Abstract
Aberrant platelet-derived growth factor receptor-α (PDGFRα) signaling is evident in a subset of hepatocellular cancers (HCCs). However, its role and regulation in hepatic physiology remains elusive. In the current study, we examined PDGFRα signaling in liver development and regeneration. We identified notable PDGFRα activation in hepatic morphogenesis that, when interrupted by PDGFRα-blocking antibody, led to decreased hepatoblast proliferation and survival in embryonic liver cultures. We also identified temporal PDGFRα overexpression, which is regulated by epidermal growth factor (EGF) and tumor necrosis factor α, and its activation at 3 to 24 hours after partial hepatectomy. Through generation of hepatocyte-specific PDGFRA knockout (KO) mice that lack an overt phenotype, we show that absent PDGFRα compromises extracelluar signal-regulated kinases and AKT activation 3 hours after partial hepatectomy, which, however, is alleviated by temporal compensatory increases in the EGF receptor (EGFR) and the hepatocyte growth factor receptor (Met) expression and activation along with rebound activation of extracellular signal-regulated kinases and AKT at 24 hours. These untimely increases in EGFR and Met allow for normal hepatocyte proliferation at 48 hours in KO, which, however, are aberrantly prolonged up to 72 hours. Intriguingly, such compensation also was visible in primary KO hepatocyte cultures but not in HCC cells after siRNA-mediated PDGFRα knockdown. Thus, temporal activation of PDGFRα in liver development is important in hepatic morphogenesis. In liver regeneration, despite increased signaling, PDGFRα is dispensable owing to EGFR and Met compensation, which is unique to normal hepatocytes but not HCC cells.
Platelet-derived growth factor receptor-α (PDGFRα) is a receptor tyrosine kinase (RTK) expressed chiefly on mesenchymal cells including fibroblasts and smooth muscle cells.1–3 In addition, it also is expressed on other cell types including neurons and endothelial cells. Its activation is elicited by PDGFs, especially AA and CC, which induce effects on growth, motility, and survival, thus regulating the function of these cells.4 After engagement, PDGFRα tyrosine phosphorylation can occur at diverse residues to elicit activation of distinct downstream effectors. Specifically relevant are downstream activation of phosphatidylinositide 3-kinases and AKT, as well as ERK signaling.5–7
Based on gene array studies using RNA from livers at different stages of gestational development in mice, our laboratory previously reported that PDGFRα expression was at its highest during early stages, especially around embryonic day 10 (E10) to E12. This coincided with the time of peak hepatoblast proliferation, after which the PDGFRα levels gradually decreased to low levels.8 In an adult liver, only low PDGFRα expression is evident, however, its expression is increased dramatically in a significant subset of hepatocellular carcinomas (HCCs) and its inhibition in human HCC cells leads to reduced tumor cell proliferation and viability.8,9
It is thus pertinent to investigate further the role and regulation of PDGFRα in liver growth. In the current study, we investigated the role of this RTK in two major models of hepatic growth. Liver development was characterized by regulated hepatic growth and differentiation of bipotential progenitors or hepatoblasts that compose the early hepatic bud at approximately E9.5 in mice showed temporal proliferation and resistance to apoptosis, eventually leading to expansion of the primitive liver bud.10 During the later stages of hepatic morphogenesis, additional molecular cues direct the differentiation of hepatic progenitors to either hepatocytes or cholangiocytes. Similarly, liver regeneration (LR) after partial hepatectomy (PHx) is a widely used model to study the importance of signaling molecules in hepatic growth. The process of LR requires an orderly interplay between many cell types and several signaling pathways.11,12 The cellular and molecular mechanisms responsible for LR show significant redundancy to allow completion of the process as shown by studies in genetic models or after chemical intervention.
In the current study, we used developing livers from various gestational stages to verify the expression of PDGFRα in hepatoblasts and show its activation temporally during early hepatic development owing to the presence of its ligands. By using previously characterized embryonic liver cultures,13,14 we interfered with PDGFRα signaling through the use of a recently characterized mouse-specific PDGFRα blocking antibody15 to address its role in hepatoblast proliferation and survival. We also show that adult murine hepatocytes indeed express PDGFRα, albeit at low levels. However, after PHx, we observed temporal up-regulation and activation of PDGFRα and we addressed its regulation during this process. Through generation of hepatocyte-specific PDGFRα knockout mice (KO), by interbreeding floxed PDGFRα16 and albumin-cre animals,17 we show that its conditional loss from hepatocytes is well tolerated. When subjected to PHx, LR proceeds uneventfully as a result of compensatory increases in the expression and activation of epidermal growth factor receptor (EGFR) and the hepatocyte growth factor receptor, Met. Such redundancy is unique to LR, but not HCC, growth, making PDGFRα an attractive therapeutic target. Thus, we show an important role of PDGFRα in various aspects of liver growth and development.
Materials and Methods
Generation of Pdgfra Conditional Knockout Mice
Homozygous Pdgfra floxed (exons 1 to 4) and albumin-Cre mice (both on a C57BL/6 background) were obtained from Jackson Laboratories (Bar Harbor, ME). Homozygous floxed Pdgfra mice were bred to albumin-Cre mice and the offspring carrying a floxed Pdgfra allele and albumin-Cre then were bred to the homozygous floxed Pdgfra mice. The mice with the floxed and floxed-deleted allele of Pdgfra henceforth are referred to as Pdgfraloxp/loxp;Alb-Cre+/− or KO mice and all other genotypes including Pdgfraloxp/loxp;Alb-Cre−/− and Pdgfraloxp/Wt;Alb-Cre−/− or Pdgfraloxp/loxp;Alb-Cre−/− are referred to as wild-type (WT) controls.
Animal Studies
All experiments on mice were performed under the strict guidelines of the NIH and the Institutional Animal Use and Care Committee at the University of Pittsburgh. Timed pregnant mice were used to isolate embryos from the E10 stage of gestational development. En bloc liver from each embryo was isolated by atraumatic dissection using an Olympus (Center Valley, PA) stereomicroscope and cultured as an organ as described previously.13,14 Five livers were cultured in 4% serum and five livers were cultured in the presence of 50 nmol/L 1E10-IMC, a mouse PDGFRα-blocking antibody, which was a kind gift from Dr. Nick Loizos (ImClone Systems Corp., New York, NY) and has been reported recently to block signaling at this concentration.15 Livers were cultured for 72 hours with fresh media containing blocking antibody or 4% serum alone, replacing the spent media every 24 hours. Livers were fixed in 10% formalin for paraffin embedding and histologic characterization.
Time pregnant mice also were used to obtain embryos (n = 3) from the E12 stage and for fixing in OCT compound for cryosections.
Eight- to 12-week-old female KO and littermate female controls were subjected to PHx as described previously.18,19 Mice (n > 3 per genotype) were sacrificed at specified time points after surgery as indicated in the Results section and in Figures 3, 5, and 6).
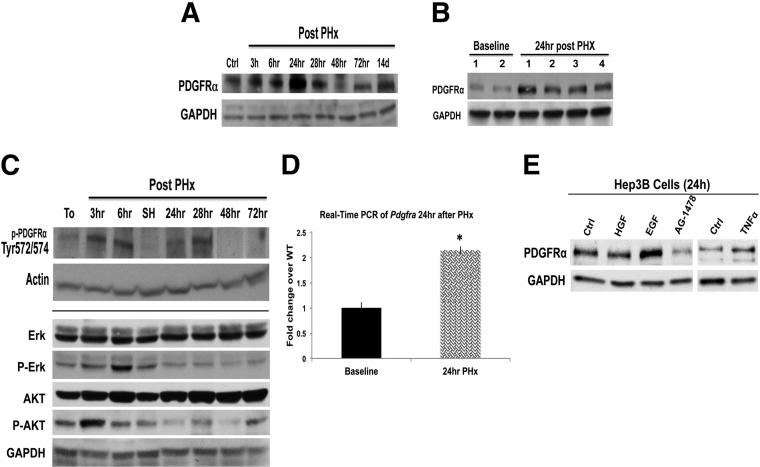
Figure 3.
