Abstract
Plant responses to insect herbivory are regulated through complex, hormone-mediated interactions. Some caterpillar species have evolved strategies to manipulate this system by inducing specific pathways that suppress plant defense responses. Effectors in the labial saliva (LS) secretions of Spodoptera exigua caterpillars are believed to induce the salicylic acid (SA) pathway to interfere with the jasmonic acid (JA) defense pathway; however, the mechanism underlying this subversion is unknown. Since noctuid caterpillar LS contains enzymes that may affect cellular redox balance, this study investigated rapid changes in cellular redox metabolites within 45 min after herbivory. Caterpillar LS is involved in suppressing the increase in oxidative stress that was observed in plants fed upon by caterpillars with impaired LS secretions. To further understand the link between cellular redox balance and plant defense responses, marker genes of SA, JA and ethylene (ET) pathways were compared in wildtype, the glutathione-compromised pad2-1 mutant and the tga2/5/6 triple mutant plants. AtPR1 and AtPDF1.2 showed LS-dependent expression that was alleviated in the pad2-1 and tga2/5/6 triple mutants. In comparison, the ET-dependent genes ERF1 expression showed LS-associated changes in both wildtype and pad2-1 mutant plants and the ORA 59 marker AtHEL had increased expression in response to herbivory, but a LS-dependent difference was not noted. These data support the model that there are SA/NPR1-, glutathione-dependent and ET-, glutathione-independent mechanisms leading to LS-associated suppression of plant induced defenses.
Keywords: Arabidopsis thaliana, caterpillar herbivory, cross-talk, induced defenses, signaling pathways, Spodoptera exigua
INTRODUCTION
As plants interact with multiple organisms, they need to prioritize their actions to respond appropriately. Plants manage this through synergistic or antagonistic interactions mediated through growth and defense hormones: a process known as cross-talk (Spoel and Dong, 2008; Robert-Seilaniantz et al., 2011). In plant–pathogen interactions, activation of the systemic acquired resistance (SAR) pathway by biotrophic pathogens may render the plant more susceptible to necrotrophic pathogens that elicit jasmonate (JA)- and ethylene (ET)-mediated responses (Glazebrook, 2005). Insect herbivores also exploit this plant hormone cross-talk to prevent the induction of defensive pathways (Felton and Korth, 2000); however, the mechanisms underlying this are not fully understood.
When tissues are damaged during caterpillar feeding, rapid changes in calcium signatures and the generation of reactive oxygen species (ROS), such as hydrogen peroxide (H2O2), leads to the induction of the JA pathway and plant defense responses (Lou and Baldwin, 2006; Arimura et al., 2011). At low, regulated concentrations, H2O2 is an important signaling molecule, however, uncontrolled levels are destructive as H2O2 readily reacts with cellular components (Schröder and Eaton, 2008; Forman et al., 2010). ROS is generated by mechanical damage but also by enzymes, such as glucose oxidase (GOX), present in the caterpillar’s labial saliva (LS; Eichenseer et al., 2010). In lima bean, the zone of H2O2 accumulation around the site of leaf damage is widened by ~500 μm by Spodoptera littoralis caterpillar herbivory compared to mechanical wounding (Maffei et al., 2006). This caterpillar LS-associated production of H2O2 is proposed to be a strategum of some insect species to interfere with induced plant defenses (Musser et al., 2002; Bede et al., 2006).
To avoid the detrimental effects of ROS, antioxidant proteins, such as superoxide dismutase, catalase, peroxidase, and the Halliwell–Asada (ascorbate/glutathione) cycle are activated to maintain cellular redox homeostasis (Noctor et al., 2012). The Halliwell–Asada cycle lowers cellular H2O2 levels through a series of redox reactions involving ascorbate and glutathione. Therefore, in response to stress, plants often alter the total glutathione pool or the ratio between oxidized to reduced glutathione (GSSG:GSH) to maintain low H2O2 levels. Recognition of biotrophic pathogen attack or salicylic acid (SA) mimic treatment may result in an increase in total glutathione levels (Fodor et al., 1997; Mou et al., 2003; Mateo et al., 2006; Mur et al., 2006). Infiltration of SA into Arabidopsis leaves initiates a transient oxidation of the glutathione pool 6 h after the time of injection (Mou et al., 2003; Mateo et al., 2006). In response to mechanical damage, the ratio of GSSG/total glutathione increases, reflecting an oxidized cellular environment, with oxidized glutathione (GSSG) positively linked to JA signaling (Mhamdi et al., 2010; Gfeller et al., 2011). Arabidopsis glutathione mutants are more susceptible to microorganism and insect attack (Ball et al., 2004; Parisy et al., 2007; Schlaeppi et al., 2008). Arabidopsis pad2-1 mutant lacks γ-glutamylcysteine synthetase that catalyzes the first step in glutathione biosynthesis (Parisy et al., 2007); therefore, glutathione levels are approximately one-fifth wildtype levels. This line is more vulnerable to S. littoralis herbivory (Schlaeppi et al., 2008; Leon-Reyes et al., 2010; Mhamdi et al., 2010; Dubreuil-Mauriza et al., 2011). As well, as glutathione pools and ratio change, related processes, such as protein glutathionylation or S-nitrosylation that are also implicated in the regulation of defense against pathogens and herbivores, are affected (Wünsche et al., 2011; Espunya et al., 2012).
In response to caterpillar herbivory, the active form of JA, (+)-7-iso-jasmonoyl-L-isoleucine, bridges JA ZIM-domain (JAZ) proteins with the E3 ubiquitin ligase SCFCOI1 complex, resulting in the proteasome-mediated degradation of JAZ and release of the basic helix-loop-helix transcription factor MYC2, responsible for the expression of JA-associated genes, such as VSP2 and LOX2 (Lorenzo et al., 2004; Dombrecht et al., 2007; Kazan and Manners, 2008; Fonseca et al., 2009; Robert-Seilaniantz et al., 2011). Caterpillar herbivory-related increases in ET biosynthesis may modulate these JA responses through cross-talk between the JA-dependent MYC2-branch and ET-dependent branches (Stotz et al., 2000; Bodenhausen and Reymond, 2007; Kazan and Manners, 2008; Diezel et al., 2009; Robert-Seilaniantz et al., 2011; Verhage et al., 2011). Two APETALA2/ERF transcription factors, ET response factor1 (ERF1) and ORA59 integrate ET cross-talk with the JA pathway (Penninckx et al., 1998; Lorenzo et al., 2003; Pré et al., 2008); though both these branches are induced by ET, evidence points to them being parallel and, perhaps, functionally redundant. Together, the MYC2 and ET pathways, ORA59/ERF1, act synergistically or antagonistically allowing the integration of temporal and spatial hormone concentrations and localization to generate a specific signal signature (Kazan and Manners, 2008; Robert-Seilaniantz et al., 2011).
Effectors in the caterpillar LS may also activate the SAR pathway leading to the attenuation of JA-dependent responses (Kazan and Manners, 2008; Weech et al., 2008; Leon-Reyes et al., 2010; Verhage et al., 2011). In recognition of attack by biotrophic pathogens, plants mount the systemic defense response, SAR, initiated by increases in cellular SA and H2O2 that positively impact each other’s production (Rao et al., 1997; Glazebrook, 2005; Mateo et al., 2006). The resultant change in glutathione redox balance results in the activation of the non-expressor of PR-genes1 (NPR1) through thioredoxin-catalyzed reduction of the disulfide bridges, changing the protein from its cytosolic oligomer form to the monomer that enters the nucleus (Spoel et al., 2009; Noctor et al., 2012). Association of NPR1 with TGA transcription factors leads to the expression of pathogenesis-related genes, such as PR1. The mechanistic basis of the antagonism between SA and JA pathways is still debated (Lorenzo and Solano, 2005). Early evidence suggests that SA interferes directly with JA biosynthesis (Doares et al., 1995; Rayapuram and Baldwin, 2007). However, NPR1 has been shown to be interfere with JA signaling downstream of JA biosynthesis (Mou et al., 2003; Spoel et al., 2003; Ndamukong et al., 2007; Koornneef and Pieterse, 2008; Spoel and Dong, 2008; Tada et al., 2008; Leon-Reyes et al., 2010). This may reflect the observation that ET modifies SA/NPR1 inhibition of JA responses such that in the presence of ET, the attenuation of JA-dependent gene expression is NPR1-independent; however, in the absence of ET, NPR1 is necessary to interfere with these responses (Leon-Reyes et al., 2009). Weech et al. (2008) used Arabidopsis mutants to show that caterpillar LS interference of JA-dependent plant defenses by activation of the SAR pathway requires an active NPR1. In addition, Diezel et al. (2009) showed that damage of wild tobacco by caterpillars of the tobacco hornworm, Manduca sexta, result in an ET burst that attenuates the SA-mediated suppression of plant defense responses. Therefore, in plant–caterpillar interactions, there appears to be extensive interplay between JA, SA, and ET pathways.
