Abstract
Background
In a prior study, we developed methods for automatically identifying associations between medications and problems using association rule mining on a large clinical data warehouse and validated these methods at a single site which used a self-developed electronic health record.
Objective
To demonstrate the generalizability of these methods by validating them at an external site.
Methods
We received data on medications and problems for 263,597 patients from the University of Texas Health Science Center at Houston Faculty Practice, an ambulatory practice that uses the Allscripts Enterprise commercial electronic health record product. We then conducted association rule mining to identify associated pairs of medications and problems and characterized these associations with five measures of interestingness: support, confidence, chi-square, interest and conviction and compared the top-ranked pairs to a gold standard.
Results
25,088 medication-problem pairs were identified that exceeded our confidence and support thresholds. An analysis of the top 500 pairs according to each measure of interestingness showed a high degree of accuracy for highly-ranked pairs.
Conclusion
The same technique was successfully employed at the University of Texas and accuracy was comparable to our previous results. Top associations included many medications that are highly specific for a particular problem as well as a large number of common, accurate medication-problem pairs that reflect practice patterns.
Keywords: Problem list, clinical decision support, data mining, automated inference, methodology
1. Background and Significance
Electronic health records (EHRs) have significant potential to improve the delivery of healthcare services, particularly if well implemented and used. Recognizing their potential, congress passed the Health Information Technology for Economic and Clinical Health (HITECH) Act as part of the American Recovery and Reinvestment Act in 2009 and developed standards for “meaningful use” of EHRs, including a requirement for maintaining an up-to-date and accurate problem list [1]. Further, federal EHR certification requirements require EHRs to support structured, coded problem lists and the Joint Commission requires clinicians to document patient problems [2].
An accurate problem list is important for several reasons. First and most directly, the problem list is a key part of the medical record, and an accurate problem list facilitates medical decision making by healthcare. Further, problem lists are also used for clinical decision support [3, 4], quality measurement and clinical research. Previous research has suggested that patients with complete problem lists are more likely to receive evidence-based care [5]. However, problem lists often remain incomplete [6], with many physicians failing to take ownership of the problem list and keep it up-to-date and accurate [7-9].
Researchers, including our own team, have explored a variety of techniques to improve the accuracy of problem list completeness. Galanter [10] and Carpenter [11] have both explored using medication orders to identify potential patient problems based on indication data. Meystre [12, 13] and others have used natural language processing and we have explored crowd-sourcing [14] and data mining techniques [6, 15, 16] to enhance problem list and, in a recent randomized trial of our data mining approach, demonstrated a three-fold increase in problem list documentation after showing physicians alerts that their patient may have an undocumented problem [16].
One particularly promising technique is the use of “proxy” data in the EHR, such as medications, laboratory results or billing data to infer a patient’s problems [15]. In a prior study, we developed a set of data mining techniques, based on association rule mining, to identify linkages between problems and medications and laboratory results [15] that we then used to infer patient problems. In brief, association rule mining looks for co-occurrence relationships between data elements [17, 18], such as between medications and laboratory results in patients’ medical records.
In our prior study, we used association rule mining to identify associations between coded problems and structured data (medications and lab results) for a sample of 100,000 patients at the Brigham and Women’s Hospital (BWH, Boston, MA). We evaluated candidate associations using five cooccurrence statistics: support, confidence, chi-square, interest, and conviction. We identified 10,735 medication-problem associations with high chi-square statistic performance and 5361 laboratory test-problem associations with high interest statistic.
An important limitation of the previously described studies, including our own, is their use at a single academic site with fairly advanced internally-developed clinical systems. However, in our paper, we posited that the techniques we developed would generalize successfully to other sites with different patient and provider populations and different EMRs [19].