Temporal increase in PDGFRα expression and activation during LR after PHx. A: Representative WB from pooled livers showed increased PDGFRα protein expression after PHx, with peak expression evident at 24 hours after PHx. GAPDH verifies comparable loading. B: WB using whole-cell lysates from 4 individual animals showed a notable increase in total PDGFRα at 24 hours after PHx as compared with normal livers at baseline, whereas GAPDH showed comparable loading. C: Representative WB shows activation of PDGFRα (phosphorylated at Tyr 572 and 574), ERK (phosphorylated at Thr202 and Tyr204 of ERK1 or Thr185 and Tyr187 of ERK2), and AKT (phosphorylated at Thr308) at various time points after PHx using a site-specific PDGFRα. SH, sham surgery sample. D: Real-time PCR showing a twofold increase in Pdgfra 24 hours after PHx (P = 0.0001). Lysates from Hep3B cells that were treated with various growth factors for 24 hours show increased PDGFRα expression in response to EGF and TNFα treatment. E: The increase by EGF was abrogated by concomitant use of EGFR inhibitor AG-1478. GAPDH depicts equal loading. Ctrl, control; GAPDH, glyceraldehyde-3-phosphate dehydrogenase. ∗P < 0.05.
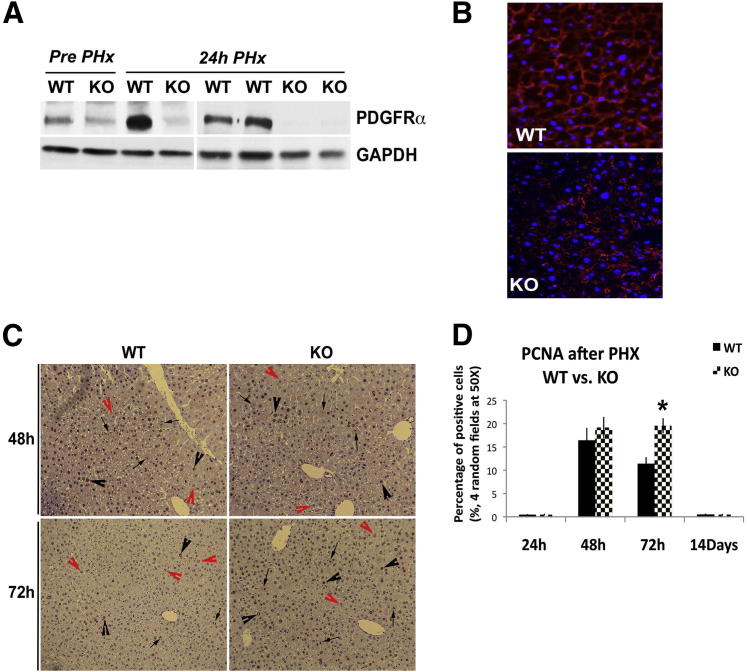
Figure 5.
Continued proliferation in PDGFRα KO during LR after PHx. A: Representative WB verifies abrogation of the surge in PDGFRα expression in KO livers at 24 hours after PHx as compared with WT. B: Immunofluorescence shows PDGFRα (red) localizing to the hepatocyte membrane at 24 hours after PHx in WT liver, whereas it is localized to nonparenchymal cells and is conspicuously absent from hepatocyte membranes in KO. Original magnification: ×400. C: PCNA IHC in WT liver and KO liver at 48 and 72 hours after PHx shows enhanced nuclear staining (black arrowheads) and mitosis (arrows) in KO, especially at 72 hours. PCNA-negative hepatocytes are indicated by red arrowheads. Original magnification: ×100. D: Bar graph of PCNA index of WT and KO livers shows comparable cells in the S-phase at 48 hours whereas a significant increase is observed in KO at 72 hours. ∗P < 0.05. GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
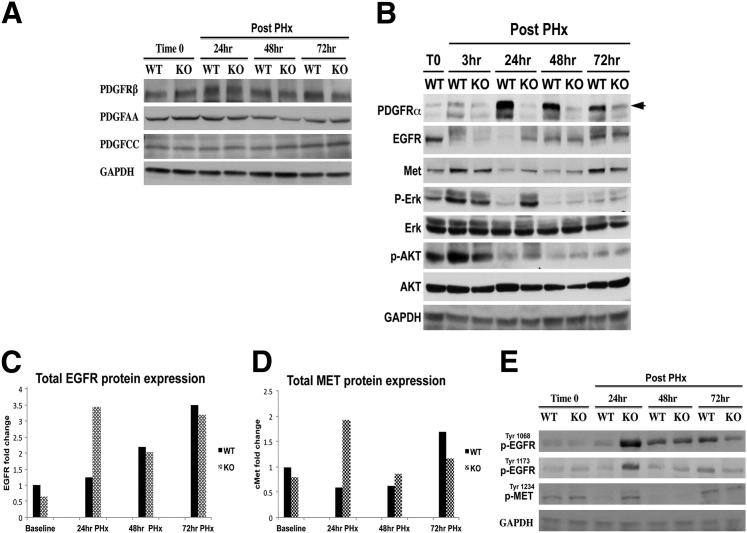
Figure 6.
Loss of hepatocyte Pdgfra induces a temporal increase in EGFR and Met expression and activation at 24 hours after PHx. A: Representative WB from pooled livers showed no changes in protein expression of PDGFRβ, PDGF-AA, or PDGF-BB at various time points after PHx in KO when compared with WT. B: Representative WB shows decreased p-AKT and p-ERK in KO compared with WT at 3 hours during LR. Enhanced EGFR and MET protein expression in KO livers at 24 hours after PHx coincides with increased p-AKT and p-ERK at this time in KO compared with WT. C: Quantification of changes in EGFR protein after PHx shows a 2.5-fold increase in KO livers at 24 hours after PHx, normalized to baseline WT. D: Quantification of changes in MET protein levels after PHx shows a twofold increase over baseline, normalized to baseline WT. E: Representative WB shows enhanced protein expression of phosphorylated forms of EGFR and MET as indicated, especially at 24 hours after PHx. GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Blood from male or female KO mice along with sex- and age-matched WT controls was collected at 3 to 10 months (Table 1) for serum biochemistry. Livers were collected from these mice for baseline analysis.
Table 1.
Serum Biochemistry from KO and WT Mice Depicts No Changes Compared with Controls
| Genotype | Sex | Age, months | Total bilirubin level, mg/dL | Aspartate transaminase level, IU/L (normal = 5–40) | Alanine transaminase level, IU/L (normal = 7–56) |
|---|---|---|---|---|---|
| WT | M | 3 | 0.3 | 70 | 31 |
| WT | F | 8 | 0.2 | 53 | 16 |
| WT | F | 8 | 0.3 | 44 | 20 |
| KO | F | 3 | 0.4 | 87 | 23 |
| KO | F | 3 | 0.2 | 82 | 25 |
| KO | M | 3 | 0.3 | 117 | 48 |
| KO | F | 10 | 0.1 | 59 | 16 |
| KO | F | 10 | 0.2 | 61 | 15 |
| KO | F | 8 | 0.3 | 160 | 30 |
F, female; M, male.
Baseline and regenerating livers were harvested and stored at −80°C until use. Livers also were fixed in 10% formalin to be used for paraffin embedding or placed in OCT compound for cryosections.
Western Blot Analysis
Multiple livers were isolated from embryos from each developmental stage [E11 to E19, postnatal day (P)1, and adult] and pooled (n > 10 for E11 to E14; n > 3 for E15 to adult) for whole-cell lysate extraction in radio immunoprecipitation assay buffer. Whole-cell lysates were prepared from regenerating liver tissue and from cell cultures and prepared from independent or pooled (n ≥ 3) livers in radio immunoprecipitation assay buffer for assessment by WB as discussed elsewhere.18 After autoradiography, the films were scanned to obtain integrated optic densitometry using NIH ImageJ software version 1.62. The average integrated optic densitometry for a protein was compared between the KO and WT groups and assessed for statistical significance by the Student’s t-test, and a P value less than 0.05 was considered significant.
