Abstract
Background
Rates of tobacco smoking are significantly higher in patients with schizophrenia compared with the general population. The underlying mechanism for this comorbidity is unclear. One hypothesis is that there are common genetic factors that predispose to both nicotine dependence (ND) and schizophrenia. To investigate this hypothesis, we examined the association of the 15q25 gene cluster, the most significant candidate region to date implicated in ND and smoking behavior, with schizophrenia and bipolar disorder.
Methods
Five variants in the 15q25 gene cluster (rs951266, rs16969968, rs1051730, rs8040868, and rs17477223) were selected to test for association with schizophrenia diagnosis, bipolar disorder diagnosis, and the presence of negative symptoms of schizophrenia. Effects of the variants on 15q25 gene expression were analyzed using publically available postmortem brain expression data.
Results
A meta-analysis revealed four markers associated with risk for schizophrenia and bipolar disorder (rs951266, rs16969968, rs8040868, and rs17477223), and with the presence of negative symptoms of schizophrenia (rs951266, rs1051730, rs8040868, and rs17477223). The associations were in the same direction as that found for ND. Gene expression analysis indicated an association between genotypes of the rs1051730 variant and CHRNA5 expression in brain and peripheral blood mononuclear cells, and with the rs16969968 and rs17477223 variants in brain.
Conclusion
Variants in the 15q25 gene cluster are associated with risk for schizophrenia/bipolar illness, negative symptoms of schizophrenia, and influence CHRNA5 expression in the brain and peripheral blood mononuclear cells. These results are consistent with the notion that there are genetic mechanisms common to schizophrenia, ND, and bipolar disorder.
Keywords: 15q25 gene cluster, bipolar disorder, CHRNA5, nicotine dependence, schizophrenia
Introduction
Diseases associated with tobacco use and dependence constitute the leading cause of preventable mortality worldwide (World Health Organization, 2008). Smoking rates are particularly high in individuals with schizophrenia, a disabling psychiatric disorder affecting ~0.6–1% of the population. While 20–30% of the general population are smokers, 80–90% of patients with schizophrenia smoke (Dalack et al., 1998; De Leon and Diaz, 2005; Kalman et al., 2005). Smokers with schizophrenia are also considered ‘heavy smokers’, taking a greater number of cigarette puffs and consuming larger total cigarette puff volumes compared with smokers in the general population (Tidey et al., 2005). The mechanisms underlying schizophrenia and smoking comorbidity are unclear. Schizophrenia patients may smoke to self-medicate, alleviating cognitive deficits and negative symptoms of the disease (Goff et al., 1992; Griffith et al., 1998; Leonard et al., 2007) or to reduce extrapyramidal side effects of antipsychotic medications (Winterer, 2010). Alternatively, common genetic factors may predispose to both schizophrenia and heavy smoking or nicotine dependence (ND). Indeed, both ND and schizophrenia are highly heritable, with heritability estimates of 50–60% in ND (Tsuang et al., 1998; Kendler et al., 2003; Li, 2006) and as high as 80–85% in schizophrenia (Sullivan et al., 2003). A discordant twin study also reported an increased prevalence of daily smoking in individuals with familial vulnerability to schizophrenia (Lyons et al., 2002).
When searching for common genes that could potentially predispose to both ND and schizophrenia, nicotinic acetylcholine receptor (nAChR) genes are prime candidates. Genome wide association studies show that variations in nAChR subunits are strongly associated with risk for smoking behavior and ND (Tobacco and Genetics (TAG) Consortium, 2010). To date, the 15q25 gene cluster, which contains the CHRNA5/CHRNA3/CHRNB4 genes, coding for the α5, α3, and β4 nAChR subunits, respectively, has shown the most robust findings as a candidate region contributing to risk of heavy smoking, ND, and smoking related diseases (Bierut, 2010; Lips et al., 2010; Liu et al., 2010; Ware et al., 2011). Interestingly, the 15q11.2–15q25 region may also harbor risk genes for schizophrenia and bipolar disorder (Gejman et al., 2001; Lewis et al., 2003). The 15q25 gene cluster is associated with cognitive performance and enhanced cognitive flexibility in current smokers in the general population (Winterer et al., 2010; Zhang et al., 2010). Variation in the CHRNA3 gene is associated with sensorimotor gating in healthy controls and schizophrenia patients, and with the number of psychotic episodes, higher doses of antipsychotics, and the presence of negative symptoms in schizophrenia patients (Petrovsky et al., 2010). Recently, rs16969968, the most consistently replicated potential causal variant in ND, located in CHRNA5, was reported to be associated with schizophrenia in nonsmokers, but not in schizophrenia smokers (Hong et al., 2011).
In the current study, we aimed to expand on recent findings, and test the hypothesis that the 15q25 gene cluster is associated with schizophrenia. In a meta-analysis of seven schizophrenia and three bipolar datasets, we have found that variants in the 15q25 gene cluster are associated with risk for schizophrenia and bipolar illness, and the presence of negative symptoms in individuals with schizophrenia. Furthermore, we have shown that variants in the 15q25 gene cluster influence CHRNA5 expression.
Methods
Datasets
All datasets used in this study were obtained under a protocol approved by the Virginia Commonwealth University Institutional Review Board and the National Institutes of Health. European and African-American participants from the Molecular Genetics of Schizophrenia GAIN (MGS-GAIN) and non-GAIN study (MGS-nonGAIN), and the Whole Genome Wide Association Study of Bipolar Disorder (GAIN-BD) were obtained from NCBI dbGaP (http://www.ncbi.nlm.nih.gov/gap). Bipolar case and control participants were obtained from the Wellcome Trust Case Control Consortium (WTCCC-BD). European and African-American participants from the Clinical Antipsychotic Trials of Intervention Effectiveness (CATIE) project were obtained from the National Institutes of Mental Health repository (http://www.nimhgenetics.org). The Irish Study of High Density Schizophrenia Families (ISHDF) is a collaborative study between Virginia Commonwealth University, the Queen’s University, Belfast, Northern Ireland, and the Health Research Board, Dublin, Ireland, that was conducted between April 1987 and November 1992, as previously described (Kendler et al., 1996). Irish schizophrenia cases and controls from the Irish Case–Control Study of Schizophrenia (ICCSS) were recruited from inpatient and outpatient psychiatric facilities in Ireland and Northern Ireland (cases) and from donors at the Northern Ireland Blood Transfusion Service, the Irish national police, and army reserve (controls) (Riley et al., 2009). Demographic information on each dataset is shown in Table 1.
Table 1.
