/s00439-011-1098-2. [DOI] [PubMed] [Google Scholar]
- 23.Peirano R.I., Goerich D.E., Riethmacher D., Wegner M. Protein zero gene expression is regulated by the glial transcription factor Sox10. Mol. Cell. Biol. 2000;20:3198–3209. doi: 10.1128/mcb.20.9.3198-3209.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chaoui A., Watanabe Y., Touraine R., Baral V., Goossens M., Pingault V., Bondurand N. Identification and functional analysis of SOX10 missense mutations in different subtypes of Waardenburg syndrome. Hum. Mutat. 2011;32:1436–1449. doi: 10.1002/humu.21583. [DOI] [PubMed] [Google Scholar]
- 25.Guex N., Peitsch M.C. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 26.Jiang L., Chen H., Jiang W., Hu Z., Mei L., Xue J., He C., Liu Y., Xia K., Feng Y. Novel mutations in the SOX10 gene in the first two Chinese cases of type IV Waardenburg syndrome. Biochem. Biophys. Res. Commun. 2011;408:620–624. doi: 10.1016/j.bbrc.2011.04.072. [DOI] [PubMed] [Google Scholar]
- 27.Sznajer Y., Coldéa C., Meire F., Delpierre I., Sekhara T., Touraine R.L. A de novo SOX10 mutation causing severe type 4 Waardenburg syndrome without Hirschsprung disease. Am. J. Med. Genet. A. 2008;146A:1038–1041. doi: 10.1002/ajmg.a.32247. [DOI] [PubMed] [Google Scholar]
- 28.Hanchate N.K., Giacobini P., Lhuillier P., Parkash J., Espy C., Fouveaut C., Leroy C., Baron S., Campagne C., Vanacker C. SEMA3A, a gene involved in axonal pathfinding, is mutated in patients with Kallmann syndrome. PLoS Genet. 2012;8:e1002896. doi: 10.1371/journal.pgen.1002896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barraud P., Seferiadis A.A., Tyson L.D., Zwart M.F., Szabo-Rogers H.L., Ruhrberg C., Liu K.J., Baker C.V. Neural crest origin of olfactory ensheathing glia. Proc. Natl. Acad. Sci. USA. 2010;107:21040–21045. doi: 10.1073/pnas.1012248107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Forni P.E., Taylor-Burds C., Melvin V.S., Williams T., Wray S. Neural crest and ectodermal cells intermix in the nasal placode to give rise to GnRH-1 neurons, sensory neurons, and olfactory ensheathing cells. J. Neurosci. 2011;31:6915–6927. doi: 10.1523/JNEUROSCI.6087-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Katoh H., Shibata S., Fukuda K., Sato M., Satoh E., Nagoshi N., Minematsu T., Matsuzaki Y., Akazawa C., Toyama Y. The dual origin of the peripheral olfactory system: placode and neural crest. Mol. Brain. 2011;4:34. doi: 10.1186/1756-6606-4-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ekberg J.A., Amaya D., Mackay-Sim A., St John J.A. The migration of olfactory ensheathing cells during development and regeneration. Neurosignals. 2012;20:147–158. doi: 10.1159/000330895. [DOI] [PubMed] [Google Scholar]
- 33.Quinton R., Duke V.M., Robertson A., Kirk J.M., Matfin G., de Zoysa P.A., Azcona C., MacColl G.S., Jacobs H.S., Conway G.S. Idiopathic gonadotrophin deficiency: genetic questions addressed through phenotypic characterization. Clin. Endocrinol. (Oxf.) 2001;55:163–174. doi: 10.1046/j.1365-2265.2001.01277.x. [DOI] [PubMed] [Google Scholar]
- 34.Dodé C., Fouveaut C., Mortier G., Janssens S., Bertherat J., Mahoudeau J., Kottler M.L., Chabrolle C., Gancel A., François I. Novel FGFR1 sequence variants in Kallmann syndrome, and genetic evidence that the FGFR1c isoform is required in olfactory bulb and palate morphogenesis. Hum. Mutat. 2007;28:97–98. doi: 10.1002/humu.9470. [DOI] [PubMed] [Google Scholar]
