Abstract
Heterochromatin is a higher order assembly that is characterized by a genomewide distribution, gene-repression, durability, and potential to spread. In this light, it is an appealing mechanism to interpret the neurobiology of complex brain disorders such as schizophrenia where down-regulation of expression appears to be the norm. H3K9 methylation (H3K9me) can initiate the seeding of a heterochromatin assembly on an inactive or poorly coordinated promoter as a consequence of a decline in transactivators either from disuse or misuse. H3K9me can extend its influence by spatial spreading through the mechanism of recursively recruiting adapters such as HP1 homodimers. HP1 itself serves as a platform for other repressive proteins such as DNA methyltransferases. In full color, heterochromatin can occupy genomewide gene networks, tissue specific ontologies, and even rearrange the nuclear architecture. Heterochromatin in the brain is modified by small molecule pharmacology, and serves a physiological role in the functioning of dopamine neurons and the construction of memory. From a therapeutic perspective, the durable nature of heterochromatin implies that it may require disassembly before the full genomic-potential of standard pharmacotherapies is achieved, especially in treatment resistant patients.
Keywords: chromosome, histone, transcription, histone methyltransferase, G9a, GLP
Introduction
Heterochromatin describes regions along the chromosome where DNA is tightly and restrictively packaged with histone and non-histone proteins. Tightly packed DNA (in the region of a promoter) is inaccessible to the free assembly and disassembly of transcriptional proteins, and consequently is less likely to be regulated or transcribed. It is our thesis that facultative heterochromatin may serve as an incubator of pathology by repressing genomewide gene networks. This outcome is a likely consequence of disease chronicity and misuse/disuse, resulting in a dearth of coordinated transcriptional activators (transactivators). Under these circumstances, the accumulation of repressive modifications may be favored for the reasons elaborated upon, resulting in a seeding, spread and encroachment by restrictive heterochromatin of previously ‘healthy’ chromosomal territory.
The process of heterochromatinization has been most thoroughly examined in non-neuronal tissue, but the key molecules involved are all expressed in the brain, suggesting neuronal function. The series of events leading to heterochromatin deposition on a promoter or DNA sequence likely depends on the biochemical context, but involves well characterized proteins and pathways. Initiation can begin with a decline of stimulatory signals and transactivators.1 This is followed by repressive histone modifications mediated primarily by histone methyltransferases (HMTs) such as G9a and GLP (see below). Methylated histones in turn serve as high affinity ligands for the attachment of adapter proteins such as heterochromatin protein 1 (HP1). At this approximate point, the chromatin-protein assembly is restrictive enough to insulate the underlying promoter from fluctuations in levels of transactivators but still susceptible to disassembly or dissolution. Either in tandem or even in parallel, the introduction of DNA methylation onto the underlying promoter sequence is now necessary, even sufficient, to hermetically seal the gene promoter. The recruitment of DNA methyltransferases can be accomplished through specific non-catalytic domains on either the HMT or the HP1 proteins.2 The finished product is a highly restrictive, highly durable, chromatin assembly. Heterochromatin deposition can be induced by acute stimuli, can be inhibited by enzyme antagonism, and can be demonstrated on a single gene promoter in primary non-dividing cells or even in euchromatic regions of the genome.3, 4 The entire scheme can be rapid and activated within one hour and has the potential to spread. These are all properties relevant towards a role in the post-mitotic neuron.
H3K9me2/3 synthesis and turnover
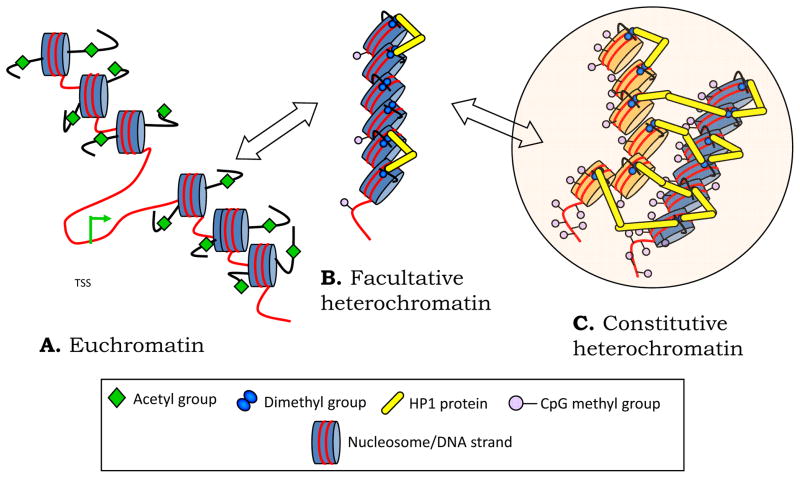
Heterochromatin is characterized by a hallmark histone signature: methylation of lysine at position 9 along the histone 3 tail. Lysine can be modified to a mono- (H3K9me1), di-(H3K9me2) or tri-methylated state (H3K9me3). H3K9me2 is of particular interest as it was originally found to associate with either ‘euchromatin’ or ‘facultative chromatin’(Figure 1).5 Unlike H3K9me3, which is primarily located to gene-poor regions of repetitive DNA, H3K9me2 can locate to gene rich areas of the genome.6 Moreover, H3K9me2 is associated quite directly with gene repression,7 and the difference between a gene that is completely shut down (hermetically closed) versus the same gene in a ‘leaky transcription’ state is often the intensity of H3K9me2 levels on the promoter.3
Figure 1.
