Abstract
Myopia, or near-sightedness, is an ocular refractive error of unfocused image quality in front of the retinal plane. Individuals with high-grade myopia (dioptric power greater than −6.00) are predisposed to ocular morbidities such as glaucoma, retinal detachment, and myopic maculopathy. Nonsyndromic, high-grade myopia is highly heritable, and to date multiple gene loci have been reported. We performed exome sequencing in 4 individuals from an 11-member family of European descent from the United States. Affected individuals had a mean dioptric spherical equivalent of −22.00 sphere. A premature stop codon mutation c.157C>T (p.Gln53*) cosegregating with disease was discovered within SCO2 that maps to chromosome 22q13.33. Subsequent analyses identified three additional mutations in three highly myopic unrelated individuals (c.341G>A, c.418G>A, and c.776C>T). To determine differential gene expression in a developmental mouse model, we induced myopia by applying a −15.00D lens over one eye. Messenger RNA levels of SCO2 were significantly downregulated in myopic mouse retinae. Immunohistochemistry in mouse eyes confirmed SCO2 protein localization in retina, retinal pigment epithelium, and sclera. SCO2 encodes for a copper homeostasis protein influential in mitochondrial cytochrome c oxidase activity. Copper deficiencies have been linked with photoreceptor loss and myopia with increased scleral wall elasticity. Retinal thinning has been reported with an SC02 variant. Human mutation identification with support from an induced myopic animal provides biological insights of myopic development.
Main Text
Myopia is a common ocular disorder primarily resulting from globe axial elongation.1,2 Its extreme form, high-grade myopia (refractive error greater than −6.00 diopters [D]) (MIM 160700, MIM 613969, MIM 60995, MIM 608367, MIM 614167, MIM 603221, MIM 608474, MIM 612554, and MIM 609994), is highly heritable and associated with ocular morbidities such as retinal detachment, maculopathy, cataracts, and glaucoma.3 Myopia prevalence rates vary worldwide. The highest prevalence rates are those in Asian countries, particularly in urban settings. Over 80% of school children in Taiwan develop myopia by adulthood, and similar rates are seen in children aged between 13 and 15 years in Hong Kong.4–6 In the United States, 33.1% of adults have some degree of myopia, and high-grade myopia affects approximately 2% of the myopic population.7,8 The economic impact of refractive error management is substantial. In the U.S., adults spend an average of $199 annually on refractive-error-correction-related costs.9 U.S. estimates in 2007 of costs associated with vision impairment exceeded 51 billion dollars annually.10 In conjunction, vision-impairment correction costs in the U.S. account for $3.8 to $7.2 billion annually.11
Multiple mapping and genome-wide association studies have identified loci and genes associated with nonsyndromic myopia.12 High-grade myopia is regarded to be distinct from low-grade myopia, as classified through thresholds in spherical refractive error and axial length measurements.13 Recently, advances in deep sequencing technology have identified mutations in genes associated with a variety of ocular disorders including myopia.14–16 In 2011, Shi et al. identified mutations in zinc finger protein 644 isoform 1 (ZNF644) in a Chinese family with autosomal-dominant high-grade myopia by using exome sequencing, which was replicated in four cases in our European descent cohort.16,17
Herein, we describe the identification of pathogenic mutations in the SCO2 cytochrome c oxidase (COX) assembly protein (SCO2) on chromosome 22q13.33 (NM_001169111.1). Gene expression studies in an experimentally induced myopic mouse model suggest that SCO2 may play a role in myopic development.
A large three-generation index family (11 members) of European descent with nine affected individuals with high-grade myopia (average spherical refractive error of −22.00D) participated in the study (Figure 1). Informed consent was obtained from all participants, with approval by the Institutional Review Board according to the principles of the Declaration of Helsinki. DNA was extracted from blood and/or saliva from all participating family members. The affected phenotype was determined as those with high myopia (refractive error greater than −6.00D) with no systemic abnormalities. To identify the genetic etiology of disease in our family, we employed exome sequencing. We selected four individuals (III:1, III:2, IV:1, IV:2) for sequencing and analysis (Figure 1). An additional 60 ethnically matched exomes and 1,172 ethnically matched controls (500 samples ascertained internally, and 672 samples purchased commercially; The Centre for Applied Genomics, The Hospital for Sick Children, Toronto, Canada) were available for subsequent validation studies to analyze allelic frequencies for candidate variants.
Figure 1.