Primary antibodies used for WB included the following: EGFR (1:200), PDGFRα (1:200), phospho-PDGFRα Tyr720 (1:100), PDGFRβ (1:300), PDGFA (1:200), PDGFB (1:200), PDGFC (1:200), Met (1:200), and glyceraldehyde-3-phosphate dehydrogenase (1:1000), all from Santa Cruz Biotechnology (Dallas, TX); phospho-PDGFRα Tyr572/574 (1:800), phospho-PDGFRα Tyr849 (1:800), and phospho-PDGFRα Tyr742 (1:800) from Invitrogen (Grand Island, NY); actin (1:2000) from Millipore (Billerica, MA); and total and p-ERK (Thr202/Tyr204 of ERK1 or Thr185/Tyr187 of ERK2) (1:1000), and total and p-AKT Thr308 (1:1000) from Cell Signaling (Danvers, MA).
Real-Time PCR
RNA was extracted from livers using TRIzol (Invitrogen) as per the manufacturer’s instructions. RNA from each sample was reverse transcribed with the SuperScript III First-Strand Synthesis System for real-time PCR with an RNase H treatment (Invitrogen). In addition, equal amounts of RNA from three age- and sex-matched KO mouse livers were pooled to make cDNA. A total of 0.1 μg cDNA along with Power SYBR-Green PCR Master Mix (Applied Biosystems; Grand Island, NY) and the appropriate primers (see following paragraph) were used for each real-time PCR reaction. The samples were analyzed for each condition in triplicate to enable statistical analysis. The Applied Biosystems StepOnePlus Real-Time PCR System was used for the transcript analysis with StepOne software version 2.1. Comparative ΔΔCT was used for data analysis, and calculations were made without the StepOne software.
The real-time PCR analysis was performed with a liver-specific reference gene (cyclophilin-A). Primer efficiencies were performed for each primer, and only similar efficiencies were used for analysis. The following real-time PCR primers were designed using Primer-BLAST (NIH): mouse Pdgfra forward: 5′-TCCTTCTACCACCTCAGCGAG-3′; mouse Pdgfra reverse: 5′-CCGGATGGTCACTCTTTAGGAAG-3′; mouse cyclophilin-A forward: 5′-CCCCACCGTGTTCTTCGACA-3′; mouse cyclophilin-A reverse: 5′-TCCAGTGCTCAGAGCTCGAAA-3′.
Histology, Immunohistochemistry, and Immunofluorescence
Four-micron sections from paraffin-embedded tissues were subjected to immunohistochemistry (IHC) for hepatocyte nuclear factor-4α (HNF4α), proliferating cell nuclear antigen (PCNA), or TUNEL, as described previously.18 PCNA-positive hepatocytes were counted using an Axioskop 40 (Carl Zeiss, Cambridge, UK) upright research microscope in four randomly selected fields per section at a magnification of ×400. PCNA counts between WT and KO livers after PHx were compared for statistical significance by the Student’s t-test, with a P value of less than 0.05 considered significant.
For immunofluorescence, cryosections were fixed for 10 minutes in 4% paraformaldehyde, washed in PBS, and blocked in 2% bovine serum albumin (BSA) for 45 minutes. Rabbit polyclonal PDGFRα antibody (1:40; Santa Cruz Biotechnology) and 1:300 mouse monoclonal anti-HNF4α antibody (Perseus Proteomics, Tokyo, Japan) in 0.5% BSA were incubated for 1 hour followed by washes in 0.5% BSA. Next, Cy3-conjugated anti-rabbit antibody and Alexa488-conjugated anti-mouse (Molecular Probes, Grand Island, NY) were applied at 1:1000 and 1:500 dilutions, respectively, in 0.5% BSA for 30 minutes. Washes were repeated in 0.5% BSA in PBS and incubated in DAPI for 45 seconds. Sections were coverslipped in Gelvatol and visualized under a Zeiss Axioscope microscope.
Cell Culture and Treatment
HBC-3, a hepatoblast cell line, was a kind gift from Dr. Leslie Rogler (Albert Einstein College of Medicine, Yeshiva University, Bronx, NY) and was cultured without inducing differentiation for obtaining whole-cell lysates as described elsewhere.20
Human hepatoma cells (Hep3B; ATCC, Manassas, VA) cultured in serum-free Eagle's minimum essential medium were treated with 20 ng/mL EGF (BD, Durham NC), 40 ng/mL hepatocyte growth factor (Snow Brand, Japan), 25 ng/mL tumor necrosis factor α (TNFα) (R&D Systems, Minneapolis MN), and 3 μmol/L/mL EGFR inhibitor (SelleckBio, Houston TX) for 24 hours. Cells were washed and lysed in radio immunoprecipitation assay buffer and assessed by WB. Hep3B cells also were used for siRNA knockdown of PDGFRα. Cells were cultured in six-well plates in Eagle's minimum essential medium with 10% fetal bovine serum. Cells were serum-starved for synchronization and transfected with 30 nmol/L PDGFRA siRNA (Ambion, Austin, TX) or control for 6 hours, followed by the addition of 1 mL Eagle's minimum essential medium media with 4% fetal bovine serum. Cells were harvested at 24 hours and 48 hours for WB.
A modified two-step collagenase perfusion was used to isolate hepatocytes from WT and KO mice for hepatocyte culture as described previously.18 Freshly isolated hepatocytes from KO and WT mice were cultured for 2 hours in MEM containing 10% serum, followed by MEM containing insulin, selenium, transferrin, dexamethasone, hepatocyte growth factor, and epidermal growth factor for 24 hours, and then harvested for whole-cell lysates in radio immunoprecipitation assay buffer for WB analysis.
Results
Increased Expression and Activation of PDGFRα during Early Hepatic Development
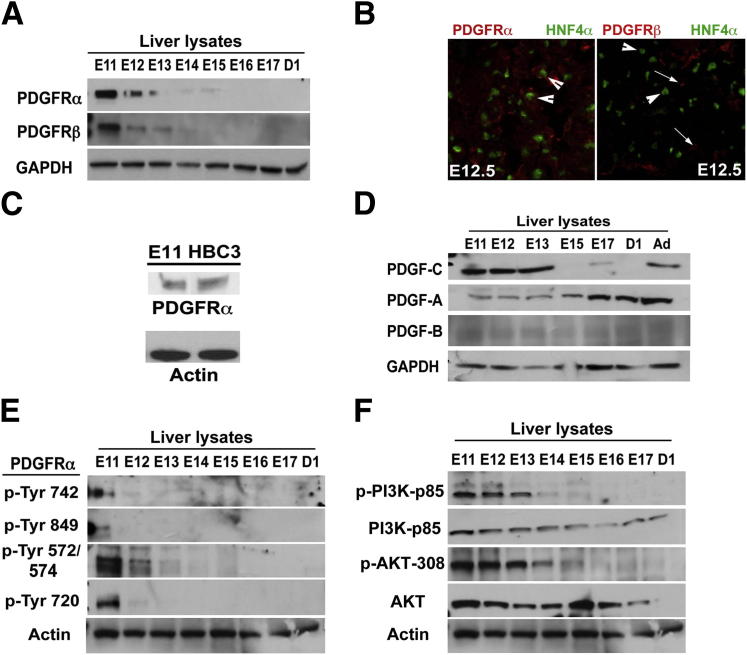
To validate previous findings,8 we used whole-cell lysates from pooled livers from various prenatal developmental stages in mice to perform WB analysis for PDGFRα and PDGFRβ. PDGFRα is expressed at high levels in E11 and E12 livers, with a gradual decrease in protein expression (Figure 1A). PDGFRβ also was expressed similarly (Figure 1A). To verify whether PDGFRα was expressed in the epithelial cell compartment of developing liver, we performed double immunofluorescence for PDGFRα and HNF4α, a known marker for both hepatoblasts and hepatocytes. Membranous and cytoplasmic localization of PDGFRα was evident in several HNF4α-positive cells at the E12.5 stage (Figure 1B). To address PDGFRβ localization, we examined its co-localization with HNF4α, but could not detect any at E12.5 (Figure 1B). To further substantiate the expression of PDGFRα in hepatoblasts, we tested HBC-3, a hepatoblast cell line, which showed comparable PDGFRα expression with E11 murine livers (Figure 1C). Next, we tested for the presence of ligands that may be responsible for engaging PDGFRα during early stages. Interestingly, although PDGF-CC was the predominant ligand at early stages, PDGF-AA became more prominent at E17 and beyond, however, PDGF-CC levels deteriorated (Figure 1D). PDGF-BB appears to be expressed minimally throughout liver development.