If successful, this generalizability would have two main benefits. First, it would allow other institutions to use the method to build problem-medication knowledge bases that reflect their own local practice and, critically, their own coding systems. Since this approach uses natively coded data, the associations produced are already encoded according to the codes in use at the institution. Second, and perhaps even more useful, there may be synergies in combining data from multiple institutions. For example, a condition that is rarely seen at one institution may not produce a strong enough association to surface during the data mining process, but if the process is repeated at another institution where the condition is more commonly seen, a reliable association may be detected. With the appropriate terminology mappings and ensemble methods for combining the mined associations, a more robust knowledge base could emerge. Moreover, these associations could also be combined with the (potentially complementary) associations found by the other approaches discussed previously, including NLP and crowdsourcing approaches.
In this study, we assess the feasibility of generalizing the approach by repeating it at the University of Texas Health Science Center at Houston Faculty Practice (UT). UT’s patient and provider population are distinct from the BWH population and, critically, UT uses a common commercially-developed EHR: the Allscripts Enterprise EHR.
2. Methods
To find associations between medications and problems at UT, we employed association rule mining, a technique which is widely used in computer science, data mining and electronic commerce [20-22]. Association rule mining assumes a database of “items” and a set of “transactions”. In the commerce scenario, the items might be products sold in a store, and the transactions might be records of purchases made in that store. In this characterization, each transaction is a set of items purchased together. In our paper, we conceive of the items as the set of medications and problems used in the Allscripts EHR at UT, and the transactions as individual patient records, so a particular patients “transaction” consists of all of his or her medications and problems.
Using this conception, an association rule, A → B, then, is a pair of items (or sets of items) that are related to each other based on their frequency of co-occurrence in patient records. For example, a hypothetical association rule might be “insulin → diabetes” with support 500 and confidence 90%. This rule would mean that 500 patients were on insulin and had diabetes on their problem list, and that 90% of patients with insulin on their medication list also had diabetes on their problem list.
Association rule mining, then, is a technique for efficiently identifying association rules with good predictive power from a dataset. Any pairwise combination of items (or sets of items) could, theoretically, form an association rule, so we employee several statistical measures of “interestingness” to differentiate rules which are likely to be predictive from those which are not. The measures we employ are:
-
•
Support: the number of patients with both the medication and problem in their record. This is a relatively crude measure, which is often useful as a threshold (e.g. we might only consider rules with a support of at least 5), but which is often high for pairs of common but unrelated items.
-
•
Confidence: the percentage of patients with the medication in their record who also have the problem. This measure is more sensitive than support, but is still subject to elevation when the problem is very common (e.g. if 15% of patients suffer from hyperlipidemia, then an association rule such as amoxicillin hyperlipidemia is likely to have 15% support, even though there is no connection between the drug and the disease, simply because 15% of patients have the disease).
-
•
Interest: the confidence divided by the baseline probability of the disease, thus correcting for the high-frequency problem issue that affects confidence.
-
•
Chi-square: its usual Pearson formulation, which has a well-known underlying probability distribution.
-
•
Conviction: the ratio of the observed association between the medication and disease and the difference expected if the association were due just to chance. This measure is discussed in elsewhere [15, 19].
Our prior paper gives more specifics on the measures and algorithms employed, as well as more formalism regarding our statistical approach [15]. The identical approach is employed in this study.
We focus, here, on validation of medication-problem associations, as these associations were much more successful in our previous study than medication-lab interactions, and because a better gold standard is available for medications. We hypothesized that frequent item set mining and association set mining approaches that we previously developed and employed at BWH would generalize successfully to UT. We identified all patients seen at UT between April, 2004 and May, 2011 with medication and problem list data available in the EHR. We used the UT Clinical Data Warehouse for our query. For each of the patients, we requested structured problems and medications that were stored in the Allscripts Enterprise EHR, a commercially developed system. The UT All-scripts EHR is an outpatient system, so data used in this study were taken from outpatient records. Problems in the UT system were coded using proprietary codes that were often mapped to ICD-9 and medications were coded using NDC codes from Medispan. When ICD-9 mappings were available, we excluded ICD-9 V codes (supplementary classification of factors influencing health status and contact with health services) from our analysis. As at Partners, the UT medication and problem list data were collected during normal clinical practice and are unaudited, so likely contain some gaps or inaccuracies, which imposes an upper bound on the accuracy of the associations found (e.g. the highest confidence for a true rule is bounded by the completeness of documentation of its member disease). Our protocol was reviewed and approved by The University of Texas Health Science Center at Houston’s Committee for the Protection of Human Subjects.