- 35.Dodé C., Levilliers J., Dupont J.M., De Paepe A., Le Dû N., Soussi-Yanicostas N., Coimbra R.S., Delmaghani S., Compain-Nouaille S., Baverel F. Loss-of-function mutations in FGFR1 cause autosomal dominant Kallmann syndrome. Nat. Genet. 2003;33:463–465. doi: 10.1038/ng1122. [DOI] [PubMed] [Google Scholar]
- 36.Falardeau J., Chung W.C., Beenken A., Raivio T., Plummer L., Sidis Y., Jacobson-Dickman E.E., Eliseenkova A.V., Ma J., Dwyer A. Decreased FGF8 signaling causes deficiency of gonadotropin-releasing hormone in humans and mice. J. Clin. Invest. 2008;118:2822–2831. doi: 10.1172/JCI34538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Massin N., Pêcheux C., Eloit C., Bensimon J.L., Galey J., Kuttenn F., Hardelin J.P., Dodé C., Touraine P. X chromosome-linked Kallmann syndrome: clinical heterogeneity in three siblings carrying an intragenic deletion of the KAL-1 gene. J. Clin. Endocrinol. Metab. 2003;88:2003–2008. doi: 10.1210/jc.2002-021981. [DOI] [PubMed] [Google Scholar]
- 38.Söderlund D., Canto P., Méndez J.P. Identification of three novel mutations in the KAL1 gene in patients with Kallmann syndrome. J. Clin. Endocrinol. Metab. 2002;87:2589–2592. doi: 10.1210/jcem.87.6.8611. [DOI] [PubMed] [Google Scholar]
- 39.Trarbach E.B., Abreu A.P., Silveira L.F., Garmes H.M., Baptista M.T., Teles M.G., Costa E.M., Mohammadi M., Pitteloud N., Mendonca B.B., Latronico A.C. Nonsense mutations in FGF8 gene causing different degrees of human gonadotropin-releasing deficiency. J. Clin. Endocrinol. Metab. 2010;95:3491–3496. doi: 10.1210/jc.2010-0176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee M.J., Calle E., Brennan A., Ahmed S., Sviderskaya E., Jessen K.R., Mirsky R. In early development of the rat mRNA for the major myelin protein P(0) is expressed in nonsensory areas of the embryonic inner ear, notochord, enteric nervous system, and olfactory ensheathing cells. Dev. Dyn. 2001;222:40–51. doi: 10.1002/dvdy.1165. [DOI] [PubMed] [Google Scholar]
- 41.Peirano R.I., Wegner M. The glial transcription factor Sox10 binds to DNA both as monomer and dimer with different functional consequences. Nucleic Acids Res. 2000;28:3047–3055. doi: 10.1093/nar/28.16.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Malki S., Boizet-Bonhoure B., Poulat F. Shuttling of SOX proteins. Int. J. Biochem. Cell Biol. 2010;42:411–416. doi: 10.1016/j.biocel.2009.09.020. [DOI] [PubMed] [Google Scholar]
- 43.Dutton K.A., Pauliny A., Lopes S.S., Elworthy S., Carney T.J., Rauch J., Geisler R., Haffter P., Kelsh R.N. Zebrafish colourless encodes sox10 and specifies non-ectomesenchymal neural crest fates. Development. 2001;128:4113–4125. doi: 10.1242/dev.128.21.4113. [DOI] [PubMed] [Google Scholar]
- 44.Kapur R.P. Early death of neural crest cells is responsible for total enteric aganglionosis in Sox10(Dom)/Sox10(Dom) mouse embryos. Pediatr. Dev. Pathol. 1999;2:559–569. doi: 10.1007/s100249900162. [DOI] [PubMed] [Google Scholar]
- 45.Paratore C., Goerich D.E., Suter U., Wegner M., Sommer L. Survival and glial fate acquisition of neural crest cells are regulated by an interplay between the transcription factor Sox10 and extrinsic combinatorial signaling. Development. 2001;128:3949–3961. doi: 10.1242/dev.128.20.3949. [DOI] [PubMed] [Google Scholar]