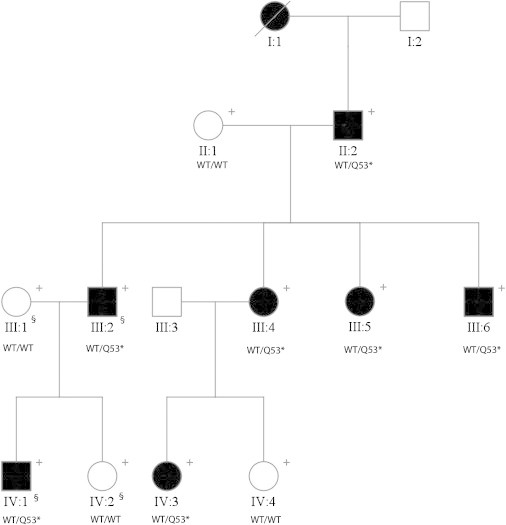
A Family with High-Grade Myopia
Segregation of SCO2 mutation (c.157C>T) in a high-grade myopia family. Abbreviations are as follows: + indicates DNA available for this study, and § indicates samples used for exome sequencing. Where available, the genotype of each individual is shown as a “WT” for the wild-type allele and as an “M” for the mutated allele.
Exome sequencing was performed by Beijing Genomics Institute (BGI), and data analyses were conducted internally. Seven micrograms (μg) of DNA were submitted with independent sample processing by using the Agilent SureSelect XT Human All Exon 38 Mb kit (Agilent, Santa Clara, CA). High throughput sequencing was performed with 91 base pair (bp) paired-end runs on a HiSeq2000 (Illumina, San Diego, CA). Read alignment was conducted by using Burrows-Wheeler Aligner (v.0.5.6) and potential duplicate reads were removed with Picard v.1.40.18 Filtering and detection of reads were conducted by using SAMtools (v.0.1.7).19 Single nucleotide variants and microindels were annotated by using the Genome Analyzer Toolkit (GATK- v1.4). We generated an average of 37.8 gigabases (Gb) of sequence and coverage of 34× for each individual. An average of 95.7% of targeted bases was covered in the four subject samples, and 87.5% of the target had at least 5× coverage (see Supplement 1 available online).
Variants in dbSNP132 with a minor allele frequency greater than 3% and those present in more than 1% of public exomes (NHLBI and 1000 Genomes) were excluded. In addition, heterozygous variants present in at least one affected individual were kept in the finalized list. In conjunction with filtering, Integrated Genome Viewer (IGV) visualization software was utilized to confirm corresponding reads and read depth to verify false positives or negatives. We identified 49 variants shared among the exome-sequenced affected members only. To minimize false positives due to batch effects, we verified all 49 variants as unique in silico relative to 60 exomes previously sequenced by our group. We performed Sanger sequencing of all 49 variants and demonstrated 100% validation (49/49) in the four index DNA samples. All microindels were eliminated based on our filtering criteria. To confirm the segregation of variants with the disease phenotype, we used Sanger sequencing for the remaining 7 family member DNA samples against all 49 variants.
A rare nonsense mutation of c.157C>T (rs74315510) within exon 2 of SCO2 cytochrome c oxidase assembly protein (SCO2, NM_001169111.1) segregated with high-grade myopia in the pedigree. The SCO2 mutation converts the amino acid glutamine to a premature stop codon on base 53 (p.Gln53*). The minor allele was not present in 1,000 control samples for the SCO2 variant. The MERLIN program using a dominant parametric model was employed to estimate the linkage of this variant.20 A two-point LOD score of 1.49 for c.157C>T was calculated for the family.20 PCR sequencing of the SCO2 coding exon 2 was conducted in an additional 140 high-grade myopia cases. The average spherical equivalent refraction of the cases was −11.00D (OD) and −10.50D (OS), respectively. We identified two additional rare variants (rs74315511 and rs8139305) and one variant in three unrelated cases that were heterozygous (Table 1). Rs74315511 (c.418G>A) is a missense mutation predicted to cause a p.Glu140Lys substitution. Rs8139305 (c.776C>T) missense mutation causes a p.Ala259Val substitution. The missense mutation (c.341G>A) causes a substitution of p.Arg114His. These variants were not seen in the same 1,000 control DNA samples. ANNOVAR was used to assess functional annotation and at least one in silico software predicted the mutations to be deleterious or damaging (Table 1).21–27 By using the Fisher exact test, the likelihood for identifying the 4 functional variants in 141 individuals with high-grade myopia relative to 1,000 nonmyopic controls was significant (p = 0.000248).
Table 1.