Number of participants with schizophrenia and bipolar disorder information, sex distribution (number of males/number of females), mean age (SD), and the number of cases and controls for all studies included
| Studies | N | Sex distribution (M/F) | Mean age (SD) | Number cases/controls |
|---|---|---|---|---|
| Molecular Genetics of Schizophrenia (GAIN) EA | 1874 | 1105/769 | 47.6 (15.1) | 1151/723 |
| Molecular Genetics of Schizophrenia (GAIN) AA | 1572 | 802/770 | 44.4 (11.9) | 915/657 |
| Molecular Genetics of Schizophrenia non-GAIN | 2115 | 1216/899 | 46.1 (15.7) | 1066/1049 |
| Clinical Antipsychotic Trials of Intervention Effectiveness-White | 732 | 630/102 | 41 (11.4) | 351/381 |
| Clinical Antipsychotic Trials of Intervention Effectiveness-Black | 177 | 128/49 | 40.9 (10.1) | 88/89 |
| Irish Case–Control Study of Schizophrenia | 1605 | 1018/587 | 42.3 (13.6) | 988/587 |
| Irish Study of High Density Schizophrenia Families | 1337 (267 families) | 769/568 | 49.5 (14.9) | 496/841 |
| Whole Genome Wide Association Study of Bipolar Disorder (GAIN-BD) EA | 1051 | 474/577 | 47.6 (15.7) | 641/410 |
| Whole Genome Association Study of Bipolar Disorder (GAIN-BD) AA | 239 | 84/155 | 44.3 (11.5) | 139/100 |
| Wellcome Trust Case Control Consortium | 5002 | 2223/2779 | NA | 1998/3004 |
Study names are listed on the left.
AA, African-American; BD, bipolar disorder; EA, European-American; F, female; M, male.
Association studies
Schizophrenia and bipolar disorder diagnosis
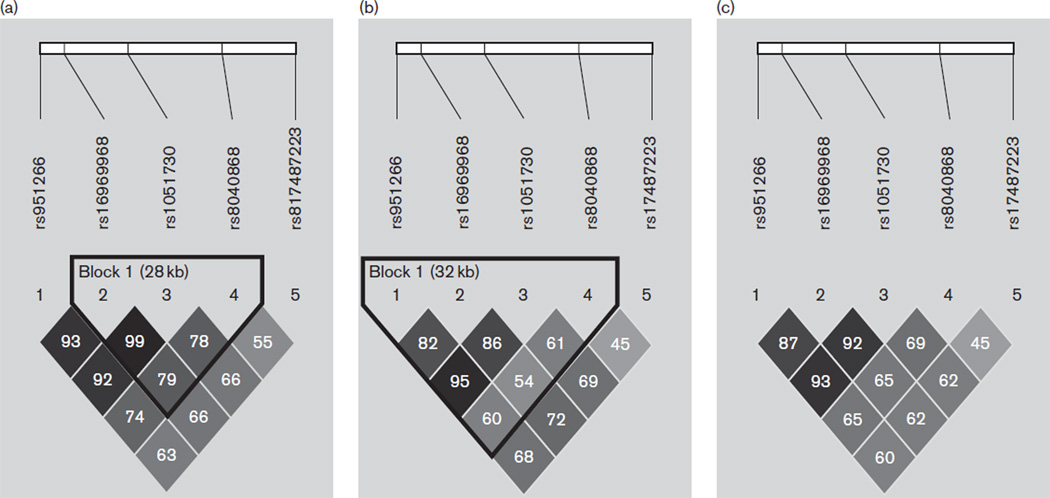
Ten datasets (seven schizophrenia and three bipolar) were used to test the association of schizophrenia and bipolar diagnosis with variants in the 15q25 gene cluster. All genotyped single nucleotide polymorphisms (SNPs) in the MGS-GAIN samples on chromosome 15 from 76 640–76 730 kb were extracted using the PLINK software (Purcell et al., 2007) for a total of 22 SNPs in the European-American sample and 28 in the African-American sample. The six unique SNPs in the African-American sample that are nonpolymorphic in European-Americans were excluded from analysis. The 22 SNPs were subsequently analyzed for association with schizophrenia and/or bipolar disorder in all datasets except the ICCSS and ISHDF datasets because only two markers, rs1051730 and rs16969968, both of which have consistently shown robust association with ND and heavy smoking, were previously genotyped in our laboratory in these datasets. Marker information and results of the 22 SNP meta-analysis are found in Supplementary Tables 1–4 (http://links.lww.com/PG/A66). Two markers, rs951266 (P = 0.04) and rs17487223 (P = 0.03), showed nominal association with the combined schizophrenia and bipolar disorder diagnosis, and rs8040868 (P = 0.06) was marginally associated. To increase the power of our meta-analysis, we incorporated the ICCSS and ISHDF datasets. On the basis of the linkage disequilibrium (LD) structure, we were able to impute rs951266, rs8040868, and rs17487223 for additional analysis in the ICCSS and ISHDF datasets. The imputation error rate for both datasets was ~6%, indicating good imputation quality. In this study, we present the results of the five markers located within the 15q25 gene cluster (rs951266, rs17487223, rs8040868, rs16969968, and rs1051730) that were analyzed in a meta-analysis using results from all datasets. Marker information and LD blocks from the combined Irish/European-American, combined African-American, and combined total sample population are shown in Table 2 and Fig. 1, respectively. Overlapping samples between the CATIE and MGS-GAIN/non-GAIN samples were removed before analyses. All controls were shared between the MGS-GAIN and GAIN-BD samples. To control for this effect, controls were split between the two samples and randomly assigned so that the ratio of cases to controls in each dataset remained consistent.
Table 2.
Chromosomal position, allele, marker location, and minor allele frequencies for the 15q25 cluster markers analyzed in this study
| SNPs | Chromosome position | Allele | Location | Freq EA | Freq AA |
|---|---|---|---|---|---|
| rs951266 | 76665596 | C/T | intron, CHRNA5 | 0.36 | 0.11 |
| rs16969968 | 76669980 | G/A | exon, CHRNA5, missense D→N | 0.35 | 0.02 |
| rs1051730 | 76681394 | G/A | exon, CHRNA3, synon Y→Y | 0.35 | 0.11 |
| rs8040868 | 76698236 | T/C | exon, CHRNA3, synon V→V | 0.40 | 0.35 |
| rs17487223 | 76711042 | C/T | intron, CHRNB4 | 0.33 | 0.12 |
The minor allele is located on the right and bolded. Minor allele frequencies for both populations are based on the combined European-American/Irish samples and the combined African-American sample.
AA, African-American; EA, European-American; SNP, single nucleotide polymorphism.
Fig. 1.
Linkage disequilibrium blocks containing r2 values for (a) the Irish/European-American datasets, (b) the combined African-American datasets, and (c) all datasets combined.
Negative symptoms
Phenotype data for the presence of negative symptoms were available for three datasets: the European-American MGS-GAIN and non-GAIN, which were analyzed as one dataset, African-American MGS-GAIN, and ISHDF. The ISHDF phenotype was based on a quantitative scale assessing the number of negative symptoms as described in detail elsewhere (Fanous et al., 2005). We converted this data into a dichotomized phenotype to coincide with the MGS phenotype data. Cases with no negative symptoms were considered ‘absent’ and cases with one or more negative symptom were considered ‘present’.
