Abstract
Recent evidence has suggested an important role of miRNAs in liver biology and diseases, although the implication of miRNAs in cholangiocarcinoma remains to be defined further. This study was designed to examine the biological function and molecular mechanism of miR-101 in cholangiocarcinogenesis and tumor progression. In situ hybridization and quantitative RT-PCR were performed to determine the expression of miR-101 in human cholangiocarcinoma tissues and cell lines. Compared with noncancerous biliary epithelial cells, the expression of miR-101 is decreased in 43.5% of human cholangiocarcinoma specimens and in all three cholangiocarcinoma cell lines used in this study. Forced overexpression of miR-101 significantly inhibited cholangiocarcinoma growth in severe combined immunodeficiency mice. miR-101-overexpressed xenograft tumor tissues showed decreased capillary densities and decreased levels of vascular endothelial growth factor (VEGF) and cyclooxygenase-2 (COX-2). The VEGF and COX-2 mRNAs were identified as the bona fide targets of miR-101 in cholangiocarcinoma cells by both computational analysis and experimental assays. miR-101 inhibits cholangiocarcinoma angiogenesis by direct targeting of VEGF mRNA 3′untranslated region and by repression of VEGF gene transcription through inhibition of COX-2. This study established a novel tumor-suppressor role of miR-101 in cholangiocarcinoma and it suggests the possibility of targeting miR-101 and related signaling pathways for future therapy.
Cholangiocarcinoma is a highly malignant cancer of the biliary tree with a dismal prognosis. The incidence and mortality of cholangiocarcinoma, especially the intrahepatic type, is increasing worldwide, and currently there is no effective chemoprevention or treatment. The tumor often arises from background conditions that cause long-standing inflammation, injury, and reparative biliary epithelial cell proliferation, such as primary sclerosing cholangitis, clonorchiasis, hepatolithiasis, or complicated fibropolycystic diseases.1–9 The pathogenesis of cholangiocarcinoma is complex and involves a number of signaling molecules, including vascular endothelial growth factor (VEGF)10,11 and cyclooxygenase-2 (COX-2).3
VEGF and COX-2 signaling pathways are known to interact with each other reciprocally and coordinately regulate cancer growth.12 VEGF is a potent angiogenic factor that binds to its receptor and stimulates cell proliferation and survival; human cholangiocarcinoma tissue samples and cell lines express VEGF.10,11 COX-2 is a rate-limiting key enzyme for the synthesis of proinflammatory and tumorigenic prostaglandins; consistent with the up-regulation of COX-2 in cholangiocarcinomas,13–17 the role of COX-2 signaling in cholangiocarcinoma growth has been well documented.3 Thus, targeting VEGF and COX-2 signaling pathways may provide effective prevention and treatment of human cholangiocarcinoma.
miRNAs are small noncoding RNAs that regulate the expression of target genes post-transcriptionally through base pairing with target mRNAs.18 In this study, we aimed to identify miRNAs that are capable of targeting key signaling pathways in cholangiocarcinogenesis. We performed a computational analysis using the algorithm provided at http://www.microrna.org,19,20 and this approach led us to identify miR-101 as a noncoding RNA that binds directly to the 3′-untranslated region (UTR) of both VEGF and COX-2 mRNAs. Although miR-101 has been shown to function as a tumor suppressor in certain cancers by targeting several molecules including enhancer of zeste homologue 2, COX-2, amyloid precursor protein, and myeloid cell leukemia sequence-1,21–25 the potential implication of miR-101 in cholangiocarcinomas remains unknown. Given that miR-101 is one of the most abundantly expressed miRNAs in the liver,26 we sought to examine the expression of miR-101 in human cholangiocarcinoma tissues and to validate the effect of miR-101 on VEGF and COX-2 in cholangiocarcinoma cells and their impact on cholangiocarcinogenesis and tumor progression.
Materials and Methods
Materials
The miR-101-1 expressed lentivirus and scramble control lentiviral, both co-expressing enhanced green fluorescent protein, were obtained from GeneCopoeia (Rockville, MD). Rabbit polyclonal antibody against CD31 and mouse monoclonal antibody against Ki-67 were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). Mouse monoclonal antibody against COX-2 was obtained from Cayman Chemical (Ann Arbor, MI). Mouse monoclonal antibody against β-actin was obtained from Sigma (St. Louis, MO). IRDye 800CW or 680LT-labeled anti-mouse or rabbit IgG secondary antibody were purchased from LI-COR Biosciences (Lincoln, NE). The VEGF promoter (from −722 to + 320) luciferase reporter construct was purchased from Switchgear (Menlo Park, CA) and the VEGF 3′UTR luciferase reporter was purchased from GeneCopoeia. Prostaglandin E2 (PGE2) was purchased from Cayman Chemical (Ann Arbor, MI). Flt-1 was purchased from ProsPec (East Brunswick, NJ). All other analytic grade chemical reagents were purchased from Sigma.
In Situ Hybridization of miRNA
Human cholangiocarcinoma tissue arrays (ISU ABXIS, Seoul, Korea) and noncholangiocarcinoma liver specimens were subjected to in situ hybridization (ISH) per institutional review board approval. ISH was performed using the miR-101 locked nucleic acid probe (5′-digoxigenin-TTCAGTTATCACAGTACTGTA-3′-digoxigenin) and the microRNA ISH Optimization Kit (Exiqon, Vedbaek, Denmark) according to the manufacturer’s instructions. Briefly, deparaffinized arrays were incubated with 15 μg/mL proteinase K at 37°C for 8 minutes. After dehydration, the slides were incubated with 40 nmol/L miR-101 probe at 50°C for 120 minutes, followed by stringent washes with 5× standard saline citrate, 1× standard saline citrate, and 0.2× standard saline citrate buffers at 50°C, digoxigenin blocking reagent (Roche, Mannheim, Germany) in maleic acid buffer containing 2% sheep serum at room temperature for 15 minutes, and alkaline phosphatase–conjugated antidigoxigenin (diluted 1:500 in blocking reagent; Roche) at room temperature for 60 minutes. Enzymatic color was developed by incubating with 4-nitro-blue tetrazolium and 5-bromo-4-chloro-3′-indolylphosphate substrate (Roche) at 30°C for 2 hours to form dark-blue 4-nitro-blue tetrazolium-formazan precipitate, followed by counterstaining with nuclear fast red solution (Vector Laboratories, Burlingame, CA). The slides then were rinsed, dehydrated, mounted, and observed under a microscope. Scrambled probe and U6 snRNA-specific probe were used as a system control. A standard four-point scale was used to evaluate the staining intensity according to the established criteria.27
Cell Culture
Three human cholangiocarcinoma cell lines (CCLP1, HuCCT1, and TFK1) and one noncancerous cholangiocyte cell line (H69) were used in this study [the CCLP1 cell line was established by Theresa Whiteside, Ph.D., at the Pittsburgh Cancer Institute (Pittsburgh, PA); the HuCCT1 and TFK1 cells were obtained from the Japanese Cancer Research Resources Bank (Osaka, Japan); and the H69 cells were kindly provided by Dr. Gregory J. Gores at the Mayo Clinic College of Medicine (Rochester, MN)]. All cells were cultured according to our previously described methods.28 The miR-101–overexpressed and scramble control stable cell lines were established by transduction with miR-101-1 lentiviral vector or miRNA-scramble control lentiviral vector, followed by selection with media containing puromycin. For VEGF induction by COX-2 overexpression, the cells transfected with the COX-2 open reading frame (without 3′UTR) plasmid were cultured in Opti-MEM medium (Invitrogen, Carlsbad, CA) containing 1% fetal bovine serum (FBS) for 24 hours and the culture media were collected for analysis. For VEGF induction by PGE2, the cells were incubated with Opti-MEM medium (Invitrogen) containing 1% FBS and 10 μmol/L PGE2 for 24 hours.
Anti-miR, miRNA Precursor, and siRNA Transfection
miR-101–specific anti-miR (Qiagen, Valencia, CA), miR-101 precursor (Ambion, Austin, TX), or COX-2 siRNA (Ambion) and control siRNA were transfected into cells using Oligofectamine (Invitrogen) per the manufacturer’s instructions. After transfection at the indicated time periods, the cell lysates or culture supernatants were obtained for analysis.