To carry out the data analysis, we used the same software that was developed for the previous study [23]. In short, the analysis software was developed in C#, compiled using Microsoft Visual Studio 2008 for the .NET 3.5 Common Language Runtime. We used the software to identify medication-problem pairs with support of at least 5 and confidence of at least 10%, meaning that at least five patients had both the medication and problem in their record and that at least 10% of patients with the medication in their record also had the problem. There was no upper bound on the support or confidence measures. We then identified the top 500 association pairs for each of the five measures of interestingness.
After the top medication-problem associations were identified, we conducted a manual review by comparing them to a gold standard: the indications listed in the Lexi-Comp drug information compendium, which contains data on both on- and off-label indications for medications. This is the same gold standard employed in our prior study. Two reviewers (a research assistant and a medical student) collaboratively reviewed each medication-problem and compared it to the gold standard, judging each pair as either indicated or not indicated.
3. Results
We received data for 263,597 UT patients, consisting of 1,182,557 coded problems and 792,962 coded medications. A total of 22,648 distinct problem codes and 16,249 distinct medication codes were represented in our data set.
After competing our association-rule mining, a total of 25,088 medication-problem pairs were identified that exceeded our confidence and support threshold. These associations were then characterized with the five measures of interestingness. ► Table 1 shows the top 50 medication-problem associations in the UT data based on the chi-square statistic. All of the top 50 associations were clinically valid when compared to the gold standard and many were very specific (often for rare diseases), such as recombinant coagulation factor IX (Benefix) for factor IV deficiency and phenylalanine-free medical food (Phenex-2) for phenylketonuria.
Table 1.
Top 50 medication-problem associations in the UT data according to the chi-square measure of interestingness.
| Rank | Medication | Problem | Support | Confidence | Chi-square | Interest |
|---|---|---|---|---|---|---|
| 1 | Permethrin | Scabies | 231 | 63.64% | 146,662.62 | 873.66 |
| 2 | Griseofulvin Microsize | Tinea Capitis | 218 | 75.17% | 84,855.09 | 222.67 |
| 3 | BeneFIX | Factor IX Deficiency | 9 | 100.00% | 81,799.76 | 2,820.90 |
| 4 | Glucagon Emergency | Type I Diabetes Mellitus – Uncontrolled | 222 | 52.86% | 72,227.04 | 320.95 |
| 5 | Levothyroxine Sodium | Hypothyroidism | 3016 | 52.67% | 64,909.58 | 20.91 |
| 6 | MetroNIDAZOLE | Bacterial Vaginosis | 1272 | 27.43% | 62,391.93 | 159.69 |
| 7 | Isoniazid | Latent Tuberculosis | 106 | 34.75% | 59,105.14 | 1,038.86 |
| 8 | AquADEKs | Pancreatic Insufficiency | 6 | 60.00% | 55,814.26 | 5,472.60 |
| 9 | LevOCARNitine | Disorder Of Mitochondrial Metabolism | 87 | 31.64% | 55,294.08 | 1,336.34 |
| 10 | Terconazole | Vaginal Candidiasis | 454 | 36.15% | 54,906.72 | 196.20 |
| 11 | Phenex-2 | Phenylketonuria | 8 | 100.00% | 54,064.82 | 1,386.44 |
| 12 | Lupron Depot-Ped | Precocious Puberty | 37 | 69.81% | 54,000.47 | 614.33 |
| 13 | Glucagon Emergency | Visit For: Fit/Adjust Insulin Pump | 119 | 28.33% | 53,420.80 | 1,138.34 |
| 14 | Pyridostigmine Bromide | Myasthenia Gravis | 45 | 58.44% | 52,869.76 | 691.21 |