Expression studies
SNPExpress dataset
The SNPExpress expression dataset was downloaded from http://compute1.lsrc.duke.edu/softwares/SNPExpress/. This project was initiated by the Duke University to investigate tissue-specific genetic regulation of splicing and expression in 93 brain samples and 80 peripheral blood mononuclear cells (PBMC). The dataset was used to determine whether variants in the 15q25 gene cluster influence expression in the region.
NCBI Gene Expression Omnibus Alzheimer’s disease dataset
Postmortem brain expression data from Alzheimer’s disease controls (n = 187) was obtained from the NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) accession number GSE15222. The original dataset was generated by the laboratory of Dr Amanda Myers at the University of Miami (Webster et al., 2009).
Stanley Medical Research Institute dataset
Postmortem brain expression data from schizophrenia, bipolar disorder, major depression, and healthy control patients were obtained from the Kato (study ID 7, frontal Brodmann area 46, n=99) and KatoB (study ID 15, prefrontal cortex, n = 46) studies through the Stanley Medical Research Institute (SMRI) (http://www.stanleyresearch.org/dnn/).
Statistical analyses
Logistic regression analysis using PLINK was performed on each dataset to test the association of schizophrenia diagnosis, bipolar disorder diagnosis, or the presence of negative symptoms. Age and sex were used as covariates for each dataset, with the exception of the WTCCC-BD dataset, where only sex information was available to us. In CATIE, seven principal components controlling for population stratification and smoking status were also included as covariates (Sullivan et al., 2008). With the exception of the CATIE datasets, smoking information was not available to us for any other dataset. For the ISHDF, PDTPHASE (pedigree disequilibrium test), implemented in the UNPHASED software (Dudbridge, 2008), was used for association analyses. Odds ratios and confidence intervals were calculated from the number of affected and unaffected siblings and trios.
Linear regression using PLINK was performed in expression analyses. Age, sex, and brain region were used as covariates in the Myers data. For the SMRI data, age, sex, brain pH, smoking status, and diagnosis were used as covariates.
Imputation was conducted using fastPHASE (Scheet and Stephens, 2006), with Hapmap CEU (Utah residents with Northern and Western European ancestry) or YOR (Yoruba in Ibadan, Nigeria) datasets as reference panels. Because rs16969968 is nonpolymorphic in African populations, the CATIE African-American dataset was used to impute this marker in the MGS-GAIN and GAIN-BD African-American datasets.
Results from all association and expression studies were used to conduct meta-analyses. Expression data were normalized from each dataset using a z-score transformation. The schizophrenia (n = 9412) and bipolar (n = 6292) datasets totaled 15 704 individuals. A total of 3 589 individuals were available for negative symptom analysis. The GWAMA program (Magi and Morris, 2010) was used for meta-analysis. Cochrane’s Q statistic P-values were calculated to measure between-study heterogeneity. Results indicated significant Q statistic P-values for some markers in the eight dataset schizophrenia meta-analysis (Supplemental Table 2, http://links.lww.com/PG/A66), indicating heterogeneity between samples, thus, for consistency a random effects meta-analysis was used to analyze all results. Q statistic P-values for all 22 SNPs are indicated in Supplemental Tables 2–4 (http://links.lww.com/PG/A66). Q statistic P-values for the five markers used in the final meta-analysis were not significant, indicating no detectable heterogeneity between datasets for these polymorphisms. Correction for multiple testing was carried out on all tests jointly for each meta-analysis using SNP spectral decomposition (Nyholt, 2004), a correction for multiple testing for SNPs in LD. On the basis of this program, the corrected P-value for the 22 SNP analysis was P-value less than 0.003, and P-value less than 0.05 for the five SNP analysis.
Results
Variants in the 15q25 gene cluster are associated with schizophrenia and bipolar disorder
Two variants, rs8040868 and rs17487223, were significantly associated with risk for schizophrenia (Table 3). Meta-analysis of the bipolar studies showed association with rs951266 and risk for bipolar disorder (Table 3). When the schizophrenia and bipolar disorder results were combined, four markers, rs951266, rs16969968, rs8040868, and rs17487223, were significantly associated with risk for schizophrenia and bipolar disorder (Table 3). All markers remained significant after correction for multiple testing.
Table 3.
Variants in the 15q25 gene cluster are associated with schizophrenia and bipolar disorder
| SNP | Allele | OR | L_95 | U_95 | P | |
|---|---|---|---|---|---|---|
| SZD | rs951266 | T | 1.05 | 1.02 | 1.18 | 0.15 |
| rs16969968 | A | 1.06 | 0.97 | 1.13 | 0.09 | |
| rs1051730 | A | 1.05 | 0.97 | 1.13 | 0.19 | |
| rs8040868 | C | 1.09 | 1.03 | 1.16 | 0.004 | |
| rs17487223 | T | 1.08 | 0.98 | 1.14 | 0.03 | |
| BD | rs951266 | T | 1.10 | 1.02 | 1.18 | 0.02 |
| rs16969968 | A | 1.05 | 0.97 | 1.13 | 0.23 | |
| rs1051730 | A | 1.05 | 0.97 | 1.13 | 0.24 | |
| rs8040868 | C | 1.03 | 0.95 | 1.11 | 0.46 | |
| rs17487223 | T | 1.06 | 0.98 | 1.14 | 0.13 | |
| SZD + BD | rs951266 | T | 1.07 | 1.02 | 1.13 | 0.009 |
| rs16969968 | A | 1.06 | 1.00 | 1.11 | 0.04 | |
| rs1051730 | A | 1.05 | 0.99 | 1.10 | 0.08 | |
| rs8040868 | C | 1.07 | 1.02 | 1.12 | 0.007 | |
| rs17487223 | T | 1.07 | 1.02 | 1.13 | 0.009 |
Columns represent the marker, minor/risk allele, odds ratio (OR), lower and upper confidence intervals (L_95; U_95) and P-values. The correction for multiple testing threshold based on single nucleotide polymorphism spectral decomposition was P < 0.05.
Significant P-values are represented in bold.
BD, bipolar disorder (SZD, n = 9412; BD = 6292; SZD + BD = 15704); SNP, single nucleotide polymorphism; SZD, schizophrenia.
Variants in the 15q25 gene cluster are associated with negative symptoms of schizophrenia
Three datasets were analyzed to test the association between variants in the 15q25 gene cluster and negative symptoms. A meta-analysis of the results revealed a significant association with the variants rs951266, rs1051730, rs8040868, and rs1748722, and the presence of negative symptoms (Table 4). Odds ratios indicate that the variants are associated with an increased risk of negative symptoms. The markers remained significant after correction for multiple testing.
Table 4.