3′UTR Luciferase Reporter Plasmid Construction
The 450-bp 3′UTR of human COX-2 was amplified from cDNA by PCR with the forward primer 5′-AAGCCTTGCCTCAGAGAGAACTGTACGGGG-3′ and reverse primer 5′-CTCGAGTGTGGGCTAGCACATAGGCCT-3′ (endonuclease restriction sites were incorporated in primers to facilitate ligation into the luciferase reporter plasmid pMIR-REP-dCMV). The correct sequence of the insert was verified by DNA sequencing. Generation of the three nucleotide mutations was achieved by site-directed mutagenesis with the QuikChange kit from Stratagene (La Jolla, CA), followed by sequence verification. The primers used for the COX-2 3′UTR mutation were as follows: 5′-CATTGTCACTGACATTTAATGGTGACGTATATTACTTAATTTATTGAAG-3′ and 5′-CTTCAATAAATTAAGTAATATACGTCACCATTAAATGTCAGTGACAATG-3′; the primers used for the VEGF 3′UTR mutation were as follows: 5′-TTTTTAATTTTAATATTTGTTATCATTTATTTATTGGTGCTCACTTTATCCGTAATAATTGTGGGGAAAAGATATAACATCACG-3′ and 5′-CGTGATGTTAATATCTTTTCCCCACAATTATTACGGATAAAGGTGAGCACCAATAAATAAATGATAACAAATATTAAAATTAAAAA-3′.
Cell Proliferation WST-1 Assay
Cells were cultured in serum-free medium for 24 hours to synchronize the cell cycle. Cells (2 × 103) were seeded onto each well of the 96-well plates in 200 μL culture medium containing 1% FBS. At the indicated time points, 90 μL of serum-free medium mixed with 10 μL WST-1 (Roche) was added to each well. After 4 hours of incubation, the absorbance of each well was measured by an enzyme-linked immunosorbent assay (ELISA) plate reader at a wavelength of 450 nm.
Luciferase Activity Assay
Twenty-four hours after transfection, cells were collected and analyzed using the Dual-Luciferase Reporter Assay System (Promega, Fitchburg, WI). Luciferase activity was measured by a Centro XS3lb 960 microplate fluorescence reader (Mandel, Ontario, Canada). The pRL-TK plasmid (Promega) with constitutive expression of Renilla luciferase was co-transfected with different firefly luciferase-based reporters as an internal control.
Quantitative RT-PCR
Total RNA was prepared using TRIzol reagent (Invitrogen). Quantitative RT-PCR (RT-qPCR) was performed using the Qiagen (Valencia, CA) miScript Kit and the miR-101 Primer Assay. U6 small nuclear 2 was used as an internal control for miR-101 amplification. The primers for COX-2 were as follows: 5′-TGAGGGATCTGTGGATGCTTCGT-3′ (forward) and 5′-AAACCCACAGTGCTTGACACAGAA-3′ (reverse); the primers for VEGF were as follows: 5′-ACACATTGTTGGAAGAAGCAGCCC-3′ (forward) and 5′-AGGAAGGTCAACCACTCACACACA-3′ (reverse); the primers for the glyceraldehyde-3-phosphate dehydrogenase control were as follows: 5′-GGCCCACATGGCCTCCAAGG-3′ (forward) and 5′-GGCAGGGACTCCCCAGCAGT-3′ (reverse).
Western Blotting
The logarithmically growing cells were washed twice with ice-cold PBS and lysed in a lysis buffer containing 50 mmol/L HEPES, 1 mmol/L EDTA (pH 8.0); one tablet of protease inhibitor cocktail (Roche) was added per 10 mL of buffer. After sonication on ice, the cell lysates were centrifuged at 12,000 × g for 20 minutes at 4°C and the samples were subjected to SDS-PAGE. The nitrocellulose membranes (Bio-Rad, Hercules, CA) were blocked in PBS with 0.1% Tween 20 containing 5% nonfat milk for 1 hour, followed by incubation with different primary antibodies (at appropriate dilutions) in 5% nonfat milk at 4°C overnight. After three washes with PBS with 0.1% Tween 20, the membranes were incubated with 1:10,000 IRDye 800CW- or IRDye 680-labeled secondary antibody (LI-COR Biosciences) for 1 hour, followed by three washes with PBS with 0.1% Tween 20. The membranes were scanned with the Odyssey Infrared Imaging System (LI-COR Biosciences).
PGE2 and VEGF Quantification
Cells with indicated treatment were incubated for 24 hours in 1 mL serum-free (for PGE2) or 1% FBS containing (for VEGF) Opti-MEM medium in 6-well plates. The supernatants were collected and centrifuged for 10 minutes at 12,000 × g to remove the floating cells and cellular debris. The amounts of PGE2 and VEGF in the spent media were measured by the Prostaglandin E2 Biotrak Enzyme Immunoassay System (GE Healthcare, Piscataway, NJ) and the Quantikine VEGF Enzyme Immunoassay kit (R&D, Minneapolis, MN), respectively.
Severe Combined Immunodeficiency Mouse Zenograft Studies
Four-week-old male nonobese diabetic CB17-Prkdc/severe combined immunodeficiency (SCID) mice were purchased from the Jackson Laboratory (Bar Harbor, ME). Subcutaneous xenografts were established by inoculating 1.5 × 106 miR-101–overexpressed or control CCLP1 or HuCCT1 cells in the flanks of mice (six mice per group). The mice were observed over 4 weeks for tumor formation. Tumor volume was measured twice a week with a caliper and calculated by using the following formula: larger diameter × (smaller diameter)2/2. At sacrifice, the tumors were recovered and the wet weight of each tumor was recorded. Separate portions of each tumor were fixed in formalin for H&E staining and immunohistochemistry, or snap frozen for Western blotting and RT-qPCR analysis.
Immunohistochemistry and Immunofluorescent Staining
Deparaffinized slides were microwaved for 15 minutes in 0.01 mol/L citrate buffer, pH 6.0, to retrieve antigen. For immunohistochemistry (IHC), slides were incubated with 0.3% H2O2 in methanol for 30 minutes to abolish endogenous peroxidase activity. The slides were blocked with 10% goat serum in PBS for 1 hour at 37°C and incubated with primary antibody at 4°C overnight, followed by three washes and a 1-hour incubation at room temperature with a secondary antibody labeled with horseradish peroxidase (for IHC) or fluorescein isothiocyanate (for immunofluorescent staining). A diaminobenzidine peroxidase substrate kit (Vector Laboratories) was used for IHC color development.
Endothelial Tube Formation Assay (in Vitro Angiogenesis)
miR-101–overexpressed or control cells (1 × 106) were incubated in 1 mL Opti-MEM medium containing 1% FBS for 24 hours and the supernatants were collected as the conditioned media. Primary human umbilical vein endothelial cells (3 × 104) in 100 μL 200 Phenol Red Free medium (Invitrogen) were mixed with 400 μL conditioned medium from the indicated cells and plated on 24-well plates coated with growth factor–reduced Matrigel (Invitrogen). Human umbilical vein endothelial cells were incubated in a humidified atmosphere of 5% CO2 at 37°C for 3 to 5 hours for tube formation. After incubation, the cells were stained with Calcein AM (Invitrogen) for 30 minutes and examined under a fluorescence microscope. At least three random images from each experiment (performed in triplicate) were analyzed to calculate the total tube length and the number of branching points using ImageJ software version 1.46 (NIH, Bethesda, MD).
Matrigel Plug Assay
Three milliliters of conditioned medium from each cell strain was condensed to 20 μL by the Centriprep Centrifugal Filter (Millipore, Billerica, MA) at 4°C. The condensed medium then was mixed with 180 μL reduced growth factor Matrigel (Invitrogen) at 0°C. The mixtures were injected in the ventral flanks of C57BL/6 mice subcutaneously using precooled syringes. Ten days after injection, the gel plugs were recovered from the mice. A portion of each plug was selected for H&E and CD31 IHC staining. Hemoglobin concentrations in the plugs were measured using a hemoglobin detection kit from ArborAssays (Ann Arbor, MI). Per H&E and CD31 IHC staining, a capillary is defined as a ring or cylindric structure with an internal endothelial cell lining (a lumen) in which red blood cells can be seen.
Statistical Analysis
Data are presented as means ± SEM or SD from a minimum of three replicates as indicated in each Figure legend. The difference between groups was evaluated by statistical software SPSS13.0 (IMB, Armonk, NY) one-way analysis of variance or repeated-measures analysis of the generalized linear model. A P value less than 0.05 was considered statistically significant.