Euchromatin and Heterochromatin. (a) Acetylated lysine 9 of histone 3 (H3K9acetyl) results in an open configuration of chromatin otherwise known as ‘euchromatin’. H3K9me2 is synthesized by histone methytransferases (HMT such as G9a, GLP, SETDB1) and initiates the assembly of a restrictive chromatin also termed ‘heterochromatin’. H3K9me2 in turn serves as a high affinity binding site for the attachment of the platform protein HP1.45 HP1 collapses the nucleosome assembly and further recruits other transcriptionally restrictive proteins to this site.46 The spatial spread of heterochromatin is a result of HP1 dimerization between nucleosomes.47 (b) The spread of heterochromatin can be contiguous as in (c) or can assemble non-contiguously (in trans, orange vs. blue colored strands) and relocate to a region of the nucleus that is restrictive to transcription. Facultative heterochromatin is an intermediate stage of restriction that retains the ‘faculty’ for promoter activity. Constitutive heterochromatin is the extreme restrictive state along this continuum; beyond a point, HDAC inhibition may be therapeutically ineffective.
HMTs are families of target-specific enzymes which can distinguish between these mono, di-, or tri-methyl states. In particular, G9a and GLP are the two HMTs dedicated towards the formation of H3K9me2, and function as a dimer; either cannot compensate for the function of the other.8 Consequently, the G9a-GLP dimer is the single protein ‘entity’ responsible for the bulk of H3K9me2 across the genome. Both G9a and GLP have an ANK domain independent of the catalytic domain. The ANK domain can attach recursively to H3K9me2 as well as recruit DNA methyltransferases (Dnmt3a or Dnmt3b) to the targeted site, inducing de-novo DNA methylation. This results in multiple layers of restrictive modifications, i.e., H3K9me2 and DNA methylation,2 which in combination greatly resist any reprogramming of the repressed gene.1
Histone lysine-methylation is a particularly durable modification with a slow turnover (compared to histone lysine-acetylation or serine-phosphorylation), and was previously believed to be irreversible.9 In dividing cells, the half-maximal turnover rate of H3K9me2/3 is about 24 hours, a period almost identical to the turnover of the bulk parent histone.10 These data suggest that in the absence of active catalytic demethylation by demethylases, H3K9 methylation can persist for the lifespan of the cell. Equivalent parameters in postmitotic neurons can be assumed to be at least of the same order of magnitude, but most likely longer, given the minimal requirement for newly synthesized histones. By comparison, turnover of histone acetylation is in the order of minutes.11, 12 For context, CpG methylation of DNA in postmitotic neurons can survive for the lifespan of the animal.13
H3K9 can also be acetylated; the switch
The H3K9 residue is a target for both acetylation and methylation; diametrically opposed functional groups as far as open (acetylation) or closed (methylation) chromatin is concerned. Acetylated H3K9 (H3K9acetyl) strongly promotes transcriptional activity, while methylated H3K9 (H3K9me2/3) strongly promotes transcriptional repression. These properties designate H3K9 as an on/off switch.