Variants in 15q25 are associated with the presence of negative symptoms of schizophrenia
| SNP | Allele | OR | L_95 | U_95 | P |
|---|---|---|---|---|---|
| rs951266 | T | 1.19 | 1.03 | 1.37 | 0.017 |
| rs16969968 | A | 1.15 | 0.99 | 1.34 | 0.07 |
| rs1051730 | A | 1.19 | 1.03 | 1.37 | 0.019 |
| rs8040868 | C | 1.16 | 1.02 | 1.31 | 0.02 |
| rs17487223 | T | 1.21 | 1.06 | 1.43 | 0.018 |
Meta-analysis results from three datasets (n = 3589) show that variants in the 15q25 gene cluster are associated with risk for the presence of negative symptoms of schizophrenia.
Significant P-values are represented in bold.
SNP, single nucleotide polymorphism.
CHRNA5 expression is associated with variants in the 15q25 gene cluster
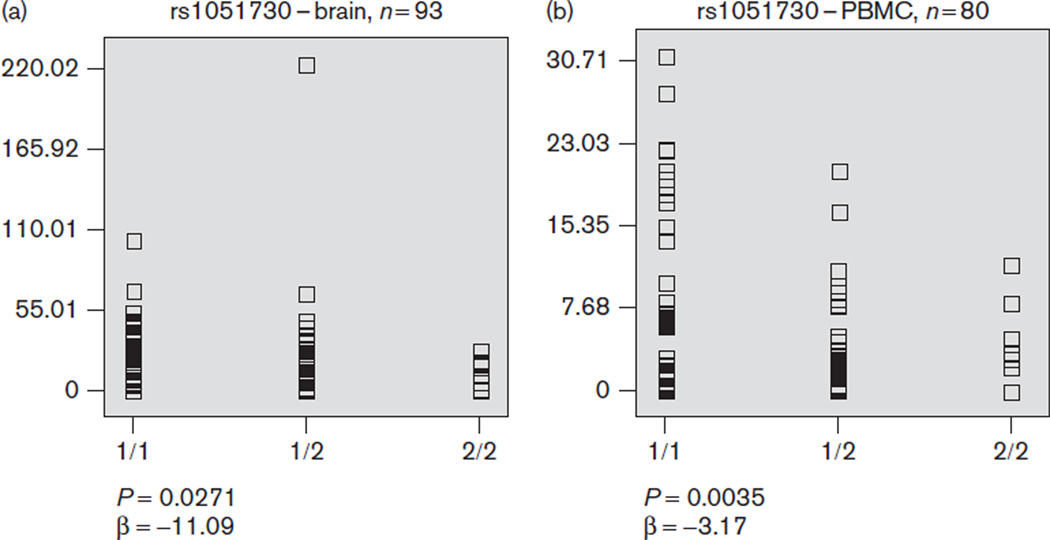
We assessed blood and PBMC expression data from the SNPExpress database. Only the variant rs1051730 was typed in this database; therefore, we tested whether its genotypes predict variation in expression of the CHRNA5, CHRNA3, and CHRNB4 genes. The minor allele homo-zygote of rs1051730 was associated with lower CHRNA5 expression in both brain (Fig. 2a) and PBMC (Fig. 2b). To follow-up these findings, we analyzed the five markers using postmortem brain expression data from Alzheimer’s disease controls acquired from the NCBI Gene Expression Ominbus and postmortem brain expression data from two SMRI studies. Consistent with preliminary results from SNPExpress, three markers, rs1051370, rs16969968, and rs17487223, were associated with lower brain CHRNA5 expression (Table 5). There was no significant association with any variant and CHRNA3 or CHRNB4 expression. Mean expression values for CHRNA5 for the three genotypes of each marker from each expression dataset are represented in Table 6.
Fig. 2.
The rs1051730 variant is associated with CHRNA5 expression. The minor allele homozygote of rs1051730, a synonymous single nucleotide polymorphism located in CHRNA3, is associated with lower brain expression of CHRNA5 in (a) brain and (b) peripheral blood mononuclear cells (PBMC). The y-axis represents relative gene expression and the x-axis represents the genotype.
Table 5.
Variants in the 15q25 gene cluster influence CHRNA5 gene expression in brain
| SNPs | Allele | β | L_95 | U_95 | P |
|---|---|---|---|---|---|
| rs951266 | T | −0.17 | −0.33 | −0.01 | 0.05 |
| rs16969968 | A | −0.33 | −0.57 | −0.09 | 0.007 |
| rs1051730 | A | −0.33 | −0.57 | −0.09 | 0.007 |
| rs8040868 | C | −0.14 | −0.29 | 0.01 | 0.07 |
| rs17487223 | T | −0.22 | −0.38 | −0.06 | 0.005 |
The minor allele is associated with lower CHRNA5 gene expression for the nonsynonymous rs16969969 variant, the synonymous rs1051730 variant, and the rs17487223 intron. Results were obtained from a meta-analysis of three expression datasets (n = 332).
Significant P-values are represented in bold.
SNP, single nucleotide polymorphism.
Table 6.
Mean expression of CHRNA5 from the three genotypes for each marker from each expression dataset
| SNP | 1/1 | 1/2 | 2/2 | |
|---|---|---|---|---|
| AD (n= 187) | rs951266 | 0.039 | 0.031 | −0.185 |
| rs16969968 | 0.206 | 0.012 | −0.095 | |
| rs1051730 | 0.206 | 0.012 | −0.095 | |
| rs8040868 | 0.206 | 0.012 | −0.095 | |
| rs17487223 | 0.039 | 0.038 | −0.205 | |
| Kato (n= 99) | rs951266 | 0.265 | −0.173 | −0.494 |
| rs16969968 | 0.265 | −0.249 | −0.336 | |
| rs1051730 | 0.265 | −0.249 | −0.215 | |
| rs8040868 | 0.259 | −0.345 | −0.215 | |
| rs17487223 | 0.315 | −0.324 | −0.367 | |
| Kato B (n = 46) | rs951266 | 1.64 | 0.112 | −0.267 |
| rs16969968 | 1.64 | 0.112 | −0.267 | |
| rs1051730 | 1.64 | 0.112 | −0.267 | |
| rs8040868 | 1.07 | 0.081 | −0.210 | |
| rs17487223 | 0.207 | 0.148 | −0.193 |
1 represents the major allele; 2 represents the minor allele.
AD, Alzheimer’s disease dataset; SNP, single nucleotide polymorphism.
Discussion
In the current study, we tested five variants in the 15q25 gene cluster, including two variants in high LD, rs16969968 and rs1051370, which have been consistently associated with risk for heavy smoking and ND, and found that variations in the 15q25 gene cluster are associated with risk for schizophrenia and bipolar disorder, and the presence of negative symptoms in patients with schizophrenia. Furthermore, we showed that three variants, rs16969968, rs1051730, and rs17487223 are associated with brain expression of CHRNA5.