Results
Expression of miR-101 Is Decreased in Human Cholangiocarcinoma Tissues and Cell Lines
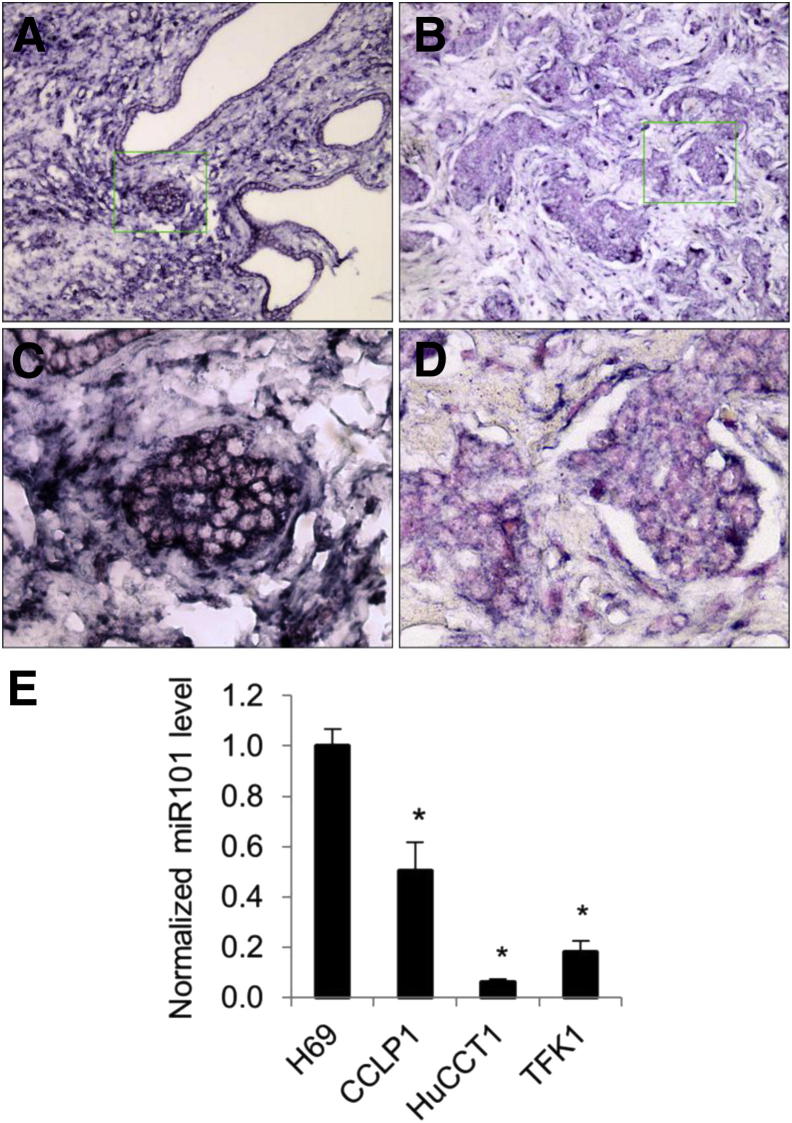
We performed ISH for miR-101 in 46 cases of human cholangiocarcinoma tissues and nine cases of noncholangiocarcinoma liver specimens by using a locked nucleic acid–modified miR-101 probe. Compared with the non-neoplastic biliary epithelium that lines bile ducts and peribiliary glands, the level of miR-101 in cholangiocarcinoma cells was decreased in 20 cases (20 of 46; 43.5%), unchanged in 26 cases (26 of 46; 56.5%), and increased in 0 cases (0 of 46; 0%). Figure 1, A–D, shows a representative image of miR-101 ISH, indicating decreased miR-101 in cholangiocarcinoma tissues in comparison with the non-neoplastic bile ducts. U6 snRNA probe and scrambled probes were hybridized with liver specimens as a systematic control under the same conditions (Supplemental Figure S1). Consistent with these observations, RT-qPCR analysis showed lower levels of miR-101 in three human cholangiocarcinoma cell lines (CCLP1, HuCCT1, and TFK1) compared with the noncancerous human cholangiocyte cell line, H69 (Figure 1E). These findings provide novel evidence for decreased miR-101 expression in human cholangiocarcinoma tissues and cell lines.
Figure 1.
Expression of miR-101 in human cholangiocarcinoma tissues and cell lines. A–D: In situ hybridization for miR-101 in human cholangiocarcinoma tissues was performed as described in Materials and Methods. Positive signals are shown as dark blue; nuclei are counterstained red. A and C: Strong staining in the bile duct epithelial cells in the non-neoplastic tissues. B and D: Weak miR-101 staining in human cholangiocarcinoma cells. Original magnification: ×400 (A); ×100 (B), from the boxed areas. E: RT-qPCR for mature miR-101 in a human cholangiocyte cell line (H69) and three human cholangiocarcinoma cell lines (CCLP1, HuCCT1, and TFK1). The results represent the average ratio between miR-101 and the internal control U6 small nuclear 2 from three experiments. ∗P < 0.01 compared with H69.
miR-101 Prevents Cholangiocarcinoma Cell Growth
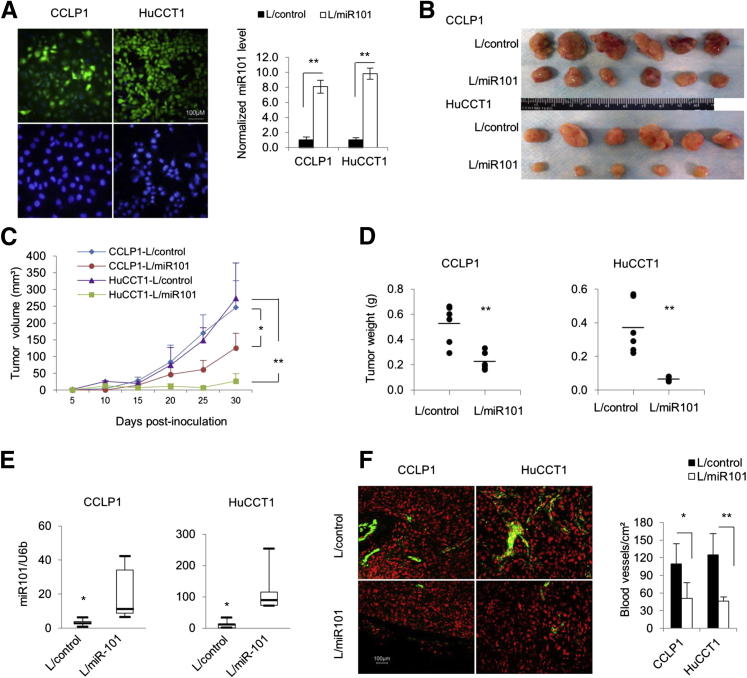
To investigate the role of miR-101 in cholangiocarcinoma cell growth, we established human cholangiocarcinoma cell lines (CCLP1 and HuCCT1) with stable overexpression of miR-101 (the cells were stably transduced with the lentivirus particles carrying the miR-101-1 gene or with the control lentivirus particle carrying the scrambled miRNA). Successful increase of miR-101 expression in miR-101 lentivirus-transduced cells was verified by RT-qPCR and by immunofluorescence for enhanced green fluorescent protein (Figure 2A). The selected stable cells with or without miR-101 overexpression then were analyzed for growth in vitro and in SCID mice. Although miR-101 overexpression slightly inhibited cholangiocarcinoma cell growth in vitro (Supplemental Figure S2), it significantly decreased tumor growth in SCID mice (Figure 2, B–D). The miR-101–overexpressed cells grown in SCID mice formed smaller tumors and had lower tumor volumes compared with the corresponding controls. The miR-101–overexpressed tumors also had decreased tumor weight compared with the vector control tumors (0.52 ± 0.15 versus 0.22 ± 0.070 g, P < 0.01 for CCLP1; and 0.37 ± 0.16 versus 0.064 ± 0.015 g, P < 0.01 for HuCCT1). RT-qPCR analysis confirmed an increased miR-101 level in the tumor tissues with stable transduction of miR-101 lentivirus (Figure 2E). We noticed that the color of miR-101–overexpressed tumors was slightly paler compared with control tumors, which likely reflects less blood/vasculature in miR-101–overexpressed tumors. Indeed, immunofluorescence staining for the vascular endothelial marker CD31 showed decreased blood vessel density in miR-101–overexpressed tumors (Figure 2F). The overall tumor cell proliferation appeared to be slightly reduced in miR-101–overexpressed tumors, as indicated by immunostaining for the proliferation marker Ki-67; however, detailed assessment for Ki-67–positive cells was impossible because of the presence of apparent tumor necrosis, which is known to interfere with immunohistochemical staining (Supplemental Figure S3A). In areas away from the necrosis, especially at tumor edges, the tumor cell morphology and the proliferation index did not appear to differ significantly between miR-101–overexpressed and control tumors (Supplemental Figure S3, B and C). Thus, the tumor volume/weight difference between the two groups most likely is caused by decreased angiogenesis in miR-101 highly expressed tumors. These findings suggest that miR-101 may prevent cholangiocarcinoma growth, at least in part, through inhibition of angiogenesis.