The present research is designed to understand the potential role of cellular redox balance in the ability of caterpillar LS to interfere with host plant defense responses. Since caterpillar LS contains redox enzymes, such as GOX that generate H2O2, caterpillar saliva should perturb the redox state or balance even more than mere wounding (Eichenseer et al., 2010; Noctor et al., 2012). By using normal caterpillars with intact LS secretions or insects where LS secretions have been impaired by cauterization of the spinneret, one can tease out the effect of LS on the modulation of plant responses. Therefore, in response to herbivory by caterpillars with intact or impaired LS secretions, the redox metabolites glutathione and ascorbate were measured to identify the impact of LS on cellular redox balance. As well, transcript responses of JA-, ET-, and SA-dependent marker genes were compared in wildtype plants and two mutant lines, pad2-1, compromised in glutathione biosynthesis, and a tga2/5/6 triple mutant that is deficient in the basic leucine zipper TGA transcription factors that interact with NPR1 (Zhang et al., 2003; Parisy et al., 2007).
MATERIALS AND METHODS
PLANTS
Arabidopsis seeds ecotype Col-0 (TAIR CS3749) and the pad2-1 mutant (TAIR CS3804) were obtained from the Arabidopsis Biological Resource Centre (Ohio State University). Seeds of the Arabidopsis tga2/5/6 triple mutant were a generous gift from Dr. Li (University of British Columbia).
For redox metabolite experiments, wildtype plants seeds were surface-sterilized by soaking them for 2 min in 70% ethanol, followed by 5 min in 50% bleach. Seeds were rinsed three times in sterile distilled water and sown in Premier Promix BS (Premier Horticulture Inc.). After cold treatment at 4°C for 3 days, seeds were transferred into a growth cabinet (light intensity 140 μEm-2s-1, 12:12 light:dark at 22°C). Plants were bottom-watered as needed, about three times per week with dilute 0.15 g/L N–P–K fertilizer.
For gene expression experiments, seeds were surface-sterilized as described above and germinated on half-strength Murashige and Skoog (MS) media with 1% agar. After cold treatment for 3 days at 4°C, seeds were placed in the growth cabinet and transferred to Agro-Mix at germination. At 5 weeks post-germination, one plant from each genotype (Col-0, pad2-1 and tga2/5/6) were transplanted into a 12.5 cm × 12 cm pot.
Approximately 6- to 7-week old plants in the late vegetative growth stage, between growth stages 3.7 and 3.9 according to Boyes et al. (2001), were used in redox metabolite or gene expression experiments.
CATERPILLARS
Beet armyworm, Spodoptera exigua (Hübner; Lepidoptera: Noctuidae), insects were reared for multiple generations from eggs purchased from Bio-Serv (Frenchtown, NJ, USA). Insects were reared under defined conditions in a growth cabinet (16:8 light:dark, RH 28-40%, temperature 28.5°C) and fed a wheat germ-based artificial diet (Bio-Serv). Adult moths were allowed to mate and the eggs collected to maintain the colony.
IMPAIRMENT OF CATERPILLAR LS SECRETIONS
Caterpillar LS is secreted through a specialized organ, the spinneret (Musser et al., 2002). To impair LS secretions, this spinneret was cauterized as previously described (Musser et al., 2002; Bede et al., 2006). Prior to the experiment, caterpillars were allowed to feed > 12 h on Arabidopsis plants to allow the insects to adjust to the plant diet.
MEASUREMENT OF REDOX METABOLITES
Leaf H2O2 levels were not measured directly due to the high variability associated with the instable nature of this compound and confounding effects by high leaf phenolic content and ascorbate (Queval et al., 2008). Therefore, other metabolites associated with the ascorbate/glutathione cycle were measured since they closely correlate with H2O2 levels (Ng et al., 2007). Six-week-old Arabidopsis plants were subject to one of three treatments: untouched (control) or subject to herbivory by 3 × 4th instar S. exigua caterpillars with intact or impaired salivary secretions. As S. exigua caterpillars feed most actively at night, experiments were performed during the dark to more accurately simulate an ecological scenario. Rosette leaves showing signs of herbivory were harvested at 5, 15, 25, 35, and 45 min and immediately frozen in N2. The experiment was repeated thrice.
At each time point, ascorbate and glutathione were measured in three to four independent samples. Plant samples were finely ground in liquid nitrogen and extracted in 0.2 N HCl at a ratio of 100 mg leaf/mL acid. This was followed by neutralization with NaOH as described in Queval and Noctor (2007). Chemicals used in redox metabolite assays were purchased from Sigma Chemical Company.
Ascorbate
Total, oxidized and reduced ascorbate from the leaf extract supernatant were determined by measuring reduced ascorbate levels spectrophotometrically at A265 using an Infinite M200 Pro microplate reader (Tecan) according to Queval and Noctor (2007). Total ascorbate was measured by converting dehydroascorbate (DHA) to the reduced form by incubating the supernatant in dithiothreitol (0.4% v/v) in 67.2 mM sodium phosphate (NaH2PO4) buffer, pH 7.5 for 30 min at room temperature. Triplicates of each sample were incubated with ascorbate oxidase (0.2 U) and reduced ascorbate was measured after an 8 min incubation. Reduced ascorbate (ASc) levels were measured by adding ascorbate oxidase to the neutralized leaf extract supernatant in 0.1 M sodium phosphate buffer, pH 5.6, incubating at room temperature for 30 min and analyzing as above. Concentrations were determined from a six-point L-ascorbate standard curve (40–240 μM). Oxidized ascorbate levels (DHA) were calculated by subtracting reduced from total ascorbate.
Glutathione
Measurement of glutathione is based on a recycling assay (Rahman et al., 2006; Queval and Noctor, 2007); glutathione reductase, in the presence of nicotinamide adenine dinucleotide phosphate (NADPH), catalyzes the reduction of GSSG to GSH that reacts with 5,5′-dithiobis(2-nitrobenzoic acid) (DTNB) forming 5-thio-2-nitrobenzoic acid (TNB) that can be measured spectrophotometrically at A412. Total glutathione was measured by incubating leaf supernatant in 0.6 mM DTNB and glutathione reductase (0.015 U) in 0.1 M sodium phosphate (NaH2PO4) buffer, pH 7.5. After the addition of 0.5 mM β-NADPH, the TNB chromophore was monitored at A412 at 5 s intervals for the first 2 min. Total glutathione concentration was calculated based on triplicate eight-point standard curve (100 nM to 60 μM). Oxidized glutathione (GSSG) was measured by removing any reduced GSH from the sample by precipitation with 2-vinylpyridine followed by conversion of GSSG to GSH and measurement using the glutathione reductase/β-NADPH/DTNB method as described above (Griffith, 1980; Rahman et al., 2006; Queval and Noctor, 2007). Briefly, leaf supernatant was incubated with 1 μl 2-vinylpyridine (approximately 10-fold above GSH levels) for 30 min at room temperature. After centrifugation at 13,000 rpm for 5 min to remove excess 2-vinylpyridine, samples were diluted in 0.1 M sodium phosphate buffer, pH 7.5 and assayed in triplicate. GSSG levels were determined from a triplicate eight-point GSSG standard curve ranging from 100 nM to 3.2 μM. Reduced GSH was calculated by subtracting 2 × GSSG from total glutathione.
GENE EXPRESSION
Three days before the herbivory experiment, clear plastic bottles were placed around the plants with mesh covering the tops. Arabidopsis plants were subject to one of three treatments: untouched (control) or subject to herbivory for 36 h by 6 × 4th instar S. exigua caterpillars with intact or impaired salivary secretions. The experiment was repeated twice; at each time point, two independent samples were taken for gene expression analysis (total n = 3–4).
RNA extraction, cDNA synthesis, and quantitative real-time polymerase chain reaction
Plants were finely ground in liquid nitrogen and total RNA was extracted using the RNeasy Mini Kit (Qiagen) following the manufacturer’s protocols. After DNase treatment (Wipeout, QuantiTect Reverse Transcription kit; Qiagen), the absence of genomic contamination was confirmed using 5′-ATG GGT CGT CAT CAG ATT CAG AGC AGA TAA-3′ and 5′-CAT ATA AGA GGT GTG TTA GAG ACA ATA ATA-3′ primers which span an intron (Weech et al., 2008). One microgram of RNA was converted to cDNA using a QuantiTect Reverse Transcription Kit following the manufacturer’s instructions.
Gene-specific primers were identified from the literature or designed using Primer3 (Table A1 in Appendix). Transcript expression was analyzed in duplicate using the Brilliant One-Step quantitative RT-PCR kit (Stratagene), according to the manufacturer’s protocol, in a Mx3000p thermocycler (Stratagene). Gene amplicon products were verified by sequencing. Each 96-well plate, contained a standard curve of the gene-of-interest, a non-template control and each sample in duplicate. Each reaction contained 1× SYBR green I, 0.375 nM ROX, 100 nM of the forward and reverse primer, mastermix that contained dNTPs, MgSO4 and Taq polymerase, and either water (non-template control), serial dilutions of PCR amplicon (standard curve) or 85 ng cDNA sample. Standard curves ensured an efficiency of between 90 and 110%. Thermocycler conditions are as follows: 95°C for 10 min; 40 cycles of denaturation at 95°C for 45 s, annealing for 1 min, and elongation at 70°C for 45 s. The annealing temperature was dependent on the primers used (Table A1 in Appendix). Dissociation curves were performed to ensure amplicon purity. Two technical plate replicates were performed.