Figure 1.
PDGFRα expression and activation in early liver development. A: A representative WB shows the highest expression of PDGFRα at E11 in liver lysates with a gradual decrease over the following days of gestational development in mice. B: Left panel: A representative double immunofluorescence with PDGFRα (red) expressed in HNF4α-positive hepatoblasts (green) cells in E12.5 murine liver (arrowheads). Right panel: PDGFRβ (red) expressed in non–HNF4α-positive cells (arrows), whereas HNF4α-positive (green) hepatoblasts (arrowheads) are negative for PDGFRβ. Original magnification: ×400. C: WB verifies PDGFRα expression in the HBC3 hepatoblast cell line and E11 liver whole-cell lysate. D: WB shows high PDGF-CC, but not PDGF-AA, during early liver development, whereas PDGF-AA levels increase at later stages. E: WB shows PDGFRα phosphorylation at multiple sites, indicating its activation in embryonic livers at early stages of hepatic development. F: WB shows tyrosine phosphorylation of phosphatidylinositide 3-kinase (PI3K)-p85 and AKT, depicting activation at corresponding early stages of hepatic development. Ad, adult; D1, postnatal day 1; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
To address whether PDGFRα signaling is ongoing during hepatic development, we tested whole- cell lysates for phosphorylation of PDGFRα at specific residues. We identified several sites to be phosphorylated in PDGFRα, prominently at Tyr-572/574, Tyr-720, and Tyr-742 at the E11 to E12 stages, whereas Tyr-849 showed only limited phosphorylation at the E11 stage (Figure 1E). To test whether some of the associated downstream signaling may be active at the corresponding times, we examined the state of phosphatidylinositide 3-kinase and AKT signaling because these have been shown to be regulated by PDGFRα, especially when they are phosphorylated at Tyr-742 and other RTKs as well.5 Although total phosphatidylinositide 3-kinase-p85 and AKT levels remained relatively unaltered during hepatic development until after birth, phospho-phosphatidylinositide 3-kinase–p85 and phospho-AKT–Thr308 levels were observed mostly in the liver lysates at the E11 to E13 stages, followed by a decrease at all later stages coinciding with the loss of PDGFRα and its phosphorylated forms (Figure 1F).
Our results therefore suggest that concomitant PDGFRα and PDGF-C expression induces activation of this RTK to contribute to hepatic morphogenesis.
Blockade of PDGFRα Signaling in Embryonic Liver Cultures Reveals Its Role in Hepatoblast Proliferation and Survival
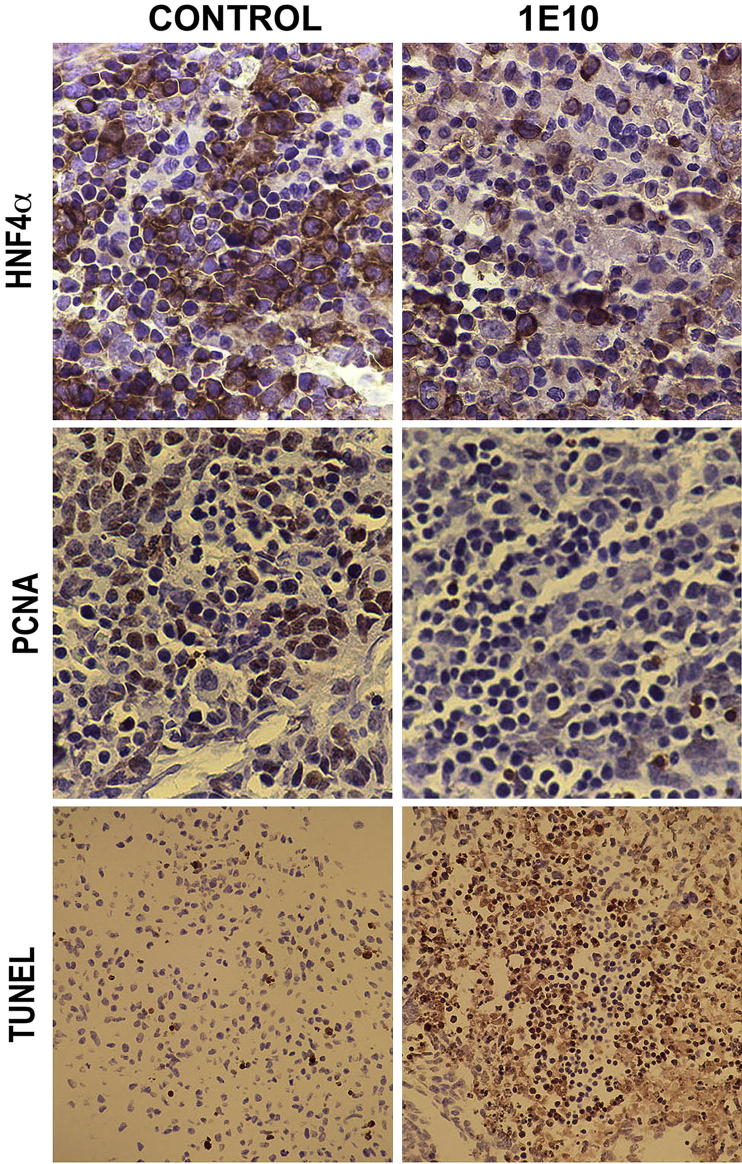
To further investigate the role of PDGFRα signaling during early liver development, we used embryonic liver cultures and PDGFRα-blocking antibody (1E10-IMC, kindly provided by ImClone), whose efficacy and specificity recently were shown.15 Livers isolated from E10 embryos and cultured in the presence of 1E10-IMC and 4% serum as compared with 4% serum alone for 72 hours showed fewer hepatoblasts when examined by IHC for HNF4α (Figure 2). To determine the mechanism of reduced hepatoblasts, we tested the organoid cultures for proliferation and any change in cell viability. The 1E10-IMC–treated embryonic livers showed dramatically fewer cells in the S-phase as indicated by IHC for PCNA, suggesting decreased cell proliferation after 72 hours of PDGFRα blockade (Figure 2). Simultaneously, these livers showed a noteworthy increase in the numbers of TUNEL-positive cells, which is indicative of ongoing apoptosis (Figure 2). Thus, based on the expression of PDGFRα mostly in hepatoblasts, it is apparent that PDGFRα signaling is one of the contributing molecular pathways in hepatoblast proliferation and viability and hence in liver growth during ex vivo hepatic morphogenesis.
Figure 2.
PDGFRα blockade in embryonic liver cultures shows its role in hepatoblast proliferation and survival. Embryonic livers cultured in PDGFRα-blocking 1E10-IMC monoclonal antibody show decreased numbers of HNF4α-positive cells by IHC. In addition, there was a notable decrease in PCNA-positive cells and increase in numbers of TUNEL-positive cells after 1E10-IMC treatment compared to cells grown in 4% serum and shown in representative photomicrographs. Original magnification: HNF4α and PCNA, ×400; TUNEL, ×200.