Figure 2.
miR-101 suppresses human cholangiocarcinoma growth in vivo. A: Construction of human cholangiocarcinoma cell lines with stable overexpression of miR-101. CCLP1 and HuCCT1 cells were stably transduced with miR-101-1 lentivirus (L/miR101) and control lentivirus (L/control), respectively. Left panel: Immunofluorescent microscopy for enhanced green fluorescent protein in cells stably transduced with the miR-101-1 lentivirus (this vector carries the enhanced green fluorescent protein gene under the control of the same cytomegalovirus promoter). Upper row: miR101-1 lentivirus-transduced cells. lower row: Noninfected control cells. Right panel: RT-qPCR analysis for miR-101 in cells infected with miR-101-1 and scramble control lentivirus. The results represent means ± SEM of the miR-101/U6 small nuclear 2 ratio normalized to the scramble control cells. B–F: miR-101–overexpressed or scramble control CCLP1 and HuCCT1 cells (2 × 106) were inoculated subcutaneously into SCID mice (n = 6). Thirty days after inoculation, the mice were sacrificed and the tumors were recovered for analyses. B: Xenograft tumor masses from SCID mice (six mice per group; no tumor development in 1 of 6 mice inoculated with miR-101–overexpressed HuCCT1 cells). C: The volume of xenograft tumors. The data represent means ± SD from six mice. D: The weight of xenograft tumors. The data represent means ± SD from six mice. The bar indicates mean weight of each group. E: The levels of miR-101 in xenograft tumor tissues as determined by RT-qPCR. The data are shown as the means ± SEM from six mice. F: CD31 immunofluorescence staining in xenograft tumor tissues. Left panel: Representative image of CD31 immunofluorescence (CD31 was shown as green and nuclei were counterstained red). Right panel: Normalized capillary numbers in the tumors of each group (the data are expressed as means ± SD from six mice). Scale bars: 100 μm (A and F). ∗P < 0.05, ∗∗P < 0.01.
VEGF Is a Direct Target of miR-101 in Human Cholangiocarcinoma Cells
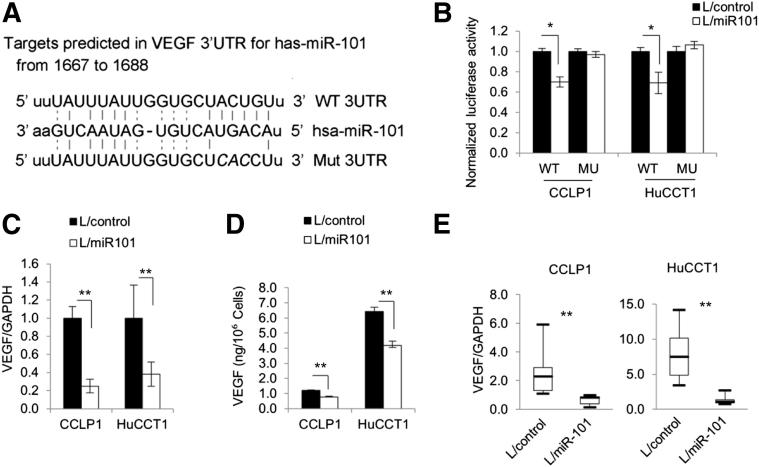
Target scan sequence analysis showed the complementary sequence of miR-101 in the 3′UTR of VEGF mRNA (Figure 3A). To verify the direct effect of miR-101 on VEGF in cholangiocarcinoma cells, CCLP1 and HuCCT1 cells were transfected with the luciferase reporter plasmid containing the 3′UTR of VEGF mRNA. As shown in Figure 3B, miR-101 overexpression decreased VEGF 3′UTR luciferase reporter activity in CCLP1 and HuCCT1 cells; this effect was abolished when the three nucleotides in the miR-101 seed-binding site of the VEGF mRNA 3′UTR were mutated. Accordingly, real-time PCR and enzyme immunoassay showed that miR-101 overexpression decreased VEGF mRNA and protein levels (Figure 3, C and D). Decreased VEGF mRNA also was observed in miR-101–overexpressed xenograft tumor tissues (Figure 3E). These results show that VEGF is a direct target of miR-101 in human cholangiocarcinoma cells.
Figure 3.
VEGF is a direct target of miR-101 in human cholangiocarcinoma cells. A: Putative miR-101 binding sequence in the 3′UTR of VEGF mRNA. Three nucleotides (italics) in the miR-101 binding site were mutated to obtain the mutation reporter construct. B: VEGF 3′UTR luciferase reporter activity assay. miR-101–overexpressed or control cells were transfected with either wild-type or mutant VEGF 3′UTR reporter plasmids (indicated as WT or MU, respectively, on the x axis). Twenty-four hours after transfection, the cell lysates were obtained to determine luciferase activity by using a luminometer. Renilla luciferase plasmid was used as the internal control. C: miR-101 reduces VEGF mRNA in cholangiocarcinoma cells. VEGF mRNA levels were determined by RT-qPCR in miR-101–overexpressed and miRNA-scramble control cells. The data are shown as means ± SEM from three independent experiments. D: miR-101 reduces VEGF production in cholangiocarcinoma cells. The level of VEGF in cell culture supernatant was determined by enzyme immunoassay. The data are shown as means ± SEM from three independent experiments. E: The levels of VEGF mRNA in xenograft tumor tissues as determined by RT-qPCR. The data are shown as means ± SEM from six mice. GAPDH, glyceraldehyde-3-phosphate dehydrogenase; L/control, control lentivirus; L/miR101, miR-101-1 lentivirus. ∗P < 0.05, ∗∗P < 0.01.
miR-101 also Targets COX-2 in Human Cholangiocarcinoma Cells
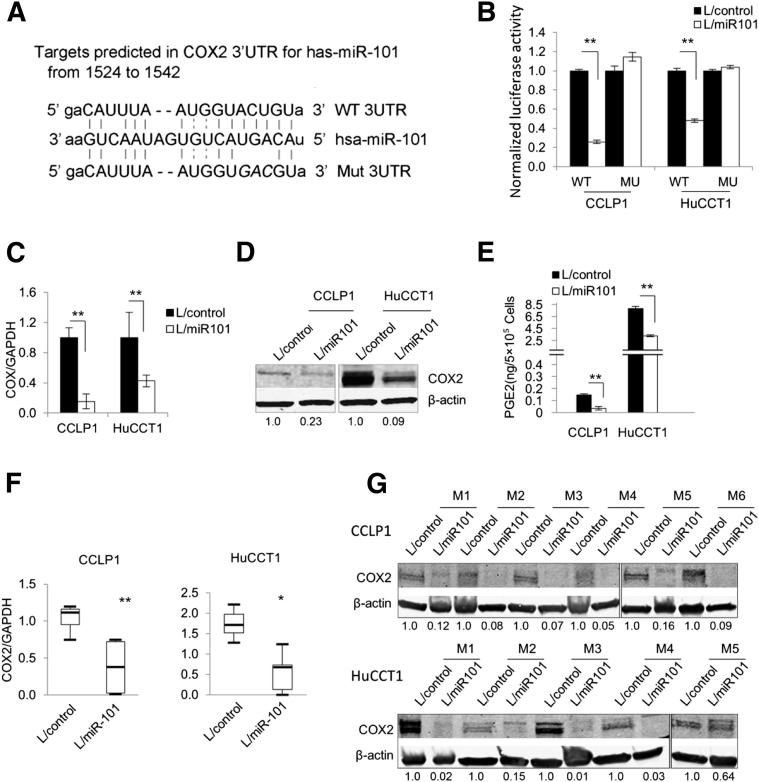
Target scan sequence analysis also revealed the presence of a miR-101 complementary sequence in the 3′UTR of COX-2 mRNA (Figure 4A). Thus, we also verified the direct effect of miR-101 on COX-2 in cholangiocarcinoma cells by transfecting CCLP1 and HuCCT1 cells with the luciferase reporter plasmid containing the 3′UTR of COX-2 mRNA. As shown in Figure 4B, miR-101 overexpression decreased the COX-2 3′UTR luciferase reporter activity and this effect was abolished when the three nucleotides in the miR-101 seed-binding site of the COX-2 3′UTR were mutated. RT-qPCR and Western blotting analyses showed that miR-101 overexpression decreased the levels of COX-2 mRNA and protein (Figure 4, C and D). In contrast, miR-101 inhibition by anti-miR increased the COX-2 protein level in CCLP1 and HuCCT1 cells (Supplemental Figure S4). Consistent with these observations, miR-101 overexpression decreased the production of PGE2 (Figure 4E). Decreased COX-2 mRNA and protein levels also were observed in miR-101–overexpressed xenograft tumor tissues recovered from the SCID mice (Figure 4, F and G).