| 15 | Levemir FlexPen | Taking Medication For Diabetes Long-term Use Of Insulin | 251 | 41.97% | 51,531.66 | 230.30 |
| 16 | Copaxone | Multiple Sclerosis | 406 | 84.06% | 50,619.37 | 34.39 |
| 17 | Truvada | HIV Infection | 230 | 65.71% | 50,189.30 | 97.14 |
| 18 | Solu-CORTEF | Congenital Adrenal Hyperplasia | 25 | 40.98% | 49,986.43 | 2,259.92 |
| 19 | Mestinon | Myasthenia Gravis | 39 | 62.90% | 49,321.72 | 599.05 |
| 20 | Viagra | Male Erectile Disorder | 528 | 36.26% | 47,376.32 | 125.76 |
| 21 | Mebendazole | Enterobiasis (Pinworm) | 27 | 23.89% | 47,210.21 | 5,491.60 |
| 22 | Nystatin | Oral Thrush | 676 | 22.21% | 46,168.09 | 249.56 |
| 23 | Methotrexate | Rheumatoid Arthritis | 479 | 41.29% | 43,468.27 | 90.07 |
| 24 | Riboflavin | Disorder Of Mitochondrial Metabolism | 37 | 55.22% | 41,071.64 | 568.33 |
| 25 | Norvir | HIV Infection | 186 | 65.49% | 40,439.78 | 78.56 |
| 26 | Elmiron | Chronic Interstitial Cystitis | 25 | 36.23% | 40,438.36 | 1,893.11 |
| 27 | NovoLOG FlexPen | Taking Medication For Diabetes Long-term Use Of Insulin | 258 | 32.09% | 40,406.22 | 236.72 |
| 28 | Synthroid | Hypothyroidism | 2154 | 45.91% | 39,694.00 | 14.93 |
| 29 | Coenzyme Q10 | Disorder Of Mitochondrial Metabolism | 53 | 34.42% | 36,636.56 | 814.09 |
| 30 | Permethrin Lice Treatment | Pediculosis Capitis | 8 | 66.67% | 36,038.43 | 1,386.44 |
| 31 | Ergocalciferol | Vitamin D Deficiency | 449 | 55.78% | 35,955.42 | 36.17 |
| 32 | Kaletra | HIV Infection | 159 | 67.66% | 35,716.97 | 67.16 |
| 33 | Griseofulvin Ultramicrosize | Tinea Capitis | 92 | 74.80% | 35,607.94 | 93.97 |
| 34 | Pen Needles 5/16” | Taking Medication For Diabetes Long-term Use Of Insulin | 192 | 37.14% | 34,822.13 | 176.16 |
| 35 | Levemir FlexPen | Diabetes Mellitus Poorly Controlled | 342 | 57.19% | 34,308.87 | 40.88 |
| 36 | Atripla | HIV Infection | 154 | 66.67% | 34,081.10 | 65.04 |
| 37 | Gris-PEG | Tinea Capitis | 88 | 73.95% | 33,671.50 | 89.89 |
| 38 | Floxin Otic | Chronic Tubotympanic Suppurative Otitis Media | 89 | 31.79% | 33,475.92 | 476.02 |
| 39 | Pen Needles 5/16” | Diabetes Mellitus Poorly Controlled | 309 | 59.77% | 32,413.11 | 36.94 |
| 40 | Reyataz | HIV Infection | 139 | 69.85% | 32,239.37 | 58.71 |
| 41 | Colistimethate Sodium | Pseudomonas Wound Infection | 12 | 17.14% | 31,883.98 | 10,945.20 |
| 42 | Methimazole | Graves’ Disease | 108 | 32.24% | 31,398.06 | 336.18 |
| 43 | Zolpidem Tartrate | Insomnia | 927 | 28.58% | 31,389.00 | 53.16 |
| 44 | Cialis | Male Erectile Disorder | 312 | 40.36% | 31,148.37 | 74.31 |
| 45 | Warfarin Sodium | Atrial Fibrillation | 695 | 28.62% | 30,999.96 | 67.78 |
| 46 | Malathion | Pediculosis Capitis | 6 | 75.00% | 30,408.46 | 1,039.83 |
| 47 | Polyethylene Glycol 3350 | Constipation | 753 | 41.93% | 29,477.84 | 26.59 |
| 48 | Carbidopa-Levodopa | Parkinson’s Disease | 186 | 46.50% | 29,062.92 | 81.00 |
| 49 | Vitamin D (Ergocalciferol) | Vitamin D Deficiency | 374 | 54.05% | 28,984.88 | 30.13 |
| 50 | NovoLOG FlexPen | Diabetes Mellitus Poorly Controlled | 361 | 44.90% | 28,299.94 | 43.15 |
► Figure 1 shows the result of evaluation of top 500 medication-problem associations according to each of the five statistics of interests to the gold standard (LexiComp drug database). As in our prior study, Chi-square had the best performance, maintaining a higher accuracy than the rest of the statistics through the top associations. On the other hand, support had the worst accuracy, starting and ending below the rest of the statistics.