A competition between these two modifications (H3K9-acetylation and H3K9-methylation), is suggested by a two-fold coordinate increase of H3K9acetyl in G9a−/− cells.5 Conversely, in G9a +/+ cells, H3K9me2 is engaged in promoter repression, causing the potent HDAC inhibitor TSA to be less effective in increasing H3K9 acetylation.5, 14 One explanation is that the G9a enzyme binds its own product and protects it from further methylation (H3K9me2 to H3K9me3 which has different functional properties; Collins and Cheng 2010), but in so doing may insulate the modification from the action of HATs and HDACs. In principle, H3K9me2 may be resistant to treatment with HDAC inhibitors. 12, 15
Acetylation is a comparatively thermo-dynamically ephemeral modification because of the presence of a keto group leading to greater macromolecular degradation, and enzymatically because of its high turnover rate (compared to the durability of the methyl modification noted above). Furthermore, histones in the proximity of actively transcribed genes undergo rapid acetylation/deacetylation of their tails and the t1/2 to equilibrium is in the order of minutes.12, 16 In the absence of transcription factors, the basal acetylation state is maintained by ambient HATs and HDACs, of which the HDACs are more efficient, possibly due to free access to the acetylated mobile histone tail.12, 17 Given this hypoacetylated state of a quiescent promoter, the equilibrium at the H3K9 switch could shift towards methylation and the seeding of a heterochromatin process as detailed below.
Heterochromatin protein 1 (HP1); spread and segregation
Heterochromatin has a propensity to spread from an initial site of nucleation such as a single promoter or even a single allele3, 18 to stretch across an entire chromosome (as in the Barr body). This spread can occur in either cis or trans, i.e. a location on one chromosome to another location on a separate chromosome.
Heterochromatin protein (HP1) attachment to H3K9me2/3 is one mechanism for propagating the spread of heterochromatin. This phenomenon is based on its chromo-shadow domain capable of homomeric multimerization.19 HP1 is family of proteins (HP1α, HP1β, HP1γ) first discovered as constituents of heterochromatin in Drosophila where they play a role in gene silencing. The N-terminus chromodomain of the HP1 protein is hydrophobic and has a high-affinity attraction to the methylated lysine residue of H3K9me2/3.20
The C-terminus of the HP1 protein contains the chromo-shadow domain noted above which is capable of homo-dimerization with a neighboring HP1.21 The simultaneous attachment of an HP1 protein at the N-terminus with an H3K9me2 histone and dimerization at the C-terminus with another HP1 protein from a neighboring nucleosome, is the basis for higher order assembly. Dimerization in-cis, (along the chromosome) could spread the heterochromatin assembly contiguously (cis-heterochromatin; ‘chromatin creep’). Possibly as a consequence of this propagation, H3K9me2 modified heterochromatin occurs in extensive contiguous regions called LOCKs, extending from 100kb to 4.9 mb.7, 22 When a tissue specific region is thus occupied, the silencing of genes tends to occur within a functional and coordinated gene cluster, such as the expression of lipase genes in the liver but not in the brain.7
These propagating characteristics notwithstanding, HP1 attachment (similar to the H3K9me2 modification) can be limited to and not extend beyond the bounds of a single regulated (repressed) promoter, and thus be capable of geographical precision.3, 23 The binding of HP1 to H3K9me2/3 is not always static or fixed, as might be anticipated from the common notion of heterochromatin as a resistant assembly, but is capable of disassembly and remains in dynamic equilibrium with unbound HP1 protein.24 HP1 disassembly kinetics are faster in euchromatic versus constitutive-heterochromatic regions along the chromosome, possibly due to the greater thermodynamic stability of the H3K9me3-HP1 complex (seen in constitutive-heterochromatin) versus the H3K9me2-HP1 complex (seen in euchromatin).20, 24
Nascent heterochromatin regions containing HP1 can be physically relocated and segregated for optimum repression in chromatin microenvironments that are tethered to the nuclear periphery by interactions between HP1 and the Lamin B receptor (Figure 1).18, 25, 26 Heterochromatin regions thus tethered are called Lamina Associated Domains (LAD) and are uniquely enriched with H3K9me2, HP1 and other repressive proteins such as HDACs.22 Dimerization of HP1 in-trans (across chromosomes) could physically relocate a cluster of H3K9me2 occupied promoters to the nuclear lamina for aggregated repression (trans-heterochromatin (Figure 1).18, 23, 27, 28 Heterochromatin interactions with the nuclear lamina have been noted in postmitotic cells.29
This genomewide spread and segregation of restrictive chromatin has functional implications if considered as a constraint on plasticity or reactivity. Highly restricted genomes may have fixed simplified and invariant transcriptional routines. In this framework, it is noteworthy that in the liver, 46% of the genome is occupied by H3K9me2/3 modifications, while in comparison, only 4% of the embryonic stem cell (ESC) and 10% of the adult neuron is similarly occupied. This would suggest that the neuron is more similar to the ESC by retaining a relatively unrestricted genome.7 Enhanced genomic plasticity as a consequence of lower H3K9me2/3 is demonstrated in nuclear transfer experiments into recipient single cell embryos; gene expression in the subsequent fetus is enhanced if the transplanted nuclei contains low levels of methylated H3K9;30 in other words nuclei containing low levels of H3Kme2/3 are easier to reprogram.