Our findings using a larger sample size support previous studies implicating the CHRNA5 gene in schizophrenia (Hong et al., 2011), and suggest a role for the 15q25 gene cluster in bipolar disorder. In the combined schizophrenia and bipolar datasets, we identified four variants, including rs16969968, the nonsynonymous SNP in CHRNA5, associated with risk for schizophrenia and bipolar disorder. Similar odds ratios and overlapping confidence intervals in individual analyses suggest commonalities between schizophrenia and bipolar disorder. Indeed, schizophrenia and bipolar disorder share a common genetic liability (see review Bramon and Sham, 2001; Lichtenstein et al., 2009), and common gene expression alterations (Shao and Vawter, 2008). While an examination of the relationship between these disorders is beyond the scope of this study, the association with the 15q25 gene cluster and both schizophrenia and bipolar disorder is of interest. Although surprisingly few studies have examined the relationship between smoking and bipolar disorder, the prevalence of smoking in bipolar disorder, though not as high as schizophrenia, has been reported between 66 and 69% (Lasser et al., 2000; Vanable et al., 2003; Diaz et al., 2009), approximately twice that of the general population (20–30%). The highest associations of comorbid substance use disorders in bipolar disorder have been reported with alcohol and cannabis, followed by cocaine and opioids (see review Cerullo and Strakowski, 2007). Interestingly, in addition to heavy smoking and ND, the 15q25 gene cluster has been associated with alcohol, cocaine, and opioid dependence (Grucza et al., 2008; Wang et al., 2009; Erlich et al., 2010; Sherva et al., 2010), suggesting a potential association with substance use disorders in general. Schizophrenia is also cormorbid with other substance abuse, though to a lesser extent than that observed with nicotine (Mueser et al., 1990; DeQuardo et al., 1994; Strakowski et al., 1994; Buckley, 1998). While a recent study investigating the relationship between the 15q25 gene cluster and psychiatric disorders commonly associated with ND found no association between the genetic variants and any comorbid psychiatric disorder assessed, schizophrenia and bipolar disorder were not tested, and limited power in the study may have impacted the ability to detect significant effects (Chen et al., 2012). Future studies with larger sample sizes may identify significant associations.
In addition to schizophrenia diagnosis, we expanded our study to assess the negative symptoms of schizophrenia, as it has been suggested that smoking in patients with schizophrenia is an attempt to alleviate cognitive deficits and negative symptoms associated with the disorder (Goff et al., 1992; Griffith et al., 1998; Leonard et al., 2007). The data suggest that variation in the 15q25 gene cluster is associated with an increased risk for the presence of negative symptoms, which include blunted affect, social withdrawal, difficulty in abstract thinking, and inappropriate affect. In previous studies, the total negative symptom score was a strong contributor to predicting smoking behavior, as more severe ND was observed in individuals with greater total number of negative symptoms (Patkar et al., 2002). Current schizophrenia smokers were also more likely to report negative symptoms than former chronic schizophrenia smokers (Hall et al., 1995). Although the current study provides evidence of a relationship between ND and negative symptoms, it is of interest in future studies to obtain additional datasets with negative symptoms measured as a quantitative phenotype. This would provide more power to the study, and allow us to assess negative symptom severity, which may be more closely associated with ND.
Expression analyses show that the minor allele of the synonymous variant rs1051730, located in CHRNA3, is associated with lower brain and PBMC expression of CHRNA5. We were only able to report expression analysis from the rs1051730 study, as this was the only marker from our study genotyped in the SNPExpress dataset. A meta-analysis of three brain datasets further supported these findings and also revealed that rs16969968, and rs17487223, located within an intron in CHRNB4, are associated with CHRNA5 brain expression. Variability in CHRNA5 gene expression in brain and whole blood is also associated with risk for ND and heavy smoking, and possibly cognitive performance (Wang et al., 2009; Winterer et al., 2010). Because we controlled for schizophrenia, bipolar disorder, major depression, and smoking in this expression analysis, the expression is not correlated with a particular phenotype; however, we cannot rule out the possibility that altered CHRNA5 brain expression is associated with ND and/or schizophrenia. In Hong et al.’s (2011) study, rs16969968 was associated with schizophrenia in nonsmokers, but not schizophrenia smokers. This may suggest a decreased function and/or expression of the CHRNA5 receptor under basal conditions in schizophrenia, which is corrected by smoking, thus obliterating differences between cases and controls. Though it is unknown whether the risk variant alters α5 nAChR subunit function under basal conditions, the rs16969968 variant, which encodes an Asp398Asn polymorphism resulting in an aspartic acid (G allele) change to asparagine (A allele), results in reduced response to agonist when the variant coassembles with the α4β2 nAChR subtype (Bierut et al., 2008). In the current study, the minor alleles of rs16969968 and rs17487223, which correlate with decreased brain expression of CHRNA5, are also associated with risk for schizophrenia and bipolar diagnosis. The variant rs1051730, though associated with CHRNA5 brain expression, did not reach significance for schizophrenia or bipolar disorder in the combined sample, but was significantly associated with risk for negative symptoms. This may be due to the allele’s smaller effect size in the combined sample, compared with the negative symptom study. Alternatively, we may be capturing a signal from rs16969968, as this marker is highly correlated with rs1051730, or vice-versa.
On the basis of the results, we propose that our data support a shared genetic component between schizophrenia and smoking. Indeed, the association with schizophrenia, bipolar disorder, and the presence of negative symptoms was in the same direction as that found for heavy smoking and ND. Future studies will attempt to verify common neurobiological pathways between schizophrenia and smoking behavior, as well as specific symptoms of schizophrenia, such as cognitive deficits and negative symptoms. A limitation to this study is that, with the exception of the CATIE sample, smoking information for the schizophrenia and bipolar disorder datasets was not available to us, thus, it is possible that the signal from our study could be driven in part by smoking, as the variants used in this study have also been implicated in risk for ND and heavy smoking (Saccone et al., 2009; Li et al., 2010). However, as mentioned, our results are consistent with a previous study implicating an association with rs16969968 and schizophrenia in nonsmokers, but not schizophrenia smokers (Hong et al., 2011). With the support of these findings, it is likely that our results provide further evidence of an association of schizophrenia with the 15q25 gene cluster.
Overall, our results implicate variants in the 15q25 gene cluster in risk for schizophrenia, the presence of negative symptoms, and bipolar disorder, and variants within the gene cluster influence CHRNA5 expression in brain and PBMC.
Supplementary Material
Acknowledgements
This study was supported in part by a research grant (07R-1770) from the Stanley Medical Research Institute and an Independent Investigator Award from NARSAD to X.C. and National Institute of Mental Health (NIMH) grant MH-020030 to K.J.J. The IHDSFand ICCSS samples were supported by grants MH41953 to K.S.K. from NIMH, and A.H.F. was supported by a grant from the Department of Veterans Affairs Merit Review Program.