From the standard curve, gene copy numbers were estimated and normalized against the constitutive reference gene AtACT2 (At3g18780). Arabidopsis AtACT2 expression was not affected by osmotic stress or when plants were treated with viral pathogens or stress-related hormones, such as methyl JA or SA, or caterpillar herbivory (Stotz et al., 2000; Dufresne et al., 2008; Weech et al., 2008). In the current study, AtACT was stably expressed within a genotype and not affected by treatment [+/+: F(2,9) = 0.26, p = 0.77; pad2-1: F(2,9) = 1.10, p = 0.37; tga2/5/6: F(2,7) = 0.42, p = 0.68; Brunner et al., 2004].
STATISTICAL ANALYSIS
For the redox experiment (repeated independently three times, n = 5–10), statistical differences (p í 0.05) in metabolite levels were determined using a two-way analysis of variance (ANOVA) using SPSS version 20 (SPSS Inc.). If a significant time × treatment factor was observed, a one-way ANOVA followed by a Tukey HSD (honestly significant difference) post hoc test was conducted to identify the significant difference. The gene expression experiment was repeated twice with two independent biological samples analyzed at each time (total n = 3–4). Within each genotype, transcript expression was analyzed by a one-way ANOVA. Statistical differences (p < 0.05) were determined using a Tukey HSD post hoc test (Rieu and Powers, 2009). Alternatively, because of the variation inherent with insect feeding studies, a greater than five-fold change in gene expression with respect to control plants was also considered significantly different. Results from statistical analyses are shown in Table A2 in Appendix.
RESULTS AND DISCUSSION
ASCORBATE–GLUTATHIONE CYCLE
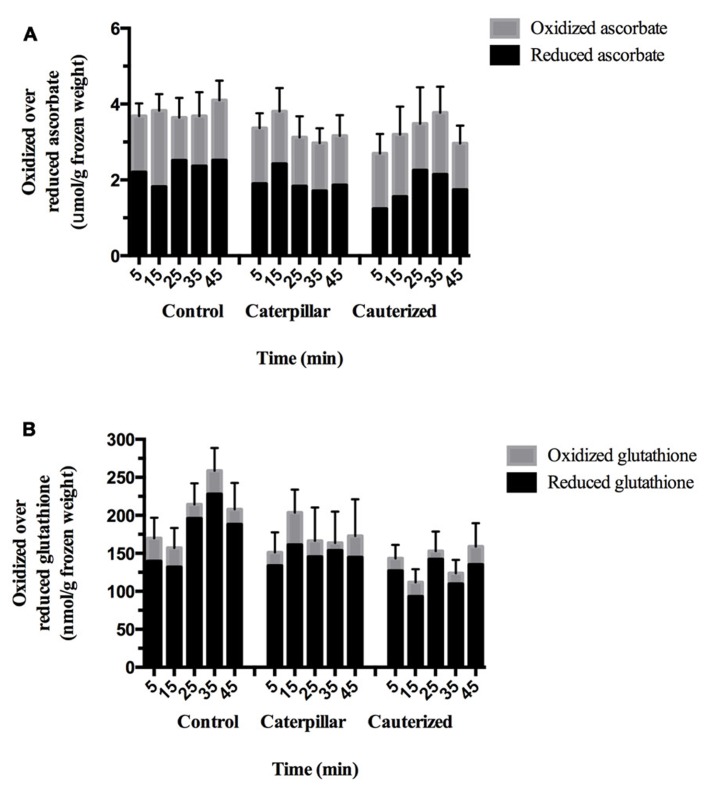
The ascorbate–glutathione cycle is critical to enable the plant to maintain cellular redox status during stresses, such as insect herbivory (Noctor et al., 2012). Oxidative stress, such as increased H2O2 levels, may result in either an increase in the levels of total glutathione (glutathione pool) or increased levels of GSSG relative to GSH (redox balance; Noctor et al., 2012). Total ascorbate levels were within the reported physiological range and did not change over the 45 min time course and was independent of treatment (Figure 1A; Table A2 in Appendix; Queval and Noctor, 2007). Oxidized and reduced ascorbate levels and the ratio of oxidized ascorbate (DHA)/reduced ascorbate (ASc) did not change in response to caterpillar herbivory. Total glutathione levels were within the expected physiological range and affected by treatment (Figure 1B; Table A2 in Appendix; Queval and Noctor, 2007). Caterpillar herbivory did not affect the oxidized GSSG/reduced GSH ratio but total glutathione levels are lower in plants infested with caterpillars with impaired salivary secretions compared to the control. This likely reflects the reduced glutathione levels found in this treatment. Caterpillar herbivory also had significantly lower oxidized GSSG levels at 35 min post-herbivory; this effect was not salivary-dependent.
FIGURE 1.
Time course of redox metabolites in Arabidopsis plants subject to caterpillar herbivory. (A) Ascorbate levels. Foliar ascorbate levels in Arabidopsis plants subject to herbivory by caterpillars with normal (caterpillar) or impaired salivary secretions (cauterized) compared to control plants. Solid bars represent reduced ascorbate (ASc) levels. Open bars represent oxidized ascorbate (dehydroascorbate, DHA) levels. Values are given in μmol/g frozen weight (FW) and represent means ± SE of three to four independent biological replications. Significant differences in ascorbate were not observed in response to caterpillar herbivory. (B) Glutathione levels. Foliar glutathione levels in Arabidopsis plants subject to herbivory by caterpillars with normal (caterpillar) or impaired salivary secretions (cauterized) compared to control plants. Solid bars represent reduced glutathione (GSH) levels. Open bars represent oxidized glutathione (GSSG) levels. Values are given in nmol/g FW and represent means ± SE of three to four independent biological replications. Significant differences were determined by two-way ANOVA (Table A2 in Appendix). At 35 min post-herbivory, a significant reduction in GSSG levels are observed in plants infested by caterpillars, both with normal or impaired salivary secretions, compared to controls. Total and reduced glutathione levels are significantly reduced in caterpillar with impaired salivary secretions compared to control levels.
Cellular glutathione–ascorbate metabolites levels and/or redox balance are involved in plant defense against pathogens or herbivores (Mou et al., 2003; Ball et al., 2004; Parisy et al., 2007; Schlaeppi et al., 2008; Wünsche et al., 2011; Espunya et al., 2012). The majority of experiments investigating changes in redox metabolites in response to stress (wound, herbivory, pathogens) characterize long-term changes in the cellular oxidative status (Fodor et al., 1997; Mou et al., 2003; Ball et al., 2004; Mateo et al., 2006; Schlaeppi et al., 2008; Gfeller et al., 2011). In this study, we are interested in early changes in cellular antioxidant levels or redox balance (ratio) to caterpillar herbivory that may lead to changes in gene expression. The difficulty in this short-term experiment is to synchronize the initiation and intensity of insect herbivory. Lou and Baldwin (2006) and this study monitored redox metabolites within the first 45 min after the initiation or simulation of herbivory. Lou and Baldwin (2006) noted an increase in H2O2 levels 30 min after wounding and application of Manduca sexta caterpillar regurgitant on Nicotiana attenuata leaves. In response to biotrophic pathogens, an increase in total or reduced glutathione levels leads to reduction and activation of NPR1 (Fodor et al., 1997; Mou et al., 2003; Fobert and Després, 2005; Mateo et al., 2006); even though SA injection into leaves shows a transient oxidation of the glutathione pool. In comparison, after wounding, the GSSG/total glutathione ratio increased leading to an activation of the JA pathway (Mhamdi et al., 2010; Gfeller et al., 2011).
Cellular redox changes occur in response to mechanical damage during insect feeding. However, noctuid caterpillar LS, that has been implicated as a stratagem to delay the induction of plant defenses, contains numerous enzymes that may affect cellular redox balance, most notably the H2O2-producing enzyme GOX (Musser et al., 2002; Weech et al., 2008; Eichenseer et al., 2010). Compared to controls, herbivory by caterpillars with intact salivary secretions did not affect cellular redox balance except for a transient decrease in oxidized GSSG at 35 min (Figure 1B). In comparison, reduced glutathione levels were lower in leaves subject to herbivory by caterpillars with impaired salivary secretion compared to controls, indicating oxidative stress. This suggests that the production of H2O2 by enzymes in the caterpillar LS may act to maintain cellular GSH levels so glutathione does not act further as a signaling molecule (Szalai et al., 2009).
TRANSCRIPT EXPRESSION IN RESPONSE TO CATERPILLAR HERBIVORY
To explore the link between cellular redox balance and plant responses to caterpillar LS, expression of JA-, ET-, and SA-dependent gene markers were analyzed in wildtype, pad2-1 mutants, that contain only about 20% of normal glutathione levels, and the tga2/5/6 triple mutant (Zhang et al., 2003; Parisy et al., 2007). Together with NPR1, TGA transcription factors are activated by a change in redox balance and responsible for SA-dependent gene expression (Després et al., 2003; Mou et al., 2003; Lindermayr et al., 2010). It must, however, be noted that the TGA transcription factors have also been shown to regulate a subset of oxylipin-dependent defensive gene expression (Mueller et al., 2008; Zander et al., 2010).