H3K9me2/3 and heterochromatin in the brain
In 2009, we reported abnormally high levels of H3K9me2 in histone extracts from peripheral blood mononuclear cells (PBMC) obtained from patients with schizophrenia.31 Interestingly, this modification was resistant in PBMC culture to treatment with HDAC inhibitors, echoing the weak chromatin response to HDAC inhibitors in schizophrenia, 32 and alluding to the ‘TSA resistance’ in H3K9me histone species.12 H3K9me2 has not been extensively studied in the human brain, and until recently, the regulation of either G9a or GLP was not known.
The distribution and architecture of H3K9me2 is described by two genomewide investigations conducted in fibroblasts and neurons. Both studies find significant enrichments with H3K9me2 across continuous chromosomal stretches (called LOCKs, noted above).7, 22 Together these studies inform us that H3K9me2 enriched domains (LOCKs) are restrictive to transcription, are the function of the G9a HMT, contain fewer genes (about 35% of the non-restricted genome), and appear to be tethered to the periphery of the nucleus by attachment to the nuclear lamina. The pattern of restriction is genomewide and distributed along tissue specific ontologies and networks; thus LOCKs within the brain will suppress liver ontologies, and vice versa.7 For example, receptor coupled G-protein signaling is suppressed in the liver, while unspecified carboxylesterase activity is suppressed in the brain. The brain distribution of H3K9me3 is the subject of another genomewide investigation on the effects of cocaine.33 As expected, H3K9me3 is enriched in areas of repetitive DNA (centromeres, pericentromeres, gene deserts), and is capable of suppressing retrotransoposon activity. But additionally and noteworthy from a therapeutic perspective, H3K9me3 can be modified by pharmacological inputs such as acute and chronic cocaine use.
The physiology of H3K9me2 in the brain has been examined by engineering a cre-mediated conditional knockout of both G9a and GLP genes in adult forebrain neurons. Knockout mice manifest significant loss of H3K9me2 levels (but not H3K9me3) specifically in euchromatic (gene rich) regions of the neuronal genome.34 Adult mice possessing G9a and GLP conditional deletions in postnatal cortical neurons do not show changes in morphology, survival or cell-specific electrophysiology34 but can be distinguished by significant behavioral deficits such as reduction in exploratory behaviors and contextual/cued fear conditioning. If G9a/GLP knockout is engineered specifically in dopaminergic spiny neurons expressing either D1 or D2 receptors, then behavioral responses dependent on these neuronal celltypes are also significantly changed. When compared to wildtype, stimulation of D1 neuronal activity by a D1 agonist (SKF 81297) in a G9a/GLP knockout mouse results in a blunted locomotor response, while depression of D2 neuronal activity using caffeine results in an amplified response.34 Curiously, these G9a/GLP knockouts manifest a derepression of early developmental genes in fully mature postmitotic neurons.34 Comparable effects are observed in pluripotent cells where inhibition of G9a by a small molecule antagonist (BIX-01294) will reprogram and derepress the expression of developmentally silenced genes.35
The interactions of H3K9me2 and dopamine signaling in neurons is independently demonstrated by Maze et al (2010) who show that G9a can be induced by acute cocaine administration (1 hour) but is decreased after chronic cocaine administration.36 Changes in H3K9me2 by cocaine are commensurate with its effects on G9a/GLP. The effect of cocaine on G9a/GLP is mediated by the immediate early gene (IEG) ΔFosB in the nucleus accumbens; a reduction in ΔFosB by a variety of genetic techniques will cause a reduction in G9a/GLP and H3K9me2. Furthermore, while dopaminemimetics such as cocaine reduce ΔFosB and consequently H3K9me2, dopamine antagonists, such as typical and atypical antipsychotics, can induce ΔFosB expression in areas known to have dopamine projections.37 Genetic manipulations of G9a in rodents can modify behavioral responses to cocaine implicating H3K9me2 in a behavioral phenotype. In summary, this series of investigations suggest that dopamine-mimetics will reduce the propensity for H3K9me2 formation, and conversely a reduction in brain H3K9me2 will alter behavioral response to an environmental input such as cocaine. A role for H3K9me2 in the creation of memory is reported by Gupta et al 2010, where global levels of H3K9me2 in the hippocampus are increased when an animal undergoes context and context-fear conditioning.38 If an HDAC inhibitor is given with context-fear conditioning, then the increase in H3K9me2 is significantly impaired, indicating that actively working promoters may be resistant to H3K9me2 deposition. On the other hand, chronic defeat stress with or without chronic imipramine treatment had no effect on H3K9me2 formation in the hippocampus even when administered for four weeks.39
Small molecule pharmacology