Fig. 1.
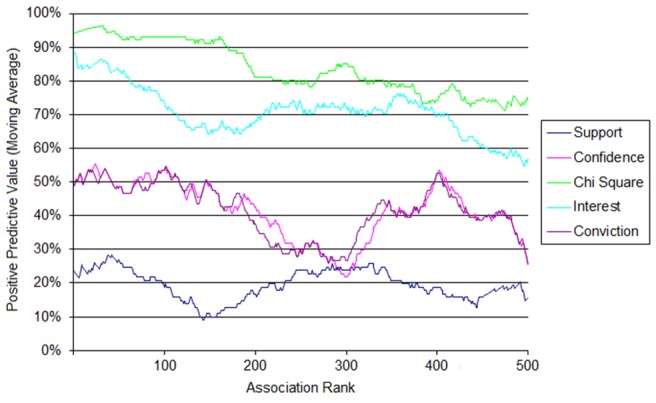
Moving average positive predictive value for the top 500 rules for each of the five measures of interestingness.
There were some important differences; however, in the top 50 medication-problem associations between BWH and UT. For example, while 17 of the top 50 medication-problem associations at BWH pertained to HIV or AIDS, only 5 out of 50 such association at University of Texas contained HIV or AIDS. Further, the UT data contains some associations involving medical supplies (particularly insulin syringes), while the Partners data does not have any top associations involving supplies. Some of these differences likely reflect differences in the underlying terminologies and dictionaries used in the system; Partners, for example, tracks insulin syringes separately from medications, while UT combines them.
4. Discussion
The technique previously employed at BWH to find medication-problem associations worked similarly on a commercially-designed EHR at The University of Texas Health Science Center at Houston. The technique achieved similar accuracy at both institutions with similar results. For example, chi-square statistic had the best performance while support statistic had the worst performance at both institutions. However, while support, confidence, chi-square, and interest statistics had similar accuracy across the top 500 medication-problem associations in the previous study at BWH, chi-square and interest statistics had visibly better accuracy than conviction and confidence statistics at UT. These results highlight the idea that statistics may not only differ on the type of data set (e.g. medication-problem associations versus laboratory results –problem associations) but also by usage patterns that may vary by institution. In practice, the best statistic (or combination of statistics) to use depends heavily on the use case being considered – for example, associations with high support are likely to be frequently occurring, so may represent high-impact targets, but associations with high interest are often more specific, and interest tends to favor less common diseases.
Our results show that the technique successfully generalized from a single academic medical center with an internally developed EHR to another center with different patient and provider populations and a commercially available EHR. The validation of the previously used technique at a different institution suggests that this method could be used on data sets with different characteristics with similar results. Importantly, using this technique offers several advantages over other knowledge-based techniques, including less manual work, which improves speed and time needed to find clinically relevant associations; ability to keep up with current information and repeat the technique as often as desired to stay up to date; ability to infer associations based on off-label use of medications; assignment of quantitative metrics to associations; and inherent use of terminologies as they are found in the EHR.