Summary of Identified SCO2 Mutations
| Family/Case Name | Ethnicity |
Refractive Error (Diopters) |
Nucleotide Change | Amino Acid Change | Chromsome 22 Location (bp)b | rs Number | LJB_PhyloP | LJB_GERP++ | AVSIFT | LJB_SIFT | LJB_PolyPhen2 | LJB_MutationTaster | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OD | OS | ||||||||||||
| CHP-6a | European descent | −22.00 | −22.00 | c.157C>T | p.Gln53* | 50962684 | rs74315510 | 0.998 | 3.13 | 0.71 | 0.911 | 0.650 | 1 |
| CHP-150 | Middle Eastern | −15.25 | −12.00 | c.341.G>A | p.Arg114His | 50962500 | NA | 0.998 | 4.44 | 0.03 | 0.86 | 0.995 | 0.930 |
| CHP-162 | European descent | −14.25 | −15.00 | c.418G>A | p.Glu140Lys | 50962423 | rs74315511 | 0.970 | 3.06 | 0 | 1 | 1 | 0.908 |
| CHP-84 | African-American | −11.00 | −10.25 | c.776C>T | p.Ala259Val | 50962065 | rs8139305 | 0.870 | 0.279 | 0.08 | 0.9 | 0.078 | 0.329 |
Annotations were derived with ANNOVAR. Evolutionary site conservation scores for PhyloP and GERP++ were calculated as described in dbNSFP and bolded when above the threshold for being classified as “conserved.” Predicted mutation effect scores are shown for SIFT, PolyPhen2, and MutationTaster. Scores above the threshold for predictions as “deleterious” are bolded if close to the threshold. OD, right eye; OS, left eye.
Refraction of CHP-6 depicts the average refractive error among the affected individuals.
GRCh37.p5.
SCO2 consists of two exons of which only the second exon is protein coding. Given the mutation location, c.157C>T truncates the protein before the catalytic domain, rendering it nonfunctional. The protein changes (p.Arg114His, p.Glu140Lys, and p.Ala259Val) by using BLAST are all located within the functional catalytic domain and are predicted to affect the protein structure. The p.Glu140Lys amino acid substitution results in removal of a salt bridge between Glu140 and Lys143 and changes the electrostatic potential of the copper binding site, which can moderately to strongly affect SCO2 function (Supplements 2 and 3). Moreover, p.Arg114His and p.Ala259Val are predicted to destabilize the structure based on FoldX, with mild-to-moderate influence on SCO2 function (Supplement 4).28
Immunohistochemical results in mouse ocular tissues confirmed SCO2 protein localization in the retina, retinal pigment epithelium (RPE), and scleral wall (Figure 2). Immunostaining intensity was reduced significantly in myopic retinal tissues of experimentally induced myopic mice compared to the nonmyopic independent controls and was significantly increased in myopic sclera (Figure 2).
Figure 2.

Immunofluorescent Labeling of Sco2 in Mouse Ocular Tissues in Induced Myopic Eyes, Fellow Eyes, and Independent Control Eyes
Immunofluorescent labeling of Sco2 in mouse retina, retinal pigment epithelium, and sclera in induced myopic eyes, fellow eyes, and independent control eyes. The florescence intensity labeled of the green color shows the localization of proteins, and blue color indicates the nuclei that were stained with DAPI. Lower level of abundance was determined for myopic retina and RPE, whereas a higher level of abundance was found in myopic sclera.
The following abbreviations represent the retinal layers: NFL, nerve fiber layer; GCL, ganglion cell layer; IPL, inner plexiform layer; INL, inner nuclear layer; OPL, outer plexiform layer; ONL, outer nuclear layer; PRL, photo receptor layer; and RPE, retinal pigment epithelium.
Sco2 expression in ocular tissues was compared between induced myopic mouse eyes relative to the control fellow eye. Ocular tissues of myopic (with spherical equivalent [SE] < −5.00D) and fellow nonoccluded eyes of the experimental mice were compared with age-matched control tissues (Supplement 6). Real-time PCR confirmed Sco2 messenger RNA (mRNA) levels to be significantly reduced in myopic retina compared to naive control retina (fold change [FC] = −8.3, p < 0.001). Increased Sco2 mRNA was detected in myopic compared to control sclera (FC = +5.6, p < 0.01) (Figure 3). Reverse transcription PCR of SCO2 expression in fetal and adult human ocular tissues confirmed expression in the choroid, sclera, retina, and RPE (Supplement 6).
Figure 3.
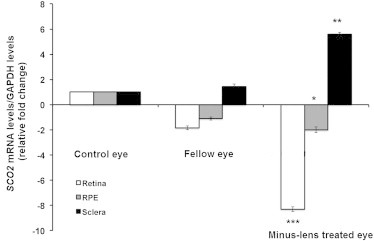
Transcription Quantification of Sco2 in Mouse Retina and Sclera in Induced Myopic Eyes, Fellow Eyes, and Independent Control Eyes
Experimental myopia was induced in B6 wild-type (WT) mice (n = 36) by applying a −15.00 D spectacle lens on the right eye (experimental eye) for 6 weeks since postnatal day 10. The left eyes were uncovered and served as contra-lateral fellow eyes. Age-matched naive mice eyes were used as independent control eyes (n = 36). Primer sequences to conduct qRT-PCR were forward 5′ ATC GCA CAG CCC TAA GTC TC 3′ and reverse 5′ CAG TAG CAT CGT GGA CCT GA 3′ (NM_001111288.1).