A small molecule antagonist (BIX-01294) of both G9a and GLP has been identified with an IC50 in the range of 1.9μM and 0.7μM respectively.40 The decrease in levels of the H3K9me2 with pharmacological inhibition of G9a/GLP is associated with only modest up-regulation of genes, but appears to facilitate the reprogramming of the genome and cell, such as after retinoic acid treatment.5,41 Modifications of the BIX backbone have produced additional antagonists, such as UNC0321 with picomolar potency.42
Another small molecular approach to the modification of the H3K9 methylation status includes the recruitment by unliganded nuclear hormone receptors, occupying their cognate binding sites along promoters of G9a/GLP. When ligand binding occurs, by either E2 or DHT, of the respective estrogen/androgen receptor, then the ligand-bound nuclear receptor in turn recruits histone demethylases such as the Lsd1 or Jmjd1/2 family, which removes the repressive H3K9me mark and facilitates transcription.43, 44 Consequently, targeted chromatin remodeling using ligand hormone receptors is a theoretical possibility.13
Heterochromatin and promoter disuse, misuse, and chronic illness; a shift in the acetylation/methylation dynamic
As reviewed here, the heterochromatin process has the capacity to repress gene activity by several mechanisms (occlude a cognate binding sequence or relocate the promoter to a repressive environment at the nucleus lamina), has the tendency to propagate spatially, is durable and may be resistant to HDAC inhibitors, and in its most aggressive form, can hermetically seal the promoter disallowing any opportunity for regulation. Heterochromatin characterized by the H3K9me2 modification, tends to occupy gene clusters, and represses genomewide tissue specific ontologies/networks. The coordination of this genomewide repression is not known, but could result from reduced availability of transcription factors for downstream nodes in a gene network.
In chronic illness, irregular transcription factor cascades could emerge as a consequence of broad psychological, cognitive or neurological restrictions and limitations. Poorly coordinated (sputtering) promoter activity is a likely result of declining transcription factors and is manifested in the commonly reported downregulation of schizophrenia candidate genes. A broad decline in transcription factors could initiate a heterochromatin process not only at the site of the original set of promoters, but across all nodes of a genomewide promoter network. Over time, this could foster the assembly of heterochromatin in widespread and disparate regions of the genome.
Therapeutic reversal of this repressive process would require interventions that could reduce or restrict the spread of heterochromatin, thereby allowing the genome to more appropriately respond to incoming stimuli, such as dopamine or serotonin signaling from the synapse (Figure 2). Furthermore, differences in network occupancy could be the focus of pathophysiological investigations attempting to distinguish diseased from normal brain.
Figure 2.
Hypothetical chromosomal dimethylated lysine 9 of histone 3 (H3K9me2) and treatment response. (a) The ‘red’ chromosome represents treatment resistant schizophrenia patient chromatin in which ‘chromatin creep’ i.e., the seeding and spread of heterochromatin assemblies across the chromosome, is represented by the color coded spots of variable sizes. Heterochromatin insulates gene rich areas from surface signals that originate at the neuronal membrane or as part of synaptic activity-where most psychotropic agents target. By contrast, the chromosome of a treatment responsive patient (blue chromosome) has less extent and intensity of this restrictive chromatin mark. This allows greater availability to signals originating at the cell membrane, such as receptor blockade or ion-channel activity capable of altering gene expression. (b) Reduced mRNA expression along regions of increased H3K9me2 occupancy is shown when comparing responder to nonresponder. The x-axis depicts mRNA expression along contiguous sections of the chromosome, moving through regions of heterochromatin. In the case of the nonresponder (red graph), the mRNA expression is reduced in regions of heterochromatin, while in the responder (blue graph), mRNA expression is increased in regions lacking heterochromatin.
Summary
Heterochromatin is a repressive and genomewide chromatin structure that starts with a single catalytic event, i.e., H3K9 methylation, which progresses to a local protein assembly, followed by a genomewide spread along gene networks. In full color, heterochromatin is capable of rearranging the nuclear architecture by chromosomal relocations. Our formulation leads to testable hypotheses (Figure 2). Is there a tendency for ‘chromatin creep’ i.e., the seeding and spread of heterochromatin assemblies across the genome, thereby insulating gene rich areas from surface signals such as dopamine antagonism? Are these changes a consequence of chronicity, drugs of abuse, or long term antipsychotic treatment? Can we release these territories with small molecular pharmacology directed at the lysine methyl modification, such as inhibitors of G9a or GLP, with or without adjunctive HDAC inhibition?