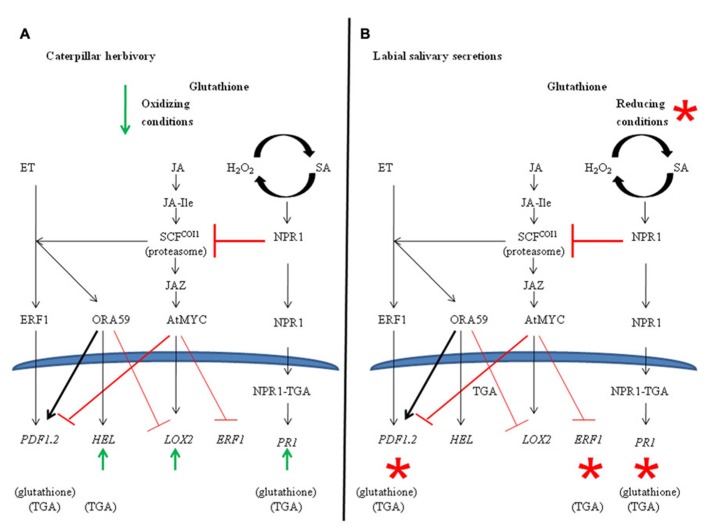
Jasmonate, SA, and ET play central roles in mediating the plant’s response to caterpillar herbivory (Figure 3; Weech et al., 2008; Diezel et al., 2009; Onkokesung et al., 2010). Pré et al. (2008) recently suggested that the transcription factors ORA59 and ERF1 act in parallel pathways to integrate these JA/ET responses. How caterpillar LS manages to manipulate these JA/ET pathways is unknown, but Weech et al. (2008) proposed that caterpillar LS requires an active SA/NPR1 pathway for this strategem. To further complicate issues, recent evidence suggests that ET potentiates SA antagonism with JA and renders it NPR1-independent (Leon-Reyes et al., 2009).
FIGURE 3.
Model of ethylene-, jasmonate-, and salicylate-dependent pathway illustrating major cross-talk signaling nodes and marker genes. (A) Changes in redox metabolites and gene expression in response to caterpillar herbivory. Illustrates caterpillar herbivory-dependent changes compared to control plants. Cellular GSSG, which is linked to the induction of JA-defenses, decrease transiently 35 min after caterpillar herbivory. Markers of the SA, ORA59, and AtMYC2 pathway, respectively AtPR1, AtHEL, and AtLOX2, are induced in response to herbivory. (B) Proposed model caterpillar labial saliva (LS) mediated suppression of jasmonate-dependent responses. Illustrates LS-associated changes between plants fed on by caterpillars with intact vs. impaired salivary secretions. Asterisks indicate LS-associated changes. Herbivory by caterpillars with impaired salivary secretions result in cellular oxidative stress (lower total and reduced cellular GSH levels) compared to controls. Induction of the AtPR1 and suppression of AtPDF1.2 and ERF1 are LS-dependent. Involvement of glutathione orTGA transcription factors are indicated in brackets underneath the marker gene.
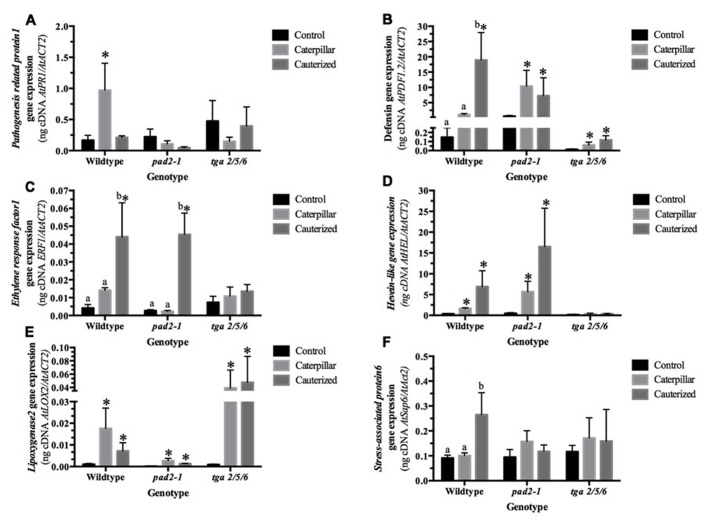
Pathogenesis-related 1 (AtPR1, At2g14610) is a SA-responsive, NPR1-dependent gene marker induced in response to biotrophic pathogen attack and aphid feeding (Glazebrook, 2005; Mur et al., 2006; Kusnierczyk et al., 2007; Walling, 2008; Zhang et al., 1999). In our study, AtPR1 gene expression was greater than fivefold higher in plants infested by caterpillars with intact LS secretions compared to caterpillars with cauterized spinnerets and control plants, indicating that caterpillar LS secretions result in the activation of SA/NPR1-dependent gene expression (Figure 2A; Table A2 in Appendix). Through activation of the SA pathway by effectors in their LS secretions, S. exigua caterpillars are believed to impair the plant’s ability to fully mount a JA-dependent defense response (Weech et al., 2008). Mewis et al. (2006) also observed AtPR1 expression in Arabidopsis response to herbivory by caterpillars of P. rapae and S. exigua; both these caterpillar LS glands contain redox enzymes, such as GOX (Eichenseer et al., 2010). The increase in AtPR1 expression was alleviated in pad2-1 and tga2/5/6 mutant plants, in line with previous studies showing that glutathione and the TGA transcription factors are upstream signals in AtPR1 expression (Després et al., 2003; Mou et al., 2003; Lindermayr et al., 2010).
FIGURE 2.
Arabidopsis transcript expression in response to caterpillar herbivory analyzed by quantitative real-time reverse transcription PCR. Seven-week-old Arabidopsis plants (Col-0, +/+), pad2-1 mutant (pad2-1), and tga2/5/6 triple mutant (tga) were untreated (control) or subject to herbivory by caterpillars with normal (intact) or impaired (cauterized) salivary secretions for 36 h. From cDNA generated from total RNA, gene-specific primers were used to determine expression levels of (A) AtPR1, (B) AtPDF1.2, (C) ERF1, (D) AtHEL, (E) AtLOX2, (F) AtSAP6. Bars represent the mean values of three to four independent replicates normalized with the reference gene AtACT2 ± SE. Within each genotype, lower case alphabetical letters indicate significant differences identified by ANOVA followed by a Tukey HSD (p ≤ 0.05; n = 3–4; Table A2 in Appendix). An asterisk denotes a fivefold or higher change in expression from control levels.
Expression of the gene encoding plant defensin, AtPDF1.2b (At2g26020), is induced by treatment of plants with JA and ET working synergistically through ORA59 (Penninckx et al., 1998; Pré et al., 2008); however, antagonism between MYC2 and ERF1 regulation of AtPDF1.2 is proposed to reflect MYC2 regulation of ERF1 expression (Dombrecht et al., 2007). As well, SA-dependent suppression of AtPDF1.2 expression requires active NPR1 and TGA transcription factors (Spoel et al., 2003; Ndamukong et al., 2007; Koornneef et al., 2008). ET modulates this SA–JA antagonism; NPR1-dependent antagonism of the expression of JA-dependent genes, such as AtPDF1.2, becomes NPR1-independent in the presence of ET (Leon-Reyes et al., 2009).
In wildtype plants, an 18-fold increase in AtPDF1.2 transcript expression is observed in response to herbivory by caterpillars with impaired salivary secretions compared to normal caterpillars or control plants, in agreement with previous studies that caterpillar LS suppresses JA-dependent plant defenses (Figure 2B; Table A2 in Appendix; Musser et al., 2002; Weech et al., 2008). In pad2-1 and tga2/5/6 mutants, LS-mediated restraint of AtPDF1.2 expression is not observed, indicating that glutathione and TGA transcription factors are required for the suppression of plant induced defenses by caterpillar herbivory. In pad2-1 mutants, a 12.5-fold increase in AtPDF1.2 levels is seen in plants infested by caterpillars compared to controls. The lower glutathione levels in the pad2-1 mutant may impair the activation of a pathway, such as the reduction of NPR1 and/or TGA transcription factors, which are needed for the LS-mediated suppression of plant defenses (Mou et al., 2003; Fobert and Després, 2005). A fivefold increase in AtPDF1.2 expression is seen in plants fed upon by caterpillars compared to controls in the tga2/5/6 mutant plants. However, it must be noted that TGA transcription factors also regulate the late expression (~48 h) of a subset of JA-dependent genes, such as AtPDF1.2 (Zander et al., 2010). Perhaps, a strong difference in gene expression between normal and cauterized caterpillars is not observed because of the requirement for TGA transcription factors, although a fivefold increase in expression is observed in caterpillar-infested tga2/5/6 mutants compared to controls. These results suggest that caterpillar LS-dependent suppression of JA-mediated activation of AtPDF1.2 gene expression is dependent on glutathione levels and, perhaps, the activation of TGA transcription factors.
In wildtype plants, results correlate with previous observations that glutathione negatively regulates AtPDF1.2 expression (Koornneef et al., 2008); we also observed that wildtype plants infested by caterpillars with impaired salivary secretions had lower reduced glutathione compared to controls and, consequently, higher AtPDF1.2 expression (Figures 1B2B). Also, the LS-associated negative regulation of AtPDF1.2 is alleviated in the pad2-1 mutant. Our observation that this LS-mediated suppression of AtPDF1.2 is lessened in the tga2/5/6 triple mutant supports observations that suppression of AtPDF1.2 gene expression requires the interaction of glutaredoxin480 with TGA transcription factors (Ndamukong et al., 2007; Koornneef and Pieterse, 2008; Zander et al., 2011). ET also plays a role in modulating the mechanism of SA/NPR1 inhibition of JA-dependent responses (Leon-Reyes et al., 2009); in the presence of ET, this suppression becomes NPR1-independent. However, given the links to glutathione and, possibly TGA transcription factors, and previous research, our data points to a LS-mediated NPR1-dependent inhibition of AtPDF1.2 gene expression (Weech et al., 2008).