The bar represents the fold changes of mRNA for Sco2 after normalization by using GAPDH as reference. The mRNA levels of sco2 in myopic and fellow retina and sclera are compared with independent controls. Relative fold change—the values are shown in log values (210). n = 36; *p < 0.05, **p < 0.01 and ***p < 0.001.
SCO2 is a copper chaperone integral to oxygen reduction catalysis by cytochrome c oxidase of the mitochondrial respiratory chain.29 The COX assembly assists in ATP metabolism, and disruptions exhibit increased intraocular oxygen levels and loss in protection to increased oxygen toxicity.30 Protein deficiency can result in reactive oxygen species increase and oxidative DNA damage.31 COX deficiencies affect organs with high energy demand.32 Because the retina is one of the most highly metabolic tissues in the body, the increased oxidative stress may alter retinal function and therefore image quality, which is essential for refractive development.31,33–41
Normal copper metabolism is essential to ocular tissue health and is associated with myopic refractive error development.42–44 As an example of trace element ocular tissue effects, copper-deficient rats exhibit a loss of conjunctival goblet cells, decrease in conjunctival and corneal microvilli and microplicae, retinal photoreceptor cell degeneration and disappearances, and degeneration and disappearance of myelin lamellae of myelinated optic nerve fibers.45,46 This implicates proper copper metabolism for cell differentiation, development, and maintenance.46 One study demonstrated the protective effect of copper supplements in individuals with myopia.47 Restoration by subTenon’s capsule injection of copper compounds resulted in increased scleral copper concentration and improved scleral tissue elasticity with cessation of myopic refractive error development.47
Reduced retinal layer thickness is correlated with higher degrees of myopia in humans with degenerative retinal changes.16,48,49 Our experimentally induced myopic mouse model demonstrated retinal thinning, corroborating previous animal studies with similar results.50,51 Tree shrews and other animal models with experimentally induced myopia demonstrated retinal ganglion cell layer thinning.51,52 Interestingly, the highly conserved paralogous SCO1 (Supplement 7) was upregulated in myopic chick retinae induced with positive (hyperopic) lens exposure.52
SCO2 mutations are associated with autosomal-recessive fatal infantile cardioencephalomyopathy (MIM 604377), COX deficiency, and milder spinal muscular atrophy-like presentations.53–57 Affected individuals harbor mutations in a compound heterozygous state, where p.Glu140Lys and an additional damaging substitution are typically present.54,58 p.Glu140Lys and p.Gln53* have been reported in cardiomyopathy patients, whereas p.Arg114His and p.Ala259Val (rs8139305) are without annotation for clinical associations.54,55 Retinal histology of a subject with cardioencephalomyopathy harboring a compound heterozygous substitution of p.Glu140Lys and p.Gln53* had retinal ganglion neuronal loss and globular distension of the retinal photoreceptors.59 Neonatal expiration precludes investigation of an associated clinical ocular phenotype such as refractive error. It is worth noting that phenotype-genotype variability does occur and has been seen in cardiovascular and ocular diseases.60,61 Within SCO2, variability in onset and systemic involvement have been reported between compound heterozygous and homozygous individuals.58 Visual impairment is often regarded as a benign disorder as a result of efficient treatment options such as glasses, contact lenses, and refractive surgery and is not always recognized as a disease phenotype in medical registries.62 The phenotypical intersection of myopia and cardioencephalomyopathy presented here must be considered exploratory and further studies are warranted.
In summary, we identified four heterozygous mutations—c.157C>T (p.Gln53*), c.341G>A (p.Arg114His), c.418G>A (p.Glu140Lys), and c.776C>T (p.Ala259Val)—in individuals and families with high-grade myopia. Investigations in silico revealed that the nonsense mutation c.157C>T truncates the protein before the catalytic domain, whereas the other three mutations are predicted to destabilize the protein structure. The destabilization of the protein may result in modulation of oxidative toxicity—particularly in the retina, leading to retinal neuronal thinning due to threshold changes in ROS.31,33–35,63 In addition, mutations can affect copper metabolism, which may result in an imbalance of copper enzymatic support activity and oxidative levels within eye tissues.
Refractive error genetics has proven to be complex, as demonstrated by mapping and large association studies. Although SCO2 did not colocalize in any reported myopia loci, our findings provide evidence that SCO2 may play an important role in eye growth and development, particularly in those who become highly myopic. A myopic phenotype should not be overlooked in studies involving a heterogeneous group of rare disorders involving SCO2.