Acknowledgments
This work was supported in part by PHS grants MH069839 (R.P.S.) and the APA AstraZeneca Young Minds in Psychiatry Award (D.P.G.).
Funding: This work was supported in part by PHS grants MH069839 (R.P.S.) and the APA AstraZeneca Young Minds in Psychiatry Award (D.P.G.).
Footnotes
Conflict of interest
The authors declare no conflict of interest.
References
- 1.Epsztejn-Litman S, Feldman N, Abu-Remaileh M, Shufaro Y, Gerson A, Ueda J, et al. De novo DNA methylation promoted by G9a prevents reprogramming of embryonically silenced genes. Nat Struct Mol Biol. 2008;15:1176–83. doi: 10.1038/nsmb.1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dong KB, Maksakova IA, Mohn F, Leung D, Appanah R, Lee S, et al. DNA methylation in ES cells requires the lysine methyltransferase G9a but not its catalytic activity. EMBO J. 2008;27:2691–701. doi: 10.1038/emboj.2008.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nguyen TT, Cho K, Stratton SA, Barton MC. Transcription factor interactions and chromatin modifications associated with p53-mediated, developmental repression of the alpha-fetoprotein gene. Mol Cell Biol. 2005;25:2147–57. doi: 10.1128/MCB.25.6.2147-2157.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.El Gazzar M, Yoza BK, Chen X, Hu J, Hawkins GA, McCall CE. G9a and HP1 couple histone and DNA methylation to TNFalpha transcription silencing during endotoxin tolerance. J Biol Chem. 2008;283:32198–208. doi: 10.1074/jbc.M803446200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tachibana M, Sugimoto K, Nozaki M, Ueda J, Ohta T, Ohki M, et al. G9a histone methyltransferase plays a dominant role in euchromatic histone H3 lysine 9 methylation and is essential for early embryogenesis. Genes Dev. 2002;16:1779–91. doi: 10.1101/gad.989402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jiang Y, Jakovcevski M, Bharadwaj R, Connor C, Schroeder FA, Lin CL, et al. Setdb1 histone methyltransferase regulates mood-related behaviors and expression of the NMDA receptor subunit NR2B. J Neurosci. 2010;30:7152–67. doi: 10.1523/JNEUROSCI.1314-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wen B, Wu H, Shinkai Y, Irizarry RA, Feinberg AP. Large histone H3 lysine 9 dimethylated chromatin blocks distinguish differentiated from embryonic stem cells. Nat Genet. 2009;41:246–50. doi: 10.1038/ng.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shinkai Y, Tachibana M. H3K9 methyltransferase G9a and the related molecule GLP. Genes Dev. 2011;25:781–8. doi: 10.1101/gad.2027411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Byvoet P, Shepherd GR, Hardin JM, Noland BJ. The distribution and turnover of labeled methyl groups in histone fractions of cultured mammalian cells. Arch Biochem Biophys. 1972;148:558–67. doi: 10.1016/0003-9861(72)90174-9. [DOI] [PubMed] [Google Scholar]
- 10.Zee BM, Levin RS, Dimaggio PA, Garcia BA. Global turnover of histone post-translational modifications and variants in human cells. Epigenetics Chromatin. 2010;3:22. doi: 10.1186/1756-8935-3-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jackson V, Shires A, Chalkley R, Granner DK. Studies on highly metabolically active acetylation and phosphorylation of histones. J Biol Chem. 1975;250:4856–63. [PubMed] [Google Scholar]
- 12.Hazzalin CA, Mahadevan LC. Dynamic acetylation of all lysine 4-methylated histone H3 in the mouse nucleus: Analysis at c-fos and c-jun. PLoS Biol. 2005;3:e393. doi: 10.1371/journal.pbio.0030393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sharma RP. Schizophrenia, epigenetics and ligand-activated nuclear receptors: A framework for chromatin therapeutics. Schizophr Res. 2005;72:79–90. doi: 10.1016/j.schres.2004.03.001. [DOI] [PubMed] [Google Scholar]
- 14.Dai B, Rasmussen TP. Global epiproteomic signatures distinguish embryonic stem cells from differentiated cells. Stem Cells. 2007;25:2567–74. doi: 10.1634/stemcells.2007-0131. [DOI] [PubMed] [Google Scholar]