Alternatively, current models propose that JA-dependent inhibition of AtPDF1.2 expression may be mediated through the negative regulation of ERF1 (At3g23240) by MYC2 (Dombrecht et al., 2007; (Zander et al., 2010). Therefore, ERF1 expression was measured to determine if it was mirrored by AtPDF1.2 expression. As seen with AtPDF1.2, a significant increase in Arabidopsis ERF1 transcript expression is observed in response to herbivory by caterpillars with impaired LS secretions compared to normal caterpillars or control plants (Figures 2B, C; Table A2 in Appendix); however, this LS-mediated suppression of ERF1 is also observed in the pad2-1 mutants. The distinct patterns between AtPDF1.2 and ERF1 expression suggest LS-mediated regulation is likely not reflective of MYC2 antagonism of ERF1; however, they suggest that there may be LS-linked, an ET, glutathione-independent mechanism of suppression. LS-suppression of ERF1 is alleviated in the tga2/5/6 triple mutant. Zander et al. (2010) found that TGA transcription factors may suppress ERF1 expression.
Hevein-like (AtHEL, PR4, At304720) gene expression is a marker of the ORA59 branch of the JA/ET signaling pathways (Potter et al., 1993; Dombrecht et al., 2007; Pré et al., 2008; Verhage et al., 2011; Zarei et al., 2011). In comparison to AtPDF1.2, suppression of JA-linked AtHEL expression by the SA pathway is NPR1-independent (Ndamukong et al., 2007). In wildtype and pad2-1 mutant plants, over a fivefold increase in gene expression is observed in plants infested by caterpillars compared with controls (Figure 2D); however, a LS effect is not observed (Table A2 in Appendix). These results support the argument that caterpillar LS-mediated suppression of induced plant defenses is glutathione- and NPR1-dependent. Unexpectedly, this caterpillar-mediated AtHEL expression was at basal levels in the tga2/5/6 triple mutant plants, suggesting that these transcription factors may be involved in regulation of AtHEL expression.
The gene encoding lipoxygenase2 (AtLOX2, At3g45410) is an early expression marker of the JA-responsive MYC2 branch (Bell et al., 1995; Dombrecht et al., 2007). As has been observed previously, AtLOX2 levels are induced sevenfold in response to insect herbivory and a LS gland-specific difference in gene expression is not observed (Figure 2E; Table A2 in Appendix; Weech et al., 2008). This same pattern was observed in pad2-1 and tga2/5/6 mutant plants. Though regulated by MYC2, the strong upregulation of this early gene occurs before SA/NPR1-mediated cross-talk (Mou et al., 2003; Spoel et al., 2003; Ndamukong et al., 2007; Koornneef and Pieterse, 2008; Spoel and Dong, 2008; Tada et al., 2008; Leon-Reyes et al., 2010). As well, LS-associated post-transcriptional modifications of LOX2 may regulate activity rather than gene expression (Thivierge et al., 2010).
The stress-associated AtSAP6 (At3g52800) was induced in plants fed upon by caterpillars with impaired LS secretions compared to controls (Figure 2F; Table A2 in Appendix). This difference was alleviated in the pad2-1 and the tga2/5/6 triple mutants indicating the possible involvement of glutathione and TGA transcription factors in the regulation of expression of this gene. AtSAP6 is strongly induced in response to numerous stresses, such as wounding and herbivory by caterpillars of the specialist P. rapae (Reymond et al., 2004; Ströher et al., 2009); however, in response to herbivory, this transcript was induced in both the wildtype and the JA-perception impaired coi1-1 gl1 mutant implying that JA signaling is not required for the expression of this gene.
CONCLUSION
Plant responses to insect herbivory are mediated through carefully regulated, complex hormone-mediated interactions. Herbivory by S. exigua caterpillars attenuate these JA-dependent plant defense responses; a mechanism believed to be related to LS-associated secretions (Musser et al., 2002; Weech et al., 2008). Given the presence of GOX in the LS of this caterpillar, the relationship between LS secretions and changes in cellular redox potential was investigated. Changes in cellular oxidative stress and, in particular, the GSSG/total glutathione ratio are signals for the induction of JA-dependent defenses (Szalai et al., 2009; Gfeller et al., 2011). Herbivory by caterpillars with intact salivary secretions did not affect cellular redox balance, except for a transient decrease in oxidized GSSG at 35 min (Figure 3). In comparison, herbivory by caterpillars with impaired salivary secretions resulted in an increase in cellular oxidative status through a decrease in reduced glutathione levels. In support of this, genes, such as AtPR1 and AtPDF1.2, showed LS-dependent transcript expression that was alleviated in the pad2-1 and tga2/5/6 triple mutant (Figures 2A, B and 3).
Increased expression of AtPR1 by herbivory using caterpillars with intact salivary secretions support the notion that LS-mediated attenuation of JA responses acts through cross-talk with the SA/NPR1 pathway (Figure 3). As well, even though AtPDF1.2 is a JA/ET marker, recent studies have shown that mid- to late-gene expression is regulated by TGA transcription factors (Zander et al., 2011). Therefore, suppression of AtPDF1.2 gene expression by caterpillar LS may also reflect cross-talk between JA- and SA/NPR1 pathways. The LS-associated modulation of ET-dependent genes, ERF1 and AtHEL, show disparate regulation since ERF1 expression shows glutathione-independent, LS-associated suppression whereas a LS-dependent difference in AtHEL is not observed (Figures 2C, D). This may support recent evidence that the ET pathway is mediated through two distinct branches regulated by ORA59 or ERF1 transcription factors (Pré et al., 2008). In fact, Pieris rapae caterpillar oral secretions, which are a mixture of gut-derived regurgitant, secretions from the ventral eversible gland and salivary secretions from the mandibular and labial glands, specifically activate the ORA59 branch of the JA/ET pathway leading to the suppression of MYC2-dependent defenses (Felton, 2008; Hogenhout and Bos, 2011; Verhage et al., 2011; Zebelo and Maffei, 2012). These caterpillars also show a feeding preference for plants that overexpress ORA59.
The mechanism behind this LS-mediated cross-talk may be explained by the model recently proposed by Van der Does et al. (2013). In an elegant set of experiments, these authors systematically demonstrated that suppression of the JA-induced pathway by the SA/NPR1 pathway occurs downstream of SCFCOI1-mediated protein degradation. Instead, the SA/NPR1 pathway negatively regulates the expression of AtPDF1.2 by affecting the accumulation of the ORA59 transcription factor. Our data from this and previous studies also show that caterpillar LS-mediated suppression of AtPDF1.2 is SA/NPR1 pathway mediated and does not appear to involve cross-talk between the ERF1 pathway (Weech et al., 2008); therefore, further studies investigating ORA59 protein levels in this plant–insect system needs to be investigated.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to Drs Guillaume Queval, Graham Noctor, and Christine Foyer for valuable discussions and advice with the optimization of experimental techniques to analyze redox metabolites. We thank two reviewers for their constructive improvements to an earlier version of this manuscript. This research was supported by operating grants from the National Sciences and Engineering Research Council to Jacqueline C. Bede.
Abbreviations
- 1D
one-dimensional
- 2D
two-dimensional
- 2,4-D
2,4-dichloroacetic acid
- 2,4-DB
2,4-dichlorobutyric acid
- ACX
acyl-CoA oxidase
- BA
benzoic acid
- BSA
bovine serum albumin
- CDS
coding sequence
- co-IP
co-immunoprecipitation
- DRP
dynamin-related protein
- ER
endoplasmic reticulum
- ERPIC
ER/peroxisome intermediate compartment
- ESI
electrospray ionization
- EST
expressed sequence tag
- FFE
free-flow electrophoresis
- FIS
fertilization-independent seed
- GFP
green fluorescent protein
- GPK
glyoxysomal protein kinase
- IAA
indole-3-acetic acid
- IBA
indole-3-butyric acid
- ICAT
isotope coded affinity tags
- ICL
isocitrate lyase
- IDHP
NADP-dependent isocitrate dehydrogenase
- IEF
iso-electric focusing
- iTRAQ
isobaric tags for relative and absolute quantitation
- LACS
long chain acyl-CoA synthetase
- MALDI
matrix-assisted laser desorption/ionization
- MFP
multi-functional protein
- MRM
multiple reaction monitoring
- MS
mass spectrometry
- MVA
mevalonic acid
- NADK
NAD kinase
- PAGE
polyacrylamide gel electrophoresis
- PMF
peptide mass fingerprint
- PTS
peroxisome targeting signal
- RNAi
ribonucleic acid interference
- RNS
reactive nitrogen species
- ROS
reactive oxygen species
- SA
salicylic acid
- SDS
sodium dodecyl sulfate
- SILAC
stable isotope labeling by amino acids in cell culture
- SRM
selected reaction monitoring
- TAP
tandem affinity purification
- TMT
tandem mass tags
APPENDIX
Table A1.