Acknowledgments
This research was funded by the National Institutes of Health Grant R01 EY014685, the Lew Wasserman Award from Research to Prevent Blindness (Chicago, IL), and a Duke-National University of Singapore core grant to T.L.Y. V.S. was supported by the Toulouse Hospital Young Researcher Fellowship, the Fondation pour la Recherche Médicale, and Fondation de France. The animal model experiments were supported by the National Medical Research Council of Singapore (NMRC/IRG/1117/2008 and NMRC/CG/T1/2010) to V.A.B. The sponsors or funding organizations had no role in the design or conduct of this research. The authors also thank Felicia Hawthorne, Xiaoyan Luo, and Kwan Jia Lin for their helpful insights regarding gene expression and immunohistochemical experiments.
Supplemental Data
Web Resources
The URLs for data presented herein are as follows:
1000 Genomes, http://browser.1000genomes.org
Broad Institute Integrated Genomics Viewer, http://www.broadinstitute.org/igv/
NHLBI Exome Sequencing Project (ESP) Exome Variant Server, http://evs.gs.washington.edu/EVS/
Online Mendelian Inheritance in Man (OMIM), http://www.omim.org/
Picard, http://picard.sourceforge.net/
PolyPhen, www.genetics.bwh.harvard.edu/pph2/
SIFT, http://sift.jcvi.org/
References
- 1.Curtin B.J. Harper & Row; Philadelphia: 1985. The Myopias: Basic Science and Clinical Management. [Google Scholar]
- 2.Curtin B.J., Karlin D.B. Axial length measurements and fundus changes of the myopic eye. Am. J. Ophthalmol. 1971;71:42–53. doi: 10.1016/0002-9394(71)91092-0. [DOI] [PubMed] [Google Scholar]
- 3.Saw S.M., Gazzard G., Shih-Yen E.C., Chua W.H. Myopia and associated pathological complications. Ophthalmic Physiol. Opt. 2005;25:381–391. doi: 10.1111/j.1475-1313.2005.00298.x. [DOI] [PubMed] [Google Scholar]
- 4.Lin L.L., Shih Y.F., Hsiao C.K., Chen C.J., Lee L.A., Hung P.T. Epidemiologic study of the prevalence and severity of myopia among schoolchildren in Taiwan in 2000. J. Formos. Med. Assoc. 2001;100:684–691. [PubMed] [Google Scholar]
- 5.Lin L.L., Shih Y.F., Tsai C.B., Chen C.J., Lee L.A., Hung P.T., Hou P.K. Epidemiologic study of ocular refraction among schoolchildren in Taiwan in 1995. Optom. Vis. Sci. 1999;76:275–281. doi: 10.1097/00006324-199905000-00013. [DOI] [PubMed] [Google Scholar]
- 6.Lam C.S., Goldschmidt E., Edwards M.H. Prevalence of myopia in local and international schools in Hong Kong. Optom. Vis. Sci. 2004;81:317–322. doi: 10.1097/01.opx.0000134905.98403.18. [DOI] [PubMed] [Google Scholar]
- 7.Vitale S., Sperduto R.D., Ferris F.L., 3rd Increased prevalence of myopia in the United States between 1971-1972 and 1999-2004. Arch. Ophthalmol. 2009;127:1632–1639. doi: 10.1001/archophthalmol.2009.303. [DOI] [PubMed] [Google Scholar]
- 8.Katz J., Tielsch J.M., Sommer A. Prevalence and risk factors for refractive errors in an adult inner city population. Invest. Ophthalmol. Vis. Sci. 1997;38:334–340. [PubMed] [Google Scholar]
- 9.Rein D.B., Zhang P., Wirth K.E., Lee P.P., Hoerger T.J., McCall N., Klein R., Tielsch J.M., Vijan S., Saaddine J. The economic burden of major adult visual disorders in the United States. Arch. Ophthalmol. 2006;124:1754–1760. doi: 10.1001/archopht.124.12.1754. [DOI] [PubMed] [Google Scholar]
- 10.Frick K.D., Gower E.W., Kempen J.H., Wolff J.L. Economic impact of visual impairment and blindness in the United States. Arch. Ophthalmol. 2007;125:544–550. doi: 10.1001/archopht.125.4.544. [DOI] [PubMed] [Google Scholar]
- 11.Vitale S., Cotch M.F., Sperduto R., Ellwein L. Costs of refractive correction of distance vision impairment in the United States, 1999-2002. Ophthalmology. 2006;113:2163–2170. doi: 10.1016/j.ophtha.2006.06.033. [DOI] [PubMed] [Google Scholar]
- 12.Young T.L. Molecular genetics of human myopia: an update. Optom. Vis. Sci. 2009;86:E8–E22. doi: 10.1097/OPX.0b013e3181940655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Young T.L. Dissecting the genetics of human high myopia: a molecular biologic approach. Trans. Am. Ophthalmol. Soc. 2004;102:423–445. [PMC free article] [PubMed] [Google Scholar]