Our study has some limitations. First, we limited our validation to only one type of association (medication-problems); therefore, we do not know whether we would have had the same success with laboratory-problem associations. Nevertheless, since the medication-problem associations worked with such similar efficacy at both institutions, we believe that we would achieve the same results with the other type of association. We also did not employ other types of data, including unstructured free text or procedure histories, which could be used in the future to complement our results in this study.
One key future direction for research is assessing the reasons for differences in the associations found at different institutions. Although these differences likely reflect actual differences in the patient and provider populations, clinical practices and use of the EHR, adding a single additional site, as we did, does not provide sufficient data for a detailed analysis of the causes and implications of these differences. If this analysis were repeated at a larger number of sites, it might be possible to more fully understand the drivers of such differences. Even more critically, such a multi-site study would allow for a more thorough assessment of the extent to which the associations found in heterogeneous institutions’ knowledge bases are complementary, and whether patterns of associations could be useful for assessing their strength. It is likely that ensemble methods could be employed to develop a more complete and accurate knowledge base by combining the associations from many sites, assuming that terminology issues could be resolved appropriately.
Another future avenue for research is exploring the utility of these associations for improving problem list completeness. As discussed in the background, we used the associations found at BWH as part of a clinical decision support intervention to improve problem list completeness [16]. Although the intervention was successful, considerable manual work was required to convert the associations into a reliable, implementable knowledge base. Future research should study whether the larger and more robust knowledge bases that may be attainable through more widespread application of this data mining approach would yield sufficient accuracy that manual knowledge review and tuning would not be needed, or would at least be significantly reduced. If successful, this would speed the path from mined associations to a working intervention considerably.
5. Clinical Relevance Statement
An accurate knowledge base of medication-problem relationships would have important clinical advantages. For example, such a database could facilitate reconciliation of data in electronic health records, enable indication-based prescribing or allow the user to more easily identify untreated (or overtreated) conditions. In this paper, we validated a data mining method for developing such a knowledge base which had previously been developed at an academic medical center with a locally-developed EHR and showed that it readily translates to a commercially-developed EHR, paving the way for more widespread adoption of the technique.
Conflicts of Interest
The authors report no conflicts of interest.
Human Subjects Protections
Our study protocol was reviewed and approved by the University of Texas at Houston Committee for the Protection of Human Subjects.
Acknowledgements
This work was supported in part by a SHARP contract from the Office of the National Coordinator for Health Information Technology (ONC #10510592).
References
- 1.Blumenthal D, Tavenner M.The “meaningful use” regulation for electronic health records. The New England journal of medicine 2010; 363(6): 501-504 Epub 2010/07/22 [DOI] [PubMed] [Google Scholar]
- 2.Information Management Processes (Standard IM 6.40): 2008 Comprehensive Accreditation Manual for Hospitals: The Official Handbook Oakbrook Terrace, Illinois: 2008 [Google Scholar]
- 3.Wright A, Goldberg H, Hongsermeier T, Middleton B. A.description and functional taxonomy of rule-based decision support content at a large integrated delivery network. Journal of the American Medical Informatics Association : JAMIA 2007; 14(4): 489-496 Epub 2007/04/27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lobach DF, Hammond WE.Computerized decision support based on a clinical practice guideline improves compliance with care standards. The American journal of medicine 1997; 102(1): 89-98 Epub 1997/01/01. [DOI] [PubMed] [Google Scholar]