Quantitative real-time polymerase chain reaction primers.
| Gene | Accession number | Annealing temperature (°C) | Forward (5′–3′) | Reverse (5′–3′) | Reference |
|---|---|---|---|---|---|
| ERF1 | At3g23240 | 62 | GAC GGA GAA TGA CCA ATA AGA AG | CCC AAA TCC TCA AAG ACA ACT AC | Swarup et al. (2007) |
| AtHEL | At304720 | 57 | CAA GTG TTT AAG GGT GAA GA | CGG TGT CTA TTT GAT TGA AC | Conn et al. (2008) |
| AtLOX2 | At3g45410 | 57 | GTC CTA CTT GCC TTC CCA AAC | ATT GTC AGG GTC ACC AAC ATC | Weech et al. (2008) |
| AtPDF1.2b | At2g26020 | 59 | CGG CAA TGG TGG AAG CA | CAT GCA TTA CTG TTT CCG CAA | Jirage et al. (2001) |
| AtPR1 | At2g14610 | 62 | CAC TAC ACT CAA GTT GTT TGG A | CAT GCA TTA CTG TTT CCG CAA A | Primer3 |
| AtSAP6 | At3g52800 | 63 | TCA ACG CAT CGA ACG GCT CTG A | GCG AAA GCG AAT CCG TTG GTG AAA | Primer3 |
| AtACT2 | At3g18780 | ACC AGC TCT TCC ATC GAG AA | GAA CCA CCG ATC CAG ACA CT | Dufresne et al. (2008) |
Table A2.
Statistical results of plant–insect experiments.
| Ascorbate | ||
|---|---|---|
| Total | Range: 2.69–3.98 μmol/g FW | Effect of treatment, F(2,103) = 1.33, p = 0.27; effect of time, F(4,103) = 0.16, p = 0.96; interaction, F(8,103) = 0.66, p = 0.73 |
| Oxidized (DHA) | Range: 1.22–1.83 μmol/g FW | Effect of treatment, F(2,100) = 0.41, p = 0.66; effect of time, F(4,100) = 0.90, p = 0.47; interaction, F(8,100) = 0.43, p = 0.90 |
| Reduced (Asc) | Range: 1.23–2.81 μmol/g FW | Effect of treatment, F(2,105) = 2.50, p = 0.09; effect of time, F(4,105) = 0.58, p = 0.68; interaction, F(8,105) = 0.97, p = 0.47 |
| Oxidized/reduced (DHA/Asc) | Effect of treatment, F(2,100) = 1.62, p = 0.21; effect of time, F(4,100) = 0.1.30, p = 0.28; interaction, F(8,100) = 1.11, p = 0.36 | |
| Glutathione | ||
| Total | Range: 151–226 nmol/g FW | Effect of treatment, F(2,109) = 3.35, p = 0.04; effect of time, F(4,109) = 0.32, p = 0.86; interaction, F(8,109) = 0.16, p = 0.99; Figure 2A) |
| Oxidized (GSSG) | Range: 9.9–42.6 nmol/g FW | Effect of treatment, F(2,89) = 3.31, p = 0.04; effect of time, F(4,89) = 2.10, p = 0.09; interaction, F(8,89) = 2.24, p = 0.03. Since interaction was significant, this was followed by a one-way ANOVA to determine the time point where there was a significant difference: 5 min: F(2,19) = 2.69, p = 0.09; 15 min: F(2,14) = 1.91, p = 0.18; 25 min: F(2,19) = 2.64, p = 0.10; 35 min: F(2,20) = 3.76, p = 0.04; 45 min: F(2,19) = 0.53, p = 0.60 |
| Reduced (GSH) | Range: 93.2–227.9 nmol/g FW | Effect of treatment, F(2,87) = 3.42, p = 0.04; effect of time, F(4,87) = 0.67, p = 0.62; interaction, F(8,87) = 0.56, p = 0.81 |
| Oxidized/reduced (GSSG/GSH) | Effect of treatment, F(2,87) = 0.99, p = 0.37; effect of time, F(4,87) = 2.18, p = 0.08; interaction, F(8,87) = 1.07, p = 0.39 | |
| Gene expression | Genotype | |
| AtPR1 | Wildtype | F(2,8) = 4.44, p = 0.05; fivefold increase in gene expression in plants attacked by caterpillars with intact labial salivary secretions compared to control plants or between control plants or plants infested by caterpillars with impaired salivary secretions |
| pad2-1 | F(2,9) = 1.23, p = 0.32 | |
| tga2/5/6 | F(2,7) = 0.35, p = 0.72 | |
| AtPDF1.2 | Wildtype | F(2,8) = 6.00, p = 0.03; 18-fold increase in gene expression in response to herbivory by caterpillars with impaired salivary secretions compared to normal caterpillars or controls |
| pad2-1 | F(2,8) = 1.50, p = 0.28; 12.5-fold increase in gene expression is seen between plants infested by caterpillars compared to controls | |
| tga2/5/6 | F(2,7) = 3.31, p = 0.10; fivefold increase in gene expression is seen in plants fed upon by caterpillars compared to controls | |
| ERF1 | Wildtype | F(2,8) = 5.07, p = 0.04; 10-fold increase in gene expression seen in plants fed upon by caterpillars with impaired salivary secretions compared to control plants |
| pad2-1 | F(2,9) = 12.83, p = 0.002; 20-fold increase in gene expression seen in plants fed upon by caterpillars with impaired salivary secretions compared to control plants or plants attacked by caterpillars with labial salivary secretions | |
| tga2/5/6 | F(2,7) = 0.61, p = 0.57 | |
| AtHEL | Wildtype | F(2,9) = 2.50, p = 0.14; fivefold increase in gene expression is observed in plants infested by caterpillars compared with control |
| pad2-1 | F(2,9) = 2.22, p = 0.17; 10-fold increase in gene expression is observed in plants infested by caterpillars compared with controls | |
| tga2/5/6 | F(2,7) = 3.31, p = 0.10 | |
| AtLOX2 | Wildtype | F(2,7) = 1.48, p = 0.29; sevenfold increase in gene expression is observed in plants infested by caterpillars compared with controls |
| pad2-1 | F(2,9) = 3.68, p = 0.07; sevenfold increase in gene expression is observed in plants infested by caterpillars compared with controls | |
| tga2/5/6 | F(2,7) = 1.16, p = 0.37; 40-fold increase in gene expression is observed in plants infested by caterpillars compared with controls | |
| AtSAP6 | Wildtype | F(2,8) = 5.02, p = 0.04 |
| pad2-1 | F(2,9) = 0.85, p = 0.46 | |
| tga2/5/6 | F(2,7) = 0.14, p = 0.87 |
A two-way ANOVA followed by a Tukey HSD was used to evaluate redox metabolite levels over a 45-min time course. A one-way ANOVA was used to evaluate differences in gene expression within each genotype. A fivefold or higher difference in gene expression is also indicated.