The authors thank Cuie Sun for her role in genotyping the ICCSS and IDHFS datasets. Clinical Antipsychotic Trials of Intervention Effectiveness (CATIE): the principal investigators of the CATIE trial were Jeffrey A. Lieberman, T. Scott Stroup, and Joseph P. McEvoy. The CATIE trial was funded by a grant from the National Institute of Mental Health (N01 MH900001) along with MH074027 (PI PF Sullivan). Genotyping for this study was funded by Eli Lilly and Company. Genome-Wide Association Study of Schizophrenia: funding support for the Genome-Wide Association of Schizophrenia Study was provided by the National Institute of Mental Health (R01 MH67257, R01 MH59588, R01 MH59571, R01 MH59565, R01 MH59587, R01 MH60870, R01 MH59566, R01 MH59586, R01 MH61675, R01 MH60879, R01 MH81800, U01 MH46276, U01 MH46289 U01 MH46318, U01 MH79469, and U01 MH79470) and the genotyping of samples was provided through the Genetic Association Information Network (GAIN). The datasets used for the analyses described in this manuscript were obtained from the database of Genotypes and Phenotypes (dbGaP) found at http://www.ncbi.nlm.nih.gov/gap through dbGaP accession number phs00013 and phg00013. Samples and associated phenotype data for the Genome-Wide Association of Schizophrenia Study were provided by the Molecular Genetics of Schizophrenia Collaboration (PI: Pablo V. Gejman, Evanston Northwestern Healthcare (ENH) and Northwestern University, Evanston, IL, USA).
Whole Genome-Wide Association Study of Bipolar Disorder: funding support for the Whole Genome Association Study of Bipolar Disorder was provided by the NIMH and the genotyping of samples was provided through GAIN. The datasets used for the analyses described in this manuscript were obtained from dbGaP found at http://www.ncbi.nlm.nih.gov/gap through dbGaP accession number phs000017.v3.p1.
Samples and associated phenotype data for the Collaborative Genomic Study of Bipolar Disorder were provided by the NIMH Genetics Initiative for Bipolar Disorder. Data and biomaterials were collected in four projects that participated in NIMH Bipolar Disorder Genetics Initiative. From 1991–1998, the principal investigators and coinvestigators were: Indiana University, Indianapolis, IN, U01 MH46282, John Nurnberger, MD, PhD, Marvin Miller, MD, and Elizabeth Bowman, MD; Washington University, St. Louis, MO, U01 MH46280, Theodore Reich, MD, Allison Goate, PhD, and John Rice, PhD; Johns Hopkins University, Baltimore, MD U01 MH46274, J. Raymond DePaulo, Jr, MD, Sylvia Simpson, MD, MPH, and Colin Stine, PhD; NIMH Intramural Research Program, Clinical Neurogenetics Branch, Bethesda, MD, Elliot, Gershon, MD, Diane Kazuba, BA, and Elizabeth Maxwell, MSW. Data and biomaterials were collected as part of 10 projects that participated in the NIMH Bipolar Disorder Genetics Initiative. From 1999–2003, the principal investigators and coinvestigators were: Indiana University, Indianapolis, IN, R01 MH59545, John Nurnberger, MD, PhD, Marvin J. Miller, MD, Elizabeth, S. Bowman, MD, N. Leela Rau, MD, P. Ryan Moe, MD, Nalini Samavedy, MD, Rif El-Mallakh, MD (at University of Louisville), Husseini Manji, MD (at Wayne State University), Debra A. Glitz, MD (at Wayne State University), Eric T. Meyer, MS, Carrie Smiley, RN, Tatiana Foroud, PhD, Leah Flury, MS, Danielle M. Dick, PhD, Howard Edenberg, PhD; Washington University, St. Louis, MO, R01 MH059534, John Rice, PhD, Theodore Reich, MD, Allison Goate, PhD, Laura Bierut, MD; Johns Hopkins University, Baltimore, MD, R01 MH59533, Melvin McInnis MD, J. Raymond DePaulo, Jr, MD, Dean F. MacKinnon, MD, Francis M. Mondimore, MD, James B. Potash, MD, Peter P. Zandi, PhD, Dimitrios Avramopoulos, and Jennifer Payne; University of Pennsylvania, PA, R01 MH59553, Wade Berrettini MD, PhD; University of California at Irvine, CA, R01 MH60068, William Byerley, MD, and Mark Vawter, MD, University of Iowa, IA, R01 MH059548, William Coryell, MD, and Raymond Crowe, MD; University of Chicago, IL, R01 MH59535, Elliot Gershon, MD, Judith Badner, PhD, Francis McMahon, MD, Chunyu Liu, PhD, Alan Sanders, MD, Maria Caserta, Steven Dinwiddie, MD, Tu Nguyen, Donna Harakal; University of California at San Diego, CA, R01 MH59567, John Kelsoe, MD, Rebecca McKinney, BA; Rush University, IL, R01 MH059556, William Scheftner, MD, Howard M. Kravitz, DO, MPH, Diana Marta, BS, Annette Vaughn-Brown, MSN, RN, and Laurie Bederow, MA; GAIN 1 March 2010 version page NIMH Intramural Research Program, Bethesda, MD, 1Z01MH002810–01, Francis J. McMahon, MD, Layla Kassem, PsyD, Sevilla Detera-Wadleigh, PhD, Lisa Austin, PhD, Dennis L. Murphy, MD.
Footnotes
Supplemental digital content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal’s Website (www.psychgenetics.com).
Conflicts and interest
There are no conflicts of interest.