Figure 4.
miR-101 targets COX-2 in cholangiocarcinoma cells. A: Putative miR-101 binding sequence in the 3′UTR of COX-2 mRNA. 3′UTR fragments of COX-2 containing wild-type or mutated (three mutated nucleotides are in italics) miR-101 binding sites were cloned into pMIR-REP-dCMV vector to obtain COX-2 3′UTR luciferase reporter plasmids as described in Materials and Methods. B: COX-2 3′UTR luciferase reporter activity assay. miR-101–overexpressed or control cells were transfected with either wild-type or mutant COX-2 3′UTR reporter plasmids (indicated as WT or MU, respectively, on the x axis). Twenty-four hours after transfection, the cell lysates were obtained to determine luciferase activity by using a luminometer. Renilla luciferase plasmid was used as the internal control. C: COX-2 mRNA levels in miR-101–overexpressed and control cells. COX-2 mRNA was measured by RT-qPCR in miR-101–overexpressed and miRNA-scramble control cells. The data are shown as means ± SEM from three independent experiments. D: Representative Western blots showing the COX-2 protein levels in miR-101–overexpressed and control cholangiocarcinoma cells. The numbers under the bands indicate the relative expression levels of COX-2 proteins (the levels in control cells were set as 1.0). E: PGE2 levels in miR-101–overexpressed and control cholangiocarcinoma cells. Serum-free medium of miR-101–overexpressed and control cells were collected and subjected to enzyme immunoassay analysis for PGE2. The data are presented as means ± SEM of three experiments. F: The levels of COX-2 mRNA in xenograft tumor tissues as determined by RT-qPCR. The data are shown as means ± SEM from six mice. G: Representative Western blots showing decreased COX-2 protein levels in miR-101–overexpressed tumor tissue recovered from the SCID mice in comparison with the control tumor tissues. M1 to M6 indicate different mice; the numbers under the bands indicate the relative expression levels of COX-2 proteins (the levels in control tumors were set as 1.0). GAPDH, glyceraldehyde-3-phosphate dehydrogenase; L/control, control lentivirus; L/miR101, miR-101-1 lentivirus. ∗P < 0.05, ∗∗P < 0.01.
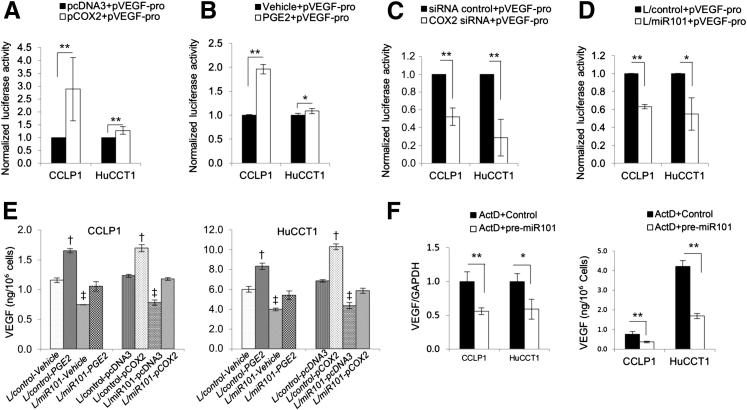
Contribution of COX-2/PGE2 in miR-101–Induced Inhibition of VEGF
Given that COX-2/PGE2 signaling is known to stimulate VEGF gene transcription in other cell types,29–34 we performed further experiments to determine whether down-regulation of COX-2/PGE2 by miR-101 might contribute to a reduction of VEGF in cholangiocarcinoma cells. As shown in Figure 5, A–C, transfection of COX-2 opening reading frame plasmid or treatment with PGE2 increased VEGF promoter luciferase reporter activity. On the other hand, depletion of COX-2 by siRNA reduced VEGF promoter luciferase reporter activity. These findings show a direct role of COX-2/PGE2 signaling in VEGF transcription in cholangiocarcinoma cells. Consistent with a reduction of COX-2/PGE2 by miR-101, miR-101 overexpression decreased VEGF promoter luciferase reporter activity in CCLP1 and HuCCT1 cells (Figure 5D). Furthermore, the production of VEGF was increased in CCLP1 and HuCCT1 cells with COX-2 overexpression or PGE2 treatment (Figure 5E). These results provide evidence for inhibition of COX-2/PGE2 by miR-101, leading to repression of VEGF gene transcription in cholangiocarcinoma cells.
Figure 5.
COX-2/PGE2 activates VEGF transcription in human cholangiocarcinoma cells. A–C: CCLP1 and HuCCT1 cells transfected with the VEGF promoter luciferase reporter (pVEGF-pro) were co-transfected with COX-2 expression plasmid [COX-2 open reading frame sequence cloned in pcDNA3.0 vector (pCOX2)] (A), treated with 10 μmol/L PGE2, (B) or co-transfected with the COX-2 siRNA. C: Twenty-four hours later, the cell lysates were obtained to measure luciferase activities. The data are presented as means ± SEM from three independent experiments. D: miR-101–overexpressed and control cells were transfected with VEGF promoter luciferase reporter plasmid. Twenty-four hours after transfection, the cell lysates were obtained to measure luciferase activities. The data are presented as means ± SEM from three independent experiments. E: VEGF in supernatants of cells with indicated treatments. The production of VEGF was increased significantly by COX-2 overexpression or 10 μmol/L PGE2 treatment. †P < 0.01 compared with vehicle or pcDNA3 control. Although miR-101–overexpression decreased VEGF production, this effect was prevented by co-treatment with PGE2 or co-expression of COX-2. ‡P < 0.01 compared with corresponding L/control cells and PEG2 treatment or pCOX2 transfected cells. The data are presented as means ± SEM from triplicate experiments. F: The cells were incubated with 1 μg/mL actinomycin D overnight, followed by transfection with miR-101 mimic or scramble control. Left panel: Twenty-four hours later, VEGF mRNA in cells were determined by RT-qPCR. Right panel: VEGF concentration in culture supernatants was determined by enzyme immunoassay. The data are shown as means ± SEM from triplicate experiments. GAPDH, glyceraldehyde-3-phosphate dehydrogenase; L/control, control lentivirus; L/miR101, miR-101-1 lentivirus. ∗P < 0.05, ∗∗P < 0.01.
We noticed that the levels of VEGF in miR-101–overexpressed and control cells continued to differ when both cells were transfected with the COX-2 open reading frame plasmid or were treated with PGE2 (Figure 5E). This finding suggests that COX-2/PGE2 signaling only partially contributes to miR-101–mediated inhibition of VEGF in cholangiocarcinoma cells. Consistent with this assertion, we noticed that when CCLP1 and HuCCT1 cells were pretreated with the transcription inhibitor actinomycin D (to eliminate COX-2/PGE2–mediated VEGF gene transcription), miR-101 continued to decrease VEGF mRNA and protein (Figure 5F). Although overexpression of miR-101 facilitated VEGF mRNA degradation at 4, 8, and 12 hours, inhibition of miR-101 by anti-miR prevented VEGF mRNA degradation at these time points (Supplemental Figure S5). Thus, miR-101 decreases VEGF production in human cholangiocarcinoma cells through targeting the VEGF mRNA 3′UTR as well as through inhibiting VEGF gene transcription via COX-2/PGE2.