REFERENCES
- Arimura G., Ozawa R., Maffei M. E. (2011). Recent advances in plant early signaling in response to herbivory. Int. J. Mol. Sci. 12 3723–3739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball L., Accotto G. P., Bechtold U., Creissen G., Funck D., Jimenez A., et al. (2004). Evidence for a direct link between glutathione biosynthesis and stress defense gene expression in Arabidopsis. Plant Cell 16 2448–2462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bede J. C., Musser R. O., Felton G. W., Korth K. L. (2006). Caterpillar herbivory and salivary enzymes decrease transcript levels of Medicago truncatula genes encoding early enzymes in terpenoid biosynthesis. Plant Mol. Biol. 60 519–531 [DOI] [PubMed] [Google Scholar]
- Bell E., Creelman R. A., Mullet J. E. (1995). A chloroplast lipoxygenase is required for wound-induced jasmonic acid accumulation in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 92 8675–8679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodenhausen N., Reymond P. (2007). Signaling pathways controlling induced resistance to insect herbivores in Arabidopsis. Mol. Plant Microbe Interact. 20 1406–1420 [DOI] [PubMed] [Google Scholar]
- Boyes D. C., Zayed A. M., Ascenzi R., McCaskill A. J., Hoffman N. E., Davis K. R., et al. (2001). Growth stage-based phenotypic analysis of Arabidopsis: a model for high throughput functional genomics in plants. Plant Cell 13 1499–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunner A. M., Yakovlev I. A., Strauss S. H. (2004). Validating internal controls for quantitative plant gene expression studies. BMC Plant Biol. 4:14 10.1186/1471-2229-4-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conn V. M., Walker A. R, Franco C. M. M. (2008). Endophytic actinobacteria induce defense pathways in Arabidopsis thaliana. Mol. Plant Microbe Interact. 21 208–218 [DOI] [PubMed] [Google Scholar]
- Després C., Chubak C., Rochon A., Clark R., Bethune T., Desveaux D., et al. (2003). The Arabidopsis NPR1 disease resistance protein is a novel cofactor that confers redox regulation of DNA binding activity to the basic domain/leucine zipper transcription factor TGA1. Plant Cell 15 2181–2191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diezel C., von Dahl C. C., Gaquerel E., Baldwin I. T. (2009). Different lepidopteran elicitors account for cross-talk in herbivory-induced phytohormone signaling. Plant Physiol. 150 1576–1587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doares S. H., Narvaez-Vasquez J., Conconi A., Ryan C. A. (1995). Salicylic acid inhibits synthesis of proteinase inhibitors in tomato leaves induced by systemin and jasmonic acid. Plant Physiol. 108 1741–1746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dombrecht B., Xue G. P., Sprague S. J., Kirkegaard J. A., Ross J. J., Reid J. B., et al. (2007). MYC2 differentially modulates diverse jasmonate-dependent functions in Arabidopsis. Plant Cell 19 2225–2245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubreuil-Mauriza C., Vitecek J., Marty L., Branciard L., Frettinger P., Wendehenne D., et al. (2011). Glutathione deficiency of the Arabidopsis mutant pad2-1 affects oxidative stress-related events, defense gene expression, and the hypersensitive response. Plant Physiol. 157 2000–2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufresne P. J., Ubalijoro E., Fortin M. G., Laliberte J.-F. (2008). Arabidopsis thaliana class II poly(A)-binding proteins are required for efficient multiplication of turnip mosaic virus. J. Gen. Virol. 89 2339–2348 [DOI] [PubMed] [Google Scholar]
- Eichenseer H., Mathews M. C., Powell J. S., Felton G. W. (2010). Survey of a salivary effector in caterpillars: glucose oxidase variation and correlation with host range. J. Chem. Ecol. 36 885–897 [DOI] [PubMed] [Google Scholar]
- Espunya M. C., De Michele R., Gómez-Cadenas A, Carmen Martïnez M. C. (2012). S-nitrosoglutathione is a component of wound- and salicylic acid-induced systemic responses in Arabidopsis thaliana. J. Exp. Bot. 63 3219–3227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felton G. W. (2008). “1Caterpillar secretions and induced plant responses,” in Induced Plant Resistance to Herbivory ed Schaller A. (Berlin: Springer-Scientific; ) 369–387 [Google Scholar]
- Felton G. W., Korth K. L. (2000). Trade-offs between pathogen and herbivore resistance. Curr. Opin. Plant Biol. 3 309–314 [DOI] [PubMed] [Google Scholar]
- Fobert P. R, Després C. (2005). Redox control of systemic acquired resistance. Curr. Opin. Plant Biol 8 378–382 [DOI] [PubMed] [Google Scholar]
- Fodor J., Gullner G., Adam A. L., Barna B., Komives T., Kiraly Z. (1997). Local and systemic responses of antioxidants to tobacco mosaic virus infection and to salicylic acid to tobacco. Plant Physiol. 114 1443–1451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca S., Chini A., Hamberg M., Adie B., Prozel A., Kramell R., et al. (2009). (+)-7-iso-Jasmonoyl-L-isoleucine is the endogenous bioactive jasmonate. Nat. Chem. Biol. 5 344–350 [DOI] [PubMed] [Google Scholar]
- Forman H. J., Maiorino M., Ursini F. (2010). Signaling functions of reactive oxygen species. Biochemistry 49 835–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gfeller A., Baerenfaller K., Loscos J., Chételat A., Baginsky S., Farmer E. E. (2011). Jasmonate controls polypeptide patterning in undamaged tissue in wounded Arabidopsis leaves. Plant Physiol. 156 1797–1807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazebrook J. (2005). Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 43 205–227 [DOI] [PubMed] [Google Scholar]
- Griffith O. W. (1980). Determination of glutathione and glutathione disulfide using glutathione reductase and 2-vinylpyridine. Anal. Biochem. 106 207–212 [DOI] [PubMed] [Google Scholar]
- Hogenhout S. A, Bos J. I. B. (2011). Effector proteins that modulate plant–insect interactions. Curr. Opin. Plant Biol. 14 422–428 [DOI] [PubMed] [Google Scholar]
- Jirage D., Zhou N., Cooper B., Clarke J. D., Dong X., Glazebrook J. (2001). Constitutive salicylic acid-dependent signaling in cpr1 and cpr6 mutants requires PAD4. Plant J. 26 395–407 [DOI] [PubMed] [Google Scholar]
- Kazan K., Manners J. M. (2008). Jasmonate signaling: toward an integrated view. Plant Physiol. 146 1459–1468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef A., Leon-Reyes A., Ritsema T., Verhage A., Den Otter F. C., Van Loon L. C., et al. (2008). Kinetics of salicylate-mediated suppression of jasmonate signaling reveal a role for redox modulation. Plant Physiol. 147 1358–1368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef A, Pieterse C. M. J. (2008). Cross talk in defense signaling. Plant Physiol. 146 839–844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusnierczyk A., Winge P., Midelfart H., Armbruster W. S., Rossiter J. T., Bones A. M. (2007). Transcriptional responses of Arabidopsis thaliana ecotypes with different glucosinolate profiles after attack by polyphagous Myzus persicae and oligophagous Brevicoryne brassicae. J. Exp. Bot. 58 2537–2552 [DOI] [PubMed] [Google Scholar]
- Leon-Reyes A., Spoel S. H., De Lange E. S., Abe H., Kobayashi M., Tsuda S., et al. (2009). Ethylene mediates the role of NONEXPRESSOR of PATHOGENESIS RELATED GENES1 in cross talk between salicylate and jasmonate signaling. Plant Physiol. 149 1797–1809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leon-Reyes A., Van der Does D., De Lange E. S., Delker C., Wasternack C., Van Wees S. C. M., et al. (2010). Salicylate-mediated suppression of jasmonate-responsive gene expression in Arabidopsis is targeted downstream of the jasmonate biosynthesis pathway. Planta 232 1423–1432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindermayr C., Sell S., Müller B., Leister D., Durner J. (2010). Redox regulation of the NPR1-TGA1 system of Arabidopsis thaliana by nitric oxide. Plant Cell 22 2894–2907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo O., Chico J. M., Sánchez-Serrano J. J., Solano R. (2004). JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell 16 1938–1950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo O., Piqueras R., Sánchez-Serrano J. J., Solano R. (2003). ETHYLENE RESPONSE FACTOR1 integrates signaling from ethylene and jasmonate pathway in plant defense. Plant Cell 15 165–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo O., Solano R. (2005). Molecular players regulating the jasmonate signalling network. Curr. Opin. Plant Biol. 8 532–540 [DOI] [PubMed] [Google Scholar]
- Lou Y., Baldwin I. T. (2006). Silencing of a germin-like gene in Nicotiana attenuata improves the performance of native herbivores. Plant Physiol. 140 1126–1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maffei M. E., Mithöfer A., Arimura G., Uchtenhagen H., Bossi S., Bertea C., et al. (2006). Effects of feeding Spodoptera littoralis on lima bean leaves. III Membrane depolarization and involvement of hydrogen peroxide. Plant Physiol. 140 1022–1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateo A., Funck D., Mühlenbock P., Kular B., Mullineaux P. M., Karpinski S. (2006). Controlled levels of salicylic acid are required for optimal photosynthesis and redox homeostasis. J. Exp. Bot. 57 1795–1807 [DOI] [PubMed] [Google Scholar]
- Mewis I., Tokuhisa J. G., Schultz J. C., Appel H. M., Ulrichs C., Gershenzon J. (2006). Gene expression and glucosinolate accumulation in Arabidopsis thaliana in response to generalist and specialist herbivores of different feeding guilds and the role of defense signaling pathways. Phytochemistry 67 2450–2462 [DOI] [PubMed] [Google Scholar]
- Mhamdi A., Hager J., Chaouch S., Queval G., Han Y., Taconnat L., et al. (2010). Arabidopsis GLUTATHIONE REDUCTASE1 plays a crucial role in leaf responses to intracellular hydrogen peroxide and in ensuring appropriate gene expression through both salicylic acid and jasmonic acid signaling pathways. Plant Physiol. 153 1144–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mou Z., Fan W., Dong X. (2003). Inducers of plant systemic acquired resistance regulate NPR1 function through redox changes. Cell 113 935–944 [DOI] [PubMed] [Google Scholar]