- 15.Collins R, Cheng X. A case study in cross-talk: The histone lysine methyltransferases G9a and GLP. Nucleic Acids Res. 2010;38:3503–11. doi: 10.1093/nar/gkq081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davie JR. Nuclear matrix, dynamic histone acetylation and transcriptionally active chromatin. Mol Biol Rep. 1997;24:197–207. doi: 10.1023/a:1006811817247. [DOI] [PubMed] [Google Scholar]
- 17.Katan-Khaykovich Y, Struhl K. Dynamics of global histone acetylation and deacetylation in vivo: Rapid restoration of normal histone acetylation status upon removal of activators and repressors. Genes Dev. 2002;16:743–52. doi: 10.1101/gad.967302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goldmit M, Ji Y, Skok J, Roldan E, Jung S, Cedar H, et al. Epigenetic ontogeny of the igk locus during B cell development. Nat Immunol. 2005;6:198–203. doi: 10.1038/ni1154. [DOI] [PubMed] [Google Scholar]
- 19.Hediger F, Gasser SM. Heterochromatin protein 1: Don’t judge the book by its cover! Curr Opin Genet Dev. 2006;16:143–50. doi: 10.1016/j.gde.2006.02.013. [DOI] [PubMed] [Google Scholar]
- 20.Jacobs SA, Khorasanizadeh S. Structure of HP1 chromodomain bound to a lysine 9-methylated histone H3 tail. Science. 2002;295:2080–3. doi: 10.1126/science.1069473. [DOI] [PubMed] [Google Scholar]
- 21.Brasher SV, Smith BO, Fogh RH, Nietlispach D, Thiru A, Nielsen PR, et al. The structure of mouse HP1 suggests a unique mode of single peptide recognition by the shadow chromo domain dimer. EMBO J. 2000;19:1587–97. doi: 10.1093/emboj/19.7.1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guelen L, Pagie L, Brasset E, Meuleman W, Faza MB, Talhout W, et al. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature. 2008;453:948–51. doi: 10.1038/nature06947. [DOI] [PubMed] [Google Scholar]
- 23.Ayyanathan K, Lechner MS, Bell P, Maul GG, Schultz DC, Yamada Y, et al. Regulated recruitment of HP1 to a euchromatic gene induces mitotically heritable, epigenetic gene silencing: A mammalian cell culture model of gene variegation. Genes Dev. 2003;17:1855–69. doi: 10.1101/gad.1102803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Festenstein R, Pagakis SN, Hiragami K, Lyon D, Verreault A, Sekkali B, et al. Modulation of heterochromatin protein 1 dynamics in primary mammalian cells. Science. 2003;299:719–21. doi: 10.1126/science.1078694. [DOI] [PubMed] [Google Scholar]
- 25.Okada Y, Suzuki T, Sunden Y, Orba Y, Kose S, Imamoto N, et al. Dissociation of heterochromatin protein 1 from lamin B receptor induced by human polyomavirus agnoprotein: Role in nuclear egress of viral particles. EMBO Rep. 2005;6:452–7. doi: 10.1038/sj.embor.7400406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kind J, van Steensel B. Genome-nuclear lamina interactions and gene regulation. Curr Opin Cell Biol. 2010;22:320–5. doi: 10.1016/j.ceb.2010.04.002. [DOI] [PubMed] [Google Scholar]
- 27.Harmon B, Sedat J. Cell-by-cell dissection of gene expression and chromosomal interactions reveals consequences of nuclear reorganization. PLoS Biol. 2005;3:e67. doi: 10.1371/journal.pbio.0030067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grewal SI, Jia S. Heterochromatin revisited. Nat Rev Genet. 2007;8:35–46. doi: 10.1038/nrg2008. [DOI] [PubMed] [Google Scholar]
- 29.Peric-Hupkes D, Meuleman W, Pagie L, Bruggeman SW, Solovei I, Brugman W, et al. Molecular maps of the reorganization of genome-nuclear lamina interactions during differentiation. Mol Cell. 2010;38:603–13. doi: 10.1016/j.molcel.2010.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baxter J, Sauer S, Peters A, John R, Williams R, Caparros ML, et al. Histone hypomethylation is an indicator of epigenetic plasticity in quiescent lymphocytes. EMBO J. 2004;23:4462–72. doi: 10.1038/sj.emboj.7600414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gavin DP, Rosen C, Chase K, Grayson DR, Tun N, Sharma RP. Dimethylated lysine 9 of histone 3 is elevated in schizophrenia and exhibits a divergent response to histone deacetylase inhibitors in lymphocyte cultures. J Psychiatry Neurosci. 2009;34:232–7. [PMC free article] [PubMed] [Google Scholar]