Temporal Increase in PDGFRα Protein and Activity after PHx
To determine whether PDGFRα may have a role in another model of regulated hepatic growth, we assessed its expression during the process of LR after PHx, especially because hepatocytes are known to secrete PDGFs during this process.21 We examined whole-cell lysates from pooled livers (n ≥ 3) at different times after PHx. A dramatic increase in the PDGFRα protein level was evident at 24 hours during LR, whereas no change in its expression was observed in samples from sham surgery (Figure 3A). To verify this observation further, PDGFRα expression was examined in individual livers from four mice harvested at 24 hours after PHx. All four samples showed a clear increase in PDGFRα expression at this time during LR (Figure 3B). Although no other phospho-specific forms of PDGFRα were detectable (data not shown), increased phosphorylation at Tyr-572/574 was observed in regenerating livers from 3 to 28 hours, whereas the 24-hour sham sample lacked such an increase (Figure 3C). Concomitant to an early increase in phospo (p)PDGFRα, there was a notable increase in both p-ERK and p-AKT, which indicates the role of PDGFRα in regulating the activity of these important mediators of LR (Figure 3C).
To determine the mechanism of enhanced protein expression of PDGFRα, we examined mRNA expression and identified a 2.0-fold increase in Pdgfra in pooled 24-hour livers as compared with prehepatectomy samples (Figure 3D). To address the molecular basis of increased PDGFRα expression after PHx, we examined the effects of known growth factors and cytokines that are known to be critical initiators of LR.12 A human hepatoma cell line, Hep3B, was treated for 24 hours with hepatocyte growth factor, EGF, and TNFα, as indicated in Materials and Methods section, and cells were tested for changes in PDGFRα protein expression by WB. EGF treatment brought about noteworthy increases in PDGFRα levels, which was abrogated by concomitant treatment of cells with an EGFR inhibitor (Figure 3E). TNFα treatment also led to a notable increase in total PDGFRα. Thus, EGF and TNFα induce PDGFRα expression during early LR.
Pdgfra Conditional KO Mice Lack any Overt Phenotype
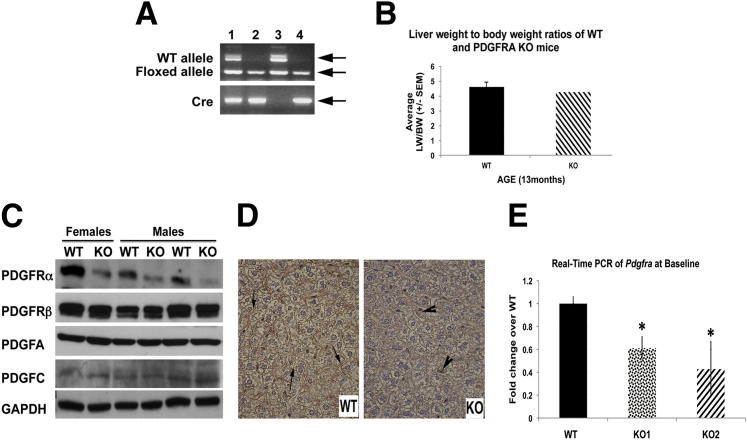
To address the function of PDGFRα during LR, we generated hepatocyte-specific conditional Pdgfra KO mice. Mice were born in a normal Mendelian ratio and did not show any apparent phenotype. Genotype was verified by the presence of a homozygous floxed PDGFRα allele and cre-recombinase by PCR (Figure 4A). The livers from KO and WT livers showed unremarkable gross differences (not shown). No differences were observed in liver weight to body weight ratios between KO and WT mice (Figure 4B), and serum biochemistry showed insignificant differences between WT and KO animals (Table 1). KO livers (≥2 months old) were used for WB, IHC, and real-time PCR. A decrease in total PDGFRα was evident in KO by WB as compared with the control livers (Figure 4C). No changes in PDGFRβ or PDGFRα ligands such as PDGF-AA or PDGF-CC were observed in the KO versus WT in the livers from age- and sex-matched littermates (Figure 4C). Because there was significant remnant expression of PDGFRα in KO livers, we next tested PDGFRα by IHC in KO and WT livers. Indeed, PDGFRα continued to be expressed in nonparenchymal cells in the KO livers as compared with WT controls because albumin-cre will specifically delete the floxed gene from hepatocytes (Figure 4D). Real-time PCR also showed an approximate 50% decrease in PDGFRA expression in KO livers, which was significantly lower than in littermate controls (Figure 4E).
Figure 4.
Generation of PDGFRα KO mice reveal a lack of overt phenotype. A: Genotyping PCR identifies KOs (lanes 2 and 4) by the presence of cre recombinase (lower panel) and floxed PDGFRα allele (242 bp) (upper panel). WT controls were identified by the absence of cre (lane 3) or the presence of cre in animals that harbor the WT PDGFRα allele (451 bp) and floxed allele (lane 1). The arrows point to the correct bands of expected sizes and represent various alleles as noted on the left side of the panel (WT allele, floxed allele, and Cre). B: Graph depicting insignificant change in liver weight to body weight (LW/BW) ratios between WT (n = 6) and KO (n = 5) mice at baseline. C: WB shows a significant decrease in total PDGFRα protein in the KO livers. No changes in the levels of PDGFRβ and PDGFRα ligands including PDGF-AA and PDGF-CC were observed. D: IHC of WT (left) livers show expression of PDGFRα at the hepatocyte membrane (arrows). PDGFRα KO livers (right) show reduced PDGFRα staining, which was localized to nonparenchymal cells (arrowheads). Original magnification: ×400. E: Real-time PCR showing a significant decrease in Pdgfra expression in KO livers (P = 0.0001). ∗P < 0.05.
Pdgfra KO Mice Show Normal Hepatocyte Proliferation after PHx
Eight-week-old or older female littermate KO and WT mice were subjected to PHx. Because peak PDGFRα expression was observed at 24 hours, we first determined these levels at this time in KO mice. KO livers lack any increase in PDGFRα at 24 hours after PHx as compared with WT, which indicates that the predominant increase in PDGFRα protein is in the parenchymal cell compartment (Figure 5A). Immunofluorescence at 24 hours during LR shows PDGFRα expression is dramatically lower in KO mice and is evident in nonparenchymal cells, whereas WT controls show strong membranous localization in hepatocytes (Figure 5B).
The KO mice were followed up after PHx for any morbidity. There appeared to be a temporal-restricted activity in the KO at approximately 24 hours after PHx, however, all KO mice recovered from surgery and were indistinguishable from WT controls at all later time points. Based on the role of PDGFRα in liver development and in HCC,8 we compared hepatocyte proliferation between the two groups of animals during LR. We hypothesized that PDGFRα induction and activation at 24 hours may be one of the upstream signaling cascades regulating hepatocyte proliferation during LR, which usually peaks approximately 48 hours after PHx in C57BL6 mice, although cell-cycle initiation occurs much earlier. However, we found comparable and high numbers of hepatocytes in the S-phase as detected by IHC for PCNA at 48 hours in KO and WT livers (Figure 5, C and D). Intriguingly, at 72 hours, although PCNA staining was expectedly lower in WT controls, KO mice continued to display several hepatocytes in the S-phase (Figure 5, C and D). However, no further growth advantage was evident at later stages and KO and WT mice had a comparable restoration in hepatic mass at 14 days after PHx.