- Mueller S., Hilbert B., Dueckershoff K., Roitsch T., Krischke M., Muelle M. J., et al. (2008). General detoxification and stress responses are mediated by oxidized lipids through TGA transcription factors in Arabidopsis. Plant Cell 20 768–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mur L. A. J., Kenton P., Atzorn R., Miersch O., Wasternack C. (2006). The outcomes of concentration-specific interactions between salicylate and jasmonate signaling include synergy, antagonism, and oxidative stress leading to cell death. Plant Physiol. 140 249–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musser R. O., Hum-Musser S. M., Eichenseer H., Peiffer M., Ervin G., Murphy J. B., et al. (2002). Caterpillar saliva beats plant defenses: a new weapon emerges in the coevolutionary race between plants and herbivores. Nature 416 599–600 [DOI] [PubMed] [Google Scholar]
- Ndamukong I., Abdallat A. A., Thurow C., Fode B., Zander M., Weigel R., et al. (2007). SA-inducible Arabidopsis glutaredoxin interacts with TGA factors and suppresses JA-responsive PDF1.2 transcription. Plant J. 50 128–139 [DOI] [PubMed] [Google Scholar]
- Ng C. F., Schafer F. Q., Buettner G. R, Rodgers V. G. J. (2007). The rate of cellular hydrogen peroxide removal shows dependency on GSH: mathematical insight into in vivo H2O2 and GPx concentrations. Free Radic. Res. 41 1201–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noctor G., Mhamdi A., Chaouch S., Han Y., Neukermans J., Marquez-Garcia B., et al. (2012). Glutathione in plants: an integrated overview. Plant Cell Environ. 35 454–484 [DOI] [PubMed] [Google Scholar]
- Onkokesung N., Baldwin I. T, Gális I. (2010). The role of jasmonic acid and ethylene crosstalk in direct defense of Nicotiana attenuata plants against chewing herbivores. Plant Signal. Behav. 5 1305–1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisy V., Poinssot B., Owsianowski L., Buchala A., Glazebrook J., Mauch F. (2007). Identification of PAD2 as a gamma-glutamylcysteine synthetase highlights the importance of glutathione in disease resistance of Arabidopsis. Plant J. 49 159–172 [DOI] [PubMed] [Google Scholar]
- Penninckx E. A. M., Thomma B. P. H. J., Buchala A., Metraux J.-P., Broekaert W. F. (1998). Concomitant activation of jasmonate and ethylene response pathway is required for induction of a plant defensin gene in Arabidopsis. Plant Cell 10 2103–2113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter S., Ukes S., Lawton K., Winter A. M., Chandler D., Di Maio J., et al. (1993). Regulation of a hevein-like gene in Arabidopsis. Mol. Plant Microbe Interact. 6 680–685 [DOI] [PubMed] [Google Scholar]
- Pré M., Atallah M., Champion A., De Vos M., Pieterse C. M. J., Memelink J. (2008). The AP2/ERF domain transcription factor ORA59 integrates jasmonic acid and ethylene signals in plant defense. Plant Physiol. 147 1347–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queval G., Hager J., Gakière B., Noctor G. (2008). Why are literature data for H2O2 contents so variable? A discussion of potential difficulties in the quantitative assay of leaf extracts. J. Exp. Bot. 59 135–146 [DOI] [PubMed] [Google Scholar]
- Queval G., Noctor G. (2007). A plate reader method for the measurement of NAD, NADP, glutathione, and ascorbate in tissue extracts: application to redox profiling during Arabidopsis rosette development. Anal. Biochem. 363 58–69 [DOI] [PubMed] [Google Scholar]
- Rahman I., Kode A., Biswas S. K. (2006). Assay for quantitative determination of glutathione and glutathione disulfide levels using enzymatic recycling method. Nat. Protoc. 1 3159–3165 [DOI] [PubMed] [Google Scholar]
- Rao M. V., Paliyath G., Ormrod D. P., Murr D. P., Watkins C. B. (1997). Influence of salicylic acid on H2O2 production, oxidative stress and H2O2-metabolizing enzymes. Plant Physiol. 115 137–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayapuram C., Baldwin I. T. (2007). Increased SA in NPR1-silenced plants antagonizes JA and JA-dependent direct and indirect defenses in herbivore-attacked Nicotiana attenuata in nature. Plant J. 52 700–715 [DOI] [PubMed] [Google Scholar]
- Reymond P., Bodenhausen N., Van Poecke R. M., Krishnamurthy V., Dicke M., Farmer E. E. (2004). A conserved transcript pattern in response to a specialist and a generalist herbivore. Plant Cell 16 3132–3147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieu I., Powers S. J. (2009). Real-time quantitative RT-PCR: design, calculations and statistics. Plant Cell 21 1031–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert-Seilaniantz A., Grant M, Jones J. D. G. (2011). Hormone crosstalk in plant disease and defense: more than just JASMONATE-SALICYLATE antagonism. Annu. Rev. Phytopathol. 49 317–343 [DOI] [PubMed] [Google Scholar]
- Schlaeppi K., Bodenhausen N., Buchala A., Mauch F., Reymond P. (2008). The glutathione-deficient mutant pad2-1 accumulates lower amounts of glucosinolates and is more susceptible to the insect herbivore Spodoptera littoralis. Plant J. 55 774–786 [DOI] [PubMed] [Google Scholar]
- Schröder E., Eaton P. (2008). Hydrogen peroxide as an endogenous mediator and exogenous tool in cardiovascular research: issues and considerations. Curr. Opin. Pharmacol. 8 153–159 [DOI] [PubMed] [Google Scholar]
- Spoel S. H., Dong X. (2008). Making sense of hormone crosstalk during plant immune response. Cell Host Microbe 3 348–351 [DOI] [PubMed] [Google Scholar]
- Spoel S. H., Koornneef A., Claessens S. M. C., Korzellus J. P., Van Pelt J. A., Mueller J. A., et al. (2003). NPR1 modulates crosstalk between salicylate- and jasmonate-dependent defense pathways through a novel function in the cytoplasm. Plant Cell 15 760–77012615947 [Google Scholar]
- Spoel S. H., Mou Z., Tada Y., Spivey N. W., Genschik P., Dong X. (2009). Proteasome-mediated turnover of the transcriptional activator NPR1 plays a dual role in regulating plant immunity. Cell 137 860–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stotz H. U., Pittendrigh B. R., Kroymann J., Weniger K., Fritsche J., Bauke A., et al. (2000). Induced plant defense responses against chewing insects. Ethylene signaling reduces resistance of Arabidopsis against Egyptian cotton worm but not diamondback moth. Plant Physiol. 124 1007–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ströher E., Wang X.-J., Roloff N., Klein P., Husemann A., Dietz K.-J. (2009). Redox-dependent regulation of the stress-induced zinc-finger protein SAP12 in Arabidopsis thaliana. Mol. Plant 2 357–367 [DOI] [PubMed] [Google Scholar]
- Swarup R., Perry P., Hagenbeek D., Van Der Straeten D., Beemster G. T. S., Sandberg G., et al. (2007). Ethylene upregulates auxin biosynthesis in Arabidopsis seedlings to enhance inhibition of root cell elongation. Plant Cell 19 2186–2196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szalai G., Kellös T, Galiba G., Kocsy G. (2009). Glutathione as an antioxidant and regulatory molecule in plants under abiotic stress conditions. J. Plant Growth Regul. 28 66–80 [Google Scholar]
- Tada Y., Spoel S. H., Pajerowska-Mukhtar K., Mou Z., Song J., Wang C., et al. (2008). Plant immunity requires conformational changes of NPR1 via S-nitrosylation and thioredoxins. Science 321 952–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thivierge K., Prado A., Driscoll B. T., Bonneil é., Thibault P., Bede J. C. (2010). Caterpillar- and salivary-specific modification of plant proteins. J. Proteome Res. 9 5887–5895 [DOI] [PubMed] [Google Scholar]
- Van der Does D., Leon-Reyes A., Koorneef A., Van Verk M. C., Rodenburg N., Pauwels L., et al. (2013). Salicylic acid suppresses jasmonic acid signaling downstream of SCFCOI1-JAZ by targeting GCC promoter motifs via transcription factor ORA59. Plant Cell 25 744–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhage A., Vlaardingerbroek I., Raaymakers C., Van Dam N. M., Dicke M., Van Wees S. C. M., et al. (2011). Rewiring of the jasmonate signaling pathway in Arabidopsis during insect herbivory. Front. Plant Sci. 2:47 10.3389/ fpls.2011.00047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walling L. L. (2008). Avoiding effective defenses: strategies employed by phloem-feeding insects. Plant Physiol. 146 859–866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weech M.-H., Chapleau M., Pan L., Ide C., Bede J. C. (2008). Caterpillar saliva interferes with induced Arabidopsis thaliana defence responses via the systemic acquired resistance pathway. J. Exp. Bot. 59 2437–2448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wünsche H., Baldwin I. T., Wu J. (2011). S-nitrosoglutathione reductase (GSNOR) mediates the biosynthesis of jasmonic acid and ethylene is induced by feeding of the insect herbivore Manduca sexta and is important for jasmonate-elicited responses in Nicotiana attenuata. J. Exp. Bot. 62 4605–4616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zander M., Chen S., Imkampe J., Thurow C., Gatz C. (2011). Repression of the Arabidopsis thaliana jasmonic acid/ethylene-induced defense pathway by TGA interacting glutaredoxins depends on their C-terminal ALWL. Mol. Plant 5 831–840 [DOI] [PubMed] [Google Scholar]
- Zander M., La Camera S., Lamotte O., Métraux J. P., Gatz C. (2010) Arabidopsis thaliana class-II TGA transcription factors are essential activators of jasmonic acid/ethylene-induced defense responses. Plant J. 61 200–210 [DOI] [PubMed] [Google Scholar]
- Zarei A., Körbes A. P., Younessi P., Montiel G., Champion A., Memelink J. (2011). Two GCC boxes and AP2/ERF-domain transcription factor ORA59 in jasmonate/ethylene-mediated activation of PDF1.2 promoter in Arabidopsis. Plant Mol. Biol. 75 321–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zebelo S. A., Maffei M. E. (2012). The ventral eversible gland (VEG) of Spodoptera littoralis triggers early responses to herbivory in Arabidopsis thaliana. Arthropod Plant Interact. 6 543–551 [Google Scholar]
- Zhang Y., Fan W., Kinkema M., Li X., Dong X. (1999). Interaction of NPR1 with basic leucine zipper protein transcription factors that bind sequences required for salicylic acid induction of the PR-1 gene. Proc. Natl. Acad. Sci. U.S.A. 96 6523–6528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Tessaro M. J., Lassner M., Li X. (2003). Knockout analysis of Arabidopsis transcription factors TGA2, TGA5 and TGA6 reveals their redundant and essential roles in Systemic Acquired Resistance. Plant Cell 15 2647–2653 [DOI] [PMC free article] [PubMed] [Google Scholar]