References
- Bierut LJ. Convergence of genetic findings for nicotine dependence and smoking related diseases with chromosome 15q24–25. Trends Pharmacol Sci. 2010;31:46–51. doi: 10.1016/j.tips.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierut LJ, Stitzel JA, Wang JC, Hinrichs AL, Grucza RA, Xuei X, et al. Variants in nicotinic receptors and risk for nicotine dependence. Am J Psychiatry. 2008;165:1163–1171. doi: 10.1176/appi.ajp.2008.07111711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bramon E, Sham PC. The common genetic liability between schizophrenia and bipolar disorder: a review. Curr Psychiatry Rep. 2001;3:332–337. doi: 10.1007/s11920-001-0030-1. [DOI] [PubMed] [Google Scholar]
- Buckley PF. Substance abuse in schizophrenia: a review. J Clin Psychiatry. 1998;59:26–30. [PubMed] [Google Scholar]
- Cerullo MA, Strakowski SM. The prevalence and significance of substance use disorders in bipolar type I and II disorder. Subst Abuse Treat Prev Policy. 2007;2:129. doi: 10.1186/1747-597X-2-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LS, Xian H, Grucza RA, Saccone NL, Wang JC, Johnson EO, et al. Nicotine dependence and comorbid psychiatric disorders: examination of specific genetic variants in the CHRNA5-A3-B4 nicotinic receptor genes. Drug Alcohol Depend. 2012;123:S42–S51. doi: 10.1016/j.drugalcdep.2012.01.014. http://dx.doi.org.proxy.library.vcu.edu/10.1016/j.drugalcdep.2012.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalack GW, Healy DJ, Meador-Woodruff JH. Nicotine dependence and schizophrenia: clinical phenomenon and laboratory findings. Am J Psychiatry. 1998;155:1490–1501. doi: 10.1176/ajp.155.11.1490. [DOI] [PubMed] [Google Scholar]
- De Leon J, Diaz FJ. A meta-analysis of worldwide studies demonstrates an association between schizophrenia and tobacco smoking behaviors. Schi-zophr Res. 2005;76:135–157. doi: 10.1016/j.schres.2005.02.010. [DOI] [PubMed] [Google Scholar]
- DeQuardo JR, Carpenter CF, Tandon R. Patterns of substance abuse in schizophrenia: nature and significance. J Psychiatr Res. 1994;28:267–275. doi: 10.1016/0022-3956(94)90010-8. [DOI] [PubMed] [Google Scholar]
- Diaz FJ, James D, Botts S, Maw L, Susce MT, de Leon J. Tobacco smoking behaviors in bipolar disorder: a comparison of the general population, schizophrenia, and major depression. Bipolar Disord. 2009;11:154–165. doi: 10.1111/j.1399-5618.2009.00664.x. [DOI] [PubMed] [Google Scholar]
- Dudbridge F. Likelihood-based association analysis for nuclear families and unrelated subjects with missing genotype data. Hum Hered. 2008;66:87–98. doi: 10.1159/000119108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erlich PM, Hoffman SN, Rukstalis M, Han JJ, Chu X, Linda Kao WH, et al. Nicotinic acetylcholine receptor genes on chromosome 15q25.1 are associated with nicotine and opioid dependence severity. Hum Genet. 2010;128:491–499. doi: 10.1007/s00439-010-0876-6. [DOI] [PubMed] [Google Scholar]
- Fanous AH, van den Oord EJ, Riley BP, Aggen SH, Neale MC, O’Neill FA, et al. Relationship between a high-risk haplotype in the DTNBP1 (dysbindin) gene and clinical features of schizophrenia. Am J Psychiatry. 2005;162:1824–1832. doi: 10.1176/appi.ajp.162.10.1824. [DOI] [PubMed] [Google Scholar]
- Gejman PV, Sanders AR, Badner JA, Cao Q, Zhang J. Linkage analysis of schizophrenia to chromosome 15. Am J Med Genet. 2001;105:789–793. doi: 10.1002/ajmg.1552. [DOI] [PubMed] [Google Scholar]
- Goff DC, Henderson DC, Amico E. Cigarette smoking in schizophrenia: relationship to psychopathology and medication side effects. Am J Psychiatry. 1992;149:1189–1194. doi: 10.1176/ajp.149.9.1189. [DOI] [PubMed] [Google Scholar]
- Griffith JM, O’Neill JE, Petty F, Garver D, Young D, Freedman R. Nicotinic receptor desensitization and sensory gating deficits in schizophrenia. Biol Psychiatry. 1998;44:98–106. doi: 10.1016/s0006-3223(97)00362-4. [DOI] [PubMed] [Google Scholar]
- Grucza RA, Wang JC, Stitzel JA, Hinrichs AL, Saccone SF, Saccone NL, et al. A risk allele for nicotine dependence in CHRNA5 is a protective allele for cocaine dependence. Biol Psychiatry. 2008;64:922–929. doi: 10.1016/j.biopsych.2008.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall RG, Duhamel M, McClanahan R, Miles G, Nason C, Rosen S, et al. Level of functioning, severity of illness, and smoking status among chronic psychiatric patients. J Nerv Ment Dis. 1995;183:468–471. doi: 10.1097/00005053-199507000-00008. [DOI] [PubMed] [Google Scholar]
- Hong LE, Yang X, Wonodi I, Hodgkinson CA, Goldman D, Stine OC, et al. A CHRNA5 allele related to nicotine addiction and schizophrenia. Gene Brain Behav. 2011;10:530–535. doi: 10.1111/j.1601-183X.2011.00689.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalman D, Morrissette SB, George TP. Co-morbidity of smoking in patients with psychiatric and substance use disorders. Am J Addict. 2005;14:106–123. doi: 10.1080/10550490590924728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendler KS, O’Neill FA, Burke J, Murphy B, Duke F, Straub RE, et al. Irish study on high-density schizophrenia families: field methods and power to detect linkage. Am J Med Genet. 1996;67:179–190. doi: 10.1002/(SICI)1096-8628(19960409)67:2<179::AID-AJMG8>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- Kendler KS, Prescott CA, Myers J, Neale MC. The structure of genetic and environmental risk factors for common psychiatric and substance use disorders in men and women. Arch Gen Psychiatry. 2003;60:929–937. doi: 10.1001/archpsyc.60.9.929. [DOI] [PubMed] [Google Scholar]
- Lasser K, Boyd JW, Woolhandler S, Himmelstein DU, McCormick D, Bor DH. Smoking and mental illness: a population-based prevalence study. JAMA. 2000;284:2606–2610. doi: 10.1001/jama.284.20.2606. [DOI] [PubMed] [Google Scholar]
- Leonard S, Mexal S, Freedman R. Smoking, genetics, and schizophrenia: evidence for self medication. J Dual Diagn. 2007;3:43–59. doi: 10.1300/J374v03n03_05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis CM, Levinson DF, Wise LH, DeLisi LE, Straub RE, Hovatta I, et al. Genome scan meta-analysis of schizophrenia and bipolar disorder, part II: schizophrenia. Am J Hum Genet. 2003;73:34–48. doi: 10.1086/376549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MD. The genetics of nicotine dependence. Curr Psychiatry Rep. 2006;8:158–164. doi: 10.1007/s11920-006-0016-0. [DOI] [PubMed] [Google Scholar]