- 46.Sonnenberg-Riethmacher E., Miehe M., Stolt C.C., Goerich D.E., Wegner M., Riethmacher D. Development and degeneration of dorsal root ganglia in the absence of the HMG-domain transcription factor Sox10. Mech. Dev. 2001;109:253–265. doi: 10.1016/s0925-4773(01)00547-0. [DOI] [PubMed] [Google Scholar]
- 47.Southard-Smith E.M., Kos L., Pavan W.J. Sox10 mutation disrupts neural crest development in Dom Hirschsprung mouse model. Nat. Genet. 1998;18:60–64. doi: 10.1038/ng0198-60. [DOI] [PubMed] [Google Scholar]
- 48.Maka M., Stolt C.C., Wegner M. Identification of Sox8 as a modifier gene in a mouse model of Hirschsprung disease reveals underlying molecular defect. Dev. Biol. 2005;277:155–169. doi: 10.1016/j.ydbio.2004.09.014. [DOI] [PubMed] [Google Scholar]
- 49.Reiprich S., Stolt C.C., Schreiner S., Parlato R., Wegner M. SoxE proteins are differentially required in mouse adrenal gland development. Mol. Biol. Cell. 2008;19:1575–1586. doi: 10.1091/mbc.E07-08-0782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stolt C.C., Lommes P., Friedrich R.P., Wegner M. Transcription factors Sox8 and Sox10 perform non-equivalent roles during oligodendrocyte development despite functional redundancy. Development. 2004;131:2349–2358. doi: 10.1242/dev.01114. [DOI] [PubMed] [Google Scholar]
- 51.Su Z., He C. Olfactory ensheathing cells: biology in neural development and regeneration. Prog. Neurobiol. 2010;92:517–532. doi: 10.1016/j.pneurobio.2010.08.008. [DOI] [PubMed] [Google Scholar]
- 52.Wray S. From nose to brain: development of gonadotrophin-releasing hormone-1 neurones. J. Neuroendocrinol. 2010;22:743–753. doi: 10.1111/j.1365-2826.2010.02034.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Breuskin I., Bodson M., Thelen N., Thiry M., Borgs L., Nguyen L., Lefebvre P.P., Malgrange B. Sox10 promotes the survival of cochlear progenitors during the establishment of the organ of Corti. Dev. Biol. 2009;335:327–339. doi: 10.1016/j.ydbio.2009.09.007. [DOI] [PubMed] [Google Scholar]
- 54.Watanabe K., Takeda K., Katori Y., Ikeda K., Oshima T., Yasumoto K., Saito H., Takasaka T., Shibahara S. Expression of the Sox10 gene during mouse inner ear development. Brain Res. Mol. Brain Res. 2000;84:141–145. doi: 10.1016/s0169-328x(00)00236-9. [DOI] [PubMed] [Google Scholar]
- 55.Breuskin I., Bodson M., Thelen N., Thiry M., Borgs L., Nguyen L., Stolt C., Wegner M., Lefebvre P.P., Malgrange B. Glial but not neuronal development in the cochleo-vestibular ganglion requires Sox10. J. Neurochem. 2010;114:1827–1839. doi: 10.1111/j.1471-4159.2010.06897.x. [DOI] [PubMed] [Google Scholar]
- 56.Janssen N., Bergman J.E., Swertz M.A., Tranebjaerg L., Lodahl M., Schoots J., Hofstra R.M., van Ravenswaaij-Arts C.M., Hoefsloot L.H. Mutation update on the CHD7 gene involved in CHARGE syndrome. Hum. Mutat. 2012;33:1149–1160. doi: 10.1002/humu.22086. [DOI] [PubMed] [Google Scholar]
- 57.Bergman J.E., de Ronde W., Jongmans M.C., Wolffenbuttel B.H., Drop S.L., Hermus A., Bocca G., Hoefsloot L.H., van Ravenswaaij-Arts C.M. The results of CHD7 analysis in clinically well-characterized patients with Kallmann syndrome. J. Clin. Endocrinol. Metab. 2012;97:E858–E862. doi: 10.1210/jc.2011-2652. [DOI] [PubMed] [Google Scholar]
- 58.Verloes A. Updated diagnostic criteria for CHARGE syndrome: a proposal. Am. J. Med. Genet. A. 2005;133A:306–308. doi: 10.1002/ajmg.a.30559. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