Pdgfra KO Mice Show No Compensatory Changes in PDGF Signaling But Show Enhanced EGFR and MET Expression and Activation during LR
Because of a lack of any defect in cell proliferation in KO mice at 48 hours and in fact a continued proliferation at higher than normal levels at 72 hours, we wondered if any compensatory mechanisms could account for this observation. We focused on 24 to 72 hours, which represents the activation times of the cell cycle and of ongoing cell proliferation during the LR process. First, liver lysates were assessed for PDGF signaling, which was unremarkable for any differences in PDGF-AA, PDGF-CC, and PDGFRβ, which has been shown to compensate for PDGFRα loss in vitro7 (Figure 6A). We next investigated expression for RTKs, such as EGFR and MET, which are major drivers of LR and also are associated with hepatocyte proliferation.12 Interestingly, WB showed increased levels of EGFR and Met protein prominently at 24 hours in the regenerating KO livers (Figure 6B). Quantification after normalization to loading control revealed an approximately 3.5-fold increase in EGFR and a 2.5-fold increase in Met greater than the WT levels at 24 hours (Figure 6, C and D). To address the state of RTK signaling, we assessed lysates for phosphorylation of EGFR and MET. Increased levels of Tyr1068-EGFR, Tyr1173-EGFR, and Tyr1234-Met were observed in lieu of PDGFRα activation at 24 hours after PHx in KO livers (Figure 6E). Thus, the anomalous increases in EGFR and Met expression and activity at 24 hours ensure hepatocyte proliferation at 48 hours in PDGFRα KO that is comparable with WT, but the aberration eventually became apparent at 72 hours when proliferation in WT receded whereas KO hepatocytes continued to proliferate, albeit temporally and without chronic consequences.
Inhibition of PDGFRα Signaling Induces EGFR or Met Expression in Vitro in Hepatocytes But Not Hepatoma Cells
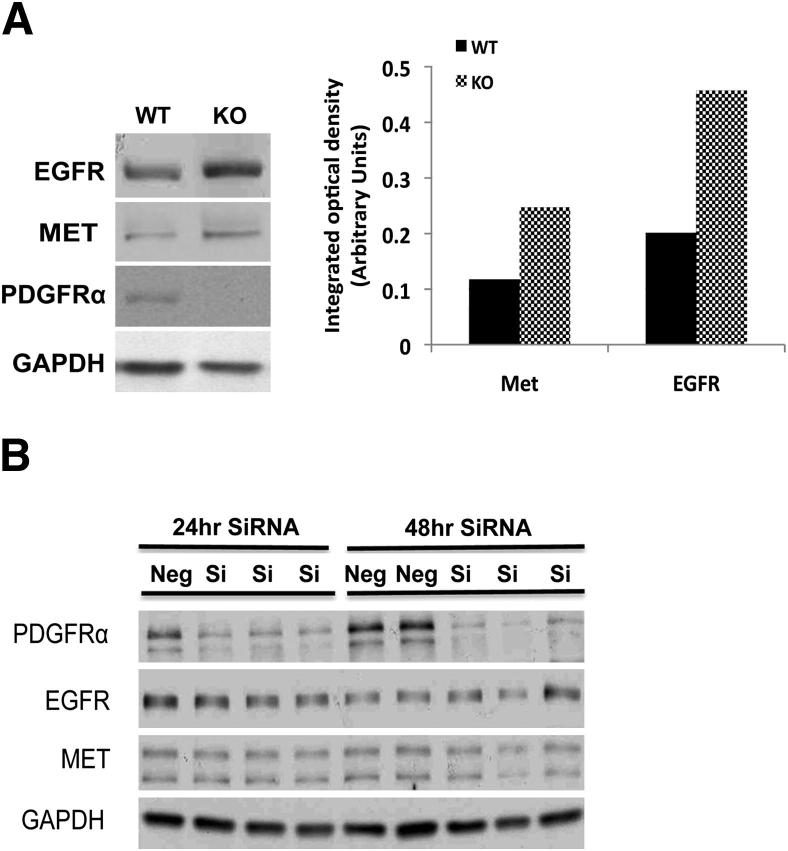
To determine whether EGFR and Met are induced globally on PDGFRα inhibition, we first tested the impact of PDGFRα loss in primary hepatocytes, which were isolated by collagenase perfusion from WT and KO mice and cultured for 24 hours as described in Materials and Methods. A notable increase in total EGFR and Met expression was evident in KO hepatocytes as compared with WT hepatocytes after 24 hours in culture (Figure 7A).
Figure 7.
EGFR and Met expression is induced after PDGFRα loss in normal hepatocytes but not in HCC cells. A: Primary hepatocytes isolated from age- and sex-matched WT and KO livers in culture for 24 hours show increased expression of EGFR and Met in KO. Densitometric analysis on the representative WB shows at least a 2.0-fold increase in Met and a 2.5-fold increase in EGFR levels in the KO hepatocytes. B: A representative WB shows that PDGFRα siRNA but not control-siRNA transfection of Hep3B cells leads to a notable decrease in PDGFRα expression, at both 24 hours and 48 hours. No changes in EGFR or Met were detectable at either time point. GAPDH, glyceraldehyde-3-phosphate dehydrogenase; Si, silencing.
Next, we tested the impact of PDGFRα knockdown in hepatoma cells on EGFR and Met expression. After transfection of PDGFRα siRNA, but not control siRNA, for either 24 hours or 48 hours, a robust PDGFRα decrease occurred in Hep3B cells (Figure 7A). However, no change in either EGFR or Met protein levels were observed at the corresponding times, indicating that only primary hepatocytes but not HCC cells had the capability to enhance the expression of the two RTKs on PDGFRα inhibition.
Discussion
PDGFRα, a traditional serum growth factor receptor for mesenchymal cells, is expressed abundantly in human HCC cells and is known to induce cell proliferation.1,3,4,6,8,9 To address the role and regulation of PDGFRα in normal liver growth, especially in epithelial cells of the liver, we investigated its status in liver development and in a model of surgically induced LR.
Previously, we reported the highest expression of the PDGFRα gene in the earliest stages of hepatic development, which was identified by microarray.8 In the current study, we verified that hepatoblasts co-express HNF4α and PDGFRα. Indeed, HNF4α is a surrogate marker of hepatoblasts and differentiating hepatocytes during hepatic development.22 However, there are other cell types in developing livers that have been reported to express PDGFRα as well. One study reported that PFGFRα is expressed in the submesothelial cell fraction, a precursor of hepatic stellate cells, and yet another group reported that it is expressed in a stromal cell that supports erythropoiesis.23,24 Intriguingly, in the latter study, the investigators identify all PDGFRα-positive cells to express β1-integrin, which also is known to be expressed in hepatocytes during early hepatic development.25 By using previously characterized embryonic liver cultures and a mouse PDGFRα blocking antibody, we showed rampant apoptosis and a decrease in cell proliferation that eventually led to lower numbers of HNF4α-positive cells in the culture, suggesting an important role of PDGFRα in hepatoblast biology. The effect likely is direct owing to RTK blockade of PDGFRα on the hepatoblasts; additional in vivo studies currently are ongoing to generate hepatoblast-specific conditional knockout using the cre-lox approach. Indeed, PDGFRα activation is associated with cell survival and proliferation in several cell types.4,5
However, conditional loss of PDGFRα in hepatocytes during adulthood was compatible with survival. Increased PDGFRα expression and signaling during liver development is highly temporal and is restricted to the early stages of hepatic development only. Based on the in vitro studies, it is apparent that loss of PDGFRα at this time remains uncompensated and yields decreased proliferation and increased apoptosis in embryonic liver cultures. The levels of PDGFRα recede with progressive hepatic development such that adult livers have low expression of PDGFRα. The lack of any overt phenotype in adults lacking hepatocyte-specific PDGFRα may in fact be because Alb-cre deletes targeted genes at 4 to 6 weeks after birth,17 a time when PDGFRα is expressed at low levels and is not actively signaling.