- 32.Sharma RP, Rosen C, Kartan S, Guidotti A, Costa E, Grayson DR, et al. Valproic acid and chromatin remodeling in schizophrenia and bipolar disorder: Preliminary results from a clinical population. Schizophr Res. 2006;88:227–31. doi: 10.1016/j.schres.2006.07.015. [DOI] [PubMed] [Google Scholar]
- 33.Maze I, Feng J, Wilkinson MB, Sun H, Shen L, Nestler EJ. Cocaine dynamically regulates heterochromatin and repetitive element unsilencing in nucleus accumbens. Proc Natl Acad Sci U S A. 2011;108:3035–40. doi: 10.1073/pnas.1015483108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schaefer A, Sampath SC, Intrator A, Min A, Gertler TS, Surmeier DJ, et al. Control of cognition and adaptive behavior by the GLP/G9a epigenetic suppressor complex. Neuron. 2009;64:678–91. doi: 10.1016/j.neuron.2009.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shi Y, Do JT, Desponts C, Hahm HS, Scholer HR, Ding S. A combined chemical and genetic approach for the generation of induced pluripotent stem cells. Cell Stem Cell. 2008;2:525–8. doi: 10.1016/j.stem.2008.05.011. [DOI] [PubMed] [Google Scholar]
- 36.Maze I, Covington HE, 3rd, Dietz DM, LaPlant Q, Renthal W, Russo SJ, et al. Essential role of the histone methyltransferase G9a in cocaine-induced plasticity. Science. 2010;327:213–6. doi: 10.1126/science.1179438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rodriguez JJ, Garcia DR, Nakabeppu Y, Pickel VM. FosB in rat striatum: Normal regional distribution and enhanced expression after 6-month haloperidol administration. Synapse. 2001;39:122–32. doi: 10.1002/1098-2396(200102)39:2<122::AID-SYN3>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 38.Gupta S, Kim SY, Artis S, Molfese DL, Schumacher A, Sweatt JD, et al. Histone methylation regulates memory formation. J Neurosci. 2010;30:3589–99. doi: 10.1523/JNEUROSCI.3732-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsankova NM, Berton O, Renthal W, Kumar A, Neve RL, Nestler EJ. Sustained hippocampal chromatin regulation in a mouse model of depression and antidepressant action. Nat Neurosci. 2006;9:519–25. doi: 10.1038/nn1659. [DOI] [PubMed] [Google Scholar]
- 40.Chang Y, Zhang X, Horton JR, Upadhyay AK, Spannhoff A, Liu J, et al. Structural basis for G9a- like protein lysine methyltransferase inhibition by BIX-01294. Nat Struct Mol Biol. 2009;16:312–7. doi: 10.1038/nsmb.1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Feldman N, Gerson A, Fang J, Li E, Zhang Y, Shinkai Y, et al. G9a-mediated irreversible epigenetic inactivation of oct-3/4 during early embryogenesis. Nat Cell Biol. 2006;8:188–94. doi: 10.1038/ncb1353. [DOI] [PubMed] [Google Scholar]
- 42.Liu F, Chen X, Allali-Hassani A, Quinn AM, Wigle TJ, Wasney GA, et al. Protein lysine methyltransferase G9a inhibitors: Design, synthesis, and structure activity relationships of 2,4-diamino-7-aminoalkoxy-quinazolines. J Med Chem. 2010;53:5844–57. doi: 10.1021/jm100478y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Garcia-Bassets I, Kwon YS, Telese F, Prefontaine GG, Hutt KR, Cheng CS, et al. Histone methylation-dependent mechanisms impose ligand dependency for gene activation by nuclear receptors. Cell. 2007;128:505–18. doi: 10.1016/j.cell.2006.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hublitz P, Albert M, Peters AH. Mechanisms of transcriptional repression by histone lysine methylation. Int J Dev Biol. 2009;53:335–54. doi: 10.1387/ijdb.082717ph. [DOI] [PubMed] [Google Scholar]
- 45.Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, et al. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–4. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- 46.Bernstein BE, Meissner A, Lander ES. The mammalian epigenome. Cell. 2007;128:669–81. doi: 10.1016/j.cell.2007.01.033. [DOI] [PubMed] [Google Scholar]
- 47.Sims RJ, 3rd, Nishioka K, Reinberg D. Histone lysine methylation: A signature for chromatin function. Trends Genet. 2003;19:629–39. doi: 10.1016/j.tig.2003.09.007. [DOI] [PubMed] [Google Scholar]