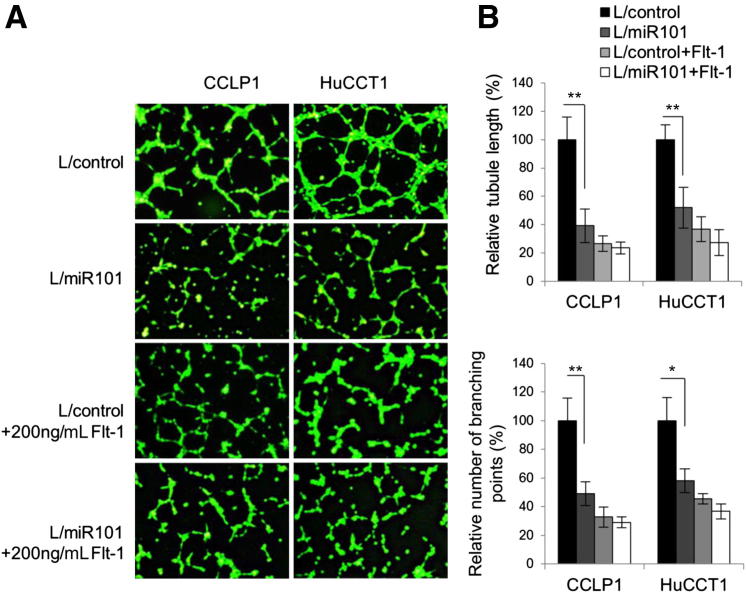
miR-101 Inhibits Cholangiocarcinoma Angiogenesis in Vitro and in Vivo
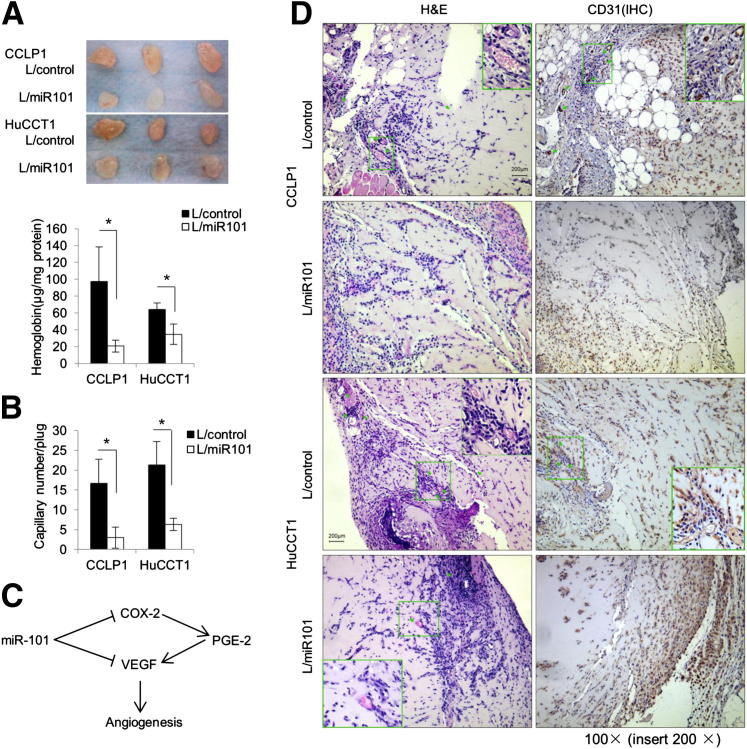
Given that miR-101 inhibits VEGF production in cholangiocarcinoma cells, we next assessed the angiogenesis capability of the conditioned media derived from miR-101–overexpressed cholangiocarcinoma cells by using complementary in vitro and in vivo assays. For the in vitro assay, human umbilical vein endothelial cells were incubated with the conditioned media collected from human cholangiocarcinoma cells transduced with the miR-101 or control lentivirus. As shown in Figure 6, the relative capillary tube length and numbers of branch points were significantly lower in human umbilical vein endothelial cells incubated with the miR-101 conditioned medium; this phenomenon no longer was observed in the presence of soluble VEGF receptor 1, Flt-1. For the in vivo assay, concentrated conditioned media of miR-101–overexpressed CCLP1 and HuCCT1 cells and their respective scrambled control cells were mixed with Matrigel and injected subcutaneously into the ventral flank of C57BL/6 mice (10 days after the injection the Matrigel plugs were recovered for further analysis). As shown in Figure 7A, the miR-101 plugs were a paler color and had lower hemoglobin concentrations compared with the control plugs. Histologic examination (H&E stain) and immunostaining for the vascular endothelial marker CD31 showed decreased capillary numbers in the miR-101 plugs compared with the control plugs (Figure 7, B and D). These results, together with the blood vessel analysis of xenograft tumor tissues from SCID mice (Figure 2F), show that miR-101 inhibits cholangiocarcinoma angiogenesis.
Figure 6.
miR-101 suppresses tube formation of human umbilical vein endothelial cells in vitro. Human umbilical vein endothelial cells (3 × 104) in 100 μL 200PRF medium were mixed with 400 μL conditional medium from indicated cells and plated in 24-well plates coated with growth factor–reduced Matrigel. The cells were incubated for 3 to 5 hours and stained with Calcein AM before immunofluorescence microscopy. A: Representative images of human umbilical vein endothelial cell tube formation. B: Upper panel: Normalized tube length of cells with indicated transduction or treatment. Lower panel: Branching points of cells with indicated transduction or treatment. The tube length and branching point of each scramble control was set as 100%. The data are expressed as means ± SEM from three independent experiments. L/control, control lentivirus; L/miR101, miR-101-1 lentivirus. ∗P < 0.05, ∗∗P < 0.01.
Figure 7.
miR-101 suppresses angiogenesis in vivo. The Matrigel plug assay was performed by s.c. injection of a 200-μL Matrigel plug into C56B/6 mice. Plugs were removed after 10 days and portions of each plug were homogenized for a hemoglobin assay or fixed for H&E or immunohistochemical staining. A: Upper panels: Matrigel plugs recovered from mice (n = 3). Lower panel: Normalized hemoglobin levels in the plugs. Data are shown as means ± SEM from three plugs. B: The average number of capillaries in plugs from each group. A qualified capillary was defined as a ring or cylindric structure with an internal endothelial cell lining (CD31+) with red blood cells in it. Data represent means ± SD from three plugs. C: Schematic representation of the mechanisms by which miR-101 regulates cholangiocarcinoma angiogenesis. D: Repetitive images of H&E stain and CD31 immunostain. Insets are the enlarged images of areas highlighted by the green rectangles; arrows indicate capillaries. L/control, control lentivirus; L/miR101, miR-101-1 lentivirus. ∗P < 0.05.
Discussion
This study showed novel evidence for the expression, function, and mechanism of miR-101 in human cholangiocarcinoma. We showed that the level of miR-101 is decreased in approximately 43% of human cholangiocarcinomas and that miR-101 inhibits cholangiocarcinogenesis and tumor progression. The anticholangiocarcinoma effect of miR-101 is mediated predominantly through the inhibition of angiogenesis rather than through inhibition of tumor cell proliferation. This assertion is supported by the following observations1: the conditioned medium from miR-101–overexpressed cholangiocarcinoma cells decreased angiogenesis in vitro and in vivo,2 miR-101 overexpression only slightly decreased cholangiocarcinoma cell proliferation, and3 miR-101–overexpressed cholangiocarcinoma in SCID mice showed decreased blood vessel density.
Our findings showed that miR-101 inhibits cholangiocarcinoma angiogenesis by targeting the 3′UTR of VEGF mRNA and by repressing VEGF gene transcription (via inhibition of COX-2/PGE2) (Figure 7C). Direct targeting of VEGF by miR-101 is indicated by the following observations1: a complementary sequence of miR-101 is present in the 3′UTR of VEGF mRNA,2 miR-101 overexpression decreases VEGF 3′UTR luciferase reporter activity in cholangiocarcinoma cells and this effect was abolished by mutation of the miR-101 binding site, and3 miR-101 overexpression decreases VEGF mRNA and protein levels in cholangiocarcinoma cells and this phenomenon persists with inhibition of transcription by actinomycin D. To our knowledge, this is the first study to show that miR-101 targets VEGF in human cells.