- Li MD, Xu Q, Lou XY, Payne TJ, Niu T, Ma JZ. Association and interaction analysis of variants in CHRNA5/CHRNA3/CHRNB4 gene cluster with nicotine dependence in African and European Americans. Am J Med Genet B Neuropsychiatr Genet. 2010;153B:745–756. doi: 10.1002/ajmg.b.31043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenstein P, Yip BH, Björk C, Pawitan Y, Cannon TD, Sullivan PF, et al. Common genetic determinants of schizophrenia and bipolar disorder in Swedish families: a population-based study. Lancet. 2009;373:234–239. doi: 10.1016/S0140-6736(09)60072-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lips EH, Gaborieau V, McKay JD, Chabrier A, Hung RJ, Boffetta P, et al. Association between a 15q25 gene variant, smoking quantity and tobacco-related cancers among 17000 individuals. Int J Epidemiol. 2010;39:563–577. doi: 10.1093/ije/dyp288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JZ, Tozzi F, Waterworth DM, Pillai SG, Muglia P, Middleton L, et al. Meta-analysis and imputation refines the association of 15q25 with smoking quantity. Nat Genet. 2010;42:436–440. doi: 10.1038/ng.572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons MJ, Bar JL, Kremen WS, Toomey R, Eisen SA, Goldberg J. Nicotine and familial vulnerability to schizophrenia: a discordant twin study. J Abnorm Psychol. 2002;111:687–693. doi: 10.1037//0021-843x.111.4.687. [DOI] [PubMed] [Google Scholar]
- Magi R, Morris AP. GWAMA: software for genome-wide association meta-analysis. BMC Bioinformatics. 2010;11:288. doi: 10.1186/1471-2105-11-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueser KT, Yarnold PR, Levinson DF, Singh H, Bellack AS, Kee K, et al. Prevalence of substance abuse in schizophrenia: demographics and clinical correlates. Schizophr Bull. 1990;16:31–54. doi: 10.1093/schbul/16.1.31. [DOI] [PubMed] [Google Scholar]
- Nyholt DR. A simple correction for multiple testing for SNPs in linkage disequilibrium with each other. Am J Hum Genet. 2004;74:765–769. doi: 10.1086/383251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patkar AA, Gopalakrishnan R, Lundy A, Leone FT, Certa KM, Weinstein SP. Relationship between tobacco smoking and positive and negative symptoms in schizophrenia. J Nerv Ment Dis. 2002;190:604–610. doi: 10.1097/00005053-200209000-00005. [DOI] [PubMed] [Google Scholar]
- Petrovsky N, Quednow BB, Ettinger U, Schmechtig A, Mössner R, Collier DA, et al. Sensorimotor gating is associated with CHRNA3 polymorphisms in schizophrenia and healthy volunteers. Neuropsychopharmacology. 2010;35:1429–1439. doi: 10.1038/npp.2010.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, et al. PLINK: a toolset for whole-genome association and population-based linkage analysis. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley B, Kuo PH, Maher BS, Fanous AH, Sun J, Wormley B, et al. The dystrobrevin binding protein 1 (DTNBP1) gene is associated with schizophrenia in the Irish Case Control Study of Schizophrenia (ICCSS) sample. Schizophr Res. 2009;115:245–253. doi: 10.1016/j.schres.2009.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccone NL, Saccone SF, Hinrichs AL, Stitzel JA, Duan W, Pergadia ML, et al. Multiple distinct risk loci for nicotine dependence identified by dense coverage of the complete family of nicotinic receptor subunit (CHRN) genes. Am J Med Genet B Neuropsychiatr Genet. 2009;150B:453–466. doi: 10.1002/ajmg.b.30828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheet P, Stephens M. A fast and flexible statistical model for large-scale population genotype data: applications to inferring missing genotypes and haplotypic phase. Am J Hum Genet. 2006;78:629–644. doi: 10.1086/502802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao L, Vawter MP. Shared gene expression alterations in schizophrenia and bipolar disorder. Biol Psychiatry. 2008;64:89–97. doi: 10.1016/j.biopsych.2007.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherva R, Kranzler HR, Yu Y, Logue MW, Poling J, Arias AJ, et al. Variation in nicotinic acetylcholine receptor genes is associated with multiple substance dependence phenotypes. Neuropsychopharmacology. 2010;35:1921–1931. doi: 10.1038/npp.2010.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strakowski SM, Tohen M, Flaum M, Amador X. Substance abuse in psychotic disorders: associations with affective syndromes. Schizophr Res. 1994;14:73–81. doi: 10.1016/0920-9964(94)90011-6. [DOI] [PubMed] [Google Scholar]
- Sullivan PF, Kendler KS, Neale MC. Schizophrenia as a complex trait: evidence from a meta analysis of twin studies. Arch Gen Psychiatry. 2003;60:1187–1192. doi: 10.1001/archpsyc.60.12.1187. [DOI] [PubMed] [Google Scholar]
- Sullivan PF, Lin D, Tzeng JY, van den OE, Perkins D, Stroup TS, et al. Genomewide association for schizophrenia in the CATIE study: results of stage 1. Mol Psychiatry. 2008;13:570–584. doi: 10.1038/mp.2008.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tidey JW, Rohsenow DJ, Kaplan GB, Swift RM. Cigarette smoking topography in smokers with schizophrenia and matched non-psychiatric controls. Drug Alcohol Depend. 2005;80:259–265. doi: 10.1016/j.drugalcdep.2005.04.002. [DOI] [PubMed] [Google Scholar]
- Tobacco and Genetics (TAG) Consortium. Genome-wide meta-analyses identify multiple loci associated with smoking behavior. Nat Genet. 2010;42:441–447. doi: 10.1038/ng.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuang MT, Lyons MJ, Meyer JM, Doyle T, Eisen SA, Goldberg J, et al. Cooccurrence of abuse of different drugs in men: the role of drug-specific and shared vulnerabilities. Arch Gen Psychiatry. 1998;55:967–972. doi: 10.1001/archpsyc.55.11.967. [DOI] [PubMed] [Google Scholar]
- Vanable PA, Carey MP, Carey KB, Maisto SA. Smoking among psychiatric outpatients: relationship to substance use, diagnosis, and illness severity. Psychol Addict Behav. 2003;17:259–265. doi: 10.1037/0893-164X.17.4.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JC, Cruchaga C, Saccone NL, Bertelsen S, Liu P, Budde JP, et al. Risk for nicotine dependence and lung cancer is conferred by mRNA expression levels and amino acid change in CHRNA5 . Hum Mol Genet. 2009;18:3125–3135. doi: 10.1093/hmg/ddp231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ware JJ, van den Bree MB, Munafò MR. Association of the CHRNA5-A3-B4 gene cluster with heaviness of smoking: a meta-analysis. Nicotine Tob Res. 2011;13:1167–1175. doi: 10.1093/ntr/ntr118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster JA, Gibbs JR, Clarke J, Ray M, Zhang W, Holmans P, et al. Genetic control of human brain transcript expression in Alzheimer disease. Am J Hum Genet. 2009;84:445–458. doi: 10.1016/j.ajhg.2009.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winterer G. Why do patients with schizophrenia smoke? Curr Opin Psychiatry. 2010;23:112–119. doi: 10.1097/YCO.0b013e3283366643. [DOI] [PubMed] [Google Scholar]
- Winterer G, Mittelstrass K, Giegling I, Lamina C, Fehr C, Brenner H, et al. Risk gene variants for nicotine dependence in the CHRNA5–CHRNA3– CHRNB4 cluster are associated with cognitive performance. Am J Med Genet B Neuropsychiatr Genet. 2010;153B:1448–1458. doi: 10.1002/ajmg.b.31126. [DOI] [PubMed] [Google Scholar]
- World Health Organization. WHO report on the global tobacco epidemic, 2008: the MPOWER package. Geneva: World Health Organization Press; 2008. [Google Scholar]
- Zhang H, Kranzler HR, Poling J, Gelernter J. Variation in the nicotinic acetylcholine receptor gene cluster CHRNA5–CHRNA3–CHRNB4 and its interaction with recent tobacco use influence cognitive flexibility. Neuropsychopharmacology. 2010;35:2211–2224. doi: 10.1038/npp.2010.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.