- 14.Nikopoulos K., Gilissen C., Hoischen A., van Nouhuys C.E., Boonstra F.N., Blokland E.A., Arts P., Wieskamp N., Strom T.M., Ayuso C. Next-generation sequencing of a 40 Mb linkage interval reveals TSPAN12 mutations in patients with familial exudative vitreoretinopathy. Am. J. Hum. Genet. 2010;86:240–247. doi: 10.1016/j.ajhg.2009.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Poulter J.A., Ali M., Gilmour D.F., Rice A., Kondo H., Hayashi K., Mackey D.A., Kearns L.S., Ruddle J.B., Craig J.E. Mutations in TSPAN12 cause autosomal-dominant familial exudative vitreoretinopathy. Am. J. Hum. Genet. 2010;86:248–253. doi: 10.1016/j.ajhg.2010.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shi Y., Li Y., Zhang D., Zhang H., Li Y., Lu F., Liu X., He F., Gong B., Cai L. Exome sequencing identifies ZNF644 mutations in high myopia. PLoS Genet. 2011;7:e1002084. doi: 10.1371/journal.pgen.1002084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tran-Viet K.N., St Germain E., Soler V., Powell C., Lim S.H., Klemm T., Saw S.M., Young T.L. Study of a US cohort supports the role of ZNF644 and high-grade myopia susceptibility. Mol. Vis. 2012;18:937–944. [PMC free article] [PubMed] [Google Scholar]
- 18.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abecasis G.R., Cherny S.S., Cookson W.O., Cardon L.R. Merlin—rapid analysis of dense genetic maps using sparse gene flow trees. Nat. Genet. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- 21.Wang K., Li M., Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pollard K.S., Hubisz M.J., Rosenbloom K.R., Siepel A. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res. 2010;20:110–121. doi: 10.1101/gr.097857.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Davydov E.V., Goode D.L., Sirota M., Cooper G.M., Sidow A., Batzoglou S. Identifying a high fraction of the human genome to be under selective constraint using GERP++ PLoS Comput. Biol. 2010;6:e1001025. doi: 10.1371/journal.pcbi.1001025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu X., Jian X., Boerwinkle E. dbNSFP: a lightweight database of human nonsynonymous SNPs and their functional predictions. Hum. Mutat. 2011;32:894–899. doi: 10.1002/humu.21517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sim N.L., Kumar P., Hu J., Henikoff S., Schneider G., Ng P.C. SIFT web server: predicting effects of amino acid substitutions on proteins. Nucleic Acids Res. 2012;40(Web Server issue) doi: 10.1093/nar/gks539. W452–W457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Adzhubei, I., Jordan, D.M., and Sunyaev, S.R. (2013). Predicting Functional Effect of Human Missense Mutations Using PolyPhen-2. Current protocols in human genetics / editorial board, Jonathan L Haines [et al] Chapter 7, Unit7 20. [DOI] [PMC free article] [PubMed]
- 27.Schwarz J.M., Rödelsperger C., Schuelke M., Seelow D. MutationTaster evaluates disease-causing potential of sequence alterations. Nat. Methods. 2010;7:575–576. doi: 10.1038/nmeth0810-575. [DOI] [PubMed] [Google Scholar]
- 28.Van Durme J., Delgado J., Stricher F., Serrano L., Schymkowitz J., Rousseau F. A graphical interface for the FoldX forcefield. Bioinformatics. 2011;27:1711–1712. doi: 10.1093/bioinformatics/btr254. [DOI] [PubMed] [Google Scholar]
- 29.Robinson N.J., Winge D.R. Copper metallochaperones. Annu. Rev. Biochem. 2010;79:537–562. doi: 10.1146/annurev-biochem-030409-143539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sung H.J., Ma W., Wang P.Y., Hynes J., O’Riordan T.C., Combs C.A., McCoy J.P., Jr., Bunz F., Kang J.G., Hwang P.M. Mitochondrial respiration protects against oxygen-associated DNA damage. Nature Communications. 2010;1:5. doi: 10.1038/ncomms1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu D.Y., Cringle S.J. Oxygen distribution and consumption within the retina in vascularised and avascular retinas and in animal models of retinal disease. Prog. Retin. Eye Res. 2001;20:175–208. doi: 10.1016/s1350-9462(00)00027-6. [DOI] [PubMed] [Google Scholar]