In parallel, our data further indicate that miR-101 also binds to the complementary sequence in the 3′UTR of COX-2 mRNA, leading to a decrease of COX-2 protein translation and a subsequent reduction of PGE2 synthesis. The latter observation is consistent with a recent study by Strillacci et al22 that showed that miR-101 targets COX-2 in human colon cancer cells. Interestingly, Strillacci et al35 also showed that COX-2/PGE2 can regulate miR-101 expression negatively in colorectal cancer cells. Thus, the interplay between miR-101 and COX-2 signaling pathways likely is important for the regulation of carcinogenesis and tumor progression. Given the documented inhibition of COX-2 by miR-101, it is conceivable that loss of miR-101 may contribute to the up-regulation and amplification of COX-2/PGE2 signaling during the multiple steps of the carcinogenic process. On the other hand, it also is possible that activation of COX-2/PGE2 signaling during carcinogenesis may represent a key mechanism for the reduction of miR-101 in cancer tissues. In this study, we showed that COX-2/PGE2 and miR-101 influenced VEGF level in cholangiocarcinoma cells through both transcriptional and post-transcriptional regulation. The role of COX-2/PGE2 signaling in VEGF gene transcription in cholangiocarcinoma cells is corroborated further by the observations that COX-2 transfection or PGE2 treatment increases VEGF promoter activity and protein synthesis whereas siRNA depletion of COX-2 reduces VEGF promoter reporter activity. Thus, miR-101 decreases VEGF transcription in cholangiocarcinoma cells, at least in part, through inhibition of COX-2/PGE2.
Su et al23 showed that miR-101 inhibits hepatocellular carcinoma growth and the effect is mediated in part through targeting Mcl-1. In our study, we observed that miR-101 did not significantly influence the level of Mcl-1 protein in human cholangiocarcinoma cells. Thus, miR-101 may inhibit hepatic cancer growth by targeting different molecules based on specific tumor types and their signaling networks.
In summary, this study provided novel evidence that miR-101 inhibits cholangiocarcinoma angiogenesis by targeting VEGF directly and indirectly via inhibition of COX-2–derived PGE2 signaling. Our findings, along with the work of Su et al,23 warrant further investigation to explore the possibility of miR-101 therapy for hepatocellular carcinoma and cholangiocarcinoma, the two most common primary cancers of the liver.
Footnotes
Supported by NIH grants CA102325, CA106280, CA134568, and DK077776 (T.W.).
Supplemental Data
U6 snRNA specific and scrambled probes were used as system control for in situ hybridization (ISH) experiments in patient slides. Positive signals were shown as dark blue; nuclei were counter stained as red. A and C: Strong U6 signal in nuclei of the bile duct epithelial cells (and adjacent inflammatory/stromal cells. B and D: Negative staining of scrambled control probe in the bile duct epithelial cells (and adjacent inflammatory/stromal cells). Original magnification, ×400 (C and D) represent high magnifications of A and B (×100), respectively.
The effect of miR-101 on human cholangiocarcinoma cell growth in vitro. Human cholangiocarcinoma cell lines (CCLP1 and HuCCT1) stably transduced with miR-101-1 lentivirus (indicated as L/miR-101) and control lentivirus (indicated as L/control) were analyzed for growth in vitro by WST-1. One thousand cells were plated in each well of 96-well plates, 100μL 1:10 diluted WST-1 reagent was added to each well at indicated time point, and the cells were incubated at 37°C for 3 hours. The absorption was measured at 450 nm. The data represent means ± SEM from three independent experiments.
Representative Ki-67 immunostain and H&E stain of xenograft cholangiocarcinoma tissues. A: Overall Ki-67 staining of miR-101 overexpressed and control xenograft tumors. Arrows indicate necrotic areas. B and C: H&E stain and Ki-67 immunostain in the tumor edge areas.
Representative Western blot for COX-2 in cholangiocarcinoma cells transfected with anti-miR101 or control (β-actin was used as loading control). Anti-miR101 increased COX-2 protein level in CCLP1 and HuCCT1 cells (note the high basal level of COX-2 expression in HuCCT1 cells).
The levels of VEGF mRNA in HuCCT1 cells with indicated treatments for different times. The cells were transfected with anti-miR101, pre-miR101, or control in medium containing 1μg/ml actinomycin D. At the indicated time points, VEGF mRNA was measured by real time-PCR assay. *P < 0.05, **P < 0.01 compared with ActD + Control group.
References
- 1.Blechacz B., Gores G.J. Cholangiocarcinoma: advances in pathogenesis, diagnosis, and treatment. Hepatology. 2008;48:308–321. doi: 10.1002/hep.22310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sirica A.E. Cholangiocarcinoma: molecular targeting strategies for chemoprevention and therapy. Hepatology. 2005;41:5–15. doi: 10.1002/hep.20537. [DOI] [PubMed] [Google Scholar]
- 3.Wu T. Cyclooxygenase-2 and prostaglandin signaling in cholangiocarcinoma. Biochim Biophys Acta. 2005;1755:135–150. doi: 10.1016/j.bbcan.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 4.Berthiaume E.P., Wands J. The molecular pathogenesis of cholangiocarcinoma. Semin Liver Dis. 2004;24:127–137. doi: 10.1055/s-2004-828890. [DOI] [PubMed] [Google Scholar]
- 5.Fava G., Lorenzini I. Molecular pathogenesis of cholangiocarcinoma. Int J Hepatol. 2012;2012:630543. doi: 10.1155/2012/630543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lazaridis K.N., Gores G.J. Primary sclerosing cholangitis and cholangiocarcinoma. Semin Liver Dis. 2006;26:42–51. doi: 10.1055/s-2006-933562. [DOI] [PubMed] [Google Scholar]
- 7.Malhi H., Gores G.J. Cholangiocarcinoma: modern advances in understanding a deadly old disease. J Hepatol. 2006;45:856–867. doi: 10.1016/j.jhep.2006.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Patel T. Cholangiocarcinoma–controversies and challenges. Nat Rev Gastroenterol Hepatol. 2011;8:189–200. doi: 10.1038/nrgastro.2011.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tyson G.L., El-Serag H.B. Risk factors for cholangiocarcinoma. Hepatology. 2011;54:173–184. doi: 10.1002/hep.24351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tang D., Nagano H., Yamamoto H., Wada H., Nakamura M., Kondo M., Ota H., Yoshioka S., Kato H., Damdinsuren B., Marubashi S., Miyamoto A., Takeda Y., Umeshita K., Dono K., Wakasa K., Monden M. Angiogenesis in cholangiocellular carcinoma: expression of vascular endothelial growth factor, angiopoietin-1/2, thrombospondin-1 and clinicopathological significance. Oncol Rep. 2006;15:525–532. [PubMed] [Google Scholar]
- 11.Glaser S.S., Gaudio E., Alpini G. Vascular factors, angiogenesis and biliary tract disease. Curr Opin Gastroenterol. 2010;26:246–250. doi: 10.1097/MOG.0b013e3283369d19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gately S. The contributions of cyclooxygenase-2 to tumor angiogenesis. Cancer Metastasis Rev. 2000;19:19–27. doi: 10.1023/a:1026575610124. [DOI] [PubMed] [Google Scholar]
- 13.Hayashi N., Yamamoto H., Hiraoka N., Dono K., Ito Y., Okami J., Kondo M., Nagano H., Umeshita K., Sakon M., Matsuura N., Nakamori S., Monden M. Differential expression of cyclooxygenase-2 (COX-2) in human bile duct epithelial cells and bile duct neoplasm. Hepatology. 2001;34:638–650. doi: 10.1053/jhep.2001.28198. [DOI] [PubMed] [Google Scholar]
- 14.Endo K., Yoon B.I., Pairojkul C., Demetris A.J., Sirica A.E. ERBB-2 overexpression and cyclooxygenase-2 up-regulation in human cholangiocarcinoma and risk conditions. Hepatology. 2002;36:439–450. doi: 10.1053/jhep.2002.34435. [DOI] [PubMed] [Google Scholar]