- 5.Szeto HC, Coleman RK, Gholami P, Hoffman BB, Goldstein MK.Accuracy of computerized outpatient diagnoses in a Veterans Affairs general medicine clinic. The American journal of managed care 2002; 8(1): 37-43 Epub 2002/01/30. [PubMed] [Google Scholar]
- 6.Wright A, Pang J, Feblowitz JC, Maloney FL, Wilcox AR, Ramelson HZ, Schneider LI, Bates DW.A method and knowledge base for automated inference of patient problems from structured data in an electronic medical record. Journal of the American Medical Informatics Association: JAMIA 2011; 18(6): 859-867 Epub 2011/05/27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wright A, Feblowitz J, Maloney FL, Henkin S, Bates DW.Use of an Electronic Problem List by Primary Care Providers and Specialists. Journal of general internal medicine 2012. Epub 2012/03/20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wright A, Maloney FL, Feblowitz JC.Clinician attitudes toward and use of electronic problem lists: a thematic analysis. BMC Med Inform Decis Mak 2011; 11: 36 Epub 2011/05/27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jao C, Hier D, Galanter W, Valenta A.Assessing physician comprehension of and attitudes toward problem list documentation. AMIA Annual Symposium proceedings / AMIA Symposium AMIA Symposium 2008: 990 Epub 2008/11/13. [PubMed] [Google Scholar]
- 10.Galanter WL, Hier DB, Jao C, Sarne D.Computerized physician order entry of medications and clinical decision support can improve problem list documentation compliance. Int J Med Inform 2010; 79(5): 332-338 Epub 2008/07/05. [DOI] [PubMed] [Google Scholar]
- 11.Carpenter JD, Gorman PN.Using medication list--problem list mismatches as markers of potential error. Proceedings / AMIA Annual Symposium AMIA Symposium 2002: 106-110 Epub 2002/12/05 [PMC free article] [PubMed] [Google Scholar]
- 12.Meystre SM, Haug PJ.Randomized controlled trial of an automated problem list with improved sensitivity. Int J Med Inform 2008; 77(9): 602-612 Epub 2008/02/19. [DOI] [PubMed] [Google Scholar]
- 13.Meystre S, Haug PJ.Automation of a problem list using natural language processing. BMC Med Inform Decis Mak 2005; 5: 30 Epub 2005/09/02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McCoy AB, Wright A, Laxmisan A, Ottosen MJ, McCoy JA, Butten D, Sittig DF.Development and evaluation of a crowdsourcing methodology for knowledge base construction: identifying relationships between clinical problems and medications. Journal of the American Medical Informatics Association: JAMIA 2012. Epub 2012/05/15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wright A, Chen ES, Maloney FL.An automated technique for identifying associations between medications, laboratory results and problems. Journal of biomedical informatics 2010; 43(6): 891-901 Epub 2010/10/05. [DOI] [PubMed] [Google Scholar]
- 16.Wright A, Pang J, Feblowitz JC, Maloney FL, Wilcox AR, McLoughlin KS, Ramelson H, Schneider L, Bates DW.Improving completeness of electronic problem lists through clinical decision support: a randomized, controlled trial. Journal of the American Medical Informatics Association: JAMIA 2012. Epub 2012/01/05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goethals B.Survey on frequent pattern mining. Univ of Helsinki. 2003. [Google Scholar]
- 18.Zhang C, Zhang S.Association rule mining: models and algorithms: Springer-Verlag; 2002 [Google Scholar]
- 19.Tan PN, Kumar V, Srivastava J, Selecting the right interestingness measure for association patterns. 2002: ACM [Google Scholar]
- 20.Hipp J, Güntzer U, Nakhaeizadeh G.Algorithms for association rule mining—a general survey and comparison. ACM SIGKDD Explorations Newsletter 2000; 2(1): 58-64 [Google Scholar]
- 21.Mobasher B, Dai H, Luo T, Nakagawa M, Effective personalization based on association rule discovery from web usage data. Proceedings of the 3rd international workshop on Web information and data management; 2001: ACM [Google Scholar]
- 22.Sarwar B, Karypis G, Konstan J, Riedl J, Analysis of recommendation algorithms for e-commerce. Proceedings of the 2nd ACM conference on Electronic commerce; 2000: ACM [Google Scholar]
- 23.Wright A CE, Maloney FL.An automated technique for identifying associations between medications, laboratory results, and problems. J Biomed Inf 2010; 43: 891-901 [DOI] [PubMed] [Google Scholar]