- 32.Zeviani M., Tiranti V., Piantadosi C. Mitochondrial disorders. Medicine (Baltimore) 1998;77:59–72. doi: 10.1097/00005792-199801000-00006. [DOI] [PubMed] [Google Scholar]
- 33.Putting B.J., Van Best J.A., Vrensen G.F., Oosterhuis J.A. Blue-light-induced dysfunction of the blood-retinal barrier at the pigment epithelium in albino versus pigmented rabbits. Exp. Eye Res. 1994;58:31–40. doi: 10.1006/exer.1994.1192. [DOI] [PubMed] [Google Scholar]
- 34.Putting B.J., van Best J.A., Zweypfenning R.C., Vrensen G.F., Oosterhuis J.A. Spectral sensitivity of the blood-retinal barrier at the pigment epithelium for blue light in the 400-500 nm range. Graefe’s archive for clinical and experimental ophthalmology = Albrecht Von Graefes Arch. Klin. Exp. Ophthalmol. 1993;231:600–606. doi: 10.1007/BF00936526. [DOI] [PubMed] [Google Scholar]
- 35.Beatty S., Koh H., Phil M., Henson D., Boulton M. The role of oxidative stress in the pathogenesis of age-related macular degeneration. Surv. Ophthalmol. 2000;45:115–134. doi: 10.1016/s0039-6257(00)00140-5. [DOI] [PubMed] [Google Scholar]
- 36.Anderson B., Jr. Ocular effects of changes in oxygen and carbon dioxide tension. Trans. Am. Ophthalmol. Soc. 1968;66:423–474. [PMC free article] [PubMed] [Google Scholar]
- 37.Stone R.A., Liu J., Sugimoto R., Capehart C., Zhu X., Pendrak K. GABA, experimental myopia, and ocular growth in chick. Invest. Ophthalmol. Vis. Sci. 2003;44:3933–3946. doi: 10.1167/iovs.02-0774. [DOI] [PubMed] [Google Scholar]
- 38.Stone R.A., McGlinn A.M., Baldwin D.A., Tobias J.W., Iuvone P.M., Khurana T.S. Image defocus and altered retinal gene expression in chick: clues to the pathogenesis of ametropia. Invest. Ophthalmol. Vis. Sci. 2011;52:5765–5777. doi: 10.1167/iovs.10-6727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guo S.S., Sivak J.G., Callender M.G., Diehl-Jones B. Retinal dopamine and lens-induced refractive errors in chicks. Curr. Eye Res. 1995;14:385–389. doi: 10.3109/02713689508999936. [DOI] [PubMed] [Google Scholar]
- 40.Troilo D., Judge S.J. Ocular development and visual deprivation myopia in the common marmoset (Callithrix jacchus) Vision Res. 1993;33:1311–1324. doi: 10.1016/0042-6989(93)90039-y. [DOI] [PubMed] [Google Scholar]
- 41.Norton T.T., Siegwart J.T. Local myopia produced by partial deprivation in tree shrew. Society for Neuroscience Abstracts. 1991;17:558. [Google Scholar]
- 42.Cai Y. Determination of select trace elements in hair of college students in Jinzhou, China. Biol. Trace Elem. Res. 2011;144:469–474. doi: 10.1007/s12011-011-9145-3. [DOI] [PubMed] [Google Scholar]
- 43.Silverstone B.Z. Effects of zinc and copper metabolism in highly myopic patients. Ciba Found. Symp. 1990;155:210–217. doi: 10.1002/9780470514023.ch12. discussion 217–221. [DOI] [PubMed] [Google Scholar]
- 44.Silverstone B., Berson D., Eilat U., Kuperman O. Copper metabolism changes in pigmentary retinopathies and high myopia. Metab. Pediatr. Ophthalmol. 1981;5:49–53. [PubMed] [Google Scholar]
- 45.Amemiya T. The eye and nutrition. Jpn. J. Ophthalmol. 2000;44:320. doi: 10.1016/s0021-5155(00)00161-1. [DOI] [PubMed] [Google Scholar]
- 46.Amemiya T. [The eye and nutrition] Nippon Ganka Gakkai Zasshi. 1999;103:829–850. [PubMed] [Google Scholar]
- 47.Avetisov E.S., Vinetskaia M.I., Iomdina E.N., Makhmudova F.R., Boltaeva Z.K., Tarutta E.P. [Copper metabolism in scleral tissue and possibilities of its correction in myopia] Vestn. Oftalmol. 1991;107:31–34. [PubMed] [Google Scholar]
- 48.Wu P.C., Chen Y.J., Chen C.H., Chen Y.H., Shin S.J., Yang H.J., Kuo H.K. Assessment of macular retinal thickness and volume in normal eyes and highly myopic eyes with third-generation optical coherence tomography. Eye (Lond.) 2008;22:551–555. doi: 10.1038/sj.eye.6702789. [DOI] [PubMed] [Google Scholar]
- 49.Chui T.Y., Yap M.K., Chan H.H., Thibos L.N. Retinal stretching limits peripheral visual acuity in myopia. Vision Res. 2005;45:593–605. doi: 10.1016/j.visres.2004.09.016. [DOI] [PubMed] [Google Scholar]