- 15.Chariyalertsak S., Sirikulchayanonta V., Mayer D., Kopp-Schneider A., Furstenberger G., Marks F., Muller-Decker K. Aberrant cyclooxygenase isozyme expression in human intrahepatic cholangiocarcinoma. Gut. 2001;48:80–86. doi: 10.1136/gut.48.1.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sirica A.E., Lai G.H., Endo K., Zhang Z., Yoon B.I. Cyclooxygenase-2 and ERBB-2 in cholangiocarcinoma: potential therapeutic targets. Semin Liver Dis. 2002;22:303–313. doi: 10.1055/s-2002-34507. [DOI] [PubMed] [Google Scholar]
- 17.Sirica A.E., Lai G.H., Zhang Z. Biliary cancer growth factor pathways, cyclo-oxygenase-2 and potential therapeutic strategies. J Gastroenterol Hepatol. 2001;16:363–372. doi: 10.1046/j.1440-1746.2001.02438.x. [DOI] [PubMed] [Google Scholar]
- 18.Valencia-Sanchez M.A., Liu J., Hannon G.J., Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2006;20:515–524. doi: 10.1101/gad.1399806. [DOI] [PubMed] [Google Scholar]
- 19.Betel D., Koppal A., Agius P., Sander C., Leslie C. Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome Biol. 2010;11:R90. doi: 10.1186/gb-2010-11-8-r90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Betel D., Wilson M., Gabow A., Marks D.S., Sander C. The microRNA.org resource: targets and expression. Nucleic Acids Res. 2008;36:D149–D153. doi: 10.1093/nar/gkm995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Au S.L., Wong C.C., Lee J.M., Fan D.N., Tsang F.H., Ng I.O., Wong C.M. Enhancer of zeste homolog 2 epigenetically silences multiple tumor suppressor microRNAs to promote liver cancer metastasis. Hepatology. 2012;56:622–631. doi: 10.1002/hep.25679. [DOI] [PubMed] [Google Scholar]
- 22.Strillacci A., Griffoni C., Sansone P., Paterini P., Piazzi G., Lazzarini G., Spisni E., Pantaleo M.A., Biasco G., Tomasi V. MiR-101 downregulation is involved in cyclooxygenase-2 overexpression in human colon cancer cells. Exp Cell Res. 2009;315:1439–1447. doi: 10.1016/j.yexcr.2008.12.010. [DOI] [PubMed] [Google Scholar]
- 23.Su H., Yang J.R., Xu T., Huang J., Xu L., Yuan Y., Zhuang S.M. MicroRNA-101, down-regulated in hepatocellular carcinoma, promotes apoptosis and suppresses tumorigenicity. Cancer Res. 2009;69:1135–1142. doi: 10.1158/0008-5472.CAN-08-2886. [DOI] [PubMed] [Google Scholar]
- 24.Thu K.L., Chari R., Lockwood W.W., Lam S., Lam W.L. miR-101 DNA copy loss is a prominent subtype specific event in lung cancer. J Thorac Oncol. 2011;6:1594–1598. doi: 10.1097/JTO.0b013e3182217d81. [DOI] [PubMed] [Google Scholar]
- 25.Vilardo E., Barbato C., Ciotti M., Cogoni C., Ruberti F. MicroRNA-101 regulates amyloid precursor protein expression in hippocampal neurons. J Biol Chem. 2010;285:18344–18351. doi: 10.1074/jbc.M110.112664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hou J., Lin L., Zhou W., Wang Z., Ding G., Dong Q., Qin L., Wu X., Zheng Y., Yang Y., Tian W., Zhang Q., Wang C., Zhuang S.M., Zheng L., Liang A., Tao W., Cao X. Identification of miRNomes in human liver and hepatocellular carcinoma reveals miR-199a/b-3p as therapeutic target for hepatocellular carcinoma. Cancer Cell. 2011;19:232–243. doi: 10.1016/j.ccr.2011.01.001. [DOI] [PubMed] [Google Scholar]
- 27.Adams E.J., Green J.A., Clark A.H., Youngson J.H. Comparison of different scoring systems for immunohistochemical staining. J Clin Pathol. 1999;52:75–77. doi: 10.1136/jcp.52.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang J., Han C., Wu T. MicroRNA-26a promotes cholangiocarcinoma growth by activating beta-catenin. Gastroenterology. 2012;143:246–256.e248. doi: 10.1053/j.gastro.2012.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang X., Klein R.D. Prostaglandin E2 induces vascular endothelial growth factor secretion in prostate cancer cells through EP2 receptor-mediated cAMP pathway. Mol Carcinog. 2007;46:912–923. doi: 10.1002/mc.20320. [DOI] [PubMed] [Google Scholar]
- 30.Bradbury D., Clarke D., Seedhouse C., Corbett L., Stocks J., Knox A. Vascular endothelial growth factor induction by prostaglandin E2 in human airway smooth muscle cells is mediated by E prostanoid EP2/EP4 receptors and SP-1 transcription factor binding sites. J Biol Chem. 2005;280:29993–30000. doi: 10.1074/jbc.M414530200. [DOI] [PubMed] [Google Scholar]
- 31.Iniguez M.A., Rodriguez A., Volpert O.V., Fresno M., Redondo J.M. Cyclooxygenase-2: a therapeutic target in angiogenesis. Trends Mol Med. 2003;9:73–78. doi: 10.1016/s1471-4914(02)00011-4. [DOI] [PubMed] [Google Scholar]
- 32.Abdel-Majid R.M., Marshall J.S. Prostaglandin E2 induces degranulation-independent production of vascular endothelial growth factor by human mast cells. J Immunol. 2004;172:1227–1236. doi: 10.4049/jimmunol.172.2.1227. [DOI] [PubMed] [Google Scholar]
- 33.Chang S.H., Liu C.H., Wu M.T., Hla T. Regulation of vascular endothelial cell growth factor expression in mouse mammary tumor cells by the EP2 subtype of the prostaglandin E2 receptor. Prostaglandins Other Lipid Mediat. 2005;76:48–58. doi: 10.1016/j.prostaglandins.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 34.Liu X.H., Kirschenbaum A., Lu M., Yao S., Dosoretz A., Holland J.F., Levine A.C. Prostaglandin E2 induces hypoxia-inducible factor-1alpha stabilization and nuclear localization in a human prostate cancer cell line. J Biol Chem. 2002;277:50081–50086. doi: 10.1074/jbc.M201095200. [DOI] [PubMed] [Google Scholar]
- 35.Strillacci A., Valerii M.C., Sansone P., Caggiano C., Sgromo A., Vittori L., Fiorentino M., Poggioli G., Rizzello F., Campieri M., Spisni E. Loss of miR-101 expression promotes Wnt/beta-catenin signalling pathway activation and malignancy in colon cancer cells. J Pathol. 2013;229:379–389. doi: 10.1002/path.4097. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
U6 snRNA specific and scrambled probes were used as system control for in situ hybridization (ISH) experiments in patient slides. Positive signals were shown as dark blue; nuclei were counter stained as red. A and C: Strong U6 signal in nuclei of the bile duct epithelial cells (and adjacent inflammatory/stromal cells. B and D: Negative staining of scrambled control probe in the bile duct epithelial cells (and adjacent inflammatory/stromal cells). Original magnification, ×400 (C and D) represent high magnifications of A and B (×100), respectively.
The effect of miR-101 on human cholangiocarcinoma cell growth in vitro. Human cholangiocarcinoma cell lines (CCLP1 and HuCCT1) stably transduced with miR-101-1 lentivirus (indicated as L/miR-101) and control lentivirus (indicated as L/control) were analyzed for growth in vitro by WST-1. One thousand cells were plated in each well of 96-well plates, 100μL 1:10 diluted WST-1 reagent was added to each well at indicated time point, and the cells were incubated at 37°C for 3 hours. The absorption was measured at 450 nm. The data represent means ± SEM from three independent experiments.
Representative Ki-67 immunostain and H&E stain of xenograft cholangiocarcinoma tissues. A: Overall Ki-67 staining of miR-101 overexpressed and control xenograft tumors. Arrows indicate necrotic areas. B and C: H&E stain and Ki-67 immunostain in the tumor edge areas.
Representative Western blot for COX-2 in cholangiocarcinoma cells transfected with anti-miR101 or control (β-actin was used as loading control). Anti-miR101 increased COX-2 protein level in CCLP1 and HuCCT1 cells (note the high basal level of COX-2 expression in HuCCT1 cells).
The levels of VEGF mRNA in HuCCT1 cells with indicated treatments for different times. The cells were transfected with anti-miR101, pre-miR101, or control in medium containing 1μg/ml actinomycin D. At the indicated time points, VEGF mRNA was measured by real time-PCR assay. *P < 0.05, **P < 0.01 compared with ActD + Control group.