The increase in PDGFRα activation and expression at 3 to 24 hours after PHx is another novel observation. There was an associated increase in AKT and ERK signaling that is known to be important in LR and also shown to be downstream effectors of PDGFRα signaling.7,26,27 The increased expression of PDGFRα at 24 hours appears to be downstream of EGF and TNFα, which are both active during the LR initiation process, especially in the PHx model.11,21 At the same time PDGFRα is activated as evident by its phosphorylation status, which may result from an autocrine mechanism because PDGFs are known to be secreted by hepatocytes during LR, although no changes in their total protein is observed during LR.21 To address its role, we generated PDGFRα-conditional KO that lacked PDGFRα in hepatocytes. These mice showed no overt phenotype and this is not surprising because of the normally low expression of PDGFRα in the liver and perhaps because of the existing redundancy in RTK signaling pathways. Also, remnant expression of PDGFRα in KO is caused by its presence in nonparenchymal cells such as endothelial cells and hepatic stellate cells.23,28 When the KO mice were subjected to PHx, we observed a notable decrease in PDGFRα expression at 3 and 24 hours, supporting the increased expression and activity of PDGFRα in hepatocytes during the early LR period. Simultaneously, a decrease in p-ERK and AKT activity was evident at 3 hours in the KO. Intriguingly, however, increased p-AKT and p-ERK levels were evident in the KO at 24 hours, which corresponded to increased EGFR and Met expression over the WT levels. Met and EGFR are paramount in normal LR and in regulating hepatocyte proliferation.29,30 This excessive EGFR and Met expression and activity in the absence of PDGFRα enabled comparable hepatocyte proliferation at 48 hours in KO and WT. These findings again highlight the existing molecular redundancy that exists in the normal LR process where blockade of a single signaling pathway is readily compensated by an alternate signaling cascade.12,31 However, this untimely and nonscheduled aberration appears to be responsible for transiently prolonging hepatocyte proliferation in PDGFRα in KO at 72 hours, as reflected by increased hepatocytes in the S-phase when compared with WT where proliferation has receded. However, all later time points in KO are unremarkable for any proliferative changes when compared with the WT. These observations also highlight a fine balance that exists between various signaling mechanisms during the process of normal LR.11,12
Intriguingly, however, the inverse relationship between EGFR and PDGFRα also was described in EGFR suppression studies. shRNA-mediated silencing of EGFR during LR led to an increase in both gene and protein expression of PDGFRα.30 Similarly, the deletion of Pdgfra lead to enhanced stimulation of the EGFR pathway in mouse embryonic fibroblasts.7 The exact mechanism of the relationship between PDGFRα and EGFR will need further studies, although EGFR and PDGFRα may be interacting physically, as shown to occur between EGFR and PDGFRβ.32–34
Hepatic fibrosis and cirrhosis as a result of chronic liver diseases are precursors of HCC. Approximately 80% of all HCCs have underlying cirrhosis.35 In fact, chronic liver injury leads to hepatocyte death, inflammation, fibrosis, oxidative stress, and hepatocyte proliferation.36 The regenerating nodules that occupy the cirrhotic liver are considered critical for maintenance of hepatic function, but proliferation in these nodules in a suboptimal environment also makes them prone to genotoxic insult, eventually evolving into dysplastic nodules and HCC. Because several signaling pathways are commonly relevant in LR and HCC, molecular targeting can be a concern in HCC in chronic liver diseases. Thus, it is critical to identify targets that may be dispensable for LR but are critical for HCC. In our current study, we show that PDGFRα signaling, which previously was shown to be a valid, biologically relevant, therapeutic target in HCC,8,9,37 may be dispensable for LR because of redundancy with other RTKs such as EGFR and Met. Intriguingly, such redundancy is unique to normal hepatocytes and not HCC cells because siRNA-mediated PDGFRα knockdown in Hep3B cells did not induce the expression of these RTKs. Although additional studies will be necessary, we propose that selective targeting of PDGFRα in eligible HCC patients may be efficacious, even in chronic liver diseases, without concern that hepatic functions will be impaired. Humanized PDGFRα-blocking antibody is in fact on the horizon for the treatment of various cancers.15,38,39
Footnotes
Supported by NIH grant 1R01DK62277 (S.P.S.M.) and an Endowed Chair for Experimental Pathology (S.P.S.M.), by a Cellular Approaches to Tissue Engineering and Regeneration fellowship (5T32EB001026 to P.K.A.), and by F31 (F31-DK094611 to P.K.A.).
References
- 1.Andrae J., Gallini R., Betsholtz C. Role of platelet-derived growth factors in physiology and medicine. Genes Dev. 2008;22:1276–1312. doi: 10.1101/gad.1653708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Claesson-Welsh L. Platelet-derived growth factor receptor signals. J Biol Chem. 1994;269:32023–32026. [PubMed] [Google Scholar]
- 3.Hoch R.V., Soriano P. Roles of PDGF in animal development. Development. 2003;130:4769–4784. doi: 10.1242/dev.00721. [DOI] [PubMed] [Google Scholar]
- 4.Heldin C.H., Ostman A., Ronnstrand L. Signal transduction via platelet-derived growth factor receptors. Biochim Biophys Acta. 1998;1378:F79–F113. doi: 10.1016/s0304-419x(98)00015-8. [DOI] [PubMed] [Google Scholar]
- 5.Franke T.F., Yang S.I., Chan T.O., Datta K., Kazlauskas A., Morrison D.K., Kaplan D.R., Tsichlis P.N. The protein kinase encoded by the Akt proto-oncogene is a target of the PDGF-activated phosphatidylinositol 3-kinase. Cell. 1995;81:727–736. doi: 10.1016/0092-8674(95)90534-0. [DOI] [PubMed] [Google Scholar]
- 6.Heldin C.H., Westermark B. Mechanism of action and in vivo role of platelet-derived growth factor. Physiol Rev. 1999;79:1283–1316. doi: 10.1152/physrev.1999.79.4.1283. [DOI] [PubMed] [Google Scholar]
- 7.Wu E., Palmer N., Tian Z., Moseman A.P., Galdzicki M., Wang X., Berger B., Zhang H., Kohane I.S. Comprehensive dissection of PDGF-PDGFR signaling pathways in PDGFR genetically defined cells. PLoS One. 2008;3:e3794. doi: 10.1371/journal.pone.0003794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stock P., Monga D., Tan X., Micsenyi A., Loizos N., Monga S.P. Platelet-derived growth factor receptor-alpha: a novel therapeutic target in human hepatocellular cancer. Mol Cancer Ther. 2007;6:1932–1941. doi: 10.1158/1535-7163.MCT-06-0720. [DOI] [PubMed] [Google Scholar]
- 9.Oseini A.M., Roberts L.R. PDGFRalpha: a new therapeutic target in the treatment of hepatocellular carcinoma? Expert Opin Ther Targets. 2009;13:443–454. doi: 10.1517/14728220902719233. [DOI] [PubMed] [Google Scholar]
- 10.Si-Tayeb K., Lemaigre F.P., Duncan S.A. Organogenesis and development of the liver. Dev Cell. 2010;18:175–189. doi: 10.1016/j.devcel.2010.01.011. [DOI] [PubMed] [Google Scholar]
- 11.Fausto N., Campbell J.S., Riehle K.J. Liver regeneration. Hepatology. 2006;43:S45–S53. doi: 10.1002/hep.20969. [DOI] [PubMed] [Google Scholar]
- 12.Michalopoulos G.K. Liver regeneration. J Cell Physiol. 2007;213:286–300. doi: 10.1002/jcp.21172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Monga S.P., Monga H.K., Tan X., Mule K., Pediaditakis P., Michalopoulos G.K. Beta-catenin antisense studies in embryonic liver cultures: role in proliferation, apoptosis, and lineage specification. Gastroenterology. 2003;124:202–216. doi: 10.1053/gast.2003.50000. [DOI] [PubMed] [Google Scholar]