- 50.Beresford J.A., Crewther S.G., Crewther D.P. Anatomical correlates of experimentally induced myopia. Aust. N. Z. J. Ophthalmol. 1998;26(Suppl 1):S84–S87. doi: 10.1111/j.1442-9071.1998.tb01383.x. [DOI] [PubMed] [Google Scholar]
- 51.Abbott C.J., Grünert U., Pianta M.J., McBrien N.A. Retinal thinning in tree shrews with induced high myopia: optical coherence tomography and histological assessment. Vision Res. 2011;51:376–385. doi: 10.1016/j.visres.2010.12.005. [DOI] [PubMed] [Google Scholar]
- 52.Feldkaemper M.P., Wang H.Y., Schaeffel F. Changes in retinal and choroidal gene expression during development of refractive errors in chicks. Invest. Ophthalmol. Vis. Sci. 2000;41:1623–1628. [PubMed] [Google Scholar]
- 53.Tay S.K., Shanske S., Kaplan P., DiMauro S. Association of mutations in SCO2, a cytochrome c oxidase assembly gene, with early fetal lethality. Arch. Neurol. 2004;61:950–952. doi: 10.1001/archneur.61.6.950. [DOI] [PubMed] [Google Scholar]
- 54.Verdijk R.M., de Krijger R., Schoonderwoerd K., Tiranti V., Smeets H., Govaerts L.C., de Coo R. Phenotypic consequences of a novel SCO2 gene mutation. Am. J. Med. Genet. A. 2008;146A:2822–2827. doi: 10.1002/ajmg.a.32523. [DOI] [PubMed] [Google Scholar]
- 55.Jaksch M., Ogilvie I., Yao J., Kortenhaus G., Bresser H.G., Gerbitz K.D., Shoubridge E.A. Mutations in SCO2 are associated with a distinct form of hypertrophic cardiomyopathy and cytochrome c oxidase deficiency. Hum. Mol. Genet. 2000;9:795–801. doi: 10.1093/hmg/9.5.795. [DOI] [PubMed] [Google Scholar]
- 56.Papadopoulou L.C., Sue C.M., Davidson M.M., Tanji K., Nishino I., Sadlock J.E., Krishna S., Walker W., Selby J., Glerum D.M. Fatal infantile cardioencephalomyopathy with COX deficiency and mutations in SCO2, a COX assembly gene. Nat. Genet. 1999;23:333–337. doi: 10.1038/15513. [DOI] [PubMed] [Google Scholar]
- 57.Pronicki M., Kowalski P., Piekutowska-Abramczuk D., Taybert J., Karkucinska-Wieckowska A., Szymanska-Debinska T., Karczmarewicz E., Pajdowska M., Migdal M., Milewska-Bobula B. A homozygous mutation in the SCO2 gene causes a spinal muscular atrophy like presentation with stridor and respiratory insufficiency. European journal of paediatric neurology: EJPN: official journal of the European Paediatric Neurology Society. 2010;14:253–260. doi: 10.1016/j.ejpn.2009.09.008. [DOI] [PubMed] [Google Scholar]
- 58.Jaksch M., Horvath R., Horn N., Auer D.P., Macmillan C., Peters J., Gerbitz K.D., Kraegeloh-Mann I., Muntau A., Karcagi V. Homozygosity (E140K) in SCO2 causes delayed infantile onset of cardiomyopathy and neuropathy. Neurology. 2001;57:1440–1446. doi: 10.1212/wnl.57.8.1440. [DOI] [PubMed] [Google Scholar]
- 59.Vesela K., Hulkova H., Hansikova H., Zeman J., Elleder M. Structural analysis of tissues affected by cytochrome C oxidase deficiency due to mutations in the SCO2 gene. APMIS: acta pathologica, microbiologica, et immunologica Scandinavica. 2008;116:41–49. doi: 10.1111/j.1600-0463.2008.00772.x. [DOI] [PubMed] [Google Scholar]
- 60.Maugeri A., van Driel M.A., van de Pol D.J., Klevering B.J., van Haren F.J., Tijmes N., Bergen A.A., Rohrschneider K., Blankenagel A., Pinckers A.J. The 2588G—>C mutation in the ABCR gene is a mild frequent founder mutation in the Western European population and allows the classification of ABCR mutations in patients with Stargardt disease. Am. J. Hum. Genet. 1999;64:1024–1035. doi: 10.1086/302323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kelly M., Semsarian C. Multiple mutations in genetic cardiovascular disease: a marker of disease severity? Circulation Cardiovascular genetics. 2009;2:182–190. doi: 10.1161/CIRCGENETICS.108.836478. [DOI] [PubMed] [Google Scholar]
- 62.Morgan I.G., Ohno-Matsui K., Saw S.M. Myopia. Lancet. 2012;379:1739–1748. doi: 10.1016/S0140-6736(12)60272-4. [DOI] [PubMed] [Google Scholar]
- 63.Gidalevitz T., Krupinski T., Garcia S., Morimoto R.I. Destabilizing protein polymorphisms in the genetic background direct phenotypic expression of mutant SOD1 toxicity. PLoS Genet. 2009;5:e1000399. doi: 10.1371/journal.pgen.1000399. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