- 14.Monga S.P., Tang Y., Candotti F., Rashid A., Wildner O., Mishra B., Iqbal S., Mishra L. Expansion of hepatic and hematopoietic stem cells utilizing mouse embryonic liver explants. Cell Transplant. 2001;10:81–89. [PubMed] [Google Scholar]
- 15.Gerber D.E., Gupta P., Dellinger M.T., Toombs J.E., Peyton M., Duignan I., Malaby J., Bailey T., Burns C., Brekken R.A., Loizos N. Stromal platelet-derived growth factor receptor alpha (PDGFRalpha) provides a therapeutic target independent of tumor cell PDGFRalpha expression in lung cancer xenografts. Mol Cancer Ther. 2012;11:2473–2482. doi: 10.1158/1535-7163.MCT-12-0431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tallquist M.D., Soriano P. Cell autonomous requirement for PDGFRalpha in populations of cranial and cardiac neural crest cells. Development. 2003;130:507–518. doi: 10.1242/dev.00241. [DOI] [PubMed] [Google Scholar]
- 17.Postic C., Magnuson M.A. DNA excision in liver by an albumin-Cre transgene occurs progressively with age. Genesis. 2000;26:149–150. doi: 10.1002/(sici)1526-968x(200002)26:2<149::aid-gene16>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 18.Nejak-Bowen K.N., Thompson M.D., Singh S., Bowen W.C., Jr., Dar M.J., Khillan J., Dai C., Monga S.P. Accelerated liver regeneration and hepatocarcinogenesis in mice overexpressing serine-45 mutant beta-catenin. Hepatology. 2010;51:1603–1613. doi: 10.1002/hep.23538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tan X., Behari J., Cieply B., Michalopoulos G.K., Monga S.P. Conditional deletion of beta-catenin reveals its role in liver growth and regeneration. Gastroenterology. 2006;131:1561–1572. doi: 10.1053/j.gastro.2006.08.042. [DOI] [PubMed] [Google Scholar]
- 20.Rogler L.E. Selective bipotential differentiation of mouse embryonic hepatoblasts in vitro. Am J Pathol. 1997;150:591–602. [PMC free article] [PubMed] [Google Scholar]
- 21.Michalopoulos G.K., DeFrances M. Liver regeneration. Adv Biochem Eng Biotechnol. 2005;93:101–134. doi: 10.1007/b99968. [DOI] [PubMed] [Google Scholar]
- 22.Parviz F., Matullo C., Garrison W.D., Savatski L., Adamson J.W., Ning G., Kaestner K.H., Rossi J.M., Zaret K.S., Duncan S.A. Hepatocyte nuclear factor 4alpha controls the development of a hepatic epithelium and liver morphogenesis. Nat Genet. 2003;34:292–296. doi: 10.1038/ng1175. [DOI] [PubMed] [Google Scholar]
- 23.Asahina K., Tsai S.Y., Li P., Ishii M., Maxson R.E., Jr., Sucov H.M., Tsukamoto H. Mesenchymal origin of hepatic stellate cells, submesothelial cells, and perivascular mesenchymal cells during mouse liver development. Hepatology. 2009;49:998–1011. doi: 10.1002/hep.22721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li W.L., Yamada Y., Ueno M., Nishikawa S., Takakura N. Platelet derived growth factor receptor alpha is essential for establishing a microenvironment that supports definitive erythropoiesis. J Biochem. 2006;140:267–273. doi: 10.1093/jb/mvj151. [DOI] [PubMed] [Google Scholar]
- 25.Weinstein M., Monga S.P., Liu Y., Brodie S.G., Tang Y., Li C., Mishra L., Deng C.X. Smad proteins and hepatocyte growth factor control parallel regulatory pathways that converge on beta1-integrin to promote normal liver development. Mol Cell Biol. 2001;21:5122–5131. doi: 10.1128/MCB.21.15.5122-5131.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bard-Chapeau E.A., Yuan J., Droin N., Long S., Zhang E.E., Nguyen T.V., Feng G.S. Concerted functions of Gab1 and Shp2 in liver regeneration and hepatoprotection. Mol Cell Biol. 2006;26:4664–4674. doi: 10.1128/MCB.02253-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Borowiak M., Garratt A.N., Wustefeld T., Strehle M., Trautwein C., Birchmeier C. Met provides essential signals for liver regeneration. Proc Natl Acad Sci U S A. 2004;101:10608–10613. doi: 10.1073/pnas.0403412101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Onitsuka I., Tanaka M., Miyajima A. Characterization and functional analyses of hepatic mesothelial cells in mouse liver development. Gastroenterology. 2010;138:1525–1535. doi: 10.1053/j.gastro.2009.12.059. 1535.e1521-1526. [DOI] [PubMed] [Google Scholar]
- 29.Paranjpe S., Bowen W.C., Bell A.W., Nejak-Bowen K., Luo J.H., Michalopoulos G.K. Cell cycle effects resulting from inhibition of hepatocyte growth factor and its receptor c-Met in regenerating rat livers by RNA interference. Hepatology. 2007;45:1471–1477. doi: 10.1002/hep.21570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paranjpe S., Bowen W.C., Tseng G.C., Luo J.H., Orr A., Michalopoulos G.K. RNA interference against hepatic epidermal growth factor receptor has suppressive effects on liver regeneration in rats. Am J Pathol. 2010;176:2669–2681. doi: 10.2353/ajpath.2010.090605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Michalopoulos G.K. Liver regeneration: alternative epithelial pathways. Int J Biochem Cell Biol. 2011;43:173–179. doi: 10.1016/j.biocel.2009.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Betsholtz C., Karlsson L., Lindahl P. Developmental roles of platelet-derived growth factors. Bioessays. 2001;23:494–507. doi: 10.1002/bies.1069. [DOI] [PubMed] [Google Scholar]
- 33.Habib A.A., Hognason T., Ren J., Stefansson K., Ratan R.R. The epidermal growth factor receptor associates with and recruits phosphatidylinositol 3-kinase to the platelet-derived growth factor beta receptor. J Biol Chem. 1998;273:6885–6891. doi: 10.1074/jbc.273.12.6885. [DOI] [PubMed] [Google Scholar]
- 34.Saito Y., Haendeler J., Hojo Y., Yamamoto K., Berk B.C. Receptor heterodimerization: essential mechanism for platelet-derived growth factor-induced epidermal growth factor receptor transactivation. Mol Cell Biol. 2001;21:6387–6394. doi: 10.1128/MCB.21.19.6387-6394.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Villanueva A., Llovet J.M. Targeted therapies for hepatocellular carcinoma. Gastroenterology. 2011;140:1410–1426. doi: 10.1053/j.gastro.2011.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Farazi P.A., DePinho R.A. Hepatocellular carcinoma pathogenesis: from genes to environment. Nat Rev Cancer. 2006;6:674–687. doi: 10.1038/nrc1934. [DOI] [PubMed] [Google Scholar]
- 37.Campbell J.S., Johnson M.M., Bauer R.L., Hudkins K.L., Gilbertson D.G., Riehle K.J., Yeh M.M., Alpers C.E., Fausto N. Targeting stromal cells for the treatment of platelet-derived growth factor C-induced hepatocellular carcinogenesis. Differentiation. 2007;75:843–852. doi: 10.1111/j.1432-0436.2007.00235.x. [DOI] [PubMed] [Google Scholar]
- 38.Loizos N., Xu Y., Huber J., Liu M., Lu D., Finnerty B., Rolser R., Malikzay A., Persaud A., Corcoran E., Deevi D.S., Balderes P., Bassi R., Jimenez X., Joynes C.J., Mangalampalli V.R., Steiner P., Tonra J.R., Wu Y., Pereira D.S., Zhu Z., Ludwig D.L., Hicklin D.J., Bohlen P., Witte L., Kussie P. Targeting the platelet-derived growth factor receptor alpha with a neutralizing human monoclonal antibody inhibits the growth of tumor xenografts: implications as a potential therapeutic target. Mol Cancer Ther. 2005;4:369–379. doi: 10.1158/1535-7163.MCT-04-0114. [DOI] [PubMed] [Google Scholar]
- 39.Shah G.D., Loizos N., Youssoufian H., Schwartz J.D., Rowinsky E.K. Rationale for the development of IMC-3G3, a fully human immunoglobulin G subclass 1 monoclonal antibody targeting the platelet-derived growth factor receptor alpha. Cancer. 2010;116:1018–1026. doi: 10.1002/cncr.24788. [DOI] [PubMed] [Google Scholar]